Abstract
The Q gene is largely responsible for the widespread cultivation of wheat because it confers the free-threshing character. It also pleiotropically influences many other domestication-related traits such as glume shape and tenacity, rachis fragility, spike length, plant height, and spike emergence time. We isolated the Q gene and verified its identity by analysis of knockout mutants and transformation. The Q gene has a high degree of similarity to members of the AP2 family of transcription factors. The Q allele is more abundantly transcribed than q, and the two alleles differ for a single amino acid. An isoleucine at position 329 in the Q protein leads to an abundance of homodimer formation in yeast cells, whereas a valine in the q protein appears to limit homodimer formation. Ectopic expression analysis allowed us to observe both silencing and overexpression effects of Q. Rachis fragility, glume shape, and glume tenacity mimicked the q phenotype in transgenic plants exhibiting post-transcriptional silencing of the transgene and the endogenous Q gene. Variation in spike compactness and plant height were associated with the level of transgene transcription due to the dosage effects of Q. The q allele is the more primitive, and the mutation that gave rise to Q occurred only once leading to the world's cultivated wheats.
WHEAT, rice, and maize are the three major cereal crops that provide most of the calories consumed by humans. Bread (common) wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD genomes) arose ∼8000–10,000 years ago (for review see Nesbitt and Samuel 1996; Feldman 2001) from the spontaneous hybridization of the tetraploid wheat T. turgidum L. (2n = 4x = 28, AABB genomes) with the diploid goatgrass Aegilops tauschii Coss. (2n = 2x = 14, DD genomes) (Kihara 1944; McFadden and Sears 1946). Domestication of wheat resulted from mutations that gave rise to traits such as soft glumes, a nonfragile rachis, and the free-threshing character.
The Q gene governs the free-threshing character and square spike phenotype. In addition, Q pleiotropically affects a repertoire of other characters important for domestication such as rachis fragility (Leighty and Boshnakian 1921; Mackey 1954; Singh et al. 1957; Jantasuriyarat et al. 2004), glume shape and tenacity (Muramatsu 1963, 1986; Singh 1969), spike length (Muramatsu 1963; Kato et al. 1999), plant height (Muramatsu 1963; Kato et al. 1999, 2003), and spike emergence time (Kato et al. 1999). The q allele and null mutations confer a speltoid spike, which is characterized by a spear-shaped spike with an elongated rachis. Speltoid spikes generally have wild grass-like features such as tenacious glumes, fragile rachises, and nonfree-threshing seed.
The emergence of the free-threshing character concomitant with reduced rachis fragility and glume tenacity allowed early farmers to efficiently harvest the grain. The significance of the Q gene is even more greatly realized in modern agriculture because nonshattering free-threshing grain is essential to mass production and mechanical harvesting on a large scale. Historically, the importance of the Q allele has been recognized, but the nature of the gene, how it arose, and its structural and functional relationship with the q allele have long been debated.
Early experiments involving the cytogenetic analysis of aneuploids located the Q gene on chromosome 5A (Huskins 1946; Unrau et al. 1950; Sears 1952, 1954; Mackey 1954). Huskins (1946) and Sears (1952, 1954) observed the dosage effect of Q on spike morphology in the T. aestivum cv. Chinese Spring (CS) background. They observed that plants nullisomic, monosomic, disomic, trisomic, and tetrasomic for 5A had speltoid, semi-speltoid, square, subcompactoid, and compactoid spikes, respectively. Muramatsu (1963) investigated the dosage effects of the q allele using the 5A chromosomes from spelt wheat (T. aestivum ssp. spelta) in the CS background. Individuals disomic, trisomic, or tetrasomic had speltoid spikes but plants pentasomic for the spelt 5A chromosome had square spikes. On the basis of these observations, Muramatsu (1963) concluded that q is hypomorphic to Q and ∼2.5 doses of q equaled 1 dose of Q.
Using chromosome deletion lines, Endo and Gill (1996) physically mapped the Q gene to a submicroscopic deletion interval on the long arm of chromosome 5A. Q was then placed on recombination-based maps (Kato et al. 1999). High-resolution mapping (Faris and Gill 2002) followed by the construction of a T. monococcum BAC contig spanning the Q locus led to the identification of a gene with similarity to the Arabidopsis APETALA2 (AP2) gene (Faris et al. 2003). A molecular marker for the AP2-like gene cosegregated with Q in 930 gametes and was considered a likely candidate.
In this article we report the validation of the wheat AP2-like (WAP2) gene as Q; investigation of structural, transcriptional, and regulatory differences between Q and q alleles; confirmation of the dosage and pleiotropic effects of Q; and some insights into the origin of cultivated macaroni and bread wheat.
MATERIALS AND METHODS
Plant materials and Southern analysis:
The genomic sequence of WAP2 was obtained from the genotypes listed in Table 1, where GenBank accession numbers for the WAP2 sequences are also presented. The sequence of the T. monococcum WAP2 gene was identified previously (Faris et al. 2003). The CS fast neutron-induced speltoid deletion mutant fndel-143 (null for Q on chromosome 5A, but retains homeoalleles on 5B and 5D) (Faris et al. 2003) was used to ensure all PCR amplifications were specific to the chromosome 5A copy of WAP2 and to evaluate the specificity of the Taqman system (see Transcription analysis). The T. aestivum cv. “Bobwhite” (QQ genotype) was used for transformation experiments. CS homeologous chromosome group 5 nullisomic-tetrasomic (NT) lines, where a pair of missing chromosomes is partially compensated by an extra pair of homeologous chromosomes, were used to assign restriction fragments to individual chromosomes by Southern analysis. DNA extraction, digestion, Southern transfer, and hybridization were performed as previously described (Faris et al. 2000).
