Abstract
Synthetic genetic analysis was improved by eliminating leaky expression of the HIS3 reporter and gene conversion between the HIS3 reporter and his3Δ1. Leaky expression was eliminated using 3-aminotriazole and gene conversion was eliminated by using the Schizosaccharomyces pombe his5+ gene, resulting in a 5- to 10-fold improvement in the efficiency of SGA.
SYNTHETIC genetic array analysis (SGA) detects genetic interactions using the collection of deletion mutants of nonessential genes in yeast (Tong et al. 2001, 2004). We adopted SGA for use on a small scale in laboratories that do not have access to robotics. SGA performed manually had a low success rate as judged by a number of criteria. We developed a “gold standard” set of genes that were synthetic lethal with mad2 and showed that a typical SGA screen, using manual manipulations, identified <10% of the bona fide interactions. SGA efficiency improved after eliminating leaky expression of the MFA1 promoter-driven HIS3 reporter. Gene conversion between the HIS3 reporter gene and the his3Δ1 allele in strains isogenic with BY4741, BY4742, and BY4743 increased false negatives and conversion was eliminated by using the Schizosaccharomyces pombe his5+ gene.
The success of SGA depends on being able to select haploid spores after meiosis. SGA utilizes a MATα query strain that contains the mutant of interest marked by a deletion with a nourseothricin resistance marker (NAT), a fusion gene with HIS3 under the control of the MFA1 promoter and can1 (Tong et al. 2001). MFA1 is a haploid-specific gene that is expressed only in MATa cells (Michaelis and Herskowitz 1988). The query strain is mated to the collection of deletion mutants where every mutation is a deletion replaced by G418 resistance (Wach et al. 1994). Diploids are isolated and sporulated, and haploids are selected on SC–his medium containing canavanine. After meiosis, populations of spores from many meioses are transferred to the haploid selection plate; the exact number depends on the sporulation efficiency of the individual cross. The haploids are then tested on medium containing NAT and G418 to determine if double mutants are inviable or compromised for growth. We constructed a query strain with mad2∷NAT, performed SGA, and assayed for synthetic lethal interactions using manual manipulations (see supplemental material at http://www.genetics.org/supplemental/).
Three evaluation criteria indicated that our SGA screens were yielding limited success. The first was that three successive screens using the identical mad2∷NAT query strain identified <10% overlap between the screens. The second indicator was that we failed to identify mutations in the full set of genes encoding proteins of multisubunit complexes. The third indicator was that we found little linkage to MAD2 in three separate screens (Tong et al. 2001, 2004). To determine the overall efficiency of the mad2 screens, we chose 170 mutants that are annotated as affecting functions relating to microtubules, cytoskeleton, kinetochore, chromosome segregation, and cell cycle (supplemental Table 1 at http://www.genetics.org/supplemental/). We reasoned that a subset of these mutants should compromise chromosome segregation and show a synthetic phenotype in combination with mad2. The mutants were mated to mad2∷NAT and each cross was evaluated directly using random spore analysis. We identified 22 mutants that showed synthetic lethal interactions with mad2∷NAT and served as a gold standard to judge the efficiency of a given SGA screen (supplemental Table 2 at http://www.genetics.org/supplemental/).
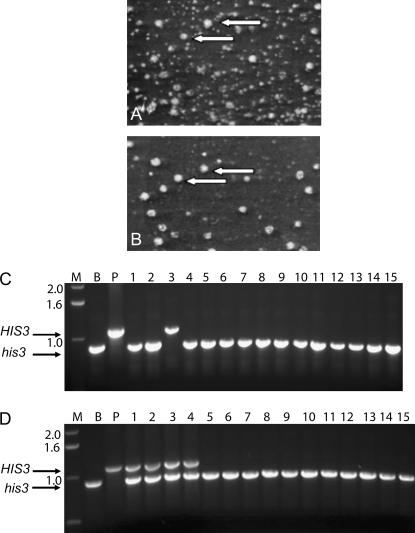
Two independent mad2 screens, performed with the entire arrays of ∼4700 mutants by manual pinning, identified only one and three of the known synthetic lethal interactions, respectively, and therefore detected 4 and 9% of the interactions. We picked 10 of the false negative colonies and in every case there were mixtures of colonies that were MATa and colonies that were sterile, suggesting they were a mixture of haploids and MATa/MATα diploids. We sporulated representative clones of the sterile colonies and the pattern of his+ segregation after meiosis suggested that there were two HIS3 genes (not shown). The diploids were arising because MATα cells were slipping through the haploid selection and then mating with the MATa cells that were properly selected. This happened because of leaky expression of His3 from PMFA1–HIS3 and by his3 reversion. The haploid query strain produced large and variable numbers of his+ revertants (Figure 1A). The small colonies were MATα and were completely eliminated by adding 2 mm 3-aminotriazole to the medium (Figure 1B). The large colonies were MATa (29/38) or MATα (9/38) and were resistant to 100 mm 3-aminotriazole. The MATa cells arise by gene conversion between HMRa and MATα. The MATα colonies arose because the his3Δ1 mutation reverted.
