Abstract
Marine mussels of the genus Mytilus have an unusual mode of mitochondrial DNA (mtDNA) transmission termed doubly uniparental inheritance (DUI). Female mussels are homoplasmic for the F mitotype, which is inherited maternally, while males are usually heteroplasmic, carrying a mixture of the maternal F mitotype and the paternally inherited M genome. Two classes of M genomes have been observed: “standard” M genomes and “recently masculinized” M genomes. The latter are more similar to F genomes at the sequence level but are transmitted paternally like standard M genomes. In this study we report the complete sequences of two standard male M. edulis and one recently masculinized male M. trossulus mitochondrial genome. A comparative analysis, including the previously sequenced M. edulis F and M. galloprovincialis F and M mtDNAs, reveals that these genomes are identical in gene order, but highly divergent in nucleotide and amino acid sequence. The large amount (>20%) of nucleotide substitutions that fall in coding regions implies that there are several amino acid replacements between the F and M genomes, which likely have an impact on the structural and functional properties of the mitochondrial proteome. Correlation of the divergence rate of different protein-coding genes indicates that mtDNA-encoded proteins of the M genome are still under selective constraints, although less highly than genes of the F genome. The mosaic F/M control region of the masculinized F genome provides evidence for lineage-specific sequences that may be responsible for the different mode of transmission genetics. This analysis shows the value of comparative genomics to better understand the mechanisms of maintenance and segregation of mtDNA sequence variants in mytilid mussels.
SEVERAL species of mollusk bivalves in the families Mytilidae, Unionidae, and Veneridae have an unusual mode of mitochondrial DNA (mtDNA) transmission termed doubly uniparental inheritance (DUI) (Zouros et al. 1992, 1994; Skibinski et al. 1994a,b; Hoeh et al. 1996; Passamonti and Scali 2001; Serb and Lydeard 2003). Instead of transmitting mtDNA exclusively through the female line as seen in most animals (uniparental inheritance), these species are characterized by the presence of two highly divergent mtDNAs: one that is inherited maternally (the F genome) and a second that is inherited paternally (the M genome). In the best-studied family Mytilidae, typical females are homoplasmic for the F mitotype throughout the entire animal, whereas males are heteroplasmic with varying mixtures of the maternal and paternal mtDNA in all tissues except the gonad, which contains almost exclusively the M-type mtDNA (Hoeh et al. 1991; Zouros et al. 1994; Stewart et al. 1995; Garrido-Ramos et al. 1998). The factors that segregate M and F mtDNAs in different tissues of heteroplasmic mussels as well as the physiological significance of this segregation are not yet understood. It was proposed that this tissue-specific segregation is a complex interaction between maternal and paternal factors of eggs and sperm (Saavedra et al. 1997; Garrido-Ramos et al. 1998). A recent report demonstrates sex-specific behavior of sperm mitochondria following fertilization (Cao et al. 2004a). In embryos from females that produce mostly sons, the sperm mitochondria tended to aggregate in a single blastomere, which is thought to go on to produce the male germline. In contrast, in embryos from females that produce only daughters, sperm mitochondria were randomly dispersed among blastomeres. This mechanism might explain the predominance but not the exclusive presence of the paternal genome in male gonads (Cao et al. 2004a,b).
The nonrandom segregation of F and M variants in heteroplasmic male mussels could also be modulated at the genome level (e.g., replication of the M-type being favored in the male gonad and that of the F-type favored elsewhere). This view draws support from the observed masculinization of F-type mtDNA. Specifically, F mtDNA seems to occasionally invade the male route of inheritance, thereby resetting the degree of sequence divergence between M and F genomes to zero (Hoeh et al. 1997; Zouros 2000). Male mussels that lack a typical M molecule thus appear to have inherited mtDNA only from their mother, and they appear to carry two F-type molecules. Such males are referred to as “recently masculinized males.” These “ancient F” or “newly masculinized” molecules behave as M mtDNA and are transmitted from generation to generation only through sperm (Hoeh et al. 1997; Saavedra et al. 1997; Zouros 2000). It has been suggested that before an F molecule can become “masculinized” (i.e., reverse its role and become transmitted paternally), it must first acquire one or more key genes from a standard M mtDNA genome (Zouros 2000). This hypothesis is consistent with the observation of mtDNA recombination between F and M molecules in mytilid mussels (Burzynski et al. 2003).
Little was known about the mtDNA genome of bivalves before the discovery of DUI. The first sequence of a nearly complete Mytilus edulis mitochondrial (mt) genome, which was published in 1992 (Hoffmann et al. 1992), was later identified as an F-type mtDNA (Skibinski et al. 1994b). (The sequence of this genome was recently completed and reported by Boore et al. 2004.) Interestingly, the gene arrangement of the M. edulis F mtDNA is considerably different from that found in any other known metazoan mtDNA. It contains an extra tRNA (trnM) and lacks the atp8 gene, a condition that is known to occur in other bivalves, including Venerupis (M. Okazaki and R. Ueshima, personal communication) and Crassostrea (S.-H. Kim, E.-Y. Je and D.-W. Park, personal communication). Recently, the complete maternal and paternal mitochondrial genomes of the Mediterranean mussel M. galloprovincialis have been reported (Mizi et al. 2005). Despite a high degree of divergence in nucleotide sequence (∼20%), both genomes retained identical gene content, gene arrangement, nucleotide composition, and codon usage bias (Mizi et al. 2005). Still, these data do not provide much insight into the role of recombination in the process of masculinization. In fact, to gain insight into DUI, we also require sequence information from recently masculinized mtDNA.
More generally, Mytilus mussels provide an excellent model for studying the evolutionary forces that operate on the mitochondrial genome, given that different levels of divergence can be studied (i.e., within and between intra- and interspecific F and M genomes). Because of the intimate interactions of nuclear and organelle genomes (Blier et al. 2001), one can predict significant correlations in the rates of amino acid substitutions between M and F mtDNAs at positions of structural or catalytic importance to enzyme function. At the same time, because the M and F mitochondrial genomes must function in different cellular environments with different physiological demands, distinct selective pressures may be operating on these molecules (Ballard and Dean 2001; Dalziel and Stewart 2002). One could argue that more similar mtDNA regions in the standard/recently masculinized male comparisons vs. female/recently masculinized male comparisons might suggest molecular adaptations over time to the functional role of male-transmitted mitochondria.
In this study, whole mtDNA genome sequencing has been undertaken on two M. edulis standard male mitotypes and one recently masculinized M. trossulus male mitotype, which have been compared with those of the published complete mtDNA sequences from other mytilid species. By a comparative genomic approach we have identified (i) the distribution of mutational differences across these genomes, (ii) evolutionary patterns from M and F genomes having similar or different nuclear backgrounds, (iii) regions that may have undergone specific adaptations as a consequence of masculinization, and (iv) gender-specific regions that may be responsible for the different transmission genetics of mtDNA. On the basis of these comparisons, we discuss mitochondrial genome diversity and evolution within mytilid mussels.
MATERIALS AND METHODS
Collection, genotyping, and mitotyping of specimens:
Mytilus mussels were collected from Chester Basin, Nova Scotia, throughout the summer of 2002. Mussels were transported on ice and housed in flowing, filtered saltwater tanks at Acadia University's Animal Care Facility. Sex was determined by microscopic examination of the gonads and only males with mature gonads were utilized for further study.