TABLE 1.
Descriptions of wheat genotypes used for sequence analysis of the WAP2 gene
| WAP2 gene source | Ploidy | Genomes | Genotype | Source designation | GenBank no. |
|---|---|---|---|---|---|
| T. aestivum ssp. aestivum (CS) | 6x | ABD | cv. Chinese Spring | AY702956 | |
| T. turgidum ssp. dicoccoides (CS-DIC 5A)a | 6x | ABD | TA3446b | AY702957 | |
| T. aestivum ssp. macha | 6x | ABD | PI 361862 | AY714342 | |
| T. aestivum ssp. spelta (Iranian)c | 6x | ABD | DS 5A Irand | AY714340 | |
| T. aestivum ssp. spelta (European)e | 6x | ABD | DS 5A Europed | AY714341 | |
| T. aestivum ssp. spelta (European) | 6x | ABD | TA2603 | AY702960 | |
| T. turgidum ssp. durum (LDN) | 4x | AB | cv. Langdon | AY702955 | |
| T. turgidum ssp. carthlicum | 4x | AB | TA2801 | AY702959 | |
| T. turgidum ssp. polonicum | 4x | AB | CItr 191826f | AY714339 | |
| T. turgidum ssp. dicoccum | 4x | AB | CItr 14621 | AY714343 | |
| T. urartu | 2x | A | TA704 | AY702958 | |
| T. monococcum | 2x | Am | cv. DV92 | AY170867 |
Disomic chromosome substitution line where a pair of 5A chromosomes from T. turgidum ssp. dicoccoides (TA106) was substituted for the native pair of 5A chromosomes in Chinese Spring by the late E. R. Sears.
Sources with TA designations are maintained by the Wheat Genetics Resource Center, Kansas State University (Manhattan, KS).
Disomic substitution line where a pair of 5A chromosomes from Iranian spelt (H. Kuckuck's accession 407a) was substituted for the native pair of 5A chromosomes in Chinese Spring by E. R. Sears as described in Luo et al. (2000). Seed provided by J. Dvorák (University of California, Davis, CA).
Designations as reported in Luo et al. (2000). Seed provided by J. Dvorák (University of California, Davis, CA).
Disomic substitution line where a pair of 5A chromosomes from European spelt (Sears' accession P78-81-1) was substituted for the native pair of 5A chromosomes in Chinese Spring by E. R. Sears as described in Luo et al. (2000). Seed provided by J. Dvorák (University of California, Davis, CA).
Sources with CItr designations are maintained by the USDA–ARS National Small Grains Collection (Aberdeen, ID).
Generation of mutants:
Mutants were generated by treating CS seeds with 0.4% ethyl methanesulfonate (EMS) in 0.05 M phosphate buffer as described (Williams et al. 1992). Selected M2 families were then planted and screened in a greenhouse for plants with speltoid spikes.
BAC sequencing and analysis:
A fragment of the T. monococcum WAP2 gene was used to screen the LDN BAC library (Cenci et al. 2003). Positive BACs were digested with HindIII and DraI, Southern blotted, and hybridized with the same probe to assign BACs to chromosome 5A or 5B on the basis of restriction fragment sizes. One BAC from chromosome 5A (376H15, 142 kb) and one from 5B (1004P5, 152 kb) were sequenced by Myriad Genetics (Salt Lake City). BAC sequences were subjected to BLASTn and BLASTx (Altshul et al. 1997) searches of the public databases to identify the WAP2 gene sequence from the BACs. The sequences were then submitted to RICEGAAS (http://ricegaas.dna.affrc.go.jp/) to identify predicted open-reading frames using multiple gene prediction programs. Further details regarding the analysis of the chromosome 5B WAP2 homeoallele are included as supplementary material available at http://www.genetics.org/supplemental/.
WAP2 sequencing and analysis:
The T. monococcum WAP2 sequence was used to design primers to amplify WAP2 from the genotypes in Table 1. The genomic WAP2 gene sequence was PCR-amplified as three separate overlapping fragments using primers AP5P.11-3 (5′-GCCCTCGCAGCCCGCGGCCACCGCGCTCCCA) or AP2startF (5′-ATGGTGCTGGATCTCAATGTGGAGTCGCCGGCGGA) in combination with AP2.8R (5′-CGCGGCCAAATCGGGGCAAAGGAATTCAAACGA) for fragment 1, WAP2.2F (5′-CACTGGATAATTTCTTCAGGTGGTTTCGACACTGC) with AP2.15R (5′-ACATGGAACCTTAATTTCAGGAACGAACTTGTCG) for fragment 2, and AP2.16F (5′-CTGCTTGGTGCGCTGCTCCACCAGCTTACTGAAA) with AP45.1R (5′-CAGAAGGCCCAACGGTTAACGCAACAATGGC) for fragment 3. PCR conditions for fragment 1 were 250 ng DraI-digested genomic DNA, 0.4 μm of each primer, 200 μm dNTP, 1× reaction buffer, 8% DMSO, and 2.5 units Herculase Hotstart DNA Polymerase (Stratagene, La Jolla, CA) in a 50-μl volume. Cycling conditions for fragment 1 amplification were the following: 98° for 3 min, 10 cycles of 98° for 40 sec, 68° for 30 sec, 72° for 2 min, 30 cycles of 98° for 40 sec, 68° for 30 sec, 72° for 2 min with extension time increasing 10 sec each cycle. One unit of Biolase DNA polymerase (Bioline USA, Randolph, MA) was added and allowed to finish cycling at 72° for 10 min. Fragments 2 and 3 were amplified using 1 μl Advantage cDNA polymerase mix (CLONTECH, Palo Alto, CA), 300 ng of DNA, 200 μm dNTP, 1× buffer, and 0.4 μm of forward and reverse primers in a 50-μl reaction. Cycling conditions for fragment 2 were the following: 94° for 4 min, 35 cycles of 94° for 40 sec, 68° for 30 sec, 72° for 3 min, and a 10-min final extension step. Cycling conditions for fragment 3 were identical to those of fragment 2 except that the annealing temperature was 72° and the extension time was reduced to 2 min.
