Abstract
The patatin multicopy gene family encodes the major storage protein in potato tubers and is organized as a single cluster in the potato genome. We sequenced a 154-kb bacterial artificial chromosome (BAC) clone containing a portion of the patatin gene cluster. Two putatively functional patatin genes were found in this BAC. These two genes are embedded within arrays of patatin pseudogenes. Using a chromatin immunoprecipitation method we demonstrate that the dramatic increase of patatin gene expression during the transition from stolons to tubers coincides with an increase of histone H4 lysine acetylation. We used 3′ rapid amplification of cDNA ends to profile expression of different patatin genes during tuber development. The profiling results revealed differential expression patterns of specific patatin gene groups throughout six different stages of tuber development. One group of patatin gene transcripts, designated patatin gene group A, was found to be the most abundant group during all stages of tuber development. Other patatin gene groups, with a 48-bp insertion in the 3′-untranslated region, are not expressed in stolons but display a gradual increase in expression level following the onset of tuberization. These results demonstrate that the patatin genes exhibit alterations in chromatin state and differential transcriptional regulation during the developmental transition from stolons into tubers, in which there is an increased demand for protein storage.
THE patatin genes in potato encode proteins that compose up to 40% of the soluble protein in the tubers (Prat et al. 1990). Thus, the patatin protein family represents the primary storage protein in potato tubers and is a critical nutritional component. Patatin proteins also exhibit enzymatic function as lipid acyl hydrolases and have been demonstrated to have both plant defense and antioxidant activities (Strickland et al. 1995; Liu et al. 2003; Rydel et al. 2003; Shewry 2003; Sharma et al. 2004).
It has been estimated that there are ∼10–18 copies of the patatin genes in each monoploid (12 chromosomes) potato genome (Twell and Ooms 1988). Genetic mapping of the patatin genes indicates that all genes map to a single locus on chromosome 8 (Ganal et al. 1991). The patatin coding regions are highly homologous with one another (in most cases, >90% nucleotide sequence identity) and most patatin proteins are immunologically and biochemically indistinguishable (Park et al. 1983; Mignery et al. 1984; Pots et al. 1999). The similarities between patatin gene copies have made it difficult to study several aspects of this multicopy gene family using traditional molecular techniques, such as gel blotting and isozyme analysis. Therefore, many questions remained about this gene family, especially those related to genomic structure, organization, and transcriptional regulation of different patatin gene loci.
Patatin genes are highly activated following tuber initiation (Hendriks et al. 1991; Bachem et al. 2000). Patatin genes are mainly expressed in tubers, with significantly lower amounts of transcripts observed in other tissues (Prat et al. 1990). However, patatin gene expression in nontuber tissues can be induced by sucrose treatment (Rocha-Sosa et al. 1989; Wenzler et al. 1989a,b). Therefore, the expression of patatin genes appears to be associated with the presence of sucrose, but sucrose is not believed to directly regulate patatin gene expression (Grierson et al. 1994).
Several studies have focused on the molecular basis of patatin gene regulation. Pikaard et al. (1987) found that two classes of patatin genes, distinguished by the presence or absence of 22 nucleotides in the 5′-untranslated region (UTR), are differentially expressed in roots and tubers. Class I transcripts (lacking the 22 nucleotides) were found to be tuber specific while class II transcripts (bearing the 22 nucleotides) were found in both tubers and roots, but at a lower abundance than class I forms. GUS reporter assays have been used to localize the expression patterns of the class I and II promoters. These assays confirmed the discovery by Pikaard et al. and further revealed that different patatin genes may be expressed in different tissue and cell types (Prat et al. 1990).
Further studies of patatin gene promoters revealed that a highly conserved 100-bp region containing two repeat domains, the A-box and the B-box, is critical for the regulation of class I patatin gene expression (Grierson et al. 1994). The A-box and the B-box were observed to bind nuclear proteins that were speculated to be transcription factors (Grierson et al. 1994). The protein believed to bind the B-box was recently identified (Zourelidou et al. 2002). This DNA-binding protein, named Storekeeper, is thought to be a regulator of patatin gene expression, possibly acting as a transcription factor (Zourelidou et al. 2002).
