Abstract
The I locus of the common bean, Phaseolus vulgaris, controls the development of four different phenotypes in response to inoculation with Bean common mosaic virus, Bean common mosaic necrosis virus, several other related potyviruses, and one comovirus. We have generated a high-resolution linkage map around this locus and have aligned it with a physical map constructed with BAC clones. These clones were obtained from a library of the cultivar “Sprite,” which carries the dominant allele at the I locus. We have identified a large cluster of TIR–NBS–LRR sequences associated within this locus, which extends over a distance >425 kb. Bean cultivars from the Andean or Mesoamerican gene pool that contain the dominant allele share the same haplotypes as revealed by gel blot hybridizations with a TIR probe. In contrast, beans with a recessive allele display simpler and variable haplotypes. A survey of wild accessions from Argentina to Mexico showed that this multigene family has expanded significantly during evolution and domestication. RNA gel blot analysis indicated that the TIR family of genes plays a role in the response to inoculations with BCMV or BCMNV.
MOLECULAR and genetic characterization of a number of disease resistance (R) genes has increased our understanding not only of their structure and possible mode of action, but also of their mode of evolution. Analysis of the deduced amino acid sequence of R genes has revealed structural motifs that are indicative of a role in signal transduction (Hammond-Kosack and Jones 1997; Ellis and Jones 1998; Dangl and Jones 2001) and that suggest the existence of conserved pathways used by plants to trigger defense responses (Bent 1996; Dangl and Jones 2001; Staskawicz et al. 2001). In addition, genetic and molecular analysis of R loci have shown varying degrees of complexity. For instance, in Arabidopsis thaliana the Rpm1 locus has one copy of a coiled-coil–nucleotide binding site–leucine rich repeat (CC–NBS–LRR) R gene in plants that are resistant to Pseudomonas syringae, but this copy is deleted in susceptible plants (Grant et al. 1995). In flax, while the L locus comprises a single copy of a Toll/interleukin-1 receptor (TIR)–NBS–LRR R gene with ∼13 alleles, the M locus encompasses ∼15 copies in tandem, but only one of these is responsible for resistance to a particular race of the rust pathogen (Anderson et al. 1997). There are six copies of a serine/threonine protein kinase sequence in the resistance allele of the Pto locus of tomato, but only five copies in the susceptible allele in which the copy that confers resistance to Pseudomonas is missing (Martin et al. 2003). Variation in copy number of R clusters and in the rate of amino acid substitutions led Michelmore and Meyers (1998) to propose a “birth-and-death” mode of evolution for R genes, similar to that observed in vertebrate gene clusters involved in immunity responses (Nei and Hughes 1992), but with emphasis on divergent selection. At the population level, the coexistence of ancient resistance and susceptibility alleles has led to the rejection of the “arms-race” hypothesis for the evolution of plant-pathogen interactions in favor of a “trench-warfare” hypothesis where these alleles are maintained by a mechanism of balancing selection (Stahl et al. 1999; Van der Hoorn et al. 2002).
The I gene of Phaseolus vulgaris, the common bean, controls resistance to Bean common mosaic virus (BCMV), a member of the Potyviridae family. This gene is widely used in bean breeding programs throughout the world. This resistance was discovered by Corbett in 1931 (Pierce 1934) and later characterized by Ali (1950) as a monogenic dominant trait. A more in-depth characterization conducted by Drijfhout (1978) revealed that the I gene confers temperature-dependent resistance to a group of BCMV strains (serotype B), but conditions development of systemic necrosis in the plant after infection with necrotic strains of BCMV (serotype A) at any temperature. Serotype B strains can induce systemic necrosis in I- genotypes at temperatures >30° or when delivered through a graft with an infected plant. On the basis of serological and sequence data, serotype A strains have been reclassified as Bean common mosaic necrosis virus (BCMNV) (McKern et al. 1992; Mink and Silbernagel 1992; Vetten et al. 1992).
In a more recent reexamination of I–BCMV interactions, Whitmer-Collmer et al. (2000) demonstrated that the I allele is incompletely dominant. At 23° and in a uniform genetic background, Whitmer-Collmer et al. (2000) showed that each of the genotypic classes displays a different phenotypic response to inoculation with a strain of BCMV: I/I shows extreme resistance or immunity; I/i, hypersensitive reaction; and i/i, systemic mosaic. The BCMV coat protein was not detected either locally or systemically when the I allele was present. However, increasing the incubation temperature to 34° induced a hypersensitive response in plants that carried the I allele and systemic necrosis in half of the homozygotes (I/I) and in all of the heterozygotes. At the higher temperature, the BCMV coat protein was detectable at the inoculation point and systemically in all I-bearing plants.
Furthermore, Fisher and Kyle (1994) have documented the broad spectrum of specificity displayed by the I locus. Three distinct responses of I- genotypes to a group of related potyviruses have been described: (a) a BCMV-like temperature-dependent resistance to the Azuki mosaic virus and Blackeye cowpea mosaic virus isolates of BCMV, Cowpea aphid-borne mosaic virus, and Watermelon mosaic virus-2; (b) a BCMNV-like temperature-independent development of systemic necrosis elicited by Soybean mosaic virus; and (c) complete local and systemic resistance against Passionfruit woodiness virus-K and Zucchini yellow mosaic virus. Dendrobium mosaic virus has also been added to the BCMV group and falls into the second group (Hu et al. 1995). Genetic analysis of 2000 F3 families has failed to detect a single recombinant among five of these specificities (Kyle and Dickson 1988; Kyle et al. 1988; Fisher and Kyle 1994). This observation has raised the question of whether the I locus has a single gene with broad spectrum resistance or represents a cluster of resistance genes with suppressed recombination. More intriguing are the reports that I-bearing bean cultivars display more severe disease symptoms, including development of necrosis, than homozygous recessive cultivars when infected with Bean severe mosaic virus (BSMV), a member of the Comoviridae family (Morales and Castaño 1992; Morales and Singh 1997).
Here we describe the construction and alignment of a high-resolution linkage map and a physical map of the region comprising the I locus. In addition, we report on a survey of cultivated and wild accessions with a probe derived from the I locus and show the apparent changes that have taken place during evolution and domestication.
MATERIALS AND METHODS
Plant material and phenotyping at the I locus:
A recombinant inbred (RI) family and an F2 population were used in this project. The RI family of 76 lines has been described elsewhere (Vallejos et al. 2000) and was used here for low-resolution mapping. The F2 population [Calima (ii) × Jamapa (II)] of 3056 individuals was analyzed for high-resolution mapping of the I locus and neighboring marker loci. Parents and F2 individuals were phenotyped on the basis of their response to inoculation with the potyvirus. Inoculation of homozygous recessive (ii) plants with BCMV causes systemic mosaic, whereas a healthy phenotype results when the dominant allele (I-) is present. However, development of systemic mosaic can take a significant period of time, and phenotypic classification can sometimes be ambiguous, requiring verification through back inoculation tests on a reporter genotype at high temperatures. To increase the efficiency and accuracy of phenotypic classification at the I locus, the F2 population was inoculated with the NL3 strain of BCMNV. This strain induces necrotic lesions and systemic necrosis within a week after inoculation of plants carrying the dominant allele (I-) (Figure 1A) and systemic mosaic in homozygous recessives (ii) (Figure 1B). The F2 population was planted in 72-well trays and germinated in Conviron E15 growth chambers (Conviron, Pembina, ND) at 25°/18° with a 14-hr photoperiod. One of the primary leaves of 2-week-old seedlings was rub inoculated with the NL3 strain of BCMNV. Plants that developed local necrotic lesions 3–4 days after inoculation and died of systemic necrosis within 10 days were assumed to be either II or Ii, while plants that survived and developed systemic mosaic were classified as ii.
Figure 1.
Phenotypes controlled by the I locus after inoculation with the NL3 strain of BNMV and genotypes of a tightly linked marker. (A) Ten-day-old seedling of Jamapa (II) showing local lesions and necrosis of the apical meristem 4 days after inoculation. (B) Three-and-a-half-week-old seedling of Calima (ii) showing symptoms of systemic mosaic 2 1/2 weeks after inoculation. (C) Ethidium-bromide-stained gel displaying the segregation of the codominant PCR-based Bng45 marker from the F2 mapping population. The higher molecular weight amplicon is the Jampa allele (J) and the low molecular weight represents the Calima (C) allele.