Products from five independent PCR reactions for each segment of the gene from each genotype were separated on 1% agarose gels and gel-purified using the QIAquick gel extraction Kit (QIAGEN, Chatsworth, CA). Fragments were cloned using the TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. The sequences were reassembled using CLUSTAL W to obtain the full-length genomic sequences.
The coding sequence of WAP2 was determined by identifying database ESTs (http://wheat.pw.usda.gov/NSF/; http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=wheat) and 5′ RACE (gene-specific primer AP2race2: 5′-GGGCGGCGACGCGGGGAAGAGCTGCCTCGTG). The 5′ UTR was obtained using the BD SMART RACE cDNA amplification kit (CLONTECH) in combination with the BD Advantage-GC 2 PCR kit (CLONTECH). Reverse transcriptase (RT)–PCR for the three EMS-induced mutants, mq36, mq125, and mq194, was done with primers flanking the regions containing single base mutations (AP45.6F: 5′-ATGGGGCAGCAGGCCCCGGCGCCTGCGCCGATGGC and AP2.13R: 5′-CTCTTGGGATCGTGCGCGGTGGGTTGCGACATC), using the TITANIUM one-step RT–PCR kit (CLONETECH).
The 5′ region extending from position −140 to −1121 from the transcription start site was PCR amplified with primers AP5P.16F: 5′-GGATCACGTGGGTGGTTCTTTGTCCATGCC and AP5P.12R: 5′-GTCGGGGAGGCCAAGGGCATCAGAGG. Cycling conditions were the following: 94° for 4 min, 35 cycles of 94° for 40 sec, 66° for 30 sec, 72° for 2 min, and a 10-min final extension step.
Transformation:
Tissue culture, particle bombardment of immature embryos, and initial screening for the bar gene were done following previously described methods (Altpeter et al. 1996; Chen et al. 1998; Huang et al. 2003). A 5.8-kb subclone containing the WAP2 gene and its native promoter from the LDN 5A BAC was ligated into the pSMART vector (Lucigen, Middleton, WI). The T. aestivum cv. Bobwhite, which is presumed to have two endogenous copies of the Q allele, was chosen for its ease of transformation. Calli were cobombarded with the subclone and the pAHC20 vector (contains the bar gene as a selectable marker driven by the maize ubiquitin promoter) using microprojectile bombardment. Positive individuals were grown to maturity and the spike morphology recorded. Selected T1 plants derived from speltoid and compactoid T0 plants were analyzed for transgene integration and transcription.
Relative quantitative-PCR:
Tissue for RNA extractions was collected, immediately frozen in liquid nitrogen, and stored at −80°. RNA was isolated using the QIAGEN RNeasy plant mini kit (QIAGEN) and on-column DNA digestion. cDNA was prepared using TaqMan RT reagents with random hexamer primers and multiScribe reverse transcriptase (Applied Biosystems, Foster City, CA) as suggested by the manufacturer. Relative quantitative (RQ)-PCR was performed on a 7500 Real Time PCR System (Applied Biosystems) using a TaqMan system for Q and either the 18S rRNA or actin genes. 18S TaqMan system (Podkowinski et al. 2003): left primer, CGGAGAATTAGGGTTCGA; right primer, CCGTGTCAGGATTGGGTA; probe, VIC-CTACCACATCCAAGGAAGGCAG-TAM. Q TaqMan system: left primer, CCCTGAATCGTCAACCACAATG; right primer, CCGTGCCATGTTGATGCA; probe, FAM-CTTCGTCCCAGTGGCCTG-NFQ. Actin Taqman system: left primer, ATGGAAGCTGCTGGAATCCAT; right primer, CCTTGCTCATACGGTCAGCAATAC; probe, VIC-CCTTCCTGATATCCACATCACACTTCATGATAGAGT-TAM. At least four replications of each reaction were performed using 1X TaqMan Universal PCR master mix (Applied Biosystems). Each reaction consisted of 5 μl of cDNA diluted 1:50, 100 mm of probe, and 200 mm of each primer. Templates to determine the amplification efficiency of 18S, actin, and Q consisted of five twofold dilutions. Raw CT values were averaged for each dilution and each sample. Amplification efficiencies of 18S, Q, and actin were tested for similarity by plotting RNA concentration vs. CT value. The difference between the slopes was <0.1 and thus considered to have similar amplification efficiencies. Sample averages were linearized using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Yeast two-hybrid analysis for detecting dimerization of Q gene products:
The yeast two-hybrid MATCHMAKER system (BD Biosciences CLONTECH, Palo Alto, CA) was used for analysis of protein-protein interactions according to the manufacturer's protocols. First, the full-length coding sequences of Q from CS and q from CS-DIC 5A were amplified from cDNA using primers Q_Nde1_F: 5′-CATATGGTGCTGGATCTCAATGTGGAGTC and Q_BH1344_R: 5′-GGATCCTCAGTTGTCCGGCGGGCGGGGGAA. Serial deletions of the coding sequences were then constructed and ligated into vectors pGBKT7 and pGADT7 to be bait and prey, respectively. Yeast Y187 cells were transformed with the bait plasmids, and AH109 cells were used for the prey plasmids. The two yeast strains containing bait and prey were mating on YPD plates for overnight, and the yeast cells were cultured in SD medium without leucine (L) and tryptophan (W) for 2 days. To test the protein interactions, the SD/-L-W cultured yeast cells were inoculated on SD plates containing X-Gal but lacking leucine, tryptophan, histidine, and adenine. The empty vectors of pGBKT7 and pGADT7 were used as a negative control, and the interaction between p53 and LTA served as a positive control.