While much has been learned about the regulation of patatin genes as a family, no known studies have addressed the question of the specific regulation of different patatin gene copies. In this study, we attempted to better define the patatin multicopy gene family in terms of gene structure, organization, and transcriptional regulation during tuber development. Using a combination of cytological and molecular techniques, we have found that patatin genes occupy a single major locus per monoploid genome that may consist of many pseudogenes as well as functional genes. The transcriptional activation of patatin genes upon tuberization is accompanied by a change in relative histone H4 lysine acetylation. We have further found that the different copies within the patatin gene family exhibit differential expression patterns dependent on both tissue type and tuber development stage.
MATERIALS AND METHODS
Plant materials:
A haploid potato clone USW1 (2n = 2x = 24) derived from potato cultivar “Katahdin” was used for bacterial artificial chromosome (BAC) library construction and fluorescence in situ hybridization (FISH) mapping. Potato cultivar “Kennebec” (2n = 4x = 48) was used for chromatin immunoprecipitation (ChIP) and all gene expression analyses. Kennebec plants were grown in a walk-in growth chamber at the University of Wisconsin (Madison, WI) Biotron. The plants were exposed to a 12:12-hr light:dark photoperiod under fluorescent and incandescent lights. Chamber temperature was 23.5° during light hours and 15.5° during dark hours. Relative humidity was held constant at 50%. Following shoot emergence, plants were given watering treatments with 0.5× strength Hoagland solution twice daily.
Sequencing and FISH mapping of BACs containing patatin genes:
A BAC library was constructed from the BamHI-digested genomic DNA of haploid clone USW1, using published protocols (Song et al. 2000). The BAC library was screened with radioactively labeled patatin cDNA clone pGM01 (Mignery et al. 1984) and patatin genomic clone PS20SB (Mignery et al. 1988). Positive clones were fingerprinted as described (Tao et al. 2001). One BAC, 14K07, was sequenced as described (Feng et al. 2002). The BAC sequence was annotated for genes using a combination of ab initio gene finders and sequence similarity evidence (Yuan et al. 2002). Gene finders used include Genemark.Hmm (Lukashin and Borodovsky 1998), Genscan, and Genscan+ (Burge and Karlin 1997). Genes with transcript evidence only were annotated as encoding “expressed proteins.” Genes predicted solely by ab initio gene finders were annotated as encoding “hypothetical proteins.” Sequence analysis and homology searches with known patatin genes were further performed using MegAlign software from the Lasergene '99 package (DNAStar, Madison, WI). FISH mapping and image processing were performed as described (Jiang et al. 1995).
RNA isolation and gel blot analysis:
Two biological replicates of 10 Kennebec plants each were grown sequentially in the same growth chamber. Plants were monitored periodically for signs of flower formation. Stolon tips and tubers were harvested from the entire population 3–5 days following the first indications of flowering in the chamber. All tissues were flash frozen in liquid nitrogen. Harvested tubers were measured in length prior to flash freezing. Tissues were divided into six groups: stolon (no tuber formation; stolon tips were harvested, including the apical tip and part of the subapical region), 1- to 5-mm tubers, 6- to 10-mm tubers, 11- to 15-mm tubers, 16- to 25-mm tubers, and 26- to 35-mm tubers. Larger tubers were not identified at the times of harvest.
Total RNA was isolated from tissue of each of the six tuber developmental stages by TRIzol extractions according to the manufacturer's instructions (Invitrogen, Carlsbad, CA) with only minor modifications. The aqueous TRIzol:chloroform extractions were precipitated with 0.5 vol isopropanol and 0.5 vol 1.2 m sodium chloride, 0.8 m sodium citrate solution. Purified RNA samples were treated with 2.5 units of DNase I and incubated at 37° for 15 min and then extracted with phenol:chloroform. Purified RNA samples were quantified and qualified using the Nanodrop spectrophotometer with version 2.5.3 software (Nanodrop Technologies, Montchanin, DE) and agarose gel electrophoresis analyses, respectively.