Genotypes that belong to each of the BCMV differentials described by Drijfhout (1978) were examined by DNA gel blot analysis. The first subgroup of seven differentials have different allelic combinations of the recessive resistance genes (bc-u, -1, -2, and -3 genes), but all have the ii genotype and develop systemic mosaic after inoculation with the NL3 strain of BCMNV; from groups 1–7, they include: Calima, Imuna, Great Northern 123, Red Mexican 34, Red Mexican 35, Pinto UI 114, and IVT 7214. The second subgroup also has different combinations of the recessive resistance genes, but has the II genotype. Jamapa and Widusa belong to group 8 and Top Crop, Amanda, and IVT7233 belong to groups 9, 10, and 11, respectively. Other genotypes subjected to DNA gel blot analysis are Stringless Green Refugee (ii), Corbett Refugee (II), BTS1 (II) and BTS2 (ii), and Sutter Pink (ii).
The wild accessions were obtained from the Plant Genetic Resources Unit of the International Center for Tropical Agriculture (CIAT). Following is a list of the accessions with the CIAT entry number, location, and the coordinates of the collection: G2771 from Jomulco, Nayarit, Mexico (M1) at 21° 10′ N and 104° 22′ W; G9989 from Mascota, Jalisco, Mexico (M2) at 20° 30′ N and 104° 49′ W; G10002 from Ixcateopan, Guerrero, Mexico (M3) at 18° 24′ N and 99° 46′ W; G11053 from Ixtlahuacan del Rio, Jalisco, Mexico (M4) at 20° 51′ N and 103° 16′ W; G19908 from San Miguel Duenas, Sacatepequez, Guatemala (G1) at 14° 32′ N and 90° 50′ W; G23438 from San Pedro Pinula, Jalapa, Guatemala (G2) at 14° 41′ N and 89° 48′ W; G21245 from San Miguel, Cajamarca, Peru (P1) at 7° 7′ S and 78° 47′ W; G23419 from Tarma, Junin, Peru (P2) at 11° 14′ S and 75° 32′ W; G23458 from Paruro, Cuzco, Peru (P3) at 13° 49′ S and 71° 51′ S; and G19894 from Trancas, Tucuman, Argentina (A) at 26° 25′ S and 65° 28′ W.
BAC library screen:
The P. vulgaris BAC library (Vanhouten and Mackenzie 1999) used in this project was constructed with genomic DNA from the cultivar “Sprite” (II). The library has 4.5 genome equivalents and an average insert size of ∼100 kb. High-density filters containing the entire BAC library were obtained directly from David Frisch (Clemson University Genomics Institute, Clemson, SC). The library was screened with 32P-labeled probes prepared using the Random Primer labeling kit from Roche Molecular Biochemicals (Indianapolis), or by PCR (dATP 70 μm, dCTP 210 μm, dGTP 210 μm, dTTP 210 μm, 50 μCi of [α-32P]dATP 3000 Ci/mmol in a 20-μl reaction volume). Immediately following labeling, probes were denatured by addition of an equal volume of freshly prepared 0.4 n NaOH, incubated for 5 min, and then added directly to the hybridization buffer. Filters were prehybridized at 65° overnight according to the protocol recommended by the Clemson University Genomics Institute. Hybridization signals were visualized by autoradiography with X-OMAT Kodak (Rochester, NY) film.
Analysis of BAC clones:
Escherichia coli strain DH10B carrying individual BAC clones was grown at 37° in LB medium supplemented with 12.5 μg/ml of chloramphenicol. BAC DNA was isolated from 6-ml overnight cultures using PSI clone BAC DNA kit (Princeton Separations, Adelphia, NJ). Purified BAC DNA was digested with NotI for 4 hr at 37°, and the insert was resolved via CHEF electrophoresis (CHEF DR III; Bio-Rad Laboratories, Hercules, CA). CHEF conditions were as follows: 1.2% agarose gels, 0.5× TBE, 14°, 6 V/cm, and a switch time of 1–10 sec for 15 hr.
Isolation and characterization of BAC ends:
The termini of BAC inserts were cloned into pBlueScript II as described elsewhere (Plyler and Vallejos 2000) and were sequenced at the DNA Sequencing Core Lab (Interdisciplinary Center of Biotechnology Research, University of Florida, Gainesville, FL) using the Applied Biosystems (Foster City, CA) model 373 system. Sequences of BAC termini were also obtained via direct BAC-end sequencing.
PCR, and DNA and RNA gel blot analyses:
Oligonucleotide primers for PCR were selected with the on-line program PRIMER 3.0 (Rozen and Skaletsky 2000; http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were synthesized by Gemini Biotech (Alachua, FL), or Integrated DNA Technologies (Coralville, IA). PCR amplifications using BAC DNA templates were conducted as described elsewhere (Plyler and Vallejos 2000) and were carried out in a GeneAmp 9600 thermocycler (Perkin-Elmer; Foster City, CA), or a DNA Engine Thermal Cycler (MJ Research, Waltham, MA). Amplicons were resolved by neutral agarose gel electrophoresis.
The insert of pBng45 was sequenced, and the following primers were selected with PRIMER 3.0: Bng45F, 5′-CAGACCCATATTTAATCGCTCC-3′; Bng45R, 5′-CAAGTGGCCAGTAGCACTAGG-3′. These primers detected codominant alleles and permitted complete genotypic classification (Figure 1C). A101400, the RAPD marker amplicon that cosegregated with the I locus, was isolated from an agarose gel, cloned into pBlueScript II, and sequenced. The following locus-specific PCR primers were designed and used for segregation analysis: A101400F, 5′-CAGAATTTTCAACCACTTAGTCTGC-3′; A101400R, 5′-TAATGACCTGATGACTTTTGAATCC-3′. The PCR-based marker for the right border of BAC12I02 was designed after a BLASTX search of GenBank revealed significant similarity between the terminal sequence and a phospholipid hydroperoxide glutathione peroxidase from several species, including Cicer arietinum and A. thaliana (S = 99–108, E = 4e−23–3e−20). To maximize the chances of detecting polymorphism between parents of the F2 population, Calima and Jamapa, PCR primers were designed from regions that flanked putative introns. The locations of introns were deduced from discontinuities in the alignment of the conceptual translation product of the bean sequence with the amino acid sequence entries and from analysis with “SplicePredictor” trained on Arabidopsis (http://deepc2.zool.iastate.edu/cgi-bin/sp.cgi). The primers PhgpF (5′-ACAGGAACCAGGAAGCAATG-3′) and PhgpR (5′ TGAACAATTCACACCCGAGA-3′) amplified a 1.1-kb fragment from both Calima and Jamapa. A cleaved amplified polymorphism (Konieczny and Ausubel 1993) between allelic amplicons was detected with PstI.
PCR-based marker analysis was conducted according to the following procedures. Prior to inoculation, leaf disks were removed by punching with the lid of microcentrifuge tubes. DNA for PCR analysis was extracted according to Edwards et al. (1991) or with the DNAzol procedure (Invitrogen, Carlsbad, CA). Amplification reactions were carried out in a 20-μl volume containing 200 μm dNTPs, 1.5 mm MgCl2, 100 pM primers, 1× reaction buffer, 0.5 units Taq DNA polymerase, and 50–100 ng of template DNA. Thermocycler conditions varied according to the primers used. Amplicons were resolved by agarose gel electrophoresis in Sunrise 96 electrophoresis equipment (Invitrogen).
DNA gel blot analysis of BAC clones was carried out exactly as described by Plyler and Vallejos (2000) and that of genomic DNA samples as described by Vallejos et al. (2000). Poly(A)+ RNA was isolated as described by Vallejos et al. (2000), and gel blot analysis was conducted according to Brown and Mackey (1997).
Segregation and linkage analysis:
The linkage relationships between the I locus and other segregating marker loci were analyzed with Mapmaker Exp. 3.0 (Lander et al. 1987; Lincoln et al. 1992).
PCR amplification and cloning of TIR–NBS–LRR sequences:
BAC terminal sequences with significant similarities to the TIR, NBS, and LRR motifs were aligned (ClustalX; Thompson et al. 1997) with the tobacco N gene (Whitham et al. 1994) and then merged in silico as a composite sequence. The terminus of the SCAR A101400 amplicon that displayed similarity to the amino terminus of the N gene was included in the composite. Primer 3 (Rozen and Skaletsky 2000) was used to design PCR primers from the composite sequence. The 5′ primer was located upstream from the putative start of translation, and the 3′ primer was selected from the most extreme region in the 3′-end of the composite.