Phylogenetic analysis:
Phylogenetic trees were constructed from CLUSTALW alignments of the complete genomic DNA sequences of Q/q using the neighbor-joining method and multiple distance-based methods available in the MacVector v7.2 software. Confidence values for nodes were calculated using 1000 bootstraps.
RESULTS
Structure and validation of Q:
We obtained the genomic sequence of the wild-type WAP2 gene from CS using PCR, and determined the full-length coding region using a combination of RT–PCR, 5′-RACE, and searches against the wheat EST databases (Figure 1A). The WAP2 gene consists of 10 exons and 9 introns, extends 3229 bp from start to stop codon, and has a GC content of 54%. The coding sequence is 1344 bp long plus a 128-bp 5′ UTR (supplemental Figure 1 at http://www.genetics.org/supplemental/) and a 255-bp 3′ UTR (supplemental Figure 2 at http://www.genetics.org/supplemental/). It encodes a protein of 447 amino acids.
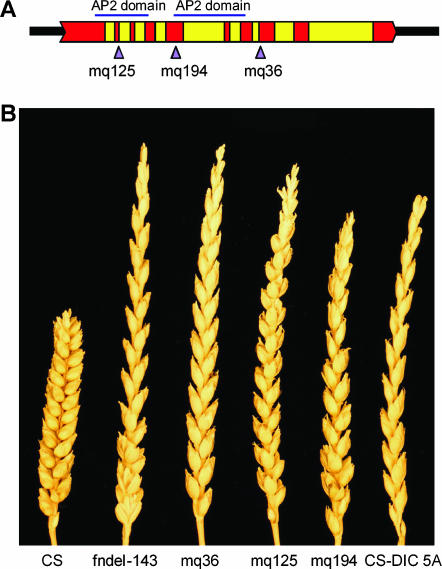
Figure 1.
Analysis of EMS mutants. (A) Illustrated structure of the Q gene. Exons are depicted in red. Arrows indicate locations of point mutations in the EMS mutants. (B) Spike morphology of CS (Q); fndel-143 (null for Q); EMS-induced mutants mq36, mq125, and mq194; and CS-DIC 5A (q).
The M2 generation of a population of CS EMS mutants was screened for the speltoid phenotype to identify putative knockouts. Three speltoid mutants harboring point mutations within WAP2 were identified (Figure 1B). Mutant mq194 had a single base substitution in an AP2 DNA binding domain (exon 5) that resulted in the change of a cysteine to a tyrosine. The mutants mq125 and mq36 had point mutations in the donor site of intron 2 and the acceptor site of intron 7, respectively. RT–PCR experiments showed that the point mutations in mq36 and mq125 resulted in larger PCR products compared to the wild-type WAP2 gene, indicating alternate splicing in these mutants (data not shown). The altered transcription of the mutated WAP2 gene in mq36 produced a predicted protein longer than the wild type due to translation of intron 7, and mq125 likely produced a truncated protein due to a stop codon encountered in the translation of intron 2. The sequence analysis of the mutants validated that WAP2 is Q and therefore, the gene will be referred to as Q hereafter.
Structural comparison of Q and q alleles:
Previous research (Kuckuck 1959) suggested that Q evolved as a duplication of q due to unequal crossing over. We performed Southern analysis by hybridizing a fragment of the Q gene with multiple nonfree-threshing and free-threshing hexaploid and tetraploid genotypes (Figure 2). Restriction patterns and fragment hybridization intensities suggested that a single copy of the Q gene is present on each of the homeologous group 5 chromosomes. In addition, sequence analysis of the 142-kb BAC harboring the Q allele from LDN indicated a single ORF corresponding in size and structure to the q ORF from T. monococcum (data not shown).
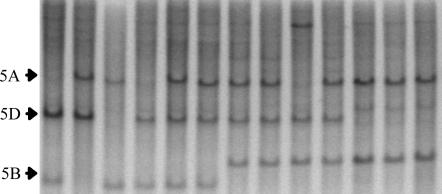
Figure 2.
Southern analysis of various wheat taxa hybridized with a fragment of WAP2 (Q). Left to right: N5AT5D, N5BT5D, N5DT5B, fndel-143 (Q-null), T. aestivum spp. aestivum cv. CS (Q), CS-DIC 5A (q), T. aestivum ssp. macha (q), European T. aestivum ssp. spelta (q; TA2603), Iranian T. aestivum ssp. spelta (Q), European T. aestivum ssp. spelta (q; DS 5A Europe), T. turgidum ssp. carthlicum (Q), T. turgidum ssp. durum cv. LDN (Q), and T. turgidum ssp. polonicum (Q).
To further investigate structural differences between Q and q alleles, we aligned the gene sequences from the 12 Triticum genotypes listed in Table 1. Comparison of the genomic sequences revealed six conserved differences between Q- and q-containing genotypes (Figure 3). Four of these differences, including a variable microsatellite, were present in introns and one was in the 3′ UTR. One conserved nucleotide difference changed a predicted amino acid where, at position 329, all Q-containing genotypes possessed an isoleucine while all q-containing genotypes possessed a valine.
Figure 3.
Locations of six conserved nucleotide differences between Q- and q-genotypes within the genomic sequence of the gene. Arrows represent single nucleotide polymorphisms between Q and q. Green, intron; red, exon; blue, 3′ UTR. The polymorphism indicated in red represents the amino acid difference between Q and q alleles at position 329 of the predicted protein. The green bar represents a variable microsatellite within intron 9.
