Abstract
Identification and characterization of the self-incompatibility genes in Brassicaceae species now allow typing of self-incompatibility haplotypes in natural populations. In this study we sampled and mapped all 88 individuals in a small population of Arabidopsis lyrata from Iceland. The self-incompatibility haplotypes at the SRK gene were typed for all the plants and some of their progeny and used to investigate the realized mating patterns in the population. The observed frequencies of haplotypes were found to change considerably from the parent generation to the offspring generation around their deterministic equilibria as determined from the known dominance relations among haplotypes. We provide direct evidence that the incompatibility system discriminates against matings among adjacent individuals. Multiple paternity is very common, causing mate availability among progeny of a single mother to be much larger than expected for single paternity.
HOMOMORPHIC self-incompatibility (SI) systems constitute invisible barriers to self-fertilization and mating among related individuals. Thus, the presence of a self-incompatibility system has a large influence on realized mating patterns in natural populations. Homomorphic SI systems are divided into gametophytic and sporophytic systems that are usually controlled by a single locus with multiple specificities (haplotypes) consisting of two (or more) genes responsible for stigma and pollen reactions. In gametophytic SI (GSI), compatibility results if the haplotype of the pollen is different from both haplotypes of the diploid stigma. When individuals share one haplotype, the reaction is compatible but only one type of pollen fertilizes; that is, an individual of genotype Si Sj can be fertilized by pollen carrying Sk from an Si Sk individual but not by pollen carrying Si. In sporophytic SI (SSI), the diploid pollen parent determines the phenotype of all pollen, and growth of the pollen is averted if this phenotype matches with the phenotype determined by the genotype of the diploid stigma. The phenotype of each sex may express either both haplotypes (codominance) or one of the haplotypes (dominance).
The strong frequency-dependent selection in both systems leads to a large number of different haplotypes maintained in populations for a very long time (Wright 1939; Schierup et al. 1997, 1998). This is because a rare haplotype has a transmission advantage since it is not denied mating opportunities by meeting its own phenotype as often as a common haplotype. At a deterministic equilibrium stabilized by selection, GSI haplotypes (and phenotypes) have equal frequencies, whereas for SSI, dominance may cause some haplotypes (the recessive ones) to have higher frequencies than other (dominant) ones (Sampson 1967; Charlesworth 1988; Schierup et al. 1997). However, more than one haplotype at a given dominance level decreases the frequency of each haplotype at that dominance level, when there is dominance in pollen and codominance in the stigma (Sampson 1974).
Mate availability for a sporophytic system, where dominance relations are the same for male and female function, can be defined as the proportion of individuals in a population that are compatible (Vekemans et al. 1998). In SSI, the mate availability is generally smaller than that in GSI. This is because matings where individuals share one haplotype usually appear compatible in GSI, even though only half the pollen can fertilize because the number of pollen in a given mating event is seldom limiting. When the number of haplotypes is small, mate availability in SSI can be very small (Byers and Meagher 1992; Vekemans et al. 1998; Glemin et al. 2005).
Recently, the main gene constituents of the S-haplotype of both the sporophytic system in Brassicaceae (Schopfer et al. 1999; Takasaki et al. 2000; Kachroo et al. 2001; Takayama et al. 2000, 2001) and the gametophytic system in Solanaceae (Lee et al. 1994; Sijacic et al. 2004) have been elucidated. In both cases, specificity is determined by the interaction of two genes underlying female and male function, respectively. Using this knowledge, it is now possible to directly study mate availability and realized mating events in natural populations by typing haplotypes.
We studied the mating patterns in an Icelandic population of Arabidopsis lyrata (Brassicaceae). A. lyrata is a perennial herb with a circumboreal distribution. It is insect pollinated, in Iceland mainly by small flies. Its sporophytic SI system was previously characterized (Charlesworth et al. 2000; Kusaba et al. 2001; Schierup et al. 2001; Mable et al. 2003, 2004), and it is orthologous to the Brassica system. A typing system that can identify most of the haplotypes has been developed (Schierup et al. 2001; Mable et al. 2003) and dominance relations between several haplotypes have been described (Schierup et al. 2001; Mable et al. 2004). Taking advantage of this, we investigate in a single population its fine-scale geographic structure, the occurrence of multiple paternity, and discrimination against matings between physically close individuals caused by restricted mate availability.