Total DNA was extracted from male mussel gonads using a modified saturated-salt extraction procedure (Miller et al. 1988; Sutherland et al. 1998). Gonad tissue was used because it consistently contains and expresses the male mitochondrial genome, whereas its expression is detected in only 50% of somatic tissues of males (Garrido-Ramos et al. 1998; Dalziel and Stewart 2002). An Ultrospec 3100 pro UV/visible spectrophotometer (Amersham Pharmacia Biotech) was used to quantify the extracted DNA, which was then stored at 4°.
Males were classified as M. edulis, M. trossulus, or hybrid, using a PCR-based assay, which targets a section of the internal transcribed spacer region between the 18S and 28S nuclear rRNA coding regions (Heath et al. 1995). After digestion with the restriction enzyme HhaI, the amplification products from M. edulis produced bands of 450 and 180 bp in length along with many smaller fragments, whereas those from M. trossulus produced bands of 280 and 180 bp as well as several smaller fragments. Hybrids possessed both bands at 450 and 280 bp. This assay appears to be highly diagnostic for these species in Nova Scotia (Garrido-Ramos et al. 1998). Only male individuals identified as “pure” species were used for subsequent analyses.
The detection of a standard male mitochondrial genome (M) in M. edulis samples was done using two levels of testing. The first involved PCR amplification of a region of the mitochondrial cytochrome oxidase III (cox3) gene using primers designed to be specific to the M mtDNA (Stewart et al. 1995; Sutherland et al. 1998). These primers correspond to the nucleotide positions 207–234 and 710–737 of the cox3 gene and amplify a 530-bp fragment only from the main M mitotype of M. edulis (M-ed 1; Stewart et al. 1995). The second level of testing was done using the primers designed by Zouros et al. (1994). This pair of primers, which corresponds to nucleotide positions 460–482 and 1326–1301 of segment 5 of the M. edulis female genome in Hoffmann et al. (1992), amplifies an 860-bp fragment of the cox3 gene from both female and male mitochondrial genomes (Zouros et al. 1994; Saavedra et al. 1997). The PCR products were then digested using HinfI: standard males produced bands of 600 and 250 bp in length, whereas those from recently masculinized males (M-A) produced bands of 375 and 275 bp with some smaller fragments. Female types (F) could also be distinguished as they produced bands of 450 and 275 bp as well as some smaller fragments. Two standard males were used for further study.
For M. trossulus samples, gender identification was performed by PCR amplification of a 700-bp fragment of the mitochondrial cytochrome oxidase subunit I (cox1) gene using the primers LCO-22MF (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO-700ME (5′-TCAGGGTGACCAAAAAATCA-3′) (J. M. Archibald and D. T. Stewart, unpublished data). PCR products were subsequently digested with AluI: standard males produced a pattern with a large fragment (∼450 bp) and several smaller fragments, whereas recently masculinized males (M-O) produced several fragments <250 bp. Female types showed patterns different from both of the M types (J. M. Archibald and D. T. Stewart, unpublished data). One recently masculinized male was used for further study.
mtDNA amplification and sequencing protocols:
The entire male M. edulis and the recently masculinized M. trossulus mitochondrial genome were amplified using species- and gender-specific primers pairs listed in Table 1. As previously reported (Garrido-Ramos et al. 1998; Dalziel and Stewart 2002), both male and female mitochondrial genomes can be present and expressed in male gonads, making difficult the direct sequencing of PCR products. To overcome this difficulty, a DNA extraction from a M. edulis female gonad, which is known to contain only the F genome, served as a control for the primers designed to amplify the standard male genome from the same species. For M. trossulus samples, DNA extractions were tested with male and female primers (designed from F, M, and M-O GenBank sequences), and individuals that were indicative of homoplasmy for either male type or female type were used for further study. Samples that tested positive for F and negative for M mtDNA were classified as recently masculinized types because they were isolated from male gonads (Saavedra et al. 1997).
TABLE 1.
Primers used in the amplification of the entire mitochondrial genome of mytilid mussels
| Region | Primer name | Oligonucleotide pair (5′–3′) | Source used for primer design |
|---|---|---|---|
| 16S-cob | em16SF | GAAGCCTCTAAGGGTAGGTCTGTTC | (AY350784–92)a |
| 2.5 kb | emUR | CTTATCAGCTCACTTGCGTACACC | (U50214)b |
| u-cox3 | emUF | GGTGTACGCAAGTGAGCTGATAAG | (U50214)b |
| 4.5 kb | emCOIIIR | CCTCGAAACCAAAATGGCGTTGGC | (AY130186–99)c |
| cox3-cox1 | emCOIIIF | GACACTTTGTGGATGTAGTATGAG | (AY130186–99)c |
| 3 kb | emCOIR | CCTACTAGAGACCTTACTCCAGCTA | (AY101397–1429)c |
| cox1-16S | emCOIF | CTGGATGAACTGTCTACCCTCCACT | (AY101397–1429)c |
| 6.8 kb | em16SR | GAACAGACCTACCCTTAGAGGCTT | (AY350784–92)a |
| 16S-cob | trm16SF | GAAGGACGACAAGACCCTATGAAG | (AF023591–99)d |
| 2.5 kb | trmUR | CATTTCAGGTCTACAGGACGACTCC | (U50213)b |
| u-cox3 | trmUF | GGAGTCGTCCTGTAGACCTGAAATG | (U50213)b |
| 4.5 kb | trmCOIIIR | CGACTACGTGCATTCCATGAAACCC | (AY130177–85)c |
| cox3-cox1 | trmCOIIIF | GTGTTCTTCCTAGTGCAACTTCGAG | (AY130177–85)c |
| 3 kb | trmCOIR | GGTCCTAGCAAAGTTAATAGCCCC | (U68773–77)e |
| cox1-16S | trmCOIF | GAAAGGAGAACGAGCGGAGCTTTA | (U68773–77)e |
| 8.7 kb | trm16SR | CCAACATCGAGGTCGCAATCTTCC | (AF023591–99)d |
em, M. edulis standard male mitochondrial genome; trm, M. trossulus recently masculinized mitochondrial genome; u, segment of the mtDNA genome (located between the cytochrome b and cytochrome c oxidase subunit II genes) for which no function has yet been assigned (Hoffmann et al. 1992).
Cao et al. (2004b).
PCR amplifications were performed in 50 μl of a solution containing ∼20 ng of total DNA, 0.4 μm of each primer, 1× Expand high-fidelity DNA polymerase buffer, 1.5 mm MgCl2, 200 μm of each dNTP, and 2.5 units Expand high-fidelity DNA polymerase (Roche Applied Science). The thermal cycling parameters were as follows: 1 min at 94° and 2 min at 88°, after which the Taq DNA polymerase was added, and then 35 cycles of 94°, 15 sec; 55°, 20 sec; and 72°, 2.30–6.30 min, depending on fragment size, followed by a termination step of 10 min at 72°. All PCR reactions (except one that yielded two bands, one stronger and of the expected size and the other weaker and of smaller size) produced a single band when visualized under UV light following ethidium bromide staining on a 0.8% agarose gel. The resulting fragments were purified either with Microcon concentrators (Millipore, Bedford, MA) or with QIAquick gel extraction kit (QIAGEN, Chatsworth, CA) in the case of the two-band product. These purified DNAs were used as template in Big Dye terminator cycle sequencing reactions according to the manufacturer's protocols (Applied Biosystem, Foster City, CA). Unincorporated dye terminator and nucleotides were removed by gel filtration (Millipore) according to the manufacturer's instructions. Readings were obtained by automated sequencing on a BaseStation DNA fragment analyzer (MJ Research, San Francisco). Sequencing of the three mtDNA molecules was completed by using the primer walking method with specific primers designed for the sequences obtained. One strand was sequenced, with several overlaps of sequences from both directions. A total of 2–14 measures were taken to assess the reliability of the sequence data, except for a few nonambiguous regions (<250 bp).