Total RNA samples from the two biological replicates of stolon and tuber tissues were used as targets for RNA gel blot analysis. RNA samples were run on a 1.2% agarose formaldehyde denaturing gel and transferred to Hybond+ nylon membrane (Amersham Biosciences, Piscataway, NJ). Patatin cDNA pGM01 (Mignery et al. 1984) and potato ubiquitin EST STMGC93 (provided by C. R. Buell at the Institute for Genomic Research) were used as probes. All probe DNA was 32P labeled using the Strip-EZ DNA labeling system, according to the manufacturer's instructions (Ambion, Austin, TX). The gel blot membrane was prewashed in 65° Church buffer (7% SDS, 0.5 m Na2HPO4, 1 mm EDTA, pH 7.2) for a minimum of 1 hr. The radioactive probes were denatured and then hybridized to the membrane overnight at 65°. The membrane was sequentially washed with 2× SSC/0.1% SDS and 0.2× SSC/0.1% SDS and exposed to X-ray film and developed. The membrane was stripped and reprobed according to the Strip-EZ manufacturer protocol (Ambion).
ChIP and real-time polymerase chain reaction:
Potato cultivar Kennebec plants were grown in walk-in growth chambers, as described above. Stolon tip and early tuber tissues (1–30 mm in length) were collected and flash frozen in liquid nitrogen. ChIP was performed essentially as described (Nagaki et al. 2003), using an antibody specific to histone H4 lysine acetylation (lys 5, 8, 12, and 16) (Upstate Biotechnology, Lake Placid, NY). Mock experiments using preimmunized rabbit serum served as nonspecific binding controls for each ChIP experiment. Real-time polymerase chain reaction (PCR) with specific primers was performed to analyze the relative enrichment of ChIPed DNA associated with histone H4 lysine acetylation in different genomic regions. All primers related to the patatin gene were designed from the BAC 14K07 sequence. Real-time PCR reactions and subsequent amplicon melting curve analyses were performed using the DyNAmo SYBR Green qPCR kit on a DNA Engine Opticon 2 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). The comparative Ct method (Livak and Schmittgen 2001) was adapted to determine the relative enrichment of each primer region. A non-patatin region within BAC 14K07 was used as the reference primer set (“PrimerReference” in the equations below), as it displayed a lower enrichment of the H4 lysine acetylation vs. mock treatment DNA in both stolons and tubers. Relative enrichment of H4 lysine acetylation for each primer set (“PrimerX” in the equations below) was calculated as follows:
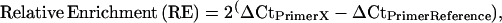 |
where
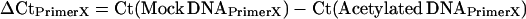 |
and
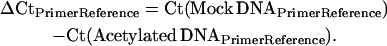 |
The ΔCtPrimerReference values were derived from averaging two experimental replicates from the same ChIP DNA samples for stolons and tubers, respectively. The same ΔCtPrimerReference values for stolons and tubers were respectively applied in all primer RE calculations. For all other primers, ΔCtPrimerX values were determined for two experimental replications from the same ChIP DNA samples for stolons and tubers, respectively. These values were independently used to calculate RE. Standard deviations for each primer set were determined from the two experimental replicates. Real-time PCR was repeated for any experimental replicate pair with a Ct difference >1.0.
Gene expression profiling using 3′ rapid amplification of cDNA ends:
A universal patatin sense primer was developed for 3′ rapid amplification of cDNA ends (RACE) analysis (details are provided in supplemental Figure 2 at http://www.genetics.org/supplemental/). This 27-bp sequence (TGTTGCTCTCATTAGGCACTGGCACTA) was identified on the sense strand in the fifth exon, 401 bp before the stop codon in patatin genes. Single-stranded 3′ RACE-ready cDNAs were generated from the total RNA of each of the six developmental stages of one bioreplicate of tuberization. The protocol for cDNA synthesis is as described by the BD SMART RACE cDNA amplification kit (BD Bioscience, Palo Alto, CA), except that Superscript III (Invitrogen) was used as the reverse transcriptase. All subsequent PCR reactions were performed with Promega (Madison, WI) Taq polymerase. PCR of the cDNAs using an intron-spanning patatin primer set (top primer, ACTGAAATGGATGATGCGTCTGAGGCTAA; bottom primer, CCAAAAGGTTACATAATCCAAGCACACAT; annealing temperature 60°) was performed to verify the quality of the cDNAs and confirm the absence of DNA templates. RACE PCR reactions were performed according to manufacturer's instructions (BD Bioscience), using the 27-bp patatin universal sense primer and the SMART RACE cDNA amplification kit universal primer A mix. Nested PCR reactions were performed on these amplicons, using the patatin universal sense primer and the nested universal primer A as specified by the manufacturer (BD Bioscience).