MATERIALS AND METHODS
Study population:
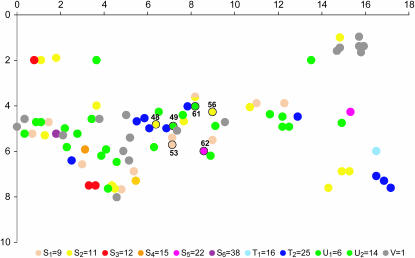
The study site (GPS coordinates 2149531, 6405485) is situated on a hill near Reykjavik, Iceland. A. lyrata grows scattered in this area with a density ≪1 individual/m2. Sampling was done August 9, 2001. An area of 30 × 50 m was searched for all individuals (88 in all), and these were located within a 10 × 20-m area central to the search area. Very few individuals occurred in a larger neighborhood of 200 × 200 m. Fresh leaf material was sampled, initially kept on ice, and frozen the same night. Seeds were sampled from all individuals with ripe seeds and kept in individual fruits. It was noted whether the individual had been flowering. The position of each plant was determined within a 10-cm grid (see Figure 1).
Figure 1.
Map of the 87 individuals sampled and typed with different colors for different haplotypes displayed in the phenotype (see inset at bottom). Axis unit is in meters. Individuals circled and numbered were used for progeny analysis.
S-haplotype typing:
DNA was extracted using a modified version of the CTAB protocol as in Bechsgaard et al. (2004). PCR was performed using three forward primers, 13seq3F, 13seq2F, and 13seq1F, and one reverse primer (SLGR) (see Schierup et al. 2001), with an annealing temperature of 50°. PCR amplification products were digested with AluI and MspI and fragments were run on a 3% agarose gel for 3 hr. From the banding patterns observed a hypothesis of the constituent SRK types was formed. Subsequently, the hypothesis was confirmed, using primers designed specifically for each SRK type (see also Mable et al. 2003, 2004 for this typing strategy and primer sequences). In each case, the procedure included negative controls (i.e., plants without the haplotype).
Investigation of progeny:
For each of six central plants (marked by their number and enclosed by a black circle in Figure 1), the seeds of three to seven fruits were germinated in small pots on the surface of a mixture of unfertilized sphagnum and fine sand (10%) and covered by punched, white plastic to ensure high humidity. All germinated seedlings were collected once the first leaves after the cotyledons appeared. Subsequently, S-haplotypes were determined as described above for the adult plants, aided by the fact that one of the haplotypes was already known from the maternal plant genotype.
Data analysis:
The S-phenotype of an individual was inferred from the genotype and from the current understanding of dominance relations among the S-haplotypes (Schierup et al. 2001; Mable et al. 2003, 2004). From the deduced phenotypes the proportion of compatible matings among the adult plants was calculated. A similar calculation was performed among progeny of a given individual, either at the fruit level or at the whole individual level. For each of the six maternal plants with typed progeny we calculated the mean distance to the possible fathers (i.e., mean distance to compatible individuals that could have provided the haplotypes received by the mother) and compared this with the mean distance to compatible fathers.
Among progeny of a given mother, the minimum number of fathers was calculated and the effective number of different haplotypes received per fruit and per plant was calculated as 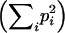 −1, where pi is the frequency of haplotype i.
−1, where pi is the frequency of haplotype i.
The proportion of compatible matings for progenies of a single fruit was calculated using all combinations of progeny. The average within-fruit compatibility for a maternal plant was weighted by the number of seeds for each fruit. The average compatibility for progeny of a maternal plant irrespective of fruit was the average of all combinations. The averages of quantities over maternal plants were weighted by the number of fruits and the number of progeny per maternal plant, respectively.