RESULTS
Complete linkage was observed between the I locus and a RAPD marker:
Previous low-resolution mapping with a recombinant inbred family of 76 lines had detected no recombinants between the I locus and either the marker locus Bng45 (Vallejos et al. 2000) or the RAPD marker A101400 (Vallejos et al. 2001). To obtain a more accurate estimate of the map distance between these loci, we analyzed an F2 population of 1659 plants arising from the cross between the susceptible Andean genotype Calima (ii) and the resistant Mesoamerican genotype Jamapa (II). Phenotypes controlled by the I locus were obtained after inoculations with the NL3 strain of BCMNV. The observed segregation ratio at this locus was 1382 I-:277 ii. The goodness-of-fit test (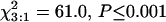 ) indicated a significant deviation from the expected Mendelian ratio with a deficit of homozygous recessives. Similar deviations have been reported for wide crosses in P. vulgaris. This phenomenon is most likely due to the partial compatibility/congruency of the Andean and Mesoamerican genetic complements interacting in the cross. Normal segregation ratios at this locus have been reported for several segregating populations obtained between closely related parents (Ali 1950; Drijfhout 1978; Kyle and Dickson 1988; Fisher and Kyle 1994).
) indicated a significant deviation from the expected Mendelian ratio with a deficit of homozygous recessives. Similar deviations have been reported for wide crosses in P. vulgaris. This phenomenon is most likely due to the partial compatibility/congruency of the Andean and Mesoamerican genetic complements interacting in the cross. Normal segregation ratios at this locus have been reported for several segregating populations obtained between closely related parents (Ali 1950; Drijfhout 1978; Kyle and Dickson 1988; Fisher and Kyle 1994).
The marker locus Bng45, originally mapped as an RFLP probe (Vallejos et al. 1992), was converted to a PCR marker to facilitate the screening of a large population (Figure 1C). As anticipated, the segregation ratio for this marker also deviated from expected Mendelian ratios: [605 JJ:778 CJ:276 CC (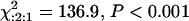 )]. A significant deficit of Calima alleles was detected. However, a contingency test indicated that the frequency of genotypic classes was a reflection of the allelic frequencies (χ2 = 0.84; d.f. = 2). The segregation distortion did not prevent us from detecting a few informative recombinants. Linkage analysis with Mapmaker 3.0 (Lander et al. 1987; Lincoln et al. 1992) and Allard's (1956) tables estimated a map distance of 0.22 ± 0.12 cM between I and Bng45.
)]. A significant deficit of Calima alleles was detected. However, a contingency test indicated that the frequency of genotypic classes was a reflection of the allelic frequencies (χ2 = 0.84; d.f. = 2). The segregation distortion did not prevent us from detecting a few informative recombinants. Linkage analysis with Mapmaker 3.0 (Lander et al. 1987; Lincoln et al. 1992) and Allard's (1956) tables estimated a map distance of 0.22 ± 0.12 cM between I and Bng45.
RAPD markers usually behave as dominant, and this was the case for the A101400 allele of Jamapa. This allelic amplicon was converted into a sequence-characterized amplified region (SCAR) marker (Paran and Michelmore 1993) and was used to screen the entire F2 population. No recombinants were detected between this SCAR marker and the I locus. The tight linkage detected between I and markers Bng45 and A101400 led us to investigate the kilobase-to-centimorgan relationship in this region.
The I locus is associated with a cluster of TIR-bearing sequences:
The average relationship between physical and map distances in the common bean has been estimated at 530 kb/cM (Vallejos et al. 1992). According to this estimate and results from our high-resolution mapping, the physical distance between Bng45 and I should be between 54 and 180 kb. Although the relationship between physical and map distances is known to vary greatly throughout the genome (Dooner and Martínez-Férez 1997; Wei et al. 1999; Fridman et al. 2000), it is also known that this relationship decreases significantly near the telomeres (Gill and Friebe 1998). Given the distal position of the I locus on chromosome 9 (formerly known as linkage group D; Pedrosa et al. 2003), the physical distance between these two loci was expected to be closer to the lower end of the estimated range. At this distance, the construction and alignment of linkage and physical maps appeared feasible.
The P. vulgaris BAC library used in this project was constructed by Vanhouten and Mackenzie (1999) from Sprite, a cultivar that carries the dominant allele I. The initial library screen with Bng45 identified two overlapping BAC clones: 12I12 (∼110 kb) and 25P23 (∼125 kb). DNA gel blot analysis confirmed that both clones shared a restriction fragment that hybridized to Bng45, a single-copy marker locus. The four termini of these BACs were subcloned and used as probes to identify the leading termini of the two-BAC contig (Plyler and Vallejos 2000) and to rescreen the BAC library. The leading terminus of 12I12 produced a hybridization pattern typical of repetitive DNA and identified a large number of clones in a BAC library screening. A probe from the leading terminus of 25P23 hybridized to a single fragment on genomic DNA gel blots and identified a single BAC clone from the library, whereas the overlapping terminus displayed a hybridization pattern typical of a multigene family on genomic blots. A similarity search in GenBank with the overlapping terminus using BLASTX (Altschul et al. 1997) revealed significant similarity (S = 76; E = 3e−21) to the LRR region of disease resistance genes. In addition, one terminus of the A101400 SCAR marker displayed significant similarity (S = 171; E = 6e−41) to the TIR motif found in several disease resistance genes, including the tobacco N gene (Whitham et al. 1994). These probes hybridized to the same three fragments in both 25P23 and 12I12. Further screening of the BAC library with these probes identified >50 clones. The majority of them were confirmed as true positives, as they revealed unique hybridization patterns in DNA gel blots.
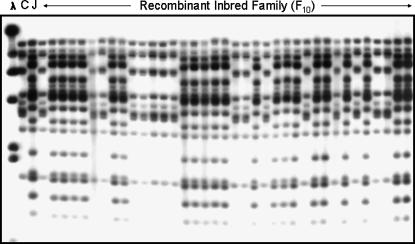
The detection of multiple cross-hybridizing BAC clones indicated the presence of a multigene family, but did not provide a clue about their genomic organization. The linkage relationships of members of this family were investigated via segregation and linkage analysis of a small RI family that was generated with the same progenitors of the large F2 population. DNA gel blot analysis of the RI family with the TIR probe revealed that Jamapa (II) has approximately twice as many fragments as Calima (ii) and that almost all of the TIR-hybridizing Jamapa fragments cosegregate (Figure 2). These results indicated that the I locus was associated with a cluster of TIR-bearing sequences.
Figure 2.
Linkage analysis of the I locus and the TIR multigene family. The DNA blot was obtained with the following samples: λ, λDNA digested with HindIII, and EcoRI digests of Calima (C), Jamapa (J), and a subset of the RI family (F10) generated from the cross [Jamapa (II) × Calima (ii)]. The blot was hybridized with a P. vulgaris-derived TIR sequence. The Jamapa haplotype shows complete cosegregation with the dominant (II) allele-controlled phenotype.
The I locus comprises a large cluster of TIR sequences and exhibits suppression of recombination:
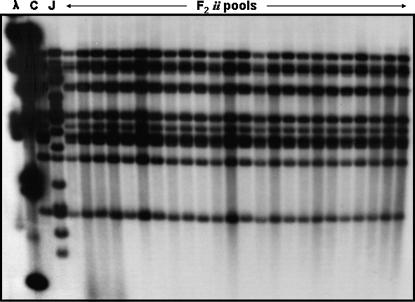
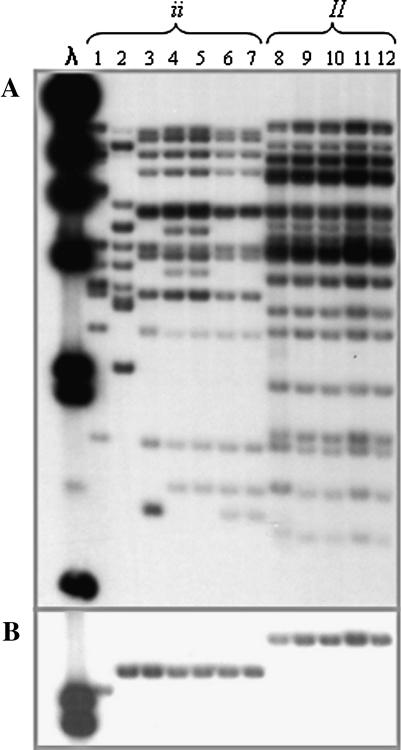
Low-resolution mapping had indicated that most TIR sequences cosegregated as a cluster, and this cluster cosegregated with the resistance phenotype. To dissect this cluster and identify informative recombinants that could provide more precise information on the location of the resistance gene, we implemented “pooled sample mapping” (Churchill et al. 1993). To implement this approach, we increased the original 1659-plant F2 population to a total of 3056 plants, assembled pools of eight recessive homozygous (ii) individuals, and analyzed them by genomic gel blot hybridization with the TIR probe. To test the limit of detection of our hybridizations, we used artificial mixtures of Calima (ii) and Jamapa (II) DNAs and were able to detect Jamapa-specific fragments in mixtures that contained 31 parts Calima DNA and 1 part Jamapa DNA. This is the equivalent of having a single recombinant individual in a pool of 16 plants. Not a single recombinant was detected after examining the 483 homozygous recessive (ii) individuals found in the 3056 F2 plants (Figure 3). These results indicated that recombination is suppressed within this cluster.