The 3′ RACE amplicons were cloned using the pGEM-T Easy Vector System II cloning kit (Promega). Clones with inserts of expected sizes (∼600–700 bp) were then used as templates in sequencing reactions. DNA sequencing templates were derived from either the colony PCR amplicons or plasmid DNA preparations. Sequence reactions using BigDye terminator chemistry (Applied Biosystems, Foster City, CA) were performed in both the forward and reverse orientation with M13 universal primers. Sequenced nucleotides were called independently by two software packages, Editview 1.0 (Applied Biosystems) and the SeqMan software from the Lasergene '99 package. Sequences and sequence assemblies were manually edited when appropriate.
The amplicon sequences were assembled into patatin groups on the basis of identical sequence matches, using MegAlign software from the Lasergene '99 package. Relative group mRNA abundance was estimated on the basis of the frequency of each group sequence per developmental stage. Sequence alignments of the groups were performed, using default settings with CLUSTAL X v.1.81 software (Thompson et al. 1997). Phylogeny of the groups was analyzed by the neighbor-joining method with CLUSTAL X v.1.81 (Saitou and Nei 1987; Thompson et al. 1997).
A PCR strategy was employed to generate sequence data for the complete coding region of some abundant patatin transcripts. A PCR primer matching a frequently observed motif in the 3′-UTR of several patatin genes was designed: ATTTACAACTACAACCCGAGACCTTGAAT. This primer and a near-universal class I patatin 5′-UTR primer (CMAACAAAATTTAAAAACAMWTTGAACATTTGC, where M = C and A and W = A and T) were used to generate PCR amplicons of the complete patatin coding regions from the 1- to 5-mm and 26- to 35-mm tuber cDNAs. Amplicons were cloned into the pGEM-T plasmid vectors as described above. Plasmids were purified and then sequenced as described above.
RESULTS AND DISCUSSION
BAC clones containing patatin genes were mapped to a single location in the potato genome:
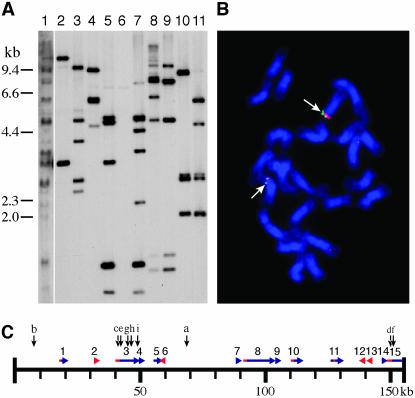
We constructed a BAC library using a haploid clone (USW1) derived from potato cultivar Katahdin. The library consists of 43,008 clones. Pulsed-field gel electrophoresis of 20 random clones indicated that the average insert size is ∼120 kb (Bresee 2002). On the basis of the clone number and average insert size, we estimated that this BAC library provides ∼5.2 times coverage of the potato genome (Bresee 2002). Sixteen positive clones were identified initially when the BAC library was screened using patatin gene plasmids pGM01 and PS20SB as probes (Figure 1A). Restriction fingerprint analysis showed that these clones fall into two contigs, consisting of 8 and 3 clones, respectively. The remaining five BACs, which showed only a single band or few weak bands in gel blot hybridization, failed to align with either contig.
Figure 1.