The expected equilibrium frequency of haplotypes was estimated in a model that is a combination of the SSIcod and SSIdom models of Schierup et al. (1997), using what is considered known about dominance classes (Table 1). Random mating and no pollen limitation are assumed; that is, only male sexual selection occurs (Christiansen and Prout 2000). Eleven haplotypes were observed. The six codominant haplotypes at the highest dominance level, S1, … , S6, cannot form homozygotes but form 15 heterozygotes SiSj where both haplotypes are expressed in the phenotype and 30 genotypes with haplotypes at the other dominance levels where only the Si haplotype is expressed in the phenotype. The two codominant haplotypes at the next level, T1 and T2, form one two-haplotype phenotype, T1T2, and eight one-haplotype phenotypes, of which two are homozygotes, T1T1 and T2T2 and the other six are heterozygotes with haplotypes at lower dominance levels. The two codominant haplotypes at the third level, U1 and U2, again form one two-haplotype phenotype, but only four one-haplotypes phenotypes. The recessive haplotype at the lowest dominance level, V, is expressed only in its homozygote, VV. Within a dominance level the haplotype frequencies are equal at equilibrium. The frequencies of two-haplotype phenotypes are thus equal, as are the homozygote frequencies. For instance, the frequency of T1T2 is expected to be higher than that of T1T1, which can be formed only if one of the parents is a heterozygote with T1 and a haplotype at the S-level. In the same way the one-haplotype phenotypes in a dominance class corresponding to a genotype with a particular haplotype at a lower dominance level have equal frequencies. The frequencies of S1T1 and S4T1 are equal and evidently equal to the frequency of S3T2. With these simplifications we need only to determine the frequency of the heterozygotes SiSj, SiTj, SiUj, SiV, TiTj, TiUj, TiV, UiUj, and UiV and the homozygotes TiTi, UiUi, and VV (where i ≠ j in the heterozygotes of haplotypes from the same dominance level). A total of 12 different equilibrium genotypic frequencies thus have to be determined, and these were found by numerically iterating the corresponding set of 12 recurrence equations of the model using Mathematica 5.0 (Wolfram Research, Champaign, IL).
TABLE 1.
Assumed dominance hierarchy of AlSRK haplotypes, and the corresponding type designations used in this article
| Dominance class | Haplotypes | Designation |
|---|---|---|
| Most dominant | 9, 11, 12, 15, 22, 38 | S1, S2, S3, S4, S5, S6 |
| Dominant | 16, 25 | T1, T2 |
| Least dominant | 6, 14 | U1, U2 |
| Recessive | 1 | V |
Codominance within each class and the same dominance hierarchy in males and females are assumed.
RESULTS
Eleven S-haplotypes were detected in the population. Table 1 shows the dominance relations among these assumed throughout this study. The experimental evidence for codominance within classes is not as strong as the evidence for dominance between classes, but the division of haplotypes into these four classes is consistent with all crossing evidence accumulated so far (Schierup et al. 2001; Mable et al. 2003, 2004) and with a phylogeny of all haplotypes where the four groups cluster with strong bootstrap support (Prigoda et al. 2005). The numbering of the haplotypes corresponds to previous reports (e.g., Schierup et al. 2001) but uses AlSRK as prefix in place of Aly13 previously used. Table 1 also serves as a translation of this numbering to the numbering system emphasizing the different dominance levels, S, T, U, and V. The genotypes of the adult plants are presented in Table 2 using the latter notation, since it is then immediately transparent which phenotype each individual expresses. The AlSRKxx notation is used in the text and in some Tables for easier comparison with previous investigations.
TABLE 2.
S-haplotypes identified in parental plants
| No. | Genotype | No. | Genotype | No. | Genotype | No. | Genotype |
|---|---|---|---|---|---|---|---|
| 1 | S3V | 23 | S3U2 | 45 | U2 | 67 | S1V |
| 2 | S2V | 24 | S3U2 | 46 | S2T2 | 68 | U2T2 |
| 3 | U2V | 25 | U2 | 47 | U2 | 69 | U2V |
| 4 | S6U1 | 26 | S2V | 48 | S2U2 | 70 | U2V |
| 5 | S1T2 | 27 | U2 | 49 | U2V | 71 | U2V |
| 6 | S2U2 | 28 | T2V | 50 | T2V | 72 | U3V |
| 7 | U2V | 29 | T2V | 51 | V | 73 | S2X |
| 8 | U2V | 30 | V | 52 | S1X | 74 | V |
| 9 | S1X | 31 | V | 53 | S1T2 | 75 | VX |
| 10 | U2V | 32 | — | 54 | U2V | 76 | VX |
| 11 | V | 33 | V | 55 | T2 | 77 | V |
| 12 | S2U1 | 34 | U2V | 56 | S2V | 78 | V |
| 13 | V | 35 | U2V | 57 | V | 79 | V |
| 14 | V | 36 | V | 58 | U2 | 80 | S2U2 |
| 15 | U1U2 | 37 | S1X | 59 | S1U2 | 81 | S2U2 |
| 16 | U2 | 38 | S3U2 | 60 | S1S2 | 82 | S2U2 |
| 17 | V | 39 | S1V | 61 | U1V | 83 | U2 |
| 18 | U2V | 40 | S2U1 | 62 | S3U2 | 84 | S5X |
| 19 | S1V | 41 | V | 63 | U2V | 85 | T1V |
| 20 | S3T2 | 42 | S2U2 | 64 | S1T2 | 86 | T2V |
| 21 | T2V | 43 | U2V | 65 | U2 | 87 | T2V |
| 22 | U2 | 44 | T2 | 66 | S2T2 | 88 | T2V |
Apparently, homozygotes with haplotypes of the most dominant class have an additional unknown AlSRKX haplotype or an even more dominant haplotype exists. The genotype of individual 32 is unknown due to poor DNA quality.