Figure 3.
Pooled sample mapping to detect recombinants within the cluster. Pools were constructed with EcoRI-digested DNA from eight F2 homozygous recessive segregants. DNA gel blots of these pools were probed with a P. vulgaris TIR probe. All 483 homozygous recessives from an F2 population of 3053 plants were screened with this procedure. No recombinants were detected.
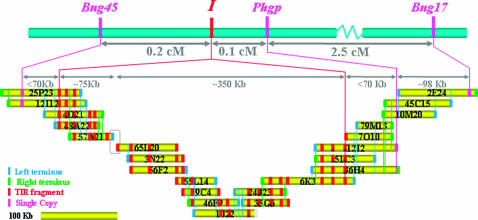
To learn about the structure of this complex locus, we arranged the selected BAC clones into contigs on the basis of shared EcoRI restriction patterns, reciprocal hybridizations with BAC-end-derived sequences, and linkage analysis of single-copy sequences. A total of five contigs were obtained. Of these, two adjacent contigs mapped to the I locus and are depicted in Figure 4. Although the BAC library was screened multiple times with the internal leading termini of the adjacent contigs, no new clones were detected. Attempts to close the gap via long-range PCR with nested primers were not productive, perhaps due to the repetitive nature of the gap termini. A first approximation estimate of the lengths of the adjacent contigs are 140 and 475 kb. The length of the gap is unknown. The other three contigs each had a single TIR-hybridizing fragment and mapped to chromosomes 4, 5, and 9 (formerly known as linkage groups A, C, and D; Pedrosa et al. 2003). The latter was distantly linked to the I locus. All of the TIR-hybridizing EcoRI fragments detected in genomic DNA blots are accounted for in the BAC clones of the five contigs.
Figure 4.
Alignment of the linkage and physical maps constituting the I locus. BAC clones in the BAC-based contig are drawn to scale. The blue and green marks indicate the left and right borders, respectively. The left and right borders correspond to the T7 and SP6 sides of the cloning site. The red marks represent TIR-hybridizing fragments, except those located terminally, where a red mark can represent a TIR-, NBS-, or LRR-related sequence. Magenta marks represent single-copy sequences. The boundaries of the I locus were established by the identification of recombination events between Bng45 and the TIR cluster on one side and between the TIR cluster and Phgp on the opposite side. Recombination was found to be suppressed within the I locus, which occupies ∼>425 kb. The empty box represents a gap in the contig.
Two common points of reference are required for a proper alignment of physical and linkage maps. Accordingly, the short 140-kb contig was aligned to the linkage map through the Bng45-I pair. The 475-kb contig contained part of the TIR cluster and required a second point of contact for proper alignment. The second point was a PCR-based marker for the Phgp locus. This marker was developed from the right terminus of BAC 12I02, a BAC clone that contained distal copies of the TIR cluster. A single recombinant plant was detected among the susceptible class of the entire F2 population, placing Phgp at 0.1 cM from the cluster (Figure 4). This map distance represents no more than 60–70 kb. To further strengthen this alignment, we included Bng17 in our analysis. This is the most distal RFLP marker on this end of chromosome 9 and had been mapped to ∼3.0 cM from Bng45 (Vallejos et al. 1992). We hybridized the contig BAC clones with Bng17. Both BAC clones 2E24 and 49O08 (not shown) displayed a single Bng17-hybridizing fragment (data not shown), indicating the inclusion of this maker locus in the contig.
Rare recombination events between Bng45 and the TIR cluster within BAC 12I12 and between the cluster and Phgp within BAC 12I2 mark the limits of the I locus. These results show that the I locus comprises a cluster of TIR sequences distributed over a distance in excess of 425 kb where recombination is suppressed. These results also raise the possibility that one of the TIR sequences in the cluster may be responsible for resistance to BCMV.
TIR sequences in the I-locus display consensus motifs of TIR–NBS–LRR disease resistance genes:
To assess the complexity of the I locus, we examined the terminal sequences of BAC clones from the I-linked contigs. These sequences were obtained by sequencing terminal subclones or by direct BAC-end sequencing. Analysis of BAC termini with BLAST 2.0 (Altschul et al. 1997) revealed significant similarities to retrotransposon-associated sequences (gag-pol, rt) or to the TIR, NBS, or LRR domains found in several R genes (Martin et al. 2003). Some termini had no match in GenBank, were AT rich, and yielded DNA gel blot hybridization patterns typical of repetitive DNA.
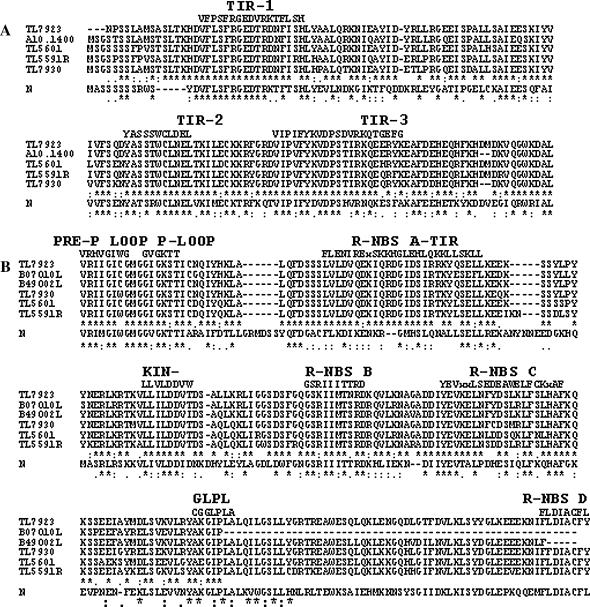
To obtain direct evidence about the structural nature of TIR-related sequences in the cluster, we used a PCR strategy (see materials and methods) to amplify and clone a few of these sequences. When individual BAC clones were used as templates in the amplification reactions, amplicons ranging in size from 3.7 to 4.5 kb were obtained. Ten clones obtained with this approach were sequenced and analyzed. Sequence comparisons identified two groups of four clones each, and two single-clone groups. Some of the redundancy is probably due to the fact that overlapping BACs were used as templates. Multiple alignment of the conceptual translation products of these sequences in the proper frame with the tobacco N gene showed that they have similar structural organization with the TIR domain at the amino terminus, followed by the NBS domain and the LRR domain at the carboxyl end (Figure 5, A–C). The TIR domains show the three consensus motifs detected in the family of TIR–NBS–LRR disease resistance genes (Meyers et al. 1999). The TL7923 clone has a deletion of 14 nucleotides that includes the apparent start of translation; however, it is possible that the next methionine at position 10 may be used as the start codon. The NBS domain of these sequences carries the three typical functional motifs (P-loop/Kin-1, Kin-2, and GLPL). Five other motifs can be recognized in the NBS domains: two TIR-specific motifs, the pre-P loop, and R-NBS-A–TIR, along with the other three motifs found in the NBS superfamily of disease resistance genes: R-NBS-B, -C, and -D. Finally, analysis with the Pfam database (Bateman et al. 2004) detected several LRR-1-type repeats in the LRR domain. This sequence analysis of contig-derived subclones provides evidence that a cluster of genes that belong to TIR–NBS–LRR family of R genes resides within the I locus.
Figure 5.