Data analysis:
Sequence readings were assembled using the XGAP package V3.6 1996 (Bonfield et al. 1995). Sequence analyses were performed on SUN workstations using software developed by Lang and Burger (1986), as well as with tools included in the Staden (1996) sequence analysis package. The BLAST network service (Altschul et al. 1990) was employed for similarity searches in GenBank at the National Center for Biotechnology Information. The genome organization, total length, and features of all protein-coding and rRNA genes of the three mtDNA molecules were identified by comparison with M. edulis and M. galloprovincialis mitochondrial genomes previously published (Table 2). Candidate tRNA genes were identified either using tRNAScan-SE version 1.21 (Lowe and Eddy 1997) or by comparison with similar positions of M. edulis and M. galloprovincialis tRNAs previously published. We also evaluated their potential to form a tRNA-like secondary structure and looked for the presence of the anticipated anticodon sequence.
TABLE 2.
Gender-associated mitochondrial genomes in mytilid mussels
| Species | Gender | Genome size (bp) | A + T content (%) | GenBank accession no. | Reference | Abbreviation |
|---|---|---|---|---|---|---|
| M. trossulus | Recently masculinized | 18,651 | 61.5 | AY823625 | This study | Mtr_RM |
| M. edulis | Male | 16,622 | 62.9 | AY823623– | This study | Med_M |
| 16,624 | AY823624 | |||||
| M. edulis | Female | 16,740 | 61.8 | AY484747 | Hoffmann et al. (1992); Boore et al. (2004) | Med_F |
| M. galloprovincialis | Male | 17,671a | 63 | AB065374 | Mizi et al. (2005) | Mga_M |
| M. galloprovincialis | Female | 16,744 | 61.8 | AB065375 | Mizi et al. (2005) | Mga_F |
This M. galloprovincialis male genome is 16,626 bp in length with a presence of a 1045-bp insertion.
The ClustalW program (Thompson et al. 1994) was used for multiple DNA and protein alignments. Sequence comparisons (nucleotide differences, synonymous and nonsynonymous substitutions, total amino acid differences and distances, and number of synonymous substitutions per synonymous site (Ks) and number of nonsynonymous substitutions per nonsynonymous site (Ka) (Nei and Gojobori 1986) were carried out with MEGA, version 2.1 (Kumar et al. 2001) and DNAsp 4.0 (Rozas and Rozas 2000). Unrooted trees were constructed for the six Mytilus mitotypes from the amino acid sequence data using the following methods: equal-weighted parsimony using a branch-and-bound search (PAUP 4.0 version b2; Swofford 1998) and neighbor-joining using a Poisson correction as the model of amino acid evolution (Saitou and Nei 1987). For both the parsimony and neighbor-joining, bootstrap support was assessed with 1000 bootstrap replicates. Sliding-window analyses were performed in PROSEQ version 2.9 (Filatov 2002).
RESULTS AND DISCUSSION
Sequence, gene content, and organization of bivalve mitochondrial genomes:
The complete mtDNA sequences of two M. edulis male and one M. trossulus recently masculinized genome have been sequenced and deposited in GenBank (accession nos. AY823623, AY823624, and AY823625, respectively). In this section, we will characterize these genomes and compare them with available data from other mytilids (Table 2). Standard male mitochondrial genomes of M. edulis are 16,622 and 16,624 bp in length with an overall A + T of 62.9%. Indels of 1 nt each in the large-subunit ribosomal RNA and in the large noncoding region account for the length difference of mtDNA between the two individuals. The length and nucleotide composition of these mtDNAs is comparable to that of other mytilids (Table 2). All Mytilus mtDNA genomes share an identical gene complement, consisting of two rRNAs, 23 tRNAs, and 12 protein-coding genes, arranged in the same order; compared to nonbivalve invertebrates, they contain an extra trnM, and atp8 is missing.
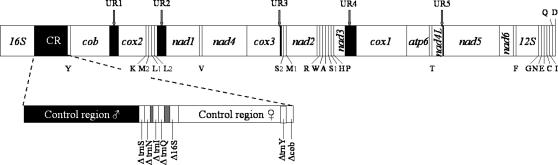
The identity of the recently masculinized M. trossulus was confirmed on the basis of a portion of the cox1 gene, whose standard M- and recently masculinized-type sequences are known from previous studies (Hoeh et al. 1997). The mitochondrial genome of the recently masculinized mtDNA is significantly longer than that of the standard male counterparts (Table 2). This gain in length is due to the presence of four additional putative tRNA genes (trnS, trnN, trnI, and trnQ) and two large noncoding regions instead of one (see below). Figure 1 compares the gene map of a typical male or female Mytilus mtDNA with that of the recently masculinized M. trossulus.
Figure 1.
Gene maps of a mitochondrial genome of Mytilus. All genes are transcribed clockwise. Names of the tRNA genes are indicated by the amino acid (one-letter code) that they specify. Solid areas indicate noncoding regions; UR1–UR5, unassigned regions 1–6. Noncoding sequences of <30 bp are not shown. 16S and 12S, large and small subunits of ribosomal RNA. The recombinant region in the recently masculinized mtDNA is shown by the dotted line. Control region ♂, corresponds to the 1265-bp male part; ΔtrnS-ΔtrnQ are full copies of the trnS, trnN, trnI, and trnQ; Δ16S corresponds to a portion sharing 89% nt identity with the normal 16S; control region ♀ corresponds to the 1340-bp female part; ΔtrnY is a full copy of trnY; Δcob corresponds to the 5′-end of the cytochrome b gene.
Mytilid mitochondria use the arthropod genetic code (Hoffmann et al. 1992): AGR codons specify serine; ATA and AAA specify methionine and lysine, respectively; and TGA is a tryptophan codon. As the presence of multiple potential start codons and often incomplete stop codons in animal mtDNAs complicates the precise inference of the coding region, we have used the same start and end points described previously for the M. edulis female mt genome (Hoffmann et al. 1992; Boore et al. 2004). Notably, some genes start with nonstandard initiation codons and some incomplete termination codons are found (Table 3), which are assumed to be completed by polyadenylation of their mRNAs (Ojala et al. 1981).
TABLE 3.
Comparisons of initiation and termination codon usage in mytilid mitochondrial genomes
| Initiation and termination codons
|
|||||
|---|---|---|---|---|---|
| Protein | Med_M | Mga_M | Med_F | Mga_F | Mtr_RM |
| cox1 | ATA/TAG | ATA/TAA | ATG/TAA | ATA/TAA | ATG/TAA |
| cox2 | ATG/TAA | ATG/TAA | ATG/TAG | ATG/TAG | ATG/TAA |
| cox3 | ATG/T** | ATG/T** | ATG/T** | ATG/T** | ATG/T** |
| Cob | ATG/TAGa | ATG/TAGa | ATG/TAA | ATG/TAA | ATG/TAAa |
| nad1 | GTG/TA* | GTG/T* | GTG/TA* | GTG/T* | GTG/TA* |
| nad2 | ATG/TAA | ATG/TAA | ATG/TAG | ATG/TAG | ATG/TAG |
| nad3 | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| nad4 | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| nad4L | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| nad5 | ATA/TA* | ATA/T* | ATA/TA* | ATA/T* | ATA/TA* |
| nad6 | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA | ATG/TAA |
| atp6 | ATG/TAA | ATG/TAA | ATG/TAG | ATG/TAG | ATG/TAG |
See Table 2 for mitochondrial genome descriptions. Asterisks indicate that the predicted stop codon is incomplete, missing either one (*) or two (**) nucleotides.