In 54 individuals, two haplotypes were identified, and in 33 individuals, only one haplotype was seen. These are thus either homozygotes for the haplotype found or heterozygotes with one haplotype failing to amplify using the PCR assay. Several attempts to identify a second haplotype in these individuals were made, e.g., by modifying PCR conditions and using different primers. Since homozygotes are readily formed in this system, many of the 33 individuals may indeed be homozygotes, and most of the plants with a single haplotype identified carry a recessive type. From the equilibrium frequencies, assuming no fine-scale population structure, a minimum of 13 individuals are expected to be homozygotes. We have direct evidence that the progeny of individuals 75 and 76 do not all inherit haplotype V (AlSRK01); thus these appear to be heterozygous for one or more unidentified haplotype(s) (termed ALSRKX or just X). Furthermore, 5 individuals appear to be homozygotes for the most dominant haplotype class, which indicates either a hidden haplotype or undetected dominance within the most dominant class of haplotypes. In the remaining analyses it is assumed that this limited nondetection of haplotypes does not severely bias estimation of mate availabilities.
Figure 1 shows a map of the distribution of phenotypes as determined from the detected haplotypes, assuming that individuals with a single detected haplotype are homozygous. There is some aggregation of phenotypes, leading to an increased probability of finding a different phenotype with physical distance (logistic regression of probability of compatibility as a function of distance <5 m, P = 0.03).
In Table 3, the observed frequency of each haplotype is calculated and compared with the expected equilibrium. In general, more recessive haplotypes are expected and indeed also found to be the most common ones, with haplotype V (AlSRK01) being observed slightly more (38%) than expected (37%). Haplotypes at the second most recessive level are found either in much higher frequency (haplotype U2 = AlSRK14) or in lower frequency (haplotype U1 = ALSRK06) than expected, but the total frequency of haplotypes at this level is close to that expected (Table 3). The more dominant haplotypes are expected to be quite rare, and, accordingly, they vary quite widely in relative frequency, but without being very common. From Table 2 the proportion of individuals that express a particular SI haplotype may be calculated. The different phenotypes are closer to having the same frequency than are the haplotypes, as expected in this system. Note that the expected equilibrium values of expression are not equal for different dominance levels due to selection occurring only through male function (Table 3). The equilibrium genotypic frequencies are given in Table 4 and compared to the observed frequencies pooled within genotypic classes. Eighty-seven observed genotypes are too few to give a detailed comparison, and the skewed frequencies of the haplotypes within dominance classes in themselves are expected to produce rather large deviations between observed frequencies and the equilibrium frequencies. The three homozygote classes (TiTi, UiUi, and VV), however, are all observed in excess. This could suggest inbreeding, but the frequency of the recessive haplotype V (AlSRK01) is fairly close to the equilibrium value. The large excesses in the two intermediate dominance classes consist only of observed homozygotes of the frequent haplotypes U2 and T2 (AlSRK14 and AlSRK25).
TABLE 3.
Observed frequency of haplotypes among 87 parents and expression of haplotypes deduced from the dominance relations of Table 1
| Haplotype (AlSRK) | Parent haplotype frequency | Equilibrium haplotype frequency | Observed expression | Equilibrium expression | |
|---|---|---|---|---|---|
| Recessive | V (1) | 0.377 | 0.370 | 0.188 | 0.128 |
| Least dominant | U1 (6) | 0.030 | 0.102 | 0.038 | 0.125 |
| U2 (14) | 0.275 | 0.102 | 0.312 | 0.125 | |
| Total | 0.305 | 0.204 | 0.350 | 0.250 | |
| Dominant | T1 (16) | 0.006 | 0.077 | 0.013 | 0.135 |
| T2 (25) | 0.108 | 0.077 | 0.125 | 0.135 | |
| Total | 0.114 | 0.154 | 0.138 | 0.270 | |
| Most dominant | S1 (9) | 0.067 | 0.045 | 0.113 | 0.108 |
| S2 (11) | 0.090 | 0.045 | 0.163 | 0.108 | |
| S3 (12) | 0.018 | 0.045 | 0.034 | 0.108 | |
| S4 (15) | 0.012 | 0.045 | 0.022 | 0.108 | |
| S5 (22) | 0.012 | 0.045 | 0.022 | 0.108 | |
| S6 (38) | 0.006 | 0.045 | 0.011 | 0.108 | |
| Total | 0.205 | 0.270 | 0.365 | 0.648 |
The sum of haplotype expressions exceeds 1 because a given individual can display two haplotypes. The equilibrium values within a dominance class are compared with the corresponding observed averages. The haplotype with unknown dominance (X in Table 2) is omitted.