Molecular and cytological characterization of BAC clones containing patatin genes. (A) DNA gel blot of HindIII restriction fragments from genomic DNA of potato haploid clone USW1 (Lane 1) and 10 BACs probed with patatin gene clone pGM01. Lane 2, 68O09; lane 3, 9E06; lane 4, 16O21; lane 5, 23P06; lane 6, 46B04 (appeared as a singleton in fingerprint analysis); lane 7, 34O11; lane 8, 14K07; lane 9, 47D05; lane 10, 55M11; lane 11, 20P13. (B) FISH analysis of BACs 34O11 (red signal) and 9E06 (green signal) on potato haploid USW1 (2n = 2x = 24). The two BACs cohybridize to the same two loci, despite being assigned to different contigs in fingerprint analysis. (C) Sequence map of BAC 14K07. Sequences similar to patatin gene exon reading frames are indicated by blue bars and sequences similar to known patatin gene promoters are indicated by red bars. Horizontal arrows indicate the directional 5′ → 3′ orientation of the reading frames and promoters, respectively. The numbers above the horizontal arrows correspond to the “Region” column in Table 1. Vertical arrows indicate the approximate positions of the primers used for ChIP analysis; the letters above the vertical arrows correspond to the “Position” column in Table 2.
The potato genome is highly heterogeneous. It is not known if the separation of the two contigs is caused by the heterozygosity of the potato genomic sequences. To better understand their relationship, one BAC from each contig was labeled for FISH analysis. BAC 34O11 associated with contig 1 and BAC 9E06 associated with contig 2 were labeled with different colors and simultaneously hybridized to the metaphase chromosomes of potato haploid clone USW1. The two BACs were mapped to the same chromosomal location (Figure 1B). This FISH result indicates that there is only one major patatin multicopy gene locus per monoploid (12-chromosome) potato genome. Contigs 1 and 2 may represent separate patatin gene clusters within the same chromosome region. This is consistent with genetic mapping experiments that concluded that the potato patatin locus is divided into discrete domains (Ganal et al. 1991).
Sequence analysis of a 154-kb BAC containing patatin genes:
To gain a better understanding of the organization of the patatin genes we sequenced a BAC clone, 14K07, selected from contig 1. BAC 14K07 was selected for sequencing due to its large insert size (154 kb) and the high number of strong bands that hybridized to patatin gene probes in DNA gel blot analysis (Figure 1A). Annotation of the sequence of BAC 14K07 identified 11 genes related to patatin-coding regions (Figure 1C; Table 1) interspersed with several regions related to transposable elements (see GenBank accession DQ274179).
TABLE 1.
Patatin genes identified in BAC 14K07
| Homology with patatin domains
|
|||||||
|---|---|---|---|---|---|---|---|
| Region | Gene ID | Status | Promotera | 5′-UTRb | Exonsc | 3′-UTR | Notes |
| 1 | StPat14K07.01 | Truncated | Partial; similarity with GenBanka | None | Exons 3–4 | None | |
| 2 | StPat14K07.02 | “Naked” partial promoter | Partial; similarity with GenBanka | None | None | None | |
| 3 | StPat14K07.03 | Intact gene | Similarity with GenBanka; A-box, B-box, CAAT box, core enhancer, TATA box conserved | Class I | Exons 1–7 | 48 bp absent | |
| 4 | StPat14K07.04 | Truncated | None | None | Exons 6–7 | 48 bp present | |
| 5 | StPat14K07.05 | Truncated | None | None | Exons 2–5 | None | |
| 6 | StPat14K07.06 | Naked partial promoter | Partial; similarity with GenBanka | None | None | None | Reverse orientation relative to others |
| 7 | StPat14K07.07 | Truncated | None | None | Exon 6 | None | |
| 8 | StPat14K07.08 | Rearranged | Partial; similarity with GenBanka; CAAT box, TATA box conserved; degenerate core enhancer | Class II | Exons 1–7 | 48 bp present | Most of exon 4 not present |
| 9 | StPat14K07.09 | Truncated | None | None | Exon 1 | None | |
| 10 | StPat14K07.10 | Truncated | Partial; similarity with GenBanka; CAAT box and core enhancer conserved; TATA box truncated | Class II | Exons 1–5 | None | |
| 11 | StPat14K07.11 | Rearranged | Partial; similarity with GenBanka; located between exons 2 and 3 | None | Exons 1–7 | 48 bp present | Premature stop codon (nt127686) |
| 12 | StPat14K07.12 | Naked partial promoter | Partial; similarity with GenBanka | None | None | None | Reverse orientation relative to others |
| 13 | StPat14K07.13 | Naked partial promoter | Partial; similarity with GenBanka | None | None | None | Reverse orientation relative to others |
| 14 | StPat14K07.14 | Truncated | None | None | Exon 7 | 48 bp absent | |
| 15 | StPat14K07.15 | Potential intact gene | Similarity to GenBanka; CAAT box, core enhancer, TATA box conserved; A-box, B-box homology ∼2 kb upstream | Class II | Exons 1–5 | NA | Interrupted in fifth intron by BAC end |
Promoter homology alignments were performed against several patatin promoters (GenBank nos. AY485645, X14483, X60396, X60397, X60399, X60398, X16046, and X17397). A-box and B-box are described by Grierson et al. (1994). Patatin promoter CAAT box, core enhancer, and TATA box features are described by Mignery et al. (1988).