Figure 1.
Sources of variability in SGA. (A) MATα cells of strain Y2454 from an overnight culture grown in YPD were spread onto an SC–his plate and incubated for three days at 30°. A small region is shown. Approximately 1000 colonies of varying sizes were evident on the entire plate. (B) The same region after replica plating to a plate containing 2 mm 3-aminotriazole, which inhibits the growth of the small colonies. Arrows point to the same colonies on both plates. (C) Haploid revertants were recovered and tested for gene conversion at the his3Δ1 locus using primers flanking the gene. Lane M is molecular weight marker, lane B is genomic DNA from BY4741 (his3Δ1) as template, lane P is plasmid pRS303 (HIS3) DNA as template, and 1–15 are DNA from the 15 revertants. All of the revertants were MATa except revertant 3, which was MATα. (D) False negative colonies were recovered from SGA after using PMFA1–HIS3 in the query strain and tested for gene conversion at the his3Δ1 locus, using primers flanking the gene. Lane M is molecular weight marker, lane B is genomic DNA from BY4741 (his3Δ1) as template, lane P is plasmid pRS303 (HIS3) DNA as template, and 1–15 are DNA from the 15 revertants. All of the revertants were MATa except 1–4, which were MATa/MATα .
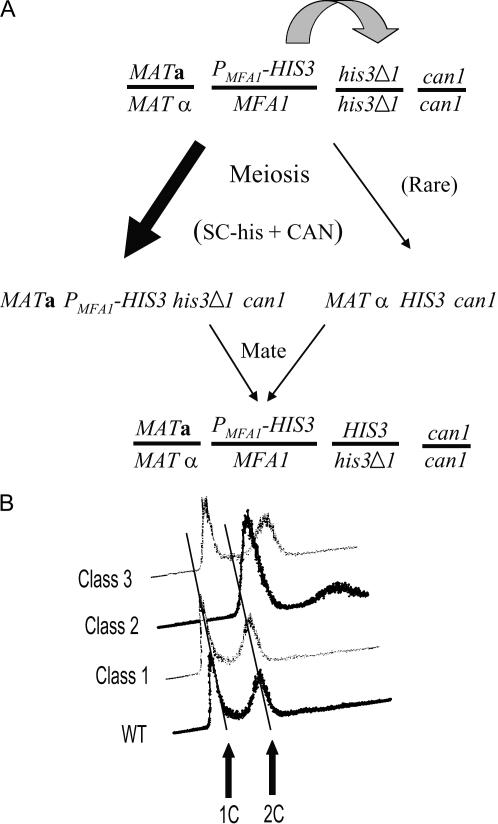
The BY4741, BY4742, and BY4743 strains have the his3Δ1 allele, a 187-bp internal deletion of the HIS3 gene. To determine if there was gene conversion of his3Δ1, we picked large colonies, extracted DNA, and performed PCR using primers that flank the HIS3 gene. There is a low frequency of ectopic mitotic gene conversion between HIS3 alleles (Figure 1C). The possibility of ectopic recombination is expected to be elevated in meiosis. It is possible that ectopic gene conversion produces MATα HIS3 spores that can mate after germination because they would germinate in the presence of a large number of MATa cells that are selected in the SGA procedure (Figure 2A). To correct the problem of gene conversion, we reconstructed the strains by substituting the S. pombe his5+ gene, which eliminated the ectopic gene conversion (data not shown).
Figure 2.