To identify regulatory elements in the promoter region of Q, we PCR amplified and sequenced a fragment extending from position −140 to −1121 from the transcription start site in the Q genotypes CS, T. turgidum ssp. carthlicum, and the Iranian spelt, and from q genotypes CS-DIC 5A, T. turgidum ssp. dicoccum, and both European spelts. We were unable to PCR amplify the 140-bp fragment immediately upstream from the transcription start site using whole genomic DNA as template. However, the sequence of this segment was available from the T. monococcum and LDN BACs, and we confirmed the sequence from these two genotypes by PCR amplification directly from the BACs followed by sequencing. We generated consensus sequences of the Q and q promoter regions on the basis of ClustalW alignments to identify conserved differences. Five conserved differences were identified (supplemental Figure 3 at http://www.genetics.org/supplemental/), but searches of plant promoter databases PlantCARE (http://intra.psb.ugent.be:8080/PlantCARE/index.html), PlantProm (http://www.softberry.com), and PLACE (http://www.dna.affrc.go.jp/PLACE/) indicated that only two of these differences are likely to be associated with putative regulatory elements.
Transcription analysis:
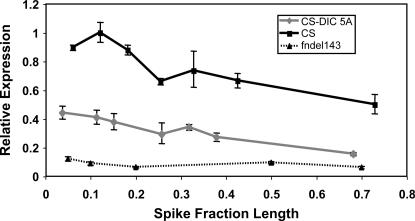
We compared the level of transcription between the hexaploids CS (Q), CS-DIC 5A (q), and fndel-143 (null for Q) in various stages of spike development. Spikes were collected and assigned fraction-length values on the basis of the length of corresponding fully matured spikes. Transcription in both CS and CS-DIC 5A peaked in the early stages of spike development and gradually declined as the spikes matured (Figure 4). While transcription patterns for both Q and q were quite similar, the transcription level of Q was consistently higher than that of q.
Figure 4.
Relative transcription levels of Q in CS and q in CS-DIC 5A during spike development. Q was consistently transcribed at higher levels than q, but transcription of Q and q followed the same trend as the spike matured. Transcription levels in fndel-143 (null for Q) are shown to indicate the specificity of the Taqman system.
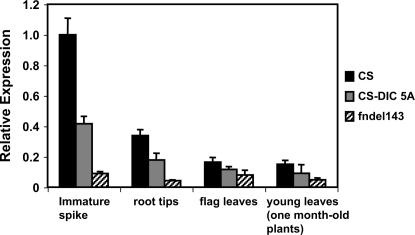
To investigate transcription levels in other tissues, we compared CS (Q), CS-DIC 5A (q), and fndel-143 (null for Q) using root tips, the youngest fully expanded leaf from 1-month-old plants (Feekes stage 4-5), and flag leaves from adult plants (Feekes stage 10). Both CS and CS-DIC 5A exhibited transcription in all tissues tested (Figure 5), but levels of transcription were less compared to immature spikes. Higher levels of transcription were observed in CS relative to CS-DIC 5A in all tissues.
Figure 5.
Transcription levels of Q from CS, q from CS-DIC 5A, and the Q null allele from fndel-143 in root tips, young leaves of 1-month-old plants, and flag leaves. The transcription levels in immature spikes (fraction length, 0.1) are shown for comparison.
Yeast two-hybrid analysis:
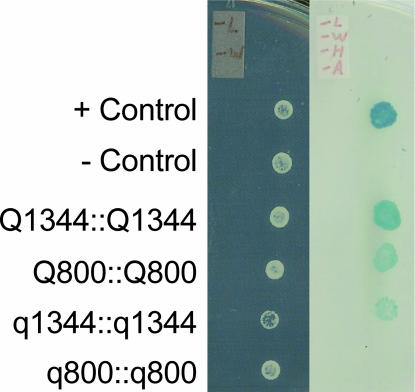
The yeast two-hybrid system was used to test for potential homodimer formation and to investigate the biological significance of the point mutation responsible for changing the valine in q to an isoleucine in Q at amino acid position 329 (I329V). The results indicated that the full-length Q protein (Q1344) has the ability to form a homodimer (Figure 6). An N-terminal deletion construct (Q800) containing the C-terminal 277 amino acids and the putative coil-forming region, which links the two AP2 domains, also had the ability to form a homodimer. Further deletions of the remaining C-terminal 247 amino acids lacking the coil region significantly decreased the level of homodimerization. The point mutation (I329V) of q greatly reduced homodimer formation of the full-length q protein (q1344), and it completely disrupted the formation of a q800 homodimer. In addition, while Q800 was able to form a homodimer, it was not able to form a dimer with q800 (data not shown).
Figure 6.
The homodimers of Q protein in yeast two-hybrid analysis. (Left) The yeast cells after mating were grown in SD/-L-W medium to confirm the cells containing both bait and prey. (Right) The protein-protein interactions were examined by the growth under the selection of SD/-L-W-H-A and in the presence of X-Gal. The cells could grow only when the combination of bait∷prey resulted in the protein interaction and therefore the activation of reporter genes (HIS, ADE2, and lacZ).
Genetic transformation and investigation of Q dosage effects:
We transformed Bobwhite with Q from the durum wheat variety LDN and obtained 17 putative transformants. We screened the T0 generation for individuals with either speltoid or compactoid spikes. One speltoid T0 plant was identified, and five T1 progeny derived from this plant were grown for further analysis. In addition to having speltoid spikes (Figure 7A), the T0 and all five T1 plants were taller than Bobwhite (Figure 7E), had keeled tenacious glumes that adhered strongly to the seed, and fragile rachises, in which the disarticulation pattern was such that the rachis broke above the junction of the rachis and rachilla leaving a portion of the rachis at the base of the spikelet (Figure 7F). This tendency mimics the disarticulation pattern of the shatter-prone wild wheats that lack the Q allele. Southern analysis showed each T1 individual harbored an abundance of transgene copies in addition to the endogenous gene (data not shown).