As interpreted, it is possible that cob of standard male and recently masculinized genomes ends on an incomplete stop codon, given that there is no similarity between the extension observed and other mtDNA sequences.
Transfer RNA and ribosomal RNA genes:
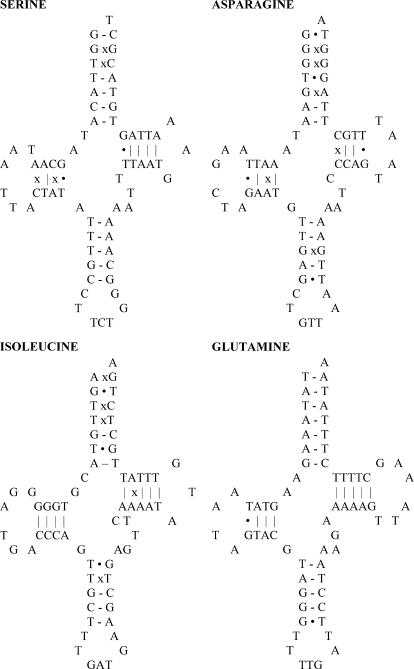
All 23 tRNA genes of the newly sequenced mtDNAs of M. edulis standard males can be folded into standard cloverleaf structures. The only exception is trnS2 (located between cox3 and trnM1), which lacks a dihydro-uridine arm. Remarkably, the M. trossulus recently masculinized mt genome contains four additional tRNA-like structures (ΔtrnS, ΔtrnN, ΔtrnI, and ΔtrnQ), inserted between the genes coding for the large-subunit ribosomal RNA (16S rRNA) and trnY. This segment normally contains the large noncoding region of the mussel mtDNA. Some of the tRNA-like structures possess several mismatched nucleotide pairs, and it remains to be determined if they are properly processed and produce functional tRNAs (Figure 2).
Figure 2.
Additional tRNA-like structures found only in M. trossulus recently masculinized mtDNA.
The boundaries of the genes coding for the large (16S)- and small (12S)-subunit ribosomal RNAs (the genes are designated rnl and rns) have been inferred by secondary structure modelling for the genomes reported here. Length variations among mussel genes range from 1243 ± 2 bp to 938 ± ∼10 bp for rnl and rns, respectively.
Noncoding sequences:
In the newly sequenced mtDNAs as well, the largest noncoding region is ∼1000 bp long and located between rnl and trnY (see Figure 1). This region likely harbors the control region of the leading strand, as inferred from the comparison with mammamlian mtDNAs, where replication and transcription initiation have been studied experimentally in great detail (Clayton 1991; Cao et al. 2004b).
Five other significant noncoding regions, designated UR1–UR5, are ∼20–500 bp long and occur across all six sequenced mytilid mtDNAs. These regions contain potential secondary structures that have been implicated in replication, transcription, or RNA processing (Clayton 1991; Mizi et al. 2005).
Comparative analysis of gender-associated mitochondrial genomes in mytilid mussels:
On the basis of the newly determined sequences, we conducted intra- and interspecific comparisons of gene sequences from standard male, recently masculinized male, and female mytilids (Table 4). Interestingly, the lowest number of nucleotide and amino acid differences in protein-coding genes is observed between the two females from different species, M. edulis and M. galloprovincialis (21 amino acid and 117 nucleotide substitutions), which is even less than the differences observed between the two standard males of M. edulis.
TABLE 4.
Intra- and interspecies comparisons for all mitochondrial genes
| Protein-coding genes
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Pairs | rRNAs | tRNAs | I | III | IV | V | Na | D |
| Med_M – Med_M | 12 (0.5) | 3 (0.2) | 17 | 4 | 3 | 5 | 29 (0.8) | 0.008 |
| Med_M – Mga_M | 130 (5.9) | 96 (6.3) | 179 | 38 | 34 | 13 | 264 (6.5) | 0.072 |
| Med_F – Mga_F | 12 (0.5) | 3 (0.2) | 16 | 2 | 3 | 0 | 21 (0.6) | 0.006 |
| Med_M – Med_F | 288 (13.1) | 150 (9.8) | 366 | 65 | 72 | 37 | 540 (14.7) | 0.157 |
| Mga_M – Mga_F | 316 (14.4) | 176 (11.5) | 357 | 52 | 85 | 32 | 526 (14.3) | 0.151 |
| Med_M – Mga_F | 283 (12.9) | 151 (9.9) | 382 | 64 | 71 | 37 | 554 (15) | 0.155 |
| Mga_M – Med_F | 319 (14.5) | 177 (11.6) | 362 | 53 | 79 | 32 | 526 (14.3) | 0.154 |
| Mtr_RM – Med_F | 166 (7.5) | 96 (6.3) | 190 | 39 | 9 | 7 | 245 (6.7) | 0.067 |
| Mtr_RM – Mga_F | 166 (7.5) | 95 (6.2) | 188 | 38 | 8 | 7 | 241 (6.6) | 0.065 |
| Mtr_RM – Med_M | 275 (12.5) | 166 (10.9) | 341 | 42 | 69 | 35 | 487 (13.2) | 0.142 |
| Mtr_RM – Mga_M | 312 (14.2) | 181 (11.9) | 385 | 62 | 76 | 31 | 554 (15) | 0.161 |
Sequence divergences for rRNAs and tRNAs are given in total number of nucleotide differences. I–V, different subunits of the same oxidative phosphorylation complex were pooled for amino acid substitutions; Na, total number of amino acid differences; D, Poisson-corrected amino acid distances for all protein-coding genes. Numbers in parentheses are percentages.
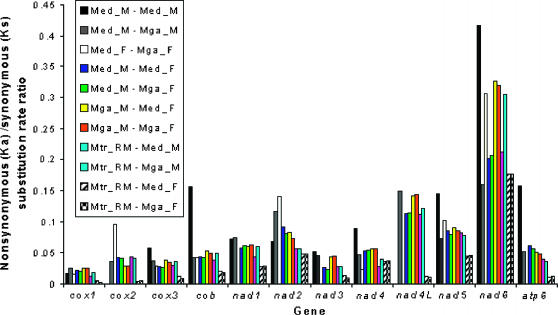
The number of nonsynonymous substitutions per nonsynonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) (Nei and Gojobori 1986) have been calculated and their ratios have been plotted for all pairwise comparisons. Figure 3 shows that Ka-to-Ks ratios are lowest for cox1 (most conserved) and highest for nad6 (least conserved). Several studies have reported significant variation in the nonsynonymous-to-synonymous substitution rate ratios for different mitochondrial genes across animals (Rand et al. 1994; Pesole et al. 1999; Ballard 2000a,b; Doiron et al. 2002). In general, animal mitochondrial genes coding for subunits of the cytochrome c oxidase complex are slowly evolving (Pesole et al. 1999; Doiron et al. 2002). Similarly, the nad6 gene appears to be weakly conserved in two other pairs of conspecific genomes (the clam Venerupis philippinarum and the freshwater mussel Inversidens japanensis) (Mizi et al. 2005), suggesting that the high rate of evolution of this gene is not a characteristic property of the M. edulis species complex.
Figure 3.