Altered chromosome transmission events in SGA. (A) Gene conversion. A diploid in meiosis can convert either of the two his3Δ1 alleles (shaded block arrow) prior to meiosis I. The vast majority of spores selected on SC–histidine containing canavanine (SC–his + CAN) after meiosis are MATa PMFA1-HIS3 his3Δ1 can1 (solid block arrow, left side). Gene conversion followed by appropriate meiotic segregation can generate rare MATα HIS3 can1 (arrow, right side). The MATα cells are surrounded by MATa cells and mate, producing diploids that escape the haploid selection and confound the SGA. (B) Flow cytometry of false negative colonies. Fifteen colonies from a bni1 × bni1 cross that mated with a MATα tester were grown to midlog, prepared for flow cytometry, and compared to BY4741. Three classes were recovered. Class 1 had an approximate haploid content of DNA, while classes 2 and 3 appeared aneuploid.
We repeated the genomewide screen to determine if including the S. pombe his5+ gene improved SGA, using our gold standard as an assay. Conventional SGA detected 9% of the “true” synthetic lethal interactions. The screen with the S. pombe his5+ gene identified 82% of the possible synthetic lethal interactions. To confirm that using PMFA1–his5+ in the query strain eliminated gene conversion we sporulated diploids derived from the mad2∷NAT query strains crossed to the cik1∷G418 strain from the haploid collection because the double mutants are synthetically lethal (supplemental Table 2 at http://www.genetics.org/supplemental/). We selected haploid spores and identified rare colonies resistant to G418 and NAT, the situation that could arise by gene conversion. We recovered rare haploids (∼10−4 of the spores) that were resistant to both drugs. When PMFA1–HIS3 was used as the query gene we recovered 4 of 15 strains that were sterile, nonmaters with copies of both his3Δ1 and HIS3 (Figure 1D). When PMFA1–his5+ was used as the query strain all of the colonies were MATa and his3Δ1 as judged by PCR (not shown). Our success with SGA was improved by eliminating gene conversion.
The improved the SGA did not completely eliminate the false negatives. The remaining false negatives (which should be MATa G418- and NAT-resistant colonies) probably arose from chromosome segregation errors in meiosis. We crossed a MATα bni1∷NAT PMFA1-HIS3 query strain to MATa bni1∷G418, sporulated the diploid, and recovered 15 rare strains that were resistant to canavanine, G418, and NAT (false positive) and mated them with a MATα tester. Given that bni1∷NAT and bni1∷G418 are alleles it should be impossible to recover haploids that contain both. We tested ploidy by two tests, flow cytometry for DNA content and pappilation to cycloheximide resistance after crossing to a cyh2 strain. The flow cytometry profiles fell into three classes (Figure 2). One class, the majority, included cells with DNA contents indistinguishable from the haploid control (12/15). The cells pappilated to cycloheximide resistance after mating to a cyh2 tester (data not shown) and therefore they had one copy of CYH2 (haploid). The second class had more than double the amount of DNA (Figure 2) and did not pappilate to cycloheximide resistance (data not shown), suggesting they were diploid or aneuploid. The final class (1/15) had a DNA content that was intermediate between haploid and diploid (Figure 2) and weakly pappilated to cycloheximide resistance (data not shown). We conclude that MATa/MATa diploids and other aneuploids can be recovered and can confound the SGA. Regardless of the origin and number of false negatives in a screen, the important consideration is that eliminating gene conversion between HIS3 alleles in the query strain and reducing leaky expression from the MFA1 promoter improves the efficiency of SGA.
Acknowledgments
We are especially thankful to the Canadian who called us (D.J.B. and D.C.A.) a “freak show” and stimulated this work. We thank Charlie Boone and Amy Tong for strains, reagents, and advice on SGA. We thank Mitch Smith, Anne Allison, and Todd Stukenberg for helpful discussions and encouragement throughout this work. We also thank members of the Burke and Stukenberg labs for helpful discussions. This work was supported by an F31 predoctoral fellowship from the National Institutes of Health (NIH) (GM65070) to J.A.D. and by RO1 grants from the NIH to D.J.B (GM40344) and D.C.A. (GM56189).
References
- Michaelis, S., and I. Herskowitz, 1988. The A-factor pheromone of Saccharomyces cerevisiae is essential for mating. Mol. Cell. Biol. 8: 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A. H. Y., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tong, A. H. Y., G. Lesage, G. D. Bader, H. Ding, H. Xu et al., 2004. Global mapping of the yeast genetic interaction network. Science 303: 808–813. [DOI] [PubMed] [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808. [DOI] [PubMed] [Google Scholar]