Figure 7.
Analysis of T1 transgenic plants. (A) T1 transgenic speltoid spike (T32). (B) Untransformed Bobwhite square spike. (C) T1 transgenic subcompactoid spike (T30). (D) T1 transgenic compactoid spike (T39). (E) Whole plant view of T1 transgenic individuals. Left to right: T32, Bobwhite, T30, and T39. (F) Differences in spikelet disarticulation pattern between untransformed Bobwhite (QQ) (left) and a T1 speltoid transgenic (T32, Q silenced) (right). In Bobwhite, Q confers a tough rachis and abscission occurs at the base of the spikelet causing the seed to be free-threshing. When Q is silenced (and in qq genotypes) abscission occurs at the junction of the rachis and rachilla causing the spikelet to shatter from the rachis and the seed to be nonfree-threshing.
Two partially sterile T0 plants with compactoid spikes were identified. Three T1 plants derived from one compactoid T0 and two T1's from a second compactoid T0 showed a range of phenotypes. One plant was of normal height and had square spikes (Figure 7B), two plants were slightly shorter than Bobwhite (Figure 7E) and had subcompactoid spikes (Figure 7C), and two were very short (Figure 7E) with compactoid spikes (Figure 7D). All plants had tough rachises and round, soft glumes that loosely held the seed, and were free-threshing (Figure 7F). Southern analysis indicated the T1 plants segregated for copy number and either were lacking the transgene (square spike) or possessed multiple copies of the transgene in addition to the endogenous gene (subcompactoid and compactoid spikes) (data not shown).
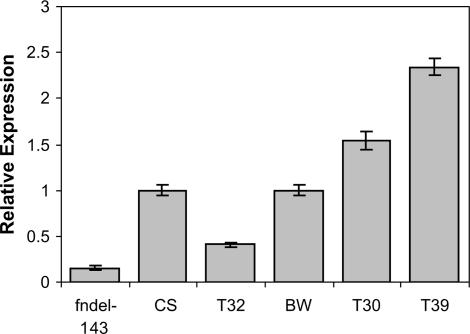
RQ-PCR indicated the level of Q transcription in the T1 speltoid plants was less than that observed in untransformed Bobwhite (Figure 8). T1 plants with subcompactoid and compactoid spikes had greater levels of Q transcription compared to Bobwhite.
Figure 8.
Relative transcription levels of the fast-neutron induced deletion mutant fndel-143 (null for Q), CS (QQ), T32 (speltoid transgenic), untransformed Bobwhite, T30 (subcompactoid transgenic), and T39 (compactoid transgenic). Transcription levels in CS (Q) and fndel-143 (null for Q) are shown for comparison. Bars represent standard error.
Phylogenetic analysis:
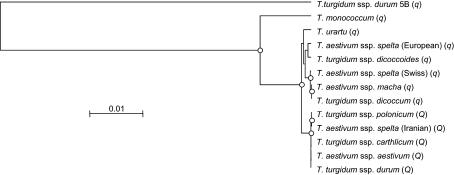
Phylogenetic analysis of the 12 Triticum genotypes indicated that all Q genotypes were highly similar, while q genotypes were more divergent (Figure 9). The genomic sequence of the AP2-like homeoallele from T. turgidum ssp. durum cv. LDN chromosome 5B was used to root the tree.
Figure 9.
Phylogenetic tree of 12 Triticum genotypes (Table 1) based on full-length genomic DNA sequences (start to stop codon) of the Q/q gene calculated by the neighbor-joining method and rooted by the q homeoallele from T. turgidum ssp. durum chromosome 5B as an outgroup. Open circles indicate nodes supported by bootstrap values >70%.
DISCUSSION
In this work, we showed that Q is a member of the AP2 class of transcription factors. AP2-like genes are characterized by having two plant-specific DNA binding motifs referred to as AP2 domains and have been implicated in a wide range of plant development roles. In Arabidopsis, AP2 is a floral homeotic gene involved in the establishment of floral meristem identity (Irish and Sussex 1990; Bowman et al. 1993), floral organ identity (Komaki et al. 1988; Bowman et al. 1989; Kunst et al. 1989; Jofuku et al. 1994), and temporal and spatial regulation of floral homeotic gene expression (Drews et al. 1991). Putative orthologs of Q have been identified in maize, rice, and barley (Faris et al. 2003). In maize, indeterminant spikelet1 (ids1) is known to determine the number of floral meristems produced (Chuck et al. 1998), but functions have not yet been assigned to the orthologs in rice and barley, and Q is so far the only AP2-like gene implicated in domestication.
Ectopic expression analysis of transgenic plants allowed us to observe both dosage and pleiotropic effects of Q. Increased transcription of Q was most obviously associated with spike compactness and reduced plant height. This observation agreed precisely with findings reported a half century ago using cytogenetic stocks (Huskins 1946; Sears 1952, 1954; Muramatsu 1963). Variation in other important morphological characters such as spike length, rachis fragility, glume shape, and glume tenacity observed in the transgenic plants agree with previous experiments that located QTL for these traits to the Q locus on chromosome 5A (Kato et al. 1999, 2003; Jantasuriyarat et al. 2004) and confirmed the pleiotropic effects of Q.