TABLE 4.
Genotypic frequencies at equilibrium and observed in the population (×1000)
| Class | SiSj | SiTj | SiUj | SiV | T1T2 | TiTi | TiUj | TiV | U1U2 | UiUi | UiV | VV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Expected | 62 | 89 | 117 | 213 | 24 | 6 | 65 | 119 | 41 | 16 | 153 | 128 |
| Observed | 12 | 75 | 175 | 88 | 0 | 25 | 12 | 100 | 12 | 113 | 200 | 188 |
Class, classes of genotypes with equal frequencies; Expected, the total expected frequency of the genotypes in the class; Observed, the observed frequency of all genotypes in the class.
The genotypes of the 87 plants may also be used to calculate the mean probability that a given haplotype will encounter a compatible mating if two individuals in the population are chosen at random. This is called the haplotype mate availability and is the main determinant of whether a given haplotype will increase or decrease through the next round of breeding by comparing it to the population-wide mate availability, which is 83%. This was partially investigated by the typing of some progeny.
A total of 227 progeny were typed from six central individuals in the population, marked in Figure 1, and termed maternal seed families. Their genotypes are found in supplemental Table S1 (http://www.genetics.org/supplemental/). Two haplotypes were found in 165 progeny and one haplotype in 62 progeny. This yields a proportion of homozygosity/nondetection of 27.5%, which is similar to what was observed for the parental generation. From these genotypes the maternal contributions were estimated. The offspring provide no information when their genotype corresponds to the maternal individual. Among informative offspring the segregation of the female haplotypes does not deviate significantly from Mendelian expectations in any of the families (total χ2(6) = 7.07) (P = 0.31).
The progeny genotypes also reveal the paternal contributions to the informative offspring. The uninformative offspring nevertheless witness that one of the maternal haplotypes was transmitted through pollen. A further uncertainty arises if the individual shows just one haplotype; thus the paternal parent has contributed either the same haplotype or a haplotype not identified. We assume, as in the analysis of the parental generation, that nondetection of haplotypes is sufficiently rare that it does not affect estimates significantly. Table 5 summarizes the pollen contributions to fruits and entire plants. This can be converted to a minimum number of fathers and the effective number of different haplotypes received, averaged over single fruits and combined over all fruits for a given mother (Table 5). Multiple paternity of single fruits is common, but pollen fertilizing a single fruit is more related than pollen fertilizing different fruits of the same mother plant. This is reflected in the differences in effective number of haplotypes received per fruit and per plant. This number is expected to be less sensitive than the pollen haplotype number to the relatively larger sample size for the whole plant as compared with the sample size of the fruit, and, consistently, it is higher (28% on average).
TABLE 5.