Class I and class II patatins are defined by the absence or presence of a 22-bp insertion in the 5′-UTR (Pikaard et al. 1987).
Patatin exon homology alignments were performed against patatin GenBank accession X01125.
Both class I and class II patatin genes were found in BAC 14K07 (Table 1). Upstream regions with similarity to known patatin gene promoters are present in 5 of the 11 patatin genes (Figure 1C; Table 1). Additionally, BAC 14K07 contains four regions with similarity to known patatin gene promoters, but they are not located immediately upstream of any patatin genes. We termed these regions “naked” patatin gene promoters. All 11 patatin genes have the same orientation (Figure 1C). However, only one of the naked patatin gene promoters is oriented in this direction; the other three naked promoters have the reverse orientation relative to the patatin genes. It is unknown if these reverse-orientated naked patatin gene promoters drive antisense patatin gene transcription.
Among the 11 patatin genes present in BAC 14K07, only 2 appear to be fully functional. One gene, StPat14K07.03 (Table 1; Figure 1C, region 3), has the appropriate patatin gene composition of seven exons and six introns (Bevan et al. 1986; Mignery et al. 1988) and does not contain any in-frame stop codons or frameshift mutations. This gene has a coding region that is nearly identical to several known patatin cDNAs. The second gene, StPat14K07.15 (Table 1; Figure 1C, region 15), is interrupted in the fifth intron by the BAC vector cloning site (Figure 1C), but contains no in-frame stop codons or frameshift mutations through the five exons present (Table 1). Both of these putative functional genes have large regions 5′ upstream of the putative transcription initiation sites that are similar to known patatin gene promoters. These promoter-similar regions include conserved CAAT boxes, core enhancers, and TATA boxes (Table 1).
The other patatin genes in BAC 14K07 appear to be structurally altered and/or truncated or contain in-frame stop codons (Table 1). These genes are incapable of encoding patatin proteins; thus we refer to them as patatin pseudogenes. The patatin multicopy gene family has long been known to contain genomic pseudogenes (Pikaard et al. 1986), which may have been functional genes prior to sequence degeneration. In general, sequences 5′ upstream to each of the patatin pseudogenes in BAC 14K07 do not appear to have intact promoters. Most 5′ upstream sequences are either unrelated to patatin gene promoters or only partially similar to known patatin gene promoters (Table 1), suggesting that the pseudogenes are not likely transcribed. The pseudogenes with the greatest similarity to the patatin gene promoter are StPat14K07.08 and StPat14K07.10 (Table 1). These two pseudogenes have 272 and 918 bp of promoter-similar sequence 5′ upstream of the putative start codons, respectively. Both of these promoter-similar regions maintain some features that may promote pseudogene transcription, such as the CAAT box, core enhancer, and TATA box. However, these features are better conserved in the two putatively functional genes StPat14K07.03 and StPat14K07.15 (Table 1). It is possible that the pseudogenes StPat14K07.08 and StPat14K07.10 were once functional patatin genes that have recently become pseudogenes. Therefore, promoter structural features are still present in these pseudogenes, which are not observed in the other patatin pseudogenes in BAC 14K07 (Table 1).