Previous research suggested that Q might have arisen through duplication of q. Evidence for this was presented by Kuckuck (1959) who reported observing occasional square-spike phenotypes in progeny derived from two speltoid parents. Also, Muramatsu (1963) suggested that approximately five doses of q conferred the same phenotype as 2 doses of Q. However, our data collected from Southern analysis and sequencing of a large BAC spanning Q indicates that Q is not a duplication of q, but most likely arose through a gain-of-function mutation. Also, we obtained no square-spike (QQ) F2 progenies from the cross CS-DIC 5A (qq)/ CS-T. spelta 5A (qq) (B. S. Gill, unpublished results).
It is clear from this and previous research (Muramatsu 1963) that both Q and q alleles are functional, but they confer different phenotypes. We found Q and q alleles to differ both in structure and in the level of transcription. An obvious and consistent difference in the level of transcription between Q and q alleles was associated with phenotypic differences from dosage effects observed in transgenic plants, and with conclusions derived from Muramatsu (1963). In addition, several bases at the DNA level defined structural differences between Q and q, but only a single putative amino acid at position 329 differentiates between the Q and q proteins. Valine and isoleucine are similar amino acids but the isoleucine in the Q protein is important for the formation of homodimers in yeast. To our knowledge, this is the first report of an AP2-like gene with the ability to form a homodimer in vitro. One might speculate that the Q homodimer may be responsible for the increased level of transcription. One or more of the conserved nucleotide differences observed in the promoter region may be critical for recognition by a Q protein homodimer complex leading to up-regulation. The q protein appears to form homodimers much less efficiently, and the q allele is transcribed at lower levels than the Q allele. This would explain the dosage effects of the q allele observed by Muramatsu (1963). Studies to determine if the Q homodimer interacts with putative regulatory elements in the promoter region, and to identify other potential interactors are under way.
It is interesting to note that AP2 in Arabidopsis is regulated at the level of translation by a microRNA, which binds to an AASSGF box (Chen 2004). The AASSGF box is conserved between Q and q genotypes, and the lack of variation would suggest that a microRNA is not responsible for governing regulation between Q and q. However, we cannot rule out the possibility of a variable microRNA existing within or near the Q gene, because some microRNAs are encoded within or very near genes (Wang et al. 2004).
It is well known that the A-genome donor of tetraploid and hexaploid wheats is T. urartu (Dvorak et al. 1993), and the D-genome donor of hexaploid wheat is Ae. tauschii (Kihara 1944; McFadden and Sears 1946). However, it is not known which AB tetraploid (qq or QQ genotype) was involved in the hybridization with Ae. tauschii (D genome) that gave rise to hexaploid wheat. And, with regard to Q, it has been a matter of speculation whether it first arose in the tetraploid progenitor of hexaploid wheat, or if it arose independently in hexaploids and tetraploids.
Our results indicate that the mutation that gave rise to the Q allele occurred only once. However, we cannot conclude whether it first arose in a tetraploid or a hexaploid. The archaeological record indicates remnants of free-threshing tetraploid and hexaploid wheats appear about the same time, and about a thousand years earlier than spelt wheats suggesting that neither Iranian spelta nor European spelta are progenitors of free-threshing hexaploid wheat (reviewed in Nesbitt and Samuel 1996; Feldman 2001). It is possible that the Q allele arose first in a Q-tetraploid similar to present day T. turgidum spp. or the extinct tetraploid naked wheat T. turgidum ssp. parvicoccum, which then hybridized with Ae. tauschii in some farmer's field to give rise to the first hexaploid. Alternatively, the Q allele may have occurred first in a hexaploid, and the present day Q-bearing tetraploids are a result of secondary hybridizations with Q-bearing hexaploids. In any case, the mutation that gave rise to the Q allele was a prominent factor in the dawn of agriculture, and it led to the rapid spread of wheat cultivation.
Acknowledgments
We thank Erik Doehler, Duane Wilson, Jacinta Kuehn, Vasu Kuraparthy, Angie Mathews, Marie Herbel, Katie Gleason, Juliane Essig, Marcy Main, and Melissa Schapaugh for technical assistance. Thanks go to J. Dvorak, University of California-Davis, for providing seed of Iranian and European spelt and their corresponding disomic substitution lines. We also thank D. Horvath, P. Gornicki, J.-M. Zhou, S. Muthukrishnan, and anonymous reviewers for critical review of the manuscript. This research was supported by USDA–ARS Current Research Information System 5442-21000-026-00D and special U.S. Department of Agriculture grant to the Wheat Genetics Resource Center at Kansas State University (to B.S.G.).
References
- Altpeter, F., V. Vasil, V. Srivastava and I. K. Vasil, 1996. Integration and expression of the high-molecular-weight glutenin subunit 1Ax1 gene into wheat. Nat. Biotech. 14: 1155–1159. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. L., D. R. Smyth and E. M. Meyerowitz, 1989. Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. L., J. Alvarez, D. Weigel, E. M. Meyerowitz and D. R. Smyth, 1993. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119: 721–743. [Google Scholar]
- Cenci, A., N. Chantret, X. Kong, Y. Gu, O. D. Anderson et al., 2003. Construction and characterization of a half million clone BAC library of durum wheat (Triticum turgidum ssp. durum). Theor. Appl. Genet. 107: 931–939. [DOI] [PubMed] [Google Scholar]
- Chen, W. P., X. Gu, G. H. Liang, S. Muthukrishnan, P. D. Chen et al., 1998. Introduction and constitutive expression of a rice chitinase gene in bread wheat using biolistic bombardment and the bar gene as a selectable marker. Theor. Appl. Genet. 97: 1296–1306. [Google Scholar]
- Chen, X., 2004. A microRNA as a translational repressor of APETALA2 in Arabidopsis. Science 303: 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., R. B. Meeley and S. Hake, 1998. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12: 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G. N., J. L. Bowman and E. M. Meyerowitz, 1991. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002. [DOI] [PubMed] [Google Scholar]
- Dvorák, J., P. di Terlizzi, H. B. Zhang and P. Resta, 1993. The evolution of polyploid wheats: identification of the A genome donor species. Genome 36: 21–31. [DOI] [PubMed] [Google Scholar]
- Endo, T. R., and B. S. Gill, 1996. The deletion stocks of common wheat. J. Hered. 87: 295–307. [Google Scholar]
- Faris, J. D., and B. S. Gill, 2002. Genomic targeting and high-resolution mapping of the domestication gene Q in wheat. Genome 45: 706–718. [DOI] [PubMed] [Google Scholar]
- Faris, J. D., K. M. Haen and B. S. Gill, 2000. Saturation mapping of a gene-rich recombination hot spot in wheat. Genetics. 154: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris, J. D., J. P. Fellers, S. A. Brooks and B. S. Gill, 2003. A bacterial artificial chromosome contig spanning the major domestication locus Q in wheat and identification of a candidate gene. Genetics 164: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M. F., 2001. Origin of cultivated wheat, pp. 1–56 in The World Wheat Book: A History of Wheat Breeding, edited by A. P. Bonjean and W. J. Angus. Lavoisier Publishing, Paris.