Multiple alignments of the deduced amino acid sequence of several P. vulgaris sequences from the contig and that from the N. glutinosa N gene (Q40392). Included in the alignments are four bean sequences (TL) derived by PCR from contig BACs (TL5591F, DQ002472; TL5591R, DQ002471; TL5601, DQ00242468; TL7923, DQ002469; and TL7930, DQ002470), a terminus of the A10.1400 SCAR amplicon (DQ00242476) and three BAC termini (B07O10L, DQ002474; B25P23R, DQ002473; and B49O02L, DQ002475). Alignments of the TIR domains (A), the NBS domains (B), and the LRR domains (C).
A calculation of the rate of synonymous (Ks) and nonsynonymous (Ka) nucleotide substitutions (K-Estimator 6.0; Comeron 1999) in deduced coding domains of the clones depicted in Figure 5 shows that the average Ka/Ks ratio is not uniform throughout the sequences. While the average ratio of the TIR domains is 0.53 [coefficient of variation (CV) = 14%], that of the NBS domains is 1.38 (CV = 23%). These ratios indicate that the TIR domains are under intense purifying selection, while the NBS domains are under diversifying selection. Ratios were not calculated for the LRR domain because they were not complete sequences and displayed several indels. Nevertheless, an extensive number of substitutions can be observed.
BCMV-resistant and susceptible bean lines display distinct haplotypes at the I locus:
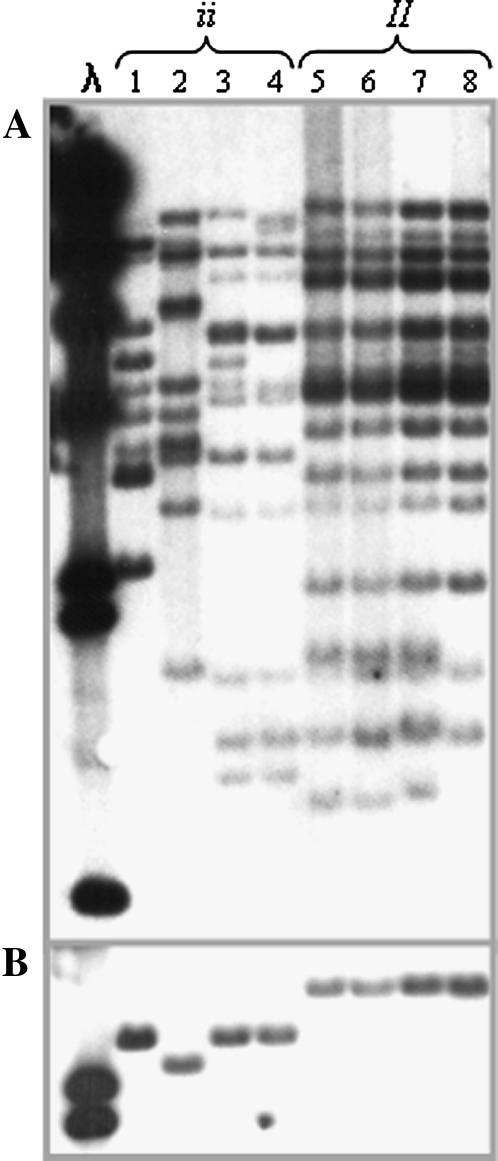
“Corbett Refugee” was the first bean line carrying the dominant allele at the I locus (Pierce 1934). This line was derived from “Stringless Green Refugee,” a line associated with the Andean gene pool. Since then, many breeding programs in the United States and around the world have incorporated that resistance into different breeding lines and cultivars. To test whether lines listed as carrying the dominant resistance possessed similar haplotypes, we selected and analyzed a representative from each of the 11 groups of BCMV differentials identified by Drijfhout (1978). These differentials have different combinations of recessive resistances and are divided into two major groups on the basis of the allele found at I. Figure 6A shows that all bean genotypes with the dominant allele at I share the same haplotype, while cultivars with a recessive allele have approximately half as many TIR-hybridizing fragments as the resistant lines and display some variation among them. In addition, Figure 6B shows that all I-bearing cultivars share the same Bng45 allele not found in any of the BCMV-susceptible cultivars. These results strongly suggest that all resistant lines ultimately received the resistance allele from a single source.
Figure 6.
DNA gel blot analysis of BCMV differentials. λ, λDNA digested with HindIII. Each lane has EcoRI-digested DNA from a representative for each of the 11 groups of differentials to BCMV strains. These differentials can be divided into two groups on the basis of the alleles that they have at the I locus. (A) TIR probe for the following bean genotypes: 1, Calima; 2, Imuna; 3, Great Northern 123; 4, Red Mexican 34; 5, Red Mexican 35; 6, Pinto UI 114; 7, IVT7214; 8, Jamapa; 9, Widusa; 10, Top Crop; 11, Amanda; 12, IVT7233. (B) Same blot as in A reprobed with Bng45.
In 1983, R. Provvidenti from Cornell University reported variation in resistance to BCMV in the Mesoamerican land race “Black Turtle Soup,” which was being grown in upstate New York. Two selections were made: BT1with a monogenic dominant resistance gene and the BMCV susceptible BT2. A test conducted by Kyle et al. (1988) showed that the resistance in BT1 and that derived from Corbett Refugee are allelic. This discovery has led bean breeders and geneticists to formulate the hypothesis that a dominant allele for resistance to BCMV has arisen independently in the Andean and Mesoamerican gene pools. To test this hypothesis, we compared the Pv–TIR hybridization profiles of these two pairs of lines (BT1/BT2 and SGR/CR). Such comparison revealed that Corbett Refugee and BT1 shared identical hybridization patterns, which were distinct from those of their susceptible counterparts, “Stringless Green Refugee” and BT2, respectively (Figure 7A). Furthermore, Sprite and Jamapa share the same haplotype as Corbett Refugee and BT1. Interestingly, Sprite, which was used for the construction of the P. vulgaris BAC library (Vanhouten and Mackenzie 1999), is associated with the Andean pool, and Jamapa, the resistant parent used in our mapping population, is a landrace associated with the Mesomerican pool. Once again, the susceptible lines showed simpler but distinct hybridization patterns. To test this hypothesis further, we compared the Bng45 marker locus alleles of lines carrying the dominant allele and found that all have the same allele, while the susceptible lines have one of two other different alleles (Figure 7B). These results strongly suggest that the dominant allele arose only once, and most likely in the Mesoamerican gene pool.
Figure 7.
DNA gel blot of analysis of putative independent sources for the I gene and their respective BCMV susceptible background. λ, λDNA digested with HindIII. Bean DNA samples were digested with EcoRI. Cultivar list: 1, Sutter Pink; 2, Calima; 3, BT2; 4, Stringless Green Refugee; 5, Corbett Refugee; 6, BT1; 7, Jamapa; 8, Sprite. DNA samples were digested with EcoRI and (A) hybridized with a Pv-TIR probe or (B) hybridized with Bng45.
The I locus has undergone tandem duplications during evolution and domestication:
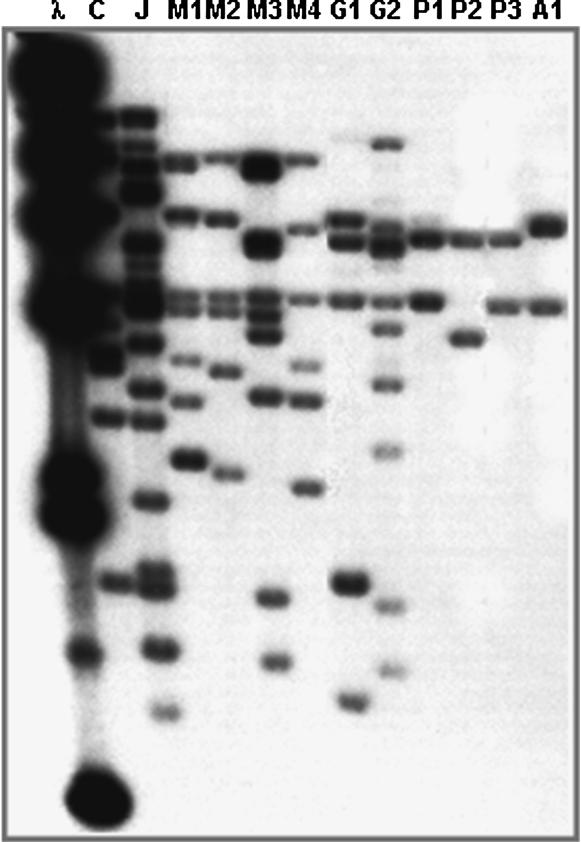
Although at present the exact identity of the resistance gene within the I locus is unknown, the number of TIR sequences within the locus could be used as an indicator of its complexity. The susceptibility alleles displayed in Figures 6 and 7 have almost half as many TIR copies as the resistance allele and therefore appear to be less complex, yet show some variation. To investigate the changes this locus may have undergone during evolution and domestication, we examined the TIR hybridization patterns of a few wild accessions collected between northern Argentina and northwestern Mexico. Figure 8 shows a latitudinal gradient in copy number with increasing complexities in accessions from more northern latitudes. Furthermore, during contig assembly of BAC clones we detected one multiple-copy locus (I) and three independent single-copy loci. The presence of only two to three TIR-hybridizing fragments in the South American accessions suggests that ectopic duplications have occurred during the evolution of this gene family. The results presented here indicate that the I locus has undergone considerable expansion through a series of tandem duplications (Leister 2004) during evolution and domestication and that this TIR family may have also increased through ectopic duplication.