Sequence analysis of BAC 14K07 revealed a unique structure of the patatin gene family in which functional patatin genes are present within regions rich in patatin pseudogenes. This structure suggests that patatin genes may evolve by a mechanism similar to the birth-and-death process described in plant pathogen resistance gene (R gene) cluster evolution (Michelmore and Meyers 1998). Patatin pseudogenes may act as a useful source of allelic variation that can become functional through interallelic recombination and gene conversion events. Interestingly, patatin proteins have been implicated in plant defense responses (Strickland et al. 1995; Sharma et al. 2004), which may explain possible structure similarities between the patatin gene family and R gene clusters. However, it remains unclear how individual patatin genes or clusters are selected for over the course of evolution and what selective pressures may dictate this process.
The abundance of patatin gene transcripts increases during tuber growth:
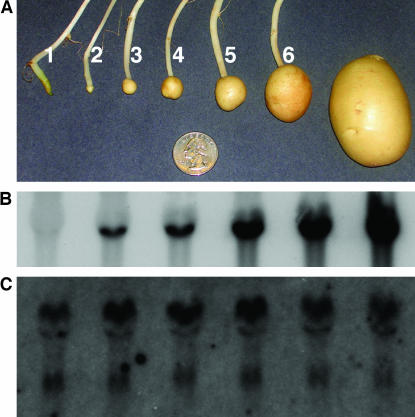
Potato variety Kennebec was used for gene expression profiling of patatin genes from pretuberizing stolons to midgrowth tubers (Figure 2A). Total RNAs from six developmental stages were collected and analyzed. RNA gel blot analysis was performed to determine the relative abundance of patatin gene transcripts during the different developmental stages of tuber growth. No visible signal was observed from the potato stolon RNA when a patatin cDNA clone pGM01 was used as a probe (Figure 2B). Reverse transcriptase PCR (RT–PCR) analyses indicated that the patatin genes were expressed in stolons (data presented below); however, the transcription level was insufficient to generate a strong visible signal on the RNA gel blot. RNA hybridization signals became visible at the onset of tuberization and increased in intensity throughout tuber growth (Figure 2B), indicating that the transcription of the patatin genes gradually increases throughout the early stages of tuber growth. Similar results, in which patatin gene expression is first detectable at tuber initiation and then subsequently increases throughout tuber growth, have previously been reported for in vitro tubers (Hendriks et al. 1991; Bachem et al. 2000). Here we have confirmed these results with tubers grown in vivo.
Figure 2.
Tuber developmental stages and patatin RNA gel blot analysis. (A) The six tuber developmental stages sampled in this study were based on tuber length (see materials and methods). Examples of sample developmental stages are shown. Stage 1, Stolon; stage 2, 1–5 mm; stage 3, 6–10 mm; stage 4, 11–15 mm; stage 5, 16–25 mm; stage 6, 26–35 mm. The diameter of the coin is 24 mm. Mature tuber is shown on the right as a developmental reference in the growth chamber environment, but was not sampled in this study. (B) Patatin gene expression level increases throughout tuber development. The patatin transcripts at the different tuber developmental stages were detected by RNA gel blot analysis using a DNA probe from patatin cDNA pGM01. (C) Equal loading of total RNA was confirmed by gel staining for rRNA with ethidium bromide (data not shown) and by reprobing with a DNA probe from ubiquitin EST STMGC93.
Patatin genes exhibit enhanced histone H4 acetylation during tuber development:
It has been recognized in numerous organisms that acetylation at the lysine residues of histone H4 is specifically associated with transcriptionally active chromatin (Strahl and Allis 2000). To reveal the H4 acetylation dynamics of the patatin genes during tuber development we conducted ChIP analysis, using an antibody specific to histone H4 lysine acetylation. Immunoprecipitated DNAs were isolated from both stolon and tuber tissues (see materials and methods). A series of PCR primers were designed from different regions of BAC 14K07, including patatin-related and non-patatin regions (Table 2). The primers were initially tested in PCR reactions on Kennebec genomic DNA templates; all primer pairs produced a single visible band of the expected size (supplemental Figure 1 at http://www.genetics.org/supplemental/). These primer pairs were then used in real-time PCR analysis of the ChIP DNA to assess the relative H4 lysine acetylation of the genomic regions in stolon and tubers, respectively.