The number of haplotypes and the effective number of haplotypes (Ne) received by six maternal plants, shown separately for each fruit and for the plant as a whole
| Mother | Offspring/fruit | Haplotypes/fruit | Ne/fruit | Haplotypes/mother | Ne/mother |
|---|---|---|---|---|---|
| 48 | 7 | 2 | 1.96 | 6 | 2.73 |
| 3 | 2 | 1.8 | |||
| 4 | 3 | 1.8 | |||
| 4 | 4 | 2 | |||
| 5 | 3 | 2.27 | |||
| 5 | 2 | 1.92 | |||
| 49 | 12 | 4 | 2.07 | 5 | 3.66 |
| 6 | 4 | 3 | |||
| 8 | 2 | 1.88 | |||
| 1 | 1 | 1 | |||
| 8 | 4 | 2.91 | |||
| 5 | 4 | 3.57 | |||
| 1 | 1 | 1 | |||
| 53 | 5 | 2 | 1.92 | 6 | 3.00 |
| 5 | 2 | 1.47 | |||
| 14 | 1 | 1 | |||
| 4 | 2 | 1.6 | |||
| 2 | 2 | 2 | |||
| 10 | 4 | 3.85 | |||
| 56 | 6 | 2 | 1.38 | 5 | 3.72 |
| 7 | 3 | 1.81 | |||
| 7 | 4 | 3.77 | |||
| 3 | 2 | 1.8 | |||
| 1 | 1 | 1 | |||
| 3 | 3 | 3 | |||
| 3 | 2 | 1.8 | |||
| 61 | 5 | 2 | 1.92 | 6 | 3.71 |
| 9 | 4 | 3.86 | |||
| 3 | 2 | 1.8 | |||
| 62 | 2 | 1 | 1 | 6 | 2.49 |
| 16 | 3 | 1.66 | |||
| 2 | 2 | 2 | |||
| 4 | 3 | 1.78 | |||
| 3 | 3 | 3 |
The frequency of haplotypes in the next generation can be deduced from the haplotypes received through pollen (not a completely unrealistic assumption since a relatively large proportion of all seeds were germinated). Interestingly, haplotypes that have a mate availability smaller than the population average are less common among successful pollen than predicted by their frequency in the paternal generation (viz. haplotypes V and U2, AlSRK01 and AlSRK14) whereas some haplotypes that have mate availabilities larger than the population average (viz. haplotypes S2 and T2, AlSRK11 and AlSRK25) have increased frequencies among successful pollen (see Table 6). The deviations from equilibrium therefore may not be larger than expected from sampling effects, caused by individuals in the parental generation contributing with a large reproductive variance to the next generation. Preliminary simulation results of the dynamics of the system are consistent with these results (M. H. Schierup and J. S. Bechsgaard, unpublished results).
TABLE 6.
Frequencies of haplotypes in parental and progeny generations
| Haplotype (AlSRK) | Parent haplotype frequency | Haplotype mate availability | Progeny haplotype frequency | Difference in haplotype frequency between generations |
|---|---|---|---|---|
| V (1) | 0.378 | 0.816 | 0.321 | −0.057 |
| U1 (6) | 0.029 | 0.989 | 0.014 | −0.015 |
| U2 (14) | 0.256 | 0.736 | 0.138 | −0.118 |
| T1 (16) | 0.006 | 1.000 | 0.005 | −0.001 |
| T2 (25) | 0.105 | 0.874 | 0.183 | 0.079 |
| S1 (9) | 0.081 | 0.897 | 0.083 | 0.001 |
| S2 (11) | 0.093 | 0.862 | 0.225 | 0.132 |
| S3 (12) | 0.017 | 0.977 | 0.009 | −0.008 |
| S4 (15) | 0.012 | 0.989 | 0.009 | −0.002 |
| S5 (22) | 0.017 | 0.989 | 0.014 | −0.004 |
| S6 (38) | 0.006 | 1.000 | 0.000 | −0.011 |
Seeds of A. lyrata have no specialized dispersal mechanism, and seeds may not spread very far. The mate availabilities within a maternal seed family might therefore affect the mate availability in a given population neighborhood in the next generation and explain why, as observed in the parental generation, mate availability increases with distance between individuals. Mate availability among progeny from the same maternal plant and from different fruits from the same maternal plants is shown in Table 7. For each maternal plant compatibility within fruits is less than that among fruits (an average of 62% of all matings between seeds from the same fruit are compatible, whereas 66% of all matings between seeds from the same maternal plant are compatible). The relatedness of seeds from a seed family results, as expected, in a lower mate availability than the population average (83%), but the multiple paternities within and between fruits of the maternal plants carry the compatibility within seed families closer to the population average.
TABLE 7.
Mate availabilities among progeny from the six mother plants at the fruit and total individual level
| Mother | MA for whole plant | Mean MA for fruits | Mean distance to compatible father | Mean distance to compatible father providing haplotype | Difference |
|---|---|---|---|---|---|
| 48 | 0.72 | 0.64 | 4.42 | 4.47 | −0.05 |
| 49 | 0.78 | 0.75 | 4.31 | 4.08 | 0.23 |
| 53 | 0.62 | 0.60 | 4.76 | 4.75 | 0.01 |
| 56 | 0.60 | 0.56 | 5.13 | 4.72 | 0.41 |
| 61 | 0.82 | 0.79 | 4.96 | 4.43 | 0.53 |
| 62 | 0.49 | 0.44 | 5.13 | 5.16 | −0.03 |
Also shown is the mean distance to fathers compatible with the maternal plant as well as the mean distance to the (compatible) fathers that could have contributed the observed paternal haplotypes, as well as their difference.