Figure 8.
DNA gel blot of analysis of wild P. vulgaris accessions. DNA samples were digested with EcoRI. The following samples were included in the blot: λ, λDNA digested with HindIII; C, Calima; J, Jamapa; M1, Nayarit, Mexico; M2, Jalisco, Mexico; M3, Guerrero, Mexico; M4, Jalisco, Mexico; G1, Sacatepequez, Guatemala; G2, Jalapa, Guatemala; P1, Cajamarca, Peru; P2, Junin, Peru; P3, Cuzco, Peru; A1, Tucuman, Argentina. All the wild accessions developed systemic mosaic after inoculation with BCMV or BCMNV.
Viral infections induce distinct patterns of I-locus gene expression in susceptible and resistant genoytpes:
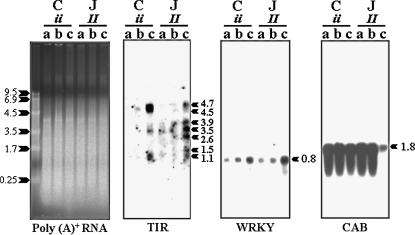
Aside from the retrotransposon related sequences, the TIR sequences were the only other coding sequences detected within the I locus via direct BAC-end sequencing or hybridization. To determine whether a member or members of the gene cluster display different patterns of gene expression between the susceptible and resistant beans, and whether the patterns change in response to inoculations with BCMV (NL1) or BCMNV (NL3), we examined the transcript levels of these sequences via RNA gel blot analysis. RNA blots were prepared with poly(A)+ RNA isolated from the primary leaves of 10-day-old seedlings 3 days after inoculation. At this time, plants with the dominant allele (Jamapa, II) were just beginning to develop necrotic lesions that were not easily recognizable. Inoculations with sterile water were used as controls.
Hybridizations with the TIR probe showed that transcripts of this gene family accumulated in response to inoculation with BCMV or BCMNV in both the susceptible and the resistant plants (Figure 9). However, BCMNV appeared to exert a stronger effect in both genotypes. The major difference between susceptible and resistant beans was in the number of different transcripts detected in the latter. This transcript diversity could be due to the expression of different family members, the products of alternative splicing, or a combination of both.
Figure 9.
RNA gel blot analysis. The primary leaves of 8-day-old seedlings of Calima (C) and Jamapa (J) were inoculated with (a) sterile water; (b) NL-1, a strain of BCMV; and (c) NL-3, a BCMNV strain. Leaf samples were harvested 3 days post-inoculation. Each lane contained 2 μg of poly(A)+ RNA. Agarose gels were stained with SYBR-gold to show approximately uniform loading of samples. Northern blots were hybridized with P. vulgaris clones of TIR and WRKY sequences and pMB123, a mung bean clone of the CAB-binding protein.
Disease resistance mediated by members of the TIR–NBS–LRR class of disease resistance genes, such as the tobacco N gene, require the expression of WRKY, a transcription factor (Chen and Chen 2000; Liu et al. 2004). Members of the WRKY superfamily of plant-specific transcription factors are known to be associated with the expression of plant defense responses (Eulgem et al. 2000). During analysis of BAC-end sequences, we came across a sequence that bore significant similarity (S = 211; E = 6e−53) to a member of the WRKY superfamily of transcription factors. This sequence was detected in a BAC clone containing a sequence for a single-copy TIR locus on chromosome 4. RNA gel blot analysis indicated that an ∼0.8-kb WRKY transcript accumulated in response to BCMV inoculation, with a stronger effect induced by BCMNV. As a point of reference, we hybridized an RNA gel blot with a mungbean chlorophyll a/b-binding (CAB) protein probe (Thompson et al. 1983). The CAB transcripts appeared to be at relatively high levels in all samples, except in the bean genotype with the dominant allele (Jamapa, II) inoculated with BCMNV. Although necrotic lesions were incipient at the time of sampling, transcripts of photosynthesis-related sequences had begun to disappear. Changes in the level of expression of TIR transcripts in response to inoculations with BCMV and BNMV and those of the related transcription factor strongly suggest that members of the TIR gene family may be involved in the defense response against the BCMV/BCMNV complex.
DISCUSSION
The I locus comprises a cluster of TIR–NBS–LRR sequences:
Aligning linkage and physical maps of the I locus and closely linked loci has shown that this resistance locus is relatively large, encompassing >450 kb. Sequence and DNA blot hybridization analyses have also shown that in addition to several retrotransposon-related sequences, multiple copies of a TIR–NBS–LRR gene family are present within the locus. This raises the possibility that a member of this gene family may be responsible for imparting resistance to BCMV. Moreover, different copies may be responsible for the different resistance specificities detected at this locus. However, even though all the TIR-hybridizing fragments detected in genomic DNA blots from Sprite are accounted for in the BAC clones of the assembled contigs, one cannot discard the possibility that the resistance phenotype(s) could be controlled by heterologous disease resistance genes present in the locus but not detected so far due to the gap in the physical map and the lack of a complete sequence of the resistance haplotype.
Comparisons among a few TIR–NBS–LRR paralogs within the I locus showed variation among them. This is in agreement with the general observation that increases in copy number of a given sequence lead to increased sequence diversity (Baumgarten et al. 2003). However, this pattern of variation was nonrandom. A gradient of substitutions and indels was detected between the amino and carboxyl ends of the deduced amino acid sequences. The average Ka/Ks ratios indicated that while the TIR domain is under purifying selection, the NBS domain is under diversifying selection. The LRR sequences were incomplete, but clearly showed a much higher degree of variation, including several indels. These results clearly point out that at least some of the TIR–NBS–LRR sequences in the locus have been under selection pressure and it is very likely that they play an active role in the defense mechanism.
Suppression of recombination at the I locus may represent an adaptive strategy:
A comparison of the linkage and physical maps shows that recombination is suppressed within the I locus and that the kilobase/centimorgan relationship fluctuates almost 20-fold within short distances in the immediate surroundings (Bng45-I: 350 kb/cM; I-Phgp: 700 kb/cM; and Phgp-Bng17: 40 kb/cM). The most likely explanations for this phenomenon are indels or inversions. For instance, it has been shown that recombination can be reduced slightly by small (1.2–3.3 kb) insertions (Dooner and Martínez-Férez 1997; Okagaki and Weil 1997) or severely by hemizygosity, as is the case for the region surrounding the apospory locus in Pennisetum squamulatum (Ozias-Akins et al. 1998; Goel et al. 2003). The disparity in the number of TIR-hybridizing fragments between the dominant (I ∼ 24) and recessive (i ∼ 12) haplotypes suggests that perhaps hemizygosity at this locus may be the cause for suppression of recombination. However, one cannot discard the role of inversions as has been documented for the complex Mla locus of barley (Wei et al. 2002). Future availability of resistance and susceptibility haplotypes may shed light on the underlying mechanism.
Suppression of recombination has been reported not only for disease resistance loci (Ganal et al. 1989; van Daelen et al. 1993; Wei et al. 1999; Chin et al. 2001), but also for natural populations of Drosophila suboscura (Navarro-Sabate et al. 2003), the mating-type chromosomes of Neurospora tetrasperma (Merino et al. 1996), and the evolution of sex chromosomes (Griffin et al. 2002). Regardless of the underlying mechanism, the latter examples have demonstrated that suppression of recombination can lead allelic haplotypes to different evolutionary paths and foster increased diversity at the affected loci.