Comparisons between the number of nonsynonymous substitutions per nonsynonymous site (Ka) and the number of synonymous substitutions per synonymous site (Ks) among mytilid mussel mtDNA-encoded protein genes.
Analysis of the expanded cox1 data set:
A broad survey of mytilid cytochrome c oxidase subunit I (Table 5) confirms earlier observations that the level of nucleotide variability is higher within the M than within the F lineage (Rawson and Hilbish 1995; Stewart et al. 1995, 1996). Several mechanisms have been suggested to explain these differences (Skibinski et al. 1994b; Stewart et al. 1996; Schmidt et al. 1997; Hasegawa et al. 1998; Ballard 2000a,b), one of which proposes a lower selective constraint for the M genome relative to its F-type (Stewart et al. 1996). Indeed, contrary to the F genome, which has to function fully in all somatic tissues and the female gonad, the male genome is expressed and serves primarily in the male gonad and only partially in somatic tissues, where it periodically occurs in conjunction with the more abundant F-type (Stewart et al. 1996; Dalziel and Stewart 2002). However, because evolutionary rates may depend on multiple overlapping variable constraints, more data are needed to draw conclusions about the factors that are affecting the levels of mtDNA polymorphism in Mytilus.
TABLE 5.
Intra- and interspecies comparisons for the cox1 mitochondrial gene
| Taxon | Nucleotide divergence (SE) |
|---|---|
| M. edulis (F genome, n = 5)a,b | 0.005 (0.002) |
| M. galloprovincialis (F genome, n = 5)b,c | 0.015 (0.005) |
| M. trossulus (F genome, n = 4)b,d | 0.009 (0.004) |
| M. edulis (M genome, n = 5)b,d | 0.080 (0.015) |
| M. galloprovincialis (M genome, n = 4)b,c | 0.093 (0.017) |
| M. trossulus (M genome, n = 5)b | 0.026 (0.007) |
| M. edulis (F/M genomes, n = 5) | 0.239 (0.034) |
| M. galloprovincialis (F/M genomes, n = 5) | 0.248 (0.037) |
| M. trossulus (F/M genomes, n = 5) | 0.219 (0.032) |
Nucleotide distances were calculated with MEGA, version 2.1 (Kumar et al. 2001) using the kimura-2 parameter model.
This study.
Evidence for nonhomologous recombination between gender-associated mtDNA lineages in M.trossulus:
Intriguingly, the major noncoding region (3061 bp) of the M. trossulus recently masculinized genome is a chimera that consists of a 1265-bp segment with high identity (93%) to the D-loop of the M. trossulus M lineage (Rawson 2005; GenBank accession nos. DQ013366–DQ013367), followed by an ∼400-bp segment having sequence similarity with several tRNA genes and the adjacent 5′-end portion of the rnl gene in the F-type M. edulis (see Figure 1). The last 1434-bp-long segment of the M. trossulus recently masculinized noncoding region contains three elements: (i) another D-loop sequence with a potential control segment having high sequence similarity (98% identity) to that of the M. trossulus F lineage (T. Barna and R. Showman, personal communication; GenBank accession no. AF188281); (ii) a repeat copy of trnY; and (iii) a 97-bp fragment of the 5′-end of F-type cob. Blast searches in GenBank indicate that similar sequences have been found recently in the F-lineage noncoding region of North American M. trossulus, but the corresponding mtDNAs have been sequenced only partially (Rawson 2005; GenBank accession nos. AY636148–AY636153). These tandem arrangements of F and M control region sequences have been fully described and designated as follows (on the basis of size): F2 (∼3060 bp; TFCR5′ for the M-type part and TFCR3′ for the F-type part of the noncoding region; without an extra trnY and 5′ cob) and F3 (∼3370 bp; with an extra trnY and 5′ cob). The F2 haplotype is nearly always present in a homoplasmic state in eggs and in a heteroplasmic state together with the standard M-type mtDNA in males. In contrast, the F3 haplotype has been found to be relatively rare and was nearly always detected in a state of heteroplasmy with the F2 haplotype. Moreover, the F2 haplotype is transmitted maternally, whereas the F3 haplotype appeared to be transmitted both maternally and paternally. Apparently, the presence of a male control region in the F-type mitochondrial genome of M. trossulus is the result of nonhomologous recombination between the M. trossulus M and F genomes.
The presence of two potential control regions and their implication in the transmission dynamics of the mitochondrial genomes in blue mussels:
The recombinant haplotype identified in our study differs from those observed previously. The product generated by PCR is 3061 bp in length, corresponding to the size of the F2 haplotype described above, but the presence of a repeat of the trnY gene and a portion from the 5′-end of cob is similar to the F3 haplotype. As we did not find any trace of standard M-type mtDNA or control region size variation in the DNA extraction from the male gonadal tissue (see materials and methods), it is reasonable to speculate that this recombinant mitotype is paternally transmitted.
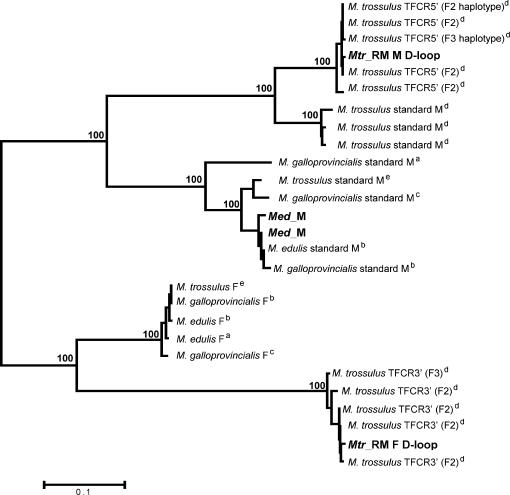
Other studies also provide evidence for mitochondrial recombination in mytilid mussels (Ladoukakis and Zouros 2001; Burzynski et al. 2003). For example, some PCR-amplified main control regions in male gonads of M. trossulus individuals from the Baltic Sea are mosaic for F- and M-like sequences (Burzynski et al. 2003). Because these recombinant variants are transmitted to sperm like the M genome, and because RFLP studies of these whole mtDNA genomes and amplified PCR products have shown high similarity to the M. edulis F genome, Burzynski et al. (2003) speculated that the M-type part of the control region confers a paternal role on mtDNA that otherwise resembles the F genome in sequence. Phylogenetic analysis confirms the high degree of similarity between the two putative M and F control regions of the recently masculinized recombinant mitotypes (TFCR5′ and TFCR3′ from the F2 and F3 haplotypes, P. D. Rawson, personal communication; male and female D-loop from the Mtr_RM mtDNA, this study) and their respective M and F control region homologs in other mytilid mussels (Figure 4). If the control region plays a role in the transmission of the mitochondrial genomes in blue mussels, then the regulation of DUI would imply at least a two-level selection model: a segregation of sperm mitochondria in the male gonad in developing embryos (Cao et al. 2004) and a replication advantage of mtDNA containing an M-type control region in male gonads.
Figure 4.
Unrooted neighbor-joining tree for the male and female D-loop portions of the recently masculinized M. trossulus mt genome (Mtr_RM in boldface type), the male and female control region portions (TFCR5′ and TFCR3′, respectively) of the M. trossulus F2 and F3 haplotypes (P. D. Rawson, personal communication), and the M- and F-type control regions for M. edulis, M. galloprovincialis, and M. trossulus (footnote a, Hoffmann et al. 1992; b, Cao et al. 2004b; c, Mizi et al. 2005; d, Rawson 2005; e, T. Barna and R. Showman, personal communication). Numbers indicate bootstrap support from 1000 replicates. Parsimony analysis yielded the same topology.