Table 7 also shows the average distances to the possible fathers of a given maternal plant, given the transmitted haplotypes and requiring the paternal plant to be compatible with the maternal plant (under the assumptions of Table 1). We calculated the average distance to all compatible fathers in the same way. This analysis was restricted to the flowering individuals only. There is a weak, but not significant, tendency that pollen has arrived from closer than average compatible fathers.
DISCUSSION
Eleven self-incompatibility haplotypes were identified in the population, but we found evidence of one more (designated X in the Table 2 legend). In this analysis we neglected this observation to focus on the haplotypes with known dominance relations. To probe the significance of this assumption we investigated the effect of more haplotypes on the deterministic equilibrium frequencies of the system and concluded that little effect of unknown haplotypes is expected. Table 8 shows some of the results, namely those where an additional haplotype is assumed in each of the dominance classes except for the most recessive one. The exercise hardly changes the frequency of a haplotype from the most dominant class; thus addition to this class results in a higher total frequency of the class. The effects on the other dominant classes are very similar. The only peculiar result is that addition of an extra haplotype to the dominant class lowers both the haplotype frequencies and the frequency of the class. We are pursuing a more thorough analysis of the dynamics of these sporophytic models with codominance among haplotypes at multiple dominance levels.
TABLE 8.
Haplotype (class) frequencies at equilibrium for different numbers of haplotypes at the four dominance levels (most dominant first)
| No. of haplotypes | S | T | U | V |
|---|---|---|---|---|
| 6221 | 0.045 (0.271) | 0.077 (0.155) | 0.102 (0.204) | 0.370 |
| 6231 | 0.043 (0.255) | 0.070 (0.140) | 0.080 (0.241) | 0.363 |
| 6321 | 0.042 (0.253) | 0.064 (0.129) | 0.099 (0.197) | 0.357 |
| 7221 | 0.042 (0.291) | 0.075 (0.151) | 0.099 (0.198) | 0.360 |
The first row shows the number of haplotypes observed in this study, and the other rows show the effect of addition of a single haplotype to each of the dominant classes.
This study is the first direct investigation of the influence of a self-incompatibility system on mating patterns in a natural population. In the parental generation, stochastic and deterministic forces may cause mate availability for the different haplotypes to deviate from equality. Stochastic factors are due to genetic drift in a subdivided population with limited dispersal and large reproductive variance. Deterministic effects are selection associated with haplotypes that are not inherent to incompatibility between individual plants. Such selection was recently demonstrated in a greenhouse population of A. lyrata originating ∼100 km from the present population (Bechsgaard et al. 2004). In controlled crosses, some haplotypes had a transmission advantage, as deduced from significant segregation distortion. This would lead to a higher frequency of the preferred haplotypes, leading to frequencies higher than those predicted at equilibrium. In Bechsgaard et al. (2004), haplotypes AlSRK01 (V) and AlSRK09 (S1) were found to be preferentially transmitted in some crosses. The present results suggest that haplotype AlSRK01 has a lower than average mate availability in the parental generation. This is not the case for haplotype AlSRK09, but note the large increase in frequency of this haplotype from the paternal generation to the progeny generation.
Self-incompatibility phenotypes were found to be slightly aggregated as evidenced by a weak, but significant, increase of mate availability with physical distance. This is consistent with limited dispersal of seeds in accordance with the seeds not having any specific dispersal mechanisms and being sufficiently heavy that they are not easily taken by the wind. The mate availability in an open-pollinated seed family is therefore of interest, since this is the main determinant of the local mate availability. The SI haplotype typing of seeds of six selected mother plants revealed that multiple paternity is common both at the level of the fruit and, more pronounced, at the level of the maternal plant. This is similar to the findings of Ellstrand (1984) in Raphanus sativus, another self-incompatible member of the Brassicaceae. The average mate availability for seeds from the same fruit is 62%, for seeds from the same plant is 66%, and for the complete population is 83%. Multiple paternity is thus important in ensuring larger mate availability at the very local level.
The fathers siring seeds appear to be somewhat closer to the maternal plant than compatible fathers in the population overall, suggesting that pollen flow is restricted even at this very short range. This is so even though the increasing mate availability with distance tends to increase the pollen dispersal distance and therefore, had A. lyrata not been self-incompatible, gene flow would probably have been even more restricted.
Acknowledgments
We thank Deborah Charlesworth and Xavier Vekemans for comments and discussions throughout this study, an anonymous reviewer for many very useful comments, staff at Paaskehojgaard experimental station for excellent facilities, Camilla Haakonsson for excellent laboratory work, and Enette B. Knudsen for editing. This study was supported by grants 00001262 from the Danish Natural Sciences Research Council and by grant no. 2052-01-0032 from the Danish Agricultural Sciences Research Council to M.H.S.