- Huang, L., S. A. Brooks, J. P. Fellers and B. S. Gill, 2003. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploidy genome of bread wheat. Genetics 164: 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huskins, C. L., 1946. Fatuoid, speltoid and related mutations of oats and wheat. Botan. Rev. 12: 457–514. [Google Scholar]
- Irish, V. F., and I. M. Sussex, 1990. Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2: 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantasuriyarat, C., M. I. Vales, C. J. W. Watson and O. Riera-Lizarazu, 2004. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 108: 261–273. [DOI] [PubMed] [Google Scholar]
- Jofuku, K. D., B. G. W. den Boer, M. Van Montagu and J. K. Okamuro, 1994. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6: 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, K., H. Miura and S. Sawada, 1999. QTL mapping of genes controlling ear emergence time and plant height on chromosome 5A of wheat. Theor. Appl. Genet. 98: 472–477. [Google Scholar]
- Kato, K., R. Sonokawa, H. Miura and S. Sawada, 2003. Dwarfing effect associated with the threshability gene Q on wheat chromosome 5A. Plant Breed. 122: 489–492. [Google Scholar]
- Kihara, H., 1944. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hortic. 19: 13–14. [Google Scholar]
- Komaki, M. K., K. Okada, E. Nishino and Y. Shimura, 1988. Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development 104: 195–203. [Google Scholar]
- Kuckuck, H., 1959. Neuere arbeiten zur Entstehung der hexaploiden Kulturweizen. Z. Pflanzenzucht 41: 205–226. [Google Scholar]
- Kunst, L., J. E. Klenz, J. Martinez-Zapater and G. W. Haughn, 1989. AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1: 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighty, C. E., and S. Boshnakian, 1921. Genetic behaviour of the spelt form in crosses between Triticum spelta and Triticum aestivum. J. Agric. Res. 7: 335–364. [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, M. C., Z. L. Yang and J. Dvorák, 2000. The Q locus of Iranian and European spelt wheat. Theor. Appl. Genet. 100: 602–606. [Google Scholar]
- MacKey, J., 1954. Neutron and X-ray experiments in wheat and a revision of the speltoid problem. Hereditas 40: 65–180. [Google Scholar]
- McFadden, E. S., and E. R. Sears, 1946. The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37: 81–89, 107–116. [DOI] [PubMed] [Google Scholar]
- Muramatsu, M., 1963. Dosage effect of the spelta gene q of hexaploid wheat. Genetics 48: 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu, M., 1986. The vulgare super gene, Q: its universality in durum wheat and its phenotypic effects in tetraploid and hexaploid wheats. Can. J. Genet. Cytol. 28: 30–41. [Google Scholar]
- Nesbitt, M., and D. Samuel, 1996. From staple crop to extinction? The archaeology and history of hulled wheats, pp. 41–100 in Hulled Wheats, Promoting the Conservation and Use of Underutilized and Neglected Crops 4: Proceedings of the First International Workshop on Hulled Wheats, edited by S. Padulosi, K. Hammer and J. Heller. Castelvecchio Pascoli, Tuscany, Italy.
- Podkowinski, J., J. Jelenska, A. Sirikhachornkit, E. Zuther, R. Haselkorn et al., 2003. Expression of cytosolic and plastid acetyl-coenzyme A carboxylase genes in young wheat plants. Plant Phys. 131: 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears, E. R., 1952. Misdivision of univalents in common wheat. Chromosoma 4: 535–550. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., 1954. The aneuploids of common wheat. MO Agr. Exp. Sta. Res. Bull. 572: 1–59. [Google Scholar]
- Singh, H. B., E. Anderson and B. P. Pal, 1957. Studies in the genetics of Triticum vavilovii Jackub. Agron. J. 49: 4–11. [Google Scholar]
- Singh, M. P., 1969. Some radiation induced changes at the ‘Q’ locus in bread wheat. Caryologia 22: 119–126. [Google Scholar]
- Unrau, J., W. E. Smith and R. C. McGinnis, 1950. Spike density, speltoidy and compactoidy in hexaploid wheat. Can. J. Res. C. 28: 273–276. [Google Scholar]
- Wang, J., H. Zhou, Y. Chen, Q. Luo and L. Qu, 2004. Identification of 20 microRNAs from Oriza sativa. Nucleic Acids Res. 32: 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N. D., J. D. Miller and D. L. Klindworth, 1992. Induced mutations of a genetic suppressor of resistance to wheat stem rust. Crop Sci. 32: 612–616. [Google Scholar]