Interestingly, examination of three other M. trossulus recently masculinized genomes, two extracted from the same male gonad sample and one from a different male gonad, indicated two size classes of PCR products after using primers trm16SF and trmUR (data not shown). Both male gonads succeeded in identifying the presence of a segment that corresponded well with the size of the product from the recently masculinized sequenced genome. The second PCR product obtained from the male gonad that generated two haplotypes was ∼2 kb shorter than the product from the sequenced genome. The presence of a male and/or a female control region was determined using the RFLP assay described by Rawson (2005): the largest PCR products contained both male and female control regions, while the smallest contained an F-type control region sequence. This suggests that “extra motifs” are not necessarily a typical feature of recently masculinized mtDNAs. However, one cannot rule out the possibility that an M-type control region is required for the transcription of a mt genome in the male gonad. At present, it is not known whether the two potential control regions are functional, whether they act in a tissue-specific manner, or what their role may be in the transmission of the mitotypes. It is premature to draw any firm conclusions about the relationship between recombination and masculinization from the available M. trossulus control region sequences. The difficulty of discerning closely related recently masculinized types and their sister F types makes it equally difficult to definitively identify the gender and tissue associations of these types. The similarity of the TFCR elements within M. trossulus also makes it difficult to reconstruct the phylogenetic relationship of these fragments (Figure 5), particularly when the nearest outgroup (e.g., the F control region segment from M. edulis) is considerably diverged from the sequences of the ingroup. A population-level study of control region sequences will be necessary to answer these questions and to determine how dynamic and frequent recombinational events are within this complex of M. trossulus genomes.
Figure 5.
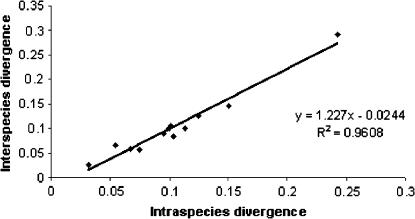
Relationships between intraspecies divergence in amino acid substitutions for protein-coding genes relative to interspecies divergences. Each diamond denotes an individual gene.
The DUI system—a network of nuclear/cytoplasmic interactions:
The high number of amino acid replacements observed between F and M genomes of a species could affect interactions with nuclear-encoded respiratory chain subunits and lead to an impairment of mitochondrial function (see Schon 2000 and Blier et al. 2001 for reviews). Indeed, as the majority of the polypeptides in the respiratory chain complexes are encoded by nuclear genes, these could function more efficiently with one or the other mtDNA genotype (Jenuth et al. 1997). If the M-type has experienced a relaxation of selection compared to the F-type, it may have accumulated more slightly deleterious mutations, and some of these mutations could alter oxidative phosphorylation in male gonad or in somatic tissues (Stewart et al. 1996). Alternatively, standard male types might have been subject to selection for particular amino acid substitutions that could improve the functional properties of mitochondrial genes in the male gonad. In this context, it is interesting to note that no significant differences have been observed in sperm swimming speeds between standard male and recently masculinized mitotypes in M. edulis, suggesting that the selective advantage of one mitotype in sperm mitochondria is not related to motility capacity (Everett et al. 2004). Swimming speed is, however, only one of the key parameters of sperm motility and fitness, and we cannot exclude the possibility of more subtle effects on mitochondrial respiratory chain function. Additional comparative analyses (e.g., measuring mitochondrial complex activities) of masculinized and standard M-type-bearing sperm are needed to clarify the potential impact associated with the amino acid substitutions observed between these mtDNAs. Such studies are currently underway in our laboratory.
The frequent hybridization between pairs of species within the M. edulis, M. trossulus, and M. galloprovincialis species complex and the retention of both species-specific M and F mtDNA in hybrid individuals (Saavedra et al. 1996; Comesaña et al. 1999) suggest a high degree of compatibility between sequence variants in the nuclear- and mtDNA-encoded subunits of the respiratory chain complexes in mytilid mussels. It was hypothesized that cytonuclear coadaptation should be evident from coordinated amino acid changes of M and F mtDNAs from the same nuclear background. Our data demonstrate a highly significant level of correlation in amino acid substitution rates for the different F and M genes irrespective of the nuclear background (Figure 5). This suggests that strong structural and/or functional constraints are gene specific rather than species or gender specific. Comparative studies of mitochondrial complex activities in natural mytilid hybrids could help to evaluate the limit of compatibility between sequence variants of nuclear- and mtDNA-encoded subunits of the respiratory chain complexes in mytilid mussels.
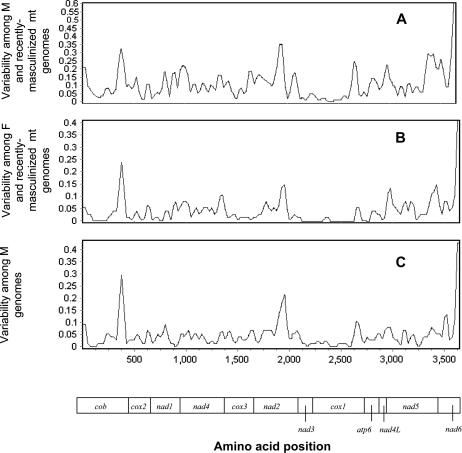
A primary motivation of this study was to present and describe the first complete sequence of a recently masculinized mitochondrial genome of a mytilid mussel. It aimed to determine: (1) candidate mitochondrial DNA regions that may demonstrate rapid adaptation associated with specific male functions and (2) distinct regions of mitochondrial DNA that may be responsible for the different transmission genetics of the gender-specific genomes. To address the first issue, a sliding-window analysis was performed to parallel the differences in amino acid substitution for all protein genes in three categories of comparisons: among standard males, between standard male and recently masculinized genomes, and between female and recently masculinized genomes (Figure 6). These pairwise comparisons showed that (i) the variability within mtDNA regions of the M lineage is highly correlated with the variability within the F lineage and (ii) the regions of mtDNA-encoded subunits that are more similar in the standard/recently masculinized males pair than in the standard female/recently masculinized male pair are situated in the NADH oxidoreductase complex (complex I) (Table 6). The degree of correlation of patterns of variability in the M and F genomes suggests that they are experiencing similar selective pressures. A comparison of mtDNA-encoded and nuclear-encoded proteins from mytilid mussels could help to determine if the subunits that directly interact with one another evolve in concert. If so, nucleotide substitutions in either an M-type or an F-type genome should constitute a selective pressure for compensatory substitutions on the opposite gender-associated genome.
Figure 6.
Variability of amino acids across the mitochondrial genome detected in sliding windows of 50 amino acids (aa) with an increment of 25 aa. Protein sequences have been concatenated as shown in the bar at the bottom. Sliding-window analyses comparing the variability among (A) male mitochondrial genomes (considering the masculinized molecule). (B) Female mitochondrial genomes (considering the masculinized molecule). (C) Male mitochondrial genomes.
TABLE 6.