References
- Bechsgaard, J., T. Bataillon and M. H. Schierup, 2004. Uneven segregation of sporophytic self-incompatibility alleles in Arabidopsis lyrata. J. Evol. Biol. 17: 554–561. [DOI] [PubMed] [Google Scholar]
- Byers, D. L., and T. R. Meagher, 1992. Mate availability in small populations of plant-species with homomorphic sporophytic self-incompatibility. Heredity 68: 353–359. [Google Scholar]
- Charlesworth, D., 1988. Evolution of homomorphic sporophytic self-incompatibility. Heredity 60: 445–453. [Google Scholar]
- Charlesworth, D., P. Awadalla, B. K. Mable and M. H. Schierup, 2000. Population-level studies of multiallelic self-incompatibility loci, with particular reference to Brassicaceae. Ann. Bot. 85: 227–239. [Google Scholar]
- Christiansen, F. B., and T. Prout, 2000. Aspects of fitness, pp. 146–156 in Evolutionary Genetics From Molecules to Morphology, edited by R. S. Singh and C. B. Krimbas. Cambridge University Press, New York.
- Ellstrand, N. C., 1984. Multiple paternity within the fruits of the wild radish, Raphanus sativus. Am. Nat. 123: 819–828. [Google Scholar]
- Glemin, S., T. Gaude, M. L. Guillemin, M. Lourmas, I. Olivieri et al., 2005. Balancing selection in the wild: testing population genetics theory of self-incompatibility in the rare species Brassica insularis. Genetics 171: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., C. R. Schopfer, M. E. Nasrallah and J. B. Nasrallah, 2001. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293: 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lee, H. S., S. S. Huang and T. H. Kao, 1994. S-proteins control rejection of incompatible pollen in Petunia inflata. Nature 367: 560–563. [DOI] [PubMed] [Google Scholar]
- Mable, B. K., M. H. Schierup and D. Charlesworth, 2003. Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity 90: 422–431. [DOI] [PubMed] [Google Scholar]
- Mable, B. K., J. Beland and C. Di Berardo, 2004. Inheritance and dominance of self-incompatibility alleles in polyploid Arabidopsis lyrata. Heredity 93: 476–486. [DOI] [PubMed] [Google Scholar]
- Prigoda, N. L., A. Nassuth and B. K. Mable, 2005. Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol. Biol. Evol. 22: 1609–1620. [DOI] [PubMed] [Google Scholar]
- Sampson, D. R., 1967. Frequency and distribution of self-incompatibility alleles in Raphanus raphanistrum. Genetics 56: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, D. R., 1974. Equilibrium frequencies of sporophytic self-incompatibility alleles. Can. J. Genet. Cytol. 16: 611–618. [Google Scholar]
- Schierup, M. H., X. Vekemans and F. B. Christiansen, 1997. Evolutionary dynamics of sporophytic self-incompatibility alleles in plants. Genetics 147: 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., X. Vekemans and F. B. Christiansen, 1998. Allelic genealogies in sporophytic self-incompatibility systems in plants. Genetics 150: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer, C. R., M. E. Nasrallah and J. B. Nasrallah, 1999. The male determinant of self-incompatibility in Brassica. Science 286: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Sijacic, P., X. Wang, A. L. Skirpan, Y. Wang, P. E. Dowd et al., 2004. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature 429: 302–305. [DOI] [PubMed] [Google Scholar]
- Takasaki, T., K. Hatakeyama, G. Suzuki, M. Watanabe, A. Isogai et al., 2000. The S receptor kinase determines self-incompatibility in Brassica stigma. Nature 403: 913–916. [DOI] [PubMed] [Google Scholar]
- Takayama, S., H. Shiba, M. Iwano, H. Shimosato, F. S. Che et al., 2000. The pollen determinant of self-incompatibility in Brassica campestris. Proc. Natl. Acad. Sci. USA 97: 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S., H. Shimosato, H. Shiba, M. Funato, F. S. Che et al., 2001. Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413: 534–538. [DOI] [PubMed] [Google Scholar]
- Vekemans, X., M. H. Schierup and F. B. Christiansen, 1998. Mate availability and fecundity selection in multi-allelic self-incompatibility systems in plants. Evolution 52: 19–29. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1939. The distribution of self-sterility alleles in populations. Genetics 24: 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]