Thus, the evolution of alleles that are unable to exchange genetic information can be seen in light of the “trench warfare” model for evolution of host-pathogen interactions proposed by Stahl et al. (1999). According to this model, natural selection maintains a dynamic equilibrium between susceptible and resistant alleles in a population. In fact, the majority of land races of Mexican black beans represent mixtures of plants with either the susceptible or the resistant allele at the I locus (J. Acosta, National Bean Breeding Program of Mexico, personal communication). This has been clearly documented by Provvidenti (1983). To understand how selection maintains both alleles in these populations, we can consider the cost of maintaining the resistance allele. While the dominant allele practically confers immunity to BCMV, it becomes a liability to the plant in the presence of BCMNV as this virus induces systemic necrosis leading to the death of the plant. In addition, the dominant allele conditions the plant to develop extreme symptoms when infected with BSMV, a member of the Comoviridae family that has severely affected bean production areas in Central America (Morales and Castaño 1992; Morales and Singh 1997). In summary, suppression of recombination at the I locus may represent an adaptive mechanism supporting balancing selection between susceptibility and resistance alleles in landraces of Mexican black beans.
The I gene likely arose within the Mesoamerican gene pool:
It is very likely that Corbett Refugee was the ultimate donor of the resistance allele found in Sprite, as is the case for almost all cultivars bred for resistance to BCMV. Thus, it is not surprising to find that Sprite and many other bean cultivars developed in the United States and Europe share similar haplotypes, as detected via TIR hybridization of genomic DNA gel blots. But it was surprising to find that the haplotype of Corbett Refugee was identical to those of Jamapa and BT1, a landrace and a selection from a landrace, respectively, both belonging to the Mesoamerican gene pool. It is widely believed in the Phaseolus community that the resistance derived from Corbett Refugee is of Andean origin because this cultivar was developed “from a single mosaic resistant plant found in a heavily infected stand of Refugee Green beans” (Pierce 1935, p. 876). The latter is associated with the Andean gene pool. The fact is that Pierce did not develop the resistant line—he simply reported that it was resistant to BCMV and that Ralph Corbett had produced that line. There is no description in the literature of how this resistant line was generated. The complexity of the haplotype of the resistant allele and the fact that all resistant beans associated with either the Andean or the Mesoamerican gene pools share the same allele at Bng45, a marker locus tightly linked to I, clearly indicate that the resistance allele originated only once, and most likely in the Mesoamerican gene pool.
From analysis of certain repeats in the phaseolin locus, Kami et al. (1995) concluded that wild P. vulgaris accessions from northern Peru and the mountainous regions of Ecuador are the most primitive accessions, as they carry the ancestral phaseolin that was present before the divergence of the Andean and Mesoamerican gene pools. G21245 was one of the accessions used by Kami et al. (1995) and was included in our analysis. This accession, along with others from Peru and Argentina, displays two TIR-hybridizing DNA fragments. The fact that we were able to detect four unlinked TIR loci in Sprite indicates that ectopic duplications have taken place during the evolution of this species. Furthermore, increasing numbers of fragments observed in northern latitudes, with a maximum observed in the resistant allele, indicate that this gene family has been in an amplification mode during both evolution and domestication. One possible mechanism for this expansion is unequal crossing over and recombination. This phenomenon has been proposed for clusters of disease resistance genes in tomato (Parniske et al. 1997; Parniske and Jones 1999) and proven to occur experimentally with transgenic plants (Jelesko et al. 1999).
The TIR family at the I locus responds to virus infections:
Regardless of the identity of the gene responsible for the resistance phenotype, our RNA gel blot analysis indicated that members of this family play an active role in the response to both BCMV and BCMNV. The accumulation of TIR-bearing transcripts in Calima (ii) and Jamapa (II) in response to inoculations with either BCMV or BCMNV indicates that no matter what the allele is at the I locus, plants can recognize and respond to the presence of the virus. The higher levels of transcript accumulation observed in response to BCMNV may imply that this strain is more aggressive. An independent line of evidence for the involvement of TIR sequences in the response to BCMV/BCMNV was provided by the accumulation of WRKY transcripts after inoculations with these viruses. These transcripts are required for the expression of N-mediated resistance to tobacco mosaic virus in tobacco (Liu et al. 2004). Our gene expression analysis has provided the strongest evidence for the role that I-locus-associated TIR sequences play in resistance to BCMV in beans.
Acknowledgments
We thank Daryl Pring, Wen Yuan Song, and L. Curtis Hannah for reviewing the manuscript and offering valuable suggestions. This work was supported in part by U.S. Department of Agriculture grant no. 97121514 and by the Florida Agricultural Experiment Station and was approved for publication as journal series no. R-08753.
References
- Ali, M. A., 1950. Genetics of resistance to the common bean mosaic virus in the bean (Phaseolus vulgaris L.). Phytopathology 40: 69–79. [Google Scholar]
- Allard, R. W., 1956. Formulas and tables to facilitate the calculation of recombination values in heredity. Hilgardia 24: 235–279. [Google Scholar]
- Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. A., G. J. Lawrence, B. C. Morrish, M. A. Ayliffe, E. J. Finnegan et al., 1997. Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9: 9641–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich et al., 2004. The Pfam protein families database. Nucleic Acids Res. 32: D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten, A., S. Cannon, R. Spangler and G. May, 2003. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A., 1996. Plant disease resistance genes: function meets structure. Plant Cell 8: 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T., and K. Mackey, 1997. Analysis of RNA by Northern and slot blot hybridization, pp. 4.9.1–16 in Current Protocols in Molecular Biology, edited by F. M. Ausubel. John Wiley & Sons, Boston. [DOI] [PubMed]
- Chen, C., and Z. Chen, 2000. Isolation and characterization of two pathogen- and salicylic acid- induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42: 387–396. [DOI] [PubMed] [Google Scholar]
- Chin, D. B., R. Arroyo-Garcia, O. E. Ochoa, R. V. Kesseli, D. O. Lavelle et al., 2001. Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157: 831–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., J. J. Giovannoni and S. D. Tanksley, 1993. Pooled sample makes high resolution mapping practical with DNA markers. Proc. Natl. Acad. Sci. USA 90: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron, J. M., 1999. K-estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15: 763–764. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. G. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., and I. M. Martínez-Férez, 1997. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9: 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drijfhout, E., 1978. Genetic interaction between Phaseolus vulgaris and bean common mosaic virus with implications for strain identification and breeding for resistance. Agricultural Research Report 872. Wageningen, The Netherlands.
- Edwards, K., C. Johnstone and C. Thompson, 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., and D. Jones, 1998. Structure and function of proteins controlling strain specific resistance in plants. Curr. Opin. Plant Biol. 1: 288–293. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., P. J. Rushton, S. Robatzek and I. E. Somssich, 2000. The WRKY superfamily of transcription factors. Trends Plant Sci. 5: 199–206. [DOI] [PubMed] [Google Scholar]
- Fisher, M. L., and M. M. Kyle, 1994. Inheritance of resistance to potyviruses in Phaseolus vulgaris L. III. Cosegregation of phenotypically similar dominant responses to nine potyviruses. Theor. Appl. Genet. 89: 818–823. [DOI] [PubMed] [Google Scholar]
- Fridman, E., T. Pleban and D. Zamir, 2000. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 97: 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal, M. W., N. D. Young and S. D. Tanksley, 1989. Pulsed field gel electrophoresis and physical mapping of the large DNA fragments in the Tm-2a region of chromosome 9 in tomato. Mol. Gen. Genet. 215: 395–400. [Google Scholar]
- Gill, B.S., and B. Friebe, 1998. Plant cytogenetics at the dawn of the 21st century. Curr. Opin. Plant Biol. 1: 109–115. [DOI] [PubMed] [Google Scholar]
- Goel, S., Z. B. Chen, J. A. Conner, Y. Akiyama, W. W. Hanna et al., 2003. Delineation by fluorescence in situ hybridization of a single hemizygous chromosomal region associated with aposporous embryo sac formation in Pennisetum squamulatum and Cenchrus ciliaris. Genetics 163: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M. R., L. Godiard, E. Straube, T. Ashfield, J. Lewald et al., 1995. Structure of the Arabidopsis Rpm1 gene enabling dual specificity disease resistance. Science 269: 843–846. [DOI] [PubMed] [Google Scholar]
- Griffin, D. K., S. C. Harvey, R. Campos-Ramos, L.-J. Ayling, N. R. Bromage et al., 2002. Early origins of the X and Y chromosomes: lessons from tilapia. Cytogenet. Genome Res. 99: 157–163. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., and J. D. G. Jones, 1997. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 575–607. [DOI] [PubMed] [Google Scholar]
- Hu, J., S. Ferreria, M. Wang, W. B. Borth, G. Mink et al., 1995. Purification, host range, serology, and partial sequencing of Dendrobium mosaic potyvirus, a new member of the bean common mosaic virus subgroup. Phytopathology 85: 542–546. [Google Scholar]
- Jelesko, J. G., R. Harper, M. Furuya and W. Gruissem, 1999. Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami, J., V. Becerra-Velázquez, D. G. Debouck and P. Gepts, 1995. Identification of presumed ancestral DNA of phaseolin in Phaseolus vulgaris. Proc. Natl. Acad. Sci. USA 92: 1101–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and F. M. Ausubel, 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Kyle, M. M., and M. H. Dickson, 1988. Linkage of hypersensitivity to five viruses with the B locus in Phaseolus vulgaris L. J. Hered. 79: 308–311. [Google Scholar]
- Kyle, M., M. H. Dickson and R. Provvidenti, 1988. The I gene and broad spectrum potyvirus resistance. Annu. Rep. Bean Improv. Coop. 31: 146–147. [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Leister, D., 2004. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 20: 116–122. [DOI] [PubMed] [Google Scholar]
- Lincoln, S. E., M. Daly and E. S. Lander, 1992. Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report, Ed. 3. Whitehead Institute, Cambridge, MA.