Gene position of amino acid substitutions that are more similar in the male/recently masculinized pairs than in the female/recently masculinized pairs
| Gene position | Med_M | Med_M | Mga_M | Mtr_RM | Med_F | Mga_F |
|---|---|---|---|---|---|---|
| cob 322…. | S | S | S | S | T | T |
| 325…. | S | S | S | S | T | T |
| 336…. | F | F | F | F | L | L |
| cox3 50…. | S | S | S | G | V | V |
| 97…. | S | S | S | S | T | T |
| 170…. | L | L | L | L | V | V |
| nad1 5…. | S | S | S | G | A | A |
| 304…. | S | S | S | S | G | G |
| nad2 79…. | F | F | F | A | S | S |
| 93…. | S | S | S | S | T | T |
| 140…. | M | M | M | M | L | L |
| 151…. | L | L | L | L | S | S |
| 220…. | G | G | G | G | S | S |
| 295…. | M | M | M | M | I | I |
| nad4 20…. | M | M | M | M | V | V |
| 36…. | T | T | T | T | A | A |
| 40…. | M | M | M | M | T | T |
| 76…. | I | I | I | I | V | V |
| 230…. | M | M | M | M | L | L |
| 246…. | M | M | M | V | S | S |
| 289…. | V | V | V | V | L | L |
| 300…. | I | I | I | I | V | V |
| 337…. | S | S | S | S | V | V |
| 414…. | V | V | V | V | L | L |
| 422…. | V | V | V | A | S | S |
| Nad4L 10…. | F | F | F | F | L | L |
| Nad5 9…. | V | V | V | I | T | T |
| 22…. | K | K | K | K | S | S |
| 166…. | T | T | T | T | M | M |
| 230…. | F | F | F | F | Y | Y |
| 450…. | L | L | L | L | F | F |
| 454…. | V | V | V | V | A | A |
| 460…. | W | W | W | W | R | R |
| 463…. | L | L | L | L | I | I |
| nad6 41…. | V | V | V | V | I | I |
| 42…. | T | T | T | T | A | A |
| 97…. | M | M | M | M | V | V |
| 105…. | T | T | T | S | V | A |
| 123…. | G | G | G | S | K | K |
Amino acid substitutions that are nonconservative are underlined.
In conclusion, data on North American M. trossulus mussels may give some insights into the mechanism of germline mitochondrial segregation and mtDNA maintenance. The observation that a recently masculinized mitotype has been found to possess a standard F control region is not consistent with the hypothesis that portions of the control region may be responsible for the different mode of transmission of the mitochondrial genomes in blue mussels (Burzynski et al. 2003; Cao et al. 2004; Mizi et al. 2005; P. D. Rawson, personal communication). Nevertheless, our observations do not directly confirm the complete absence of DUI regulation by the mtDNA itself. It is clear that gender-specific sequences can be acquired through recombination (Ladoukakis and Zouros 2001; Burzynski et al. 2003; this study). Furthermore, the requirement for recombination to introduce a critical control region or other sequence into an otherwise female type could explain why the masculinization events are absent in unionids (Mizi et al. 2005), which apparently do not possess recombinant F-M genomes (Hoeh et al. 2002).
Our data reveal individual patterns of evolution of the different mitochondrial genes—patterns that seem independent of the nuclear or tissue background. Despite a higher evolutionary rate possibly associated with relaxed or positive selection, our results suggest that the M genome is functional and under selective constraints. As proposed earlier, selective constraints on the M genome may be required to meet the specific needs of sperm (Skibinski et al. 1994b). Qualitative or quantitative differences in respiratory chain activity between the male gonad and somatic tissues may cause the biased distribution of M mtDNA in heteroplasmic males. However, cytonuclear interactions, interaction between different F and M mtDNAs, and/or differences in their replication abilities should also be important cellular and molecular mechanisms that underlie the phenomenon of doubly uniparental inheritance.
Acknowledgments
We thank members of the Burger laboratory for help with sequencing and Michael W. Gray and Murray N. Schnare, Dalhousie University, for rRNA secondary structure modelling. We are also grateful to the three anonymous reviewers for comments on the manuscript. This study was supported by research grants from the National Sciences and Engineering Research Council (NSERC) to P. Blier and D. Stewart. S. Breton was financially supported by a NSERC scholarship.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ballard, J. W. O., 2000. a Comparative genomics of mitochondrial DNA in members of the Drosophila melanogaster subgroup. J. Mol. Evol. 51: 48–63. [DOI] [PubMed] [Google Scholar]
- Ballard, J. W. O., 2000. b Comparative genomics of mitochondrial DNA in Drosophila simulans. J. Mol. Evol. 51: 64–75. [DOI] [PubMed] [Google Scholar]
- Ballard, J. W. O., and M. D. Dean, 2001. The mitochondrial genome: mutation selection and recombination. Curr. Opin. Genet. Dev. 11: 667–672. [DOI] [PubMed] [Google Scholar]
- Blier, P. U., F. Dufresne and R. S. Burton, 2001. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 17: 400–406. [DOI] [PubMed] [Google Scholar]
- Bonfield, J. K., K. F. Smith and R. Staden, 1995. A new DNA sequence assembly program. Nucleic Acids Res. 23: 4992–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L., M. Medina and L. A. Rosenberg, 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 21: 1492–1503. [DOI] [PubMed] [Google Scholar]
- Burzynski, A., M. Zbawizka, D. O. Skibinski and R. Wenne, 2003. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 20: 388–392. [DOI] [PubMed] [Google Scholar]
- Cao, L., E. Kenchington and E. Zouros, 2004. a Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis). Genetics 166: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L., E. Kenchington, E. Zouros and G. C. Rodakis, 2004. b Evidence that the large noncoding sequence is the main control region of maternally and paternally transmitted mitochondrial genomes of the marine mussel (Mytilus spp.). Genetics 167: 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, D. A., 1991. Replication and transcription of vertebrate mitochondrial DNA. Annu. Rev. Cell Biol. 7: 453–478. [DOI] [PubMed] [Google Scholar]
- Comesaña, A. S., J. E. Toro, D. J. Innes and R. J. Thompson, 1999. A molecular approach to the ecology of a mussel (Mytilus edulis–M. trossulus) hybrid zone on the east coast of Newfoundland, Canada. Mar. Biol. 133: 213–221. [Google Scholar]
- Dalziel, A. C., and D. T. Stewart, 2002. Tissue-specific expression of male-transmitted mitochondrial DNA and its implications for rates of molecular evolution in Mytilus mussels (Bivalvia: Mytilidae). Genome 45: 348–355. [DOI] [PubMed] [Google Scholar]
- Doiron, S., L. Bernatchez and P. U. Blier, 2002. A comparative mitogenomic analysis of the potential adaptative value of arctic charr mtDNA introgression in brook charr populations (Salvelinus fontinalis Mitchill). Mol. Biol. Evol. 19: 1902–1909. [DOI] [PubMed] [Google Scholar]
- Everett, E. M., P. J. Williams, G. Gibson and D. T. Stewart, 2004. Mitochondrial DNA polymorphisms and sperm motility in Mytilus edulis (Bivalvia: Mytilidae). J. Exp. Zool. 301: 906–910. [DOI] [PubMed] [Google Scholar]
- Filatov, D. A., 2002. ProSeq: a software for preparation and evolutionary analysis of DNA sequence data sets. Mol. Ecol. Notes 2: 621–624. [Google Scholar]
- Garrido-Ramos, M. A., D. T. Stewart, B. W. Sutherland and E. Zouros, 1998. The distribution of male-transmitted and female-transmitted mitochondrial types in somatic tissues of blue mussels: implications for the operation of doubly uniparental inheritance of mitochondrial DNA. Genome 41: 818–824. [Google Scholar]
- Hasegawa, M., Y. Cao and Z. Yang, 1998. Preponderance of slightly deleterious polymorphism in mitochondrial DNA: nonsynonymous/synonymous rate ratio is much higher within species than between species. Mol. Biol. Evol. 15: 1499–1505. [DOI] [PubMed] [Google Scholar]
- Heath, D. D., P. D. Rawson and T. J. Hilbish, 1995. PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can. J. Fish. Aquat. Sci. 52: 2621–2627. [Google Scholar]
- Hoeh, W. R., K. H. Blakley and W. M. Brown, 1991. Heteroplasmy suggests limited biparental inheritance of Mytilus mitochondrial DNA. Science 251: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Hoeh, W. R., D. T. Stewart, B. W. Sutherland and E. Zouros, 1996. Multiple origins of gender-associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia). Evolution 50: 2276–2286. [DOI] [PubMed] [Google Scholar]
- Hoeh, W. R., D. T. Stewart, C. Saavedra, B. W. Sutherland and E. Zouros, 1997. Phylogenetic evidence for role-reversals of gender-associated mitochondrial DNA in Mytilus (Bivalvia: Mytilidae). Mol. Biol. Evol. 14: 959–967. [DOI] [PubMed] [Google Scholar]
- Hoeh, W. R., D. T. Stewart and S. I. Guttman, 2002. High fidelity of mitochondrial genome transmission under the doubly uniparental mode of inheritance in freshwater mussels (Bivalvia: Unionoidea). Evolution 56: 2252–2261. [DOI] [PubMed] [Google Scholar]
- Hoffmann, R. J., J. L. Boore and W. M. Brown, 1992. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics 131: 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuth, J. P., A. C. Peterson and E. A. Shoubridge, 1997. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat. Genet. 16: 93–95. [DOI] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA: molecular evolutionary genetic analysis, Version 2.1. Pennsylvania State University, University Park, PA.