- Liu, Y., M. Schiff and S. P. Dinesh-Kumar, 2004. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 38: 800–809. [DOI] [PubMed] [Google Scholar]
- Martin, G. B., A. J. Bogdanove and G. Sessa, 2003. Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54: 23–61. [DOI] [PubMed] [Google Scholar]
- McKern, N. M., G. I. Mink, O. W. Barnett, A. Mishra, L. A. Whittaker et al., 1992. Isolates of bean common mosaic virus comprising two distinct potyviruses. Phytopathology 82: 923–929. [Google Scholar]
- Merino, S. T., M. A. Nelson, D. J. Jacobson and D. O. Natvig, 1996. Pseudohomothallism and evolution of the mating-type chromosome in Neurospora tetrasperma. Genetics 143: 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., A. W. Dickerman, R. W. Michelmore, S. Sivaramakrishnan, B. W. Sobral et al., 1999. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20: 317–332. [DOI] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth and death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mink, G. I., and M. J. Silbernagel, 1992. Serological and biological relationships among viruses in the bean common mosaic virus group. Arch. Virol. Suppl. 5: 397–406. [DOI] [PubMed] [Google Scholar]
- Morales, F. J., and M. Castaño, 1992. Increased disease severity induced by some comoviruses in bean genotypes possessing monogenic dominant resistance to bean common mosaic potyvirus. Plant Dis. 76: 570–573. [Google Scholar]
- Morales, F. J., and S. P. Singh, 1997. Inheritance of the mosaic and necrosis reactions induced by bean severe mosaic comoviruses in Phaseolus vulgaris L. Euphytica 93: 223–226. [Google Scholar]
- Navarro-Sabate, A., M. Aguade and C. Segarra, 2003. Excess of non-synonymous polymorphism at Acph-1 in different gene arrangements of Drosophila subobscura. Mol. Biol. Evol. 20: 1833–1843. [DOI] [PubMed] [Google Scholar]
- Nei, M., and A. L. Hughes, 1992. Balanced polymorphism and evolution by the birth-and-death process in the MHC loci, pp. 27–38 in Histocompatibility Workshop and Conference, edited by K. Tsuji, M. Aizawa and T. Sasazuki. Oxford University Press, Oxford.
- Okagaki, R. J., and C. R. Weil, 1997. Analysis of recombination sites within the maize waxy locus. Genetics 147: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozias-Akins, P., D. Roche and W. W. Hanna, 1998. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 95: 5127–5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paran, I., and R. W. Michelmore, 1993. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 85: 985–993. [DOI] [PubMed] [Google Scholar]
- Parniske, M., and J. D. G. Jones, 1999. Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., K. E. Hammond-Kosack, C. Golstein, C. M. Thomas, D. A. Jones et al., 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91: 821–832. [DOI] [PubMed] [Google Scholar]
- Pedrosa, A., C. E. Vallejos, A. Bachmair and D. Schweizer, 2003. Integration of common bean (Phaseolus vulgaris L.) linkage and chromosomal maps. Theor. Appl. Genet. 106: 205–212. [DOI] [PubMed] [Google Scholar]
- Pierce, W.H., 1934. Viroses of the bean. Phytopathology 24: 87–115. [Google Scholar]
- Pierce, W. H., 1935. The inheritance of resistance to common bean mosaic in field and garden beans. Phytopathology 25: 875–883. [Google Scholar]
- Plyler, T. R., and C. E. Vallejos, 2000. A method for cloning restriction fragments containing the termini of BAC inserts. BioTechniques 28: 1012–1016, 1018. [DOI] [PubMed] [Google Scholar]
- Provvidenti, R., 1983. Two useful selections of the bean cultivar Black Turtle Soup for viral identification. Annu. Rep. Bean Improv. Coop. 26: 73–75. [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Stahl, E. A, G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Staskawicz, B. J., M. B. Mudgett, J. L. Dangl and J. E. Galan, 2001. Common and contrasting themes of plant and animal diseases. Science 292: 2285–2289. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, W. F., M. Everett, N. O. Polans and R. A. Jorgensen, 1983. Phytochrome control of RNA levels in developing pea and mungbean leaves. Planta 158: 487–500. [DOI] [PubMed] [Google Scholar]
- Vallejos, C. E., N. S. Sakiyama and C. D. Chase, 1992. A molecular marker-based linkage map of Phaseolus vulgaris L. Genetics 131: 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejos, C. E., J. J. Malandro, K. Sheehy and M. J. Zimmermann, 2000. Detection and cloning of expressed sequences linked to a target gene. Theor. Appl. Genet. 101: 1109–1113. [Google Scholar]
- Vallejos, C. E., P. W. Skroch and J. Nienhuis, 2001. Phaseolus vulgaris: the common bean, integration of RFLP and RAPD-based linkage maps, pp. 300–317 in DNA-Based Markers in Plants, Ed. 2, edited by R. L. Phillips and I. K. Vasil. Kluwer Academic, Dordrecht, The Netherlands.
- van Daelen, R. A., F. Gerbens, F. van Ruissen, J. Aarts, J. Hontelez et al., 1993. Long-range physical maps of two loci (Aps-1 and GP79) flanking the root-knot nematode resistance gene (Mi) near the centromere of tomato chromosome 6. Plant Mol. Biol. 23: 185–192. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R. A., P. J. De Wit and M. H. Joosten, 2002. Balancing selection favors guarding resistance proteins. Trends Plant Sci. 7: 67–71. [DOI] [PubMed] [Google Scholar]
- Vanhouten, W., and S. A. Mackenzie, 1999. Construction and characterization of a common bean bacterial artificial chromosome library. Plant Mol. Biol. 40: 977–983. [DOI] [PubMed] [Google Scholar]
- Vetten, H. J., D. E. Lesemann and E. Maiss, 1992. Serotype A and B strains of bean common mosaic virus are two distinct potyviruses. Arch. Virol. Suppl. 5: 415–431. [DOI] [PubMed] [Google Scholar]
- Wei, F., K. Gobelman-Werner, S. M. Morroll, J. Kurth, L. Mao et al., 1999. The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153: 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F. S., R. A. Wong and R. P. Wise, 2002. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in Barley. Plant Cell 14: 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., S. P. Dinesh-Kumar, D. Choi, R. Hehl, C. Corr et al., 1994. The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin-1 receptor. Cell 78: 1101–1115. [DOI] [PubMed] [Google Scholar]
- Whitmer-Collmer, C., M. Fisher-Marston, J. C. Taylor and M. Jahn, 2000. The I gene of bean: a dosage dependent allele conferring extreme resistance, hypersensitive resistance, or spreading vascular necrosis in response to the potyvirus bean common mosaic virus. Mol. Plant-Microbe Interact. 11: 1266–1270. [DOI] [PubMed] [Google Scholar]