- Ladoukakis, E. D., and E. Zouros, 2001. Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18: 1168–1175. [DOI] [PubMed] [Google Scholar]
- Lang, B. F., and G. Burger, 1986. A collection of programs for nucleic acid and protein analysis, written in FORTRAN 77 for IBM-PC compatible microcomputers. Nucleic Acids Res. 14: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T. M., and S. R. Eddy, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. A., D. D. Dykes and H. F. Polesky, 1988. A simple salting out procedure for extraction DNA from human nucleated cells. Nucleic Acids Res. 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizi, A., E. Zouros, N. Moschonas and G. C. Rodakis, 2005. The complete maternal and paternal mitochondrial genomes of the Mediterranean mussel Mytilus galloprovincialis: implications for the doubly uniparental inheritances mode of mtDNA. Mol. Biol. Evol. 22: 952–967. [DOI] [PubMed] [Google Scholar]
- Nei, M., and T. Gojobori, 1986. Simple methods for estimating the number of synonymous and nonsynonymous substitutions. Mol. Biol. Evol. 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Ojala, D., J. Montoya and G. Attardi, 1981. TRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474. [DOI] [PubMed] [Google Scholar]
- Passamonti, M., and V. Scali, 2001. Gender-associated mitochondrial DNA heteroplasmy in the venerid clam Tapes philippinarum (Mollusca Bivalvia). Curr. Genet. 39: 117–124. [DOI] [PubMed] [Google Scholar]
- Pesole, G., C. Gissi, A. De Chirico and C. Saccone, 1999. Nucleotide substitution rate of mammalian mitochondrial genomes. J. Mol. Evol. 48: 427–434. [DOI] [PubMed] [Google Scholar]
- Rand, D. M., M. Dorfsman and L. M. Kann, 1994. Neutral and non-neutral evolution of Drosophila mitochondrial DNA. Genetics 138: 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson, P. D., 2005. Nonhomologous recombination between the large unassigned region of the male and female mitochondrial genomes in the mussel, Mytilus trossulus. J. Mol. Evol. 61: 717–732. [DOI] [PubMed] [Google Scholar]
- Rawson, P. D., and T. J. Hilbish, 1995. Evolutionary relationships among the male and female mitochondrial DNA lineages in the Mytilus edulis species complex. Mol. Biol. Evol. 12: 893–901. [DOI] [PubMed] [Google Scholar]
- Riginos, C., M. J. Hickerson, C. M. Henzler and C. W. Cunningham, 2004. Differential patterns of male and female mtDNA exchange across the Atlantic ocean in the blue mussel, Mytilus edulis. Evolution 58: 2438–2451. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 2000. DNA sequence polymorphism, Version 4.0. Departament de Genètica, Universitat de Barcelona, Barcelona.
- Saavedra, C., D. T. Stewart, R. R. Stanwood and E. Zouros, 1996. Species-specific segregation of gender-associated mitochondrial DNA types in an area where two mussel species (Mytilus edulis and M. trossulus) hybridize. Genetics 143: 1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra, C., M.-I. Reyero and E. Zouros, 1997. Male-dependent doubly uniparental inheritance of mitochondrial DNA and female-dependent sex-ratio in the mussel Mytilus galloprovincialis. Genetics 145: 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schmidt, T. R., S. A. Jaradat, M. Goodman, M. I. Lomax and L. I. Grossman, 1997. Molecular evolution of cytochrome c oxidase: rate variation among subunit VIa isoforms. Mol. Biol. Evol. 14: 595–601. [DOI] [PubMed] [Google Scholar]
- Schon, E. A., 2000. Mitochondrial genetics and disease. Trends Biochem. Sci. 25: 555–560. [DOI] [PubMed] [Google Scholar]
- Serb, J. M., and C. Lydeard, 2003. Complete mtDNA sequence of the North American freshwater mussel, Lampsilis ornata (Unionidae): an examination of the evolution and phylogenetic utility of mitochondrial genome organization in Bivalvia (Mollusca). Mol. Biol. Evol. 20: 1854–1866. [DOI] [PubMed] [Google Scholar]
- Skibinski, D. O. F., C. Gallagher and C. M. Beynon, 1994. a Mitochondrial DNA inheritance. Nature 368: 817–818. [DOI] [PubMed] [Google Scholar]
- Skibinski, D. O. F., C. Gallagher and C. M. Beynon, 1994. b Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 138: 801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden, R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241. [DOI] [PubMed] [Google Scholar]
- Stewart, D. T., C. Saavedra, R. R. Stanwood, A. O. Ball and E. Zouros, 1995. Male and female mitochondrial DNA lineages in the blue mussel (Mytilus edulis) species group. Mol. Biol. Evol. 12: 735–747. [DOI] [PubMed] [Google Scholar]
- Stewart, D. T., E. R. Kenchington, R. K. Singh and E. Zouros, 1996. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics 143: 1349–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, B., D. T. Stewart, E. Kenchington and E. Zouros, 1998. The fate of paternal mitochondrial DNA in developing female mussels, Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA. Genetics 148: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 1998. PAUP: Phylogenetic Analysis Using Parsimony, Version 4.0b2. Illinois Natural History Survey, Champaign, IL.
- Thompson, J. D., D. G. Higgins and T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros, E., 2000. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet. Syst. 75: 313–318. [DOI] [PubMed] [Google Scholar]
- Zouros, E., K. R. Freeman, A. O. Ball and G. H. Pogson, 1992. Direct evidence for extensive paternal mitochondrial DNA inheritances in the marine mussel Mytilus. Nature 359: 412–414. [DOI] [PubMed] [Google Scholar]
- Zouros, E., A. O. Ball, C. Saavedra and K. R. Freeman, 1994. An unusual type of mitochondrial inheritance in the blue mussel Mytilus. Proc. Natl. Acad. Sci. USA 91: 7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]