TABLE 2.
Primers for real-time PCR using ChIP DNA as templates
| Positiona | Gene specificity | Primer design template | Primer sequence | Amplicon (bp) | Annealing temperature |
|---|---|---|---|---|---|
| a | Non-patatin | BAC 14K07 | GCTAAAGGTGTTATTGAAACTGAGGAATC | 242 | 60° |
| TCTGAACTCTATGAAATGCTTGTTGAGAC | |||||
| b | Non-patatin | BAC 14K07 | GTCAGTCATTCCATTACTTGCCGAAACGA | 255 | 60° |
| CGTGTGACCACATTTTATGTAATAGGTAG | |||||
| c | Class I promoter | BAC 14K07 gene 3 | ATGGTAGATTGTTTGAGCGGATAATCTTC | 252 | 60° |
| GGTATATATAGGCACGTAAACAAGTTGAG | |||||
| d | Class II promoter | BAC 14K07 gene 15 | TGCTGCATTATTTCTTTTACATTGTTGGAT | 304 | 60° |
| ACTACAACAAGGAGTCGTTTGATAGAGTG | |||||
| e | Class I 5′-UTR— exon 1 | BAC 14K07 gene 3 | CCAACAAAATTTAAAAACACTTTGAACATTTGC | 200 | 60° |
| AGTTGTCCTTCAAGAAATTCGAGAATGGT | |||||
| f | Class II 5′-UTR— exon 1 | BAC 14K07 gene 15 | TTGAGATATCAGTTTTTATTAATTATAATCTGC | 200 | 55° |
| AGTTGTCCTTCAAGAAATTCGAGAATGGT | |||||
| g | Patatin intron 3 | BAC 14K07 gene 3 | GGTGAGATCGGACGTGTTTAGGAGTAGTT | 231 | 65° |
| AAATGAACCCATTCACCACTGAGATCCAC | |||||
| h | Patatin exon 5 | BAC 14K07 gene 3 | GATCCAGCATTTGCTTCAATTAGGTCATT | 241 | 60° |
| CCCTGAGGTAATTGTTTTGTGAATGAAGA | |||||
| i | Patatin 3′-UTR | BAC 14K07 gene 3 | TTCAAGGTCTCGGGTTGTAGTTGTAAAT | 147 | 55° |
| TCAAACATTAAGCACAACTAAAAGGTTAC | |||||
| NA | Positive control | Ubiq conj enzyme2 | AGCTTTGATGACTGGAATCTTGTTTTGTG | 269 | 65° |
| CAACTCTGAGCCCTTCACAATAAAAGCAC |
Refers to the position indicated in Figure 1C.
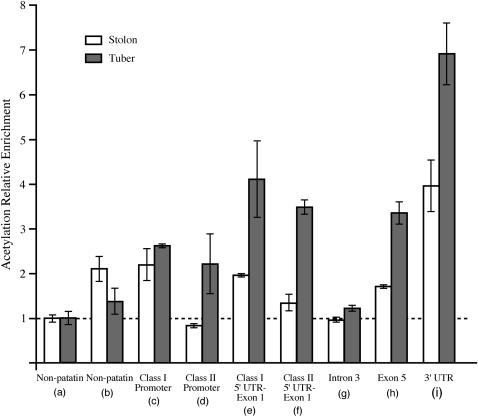
We used a non-patatin gene primer pair “a” (Figure 1C; Table 2) as a reference for all other primers because this primer represents a region that displayed a low relative histone H4 lysine acetylation in both stolons and tubers and is thus effective as a negative control. The ChIP data showed that patatin gene-related regions generally have more histone H4 lysine acetylation than the reference non-patatin gene region “a” (Figure 3). The histone H4 lysine acetylation level of patatin gene regions relative to non-patatin regions appears to be greater in tubers than in stolons (Figure 3). These results indicate that there are alterations in the histone acetylation pattern of patatin gene regions during the developmental transition from stolons to tubers. Furthermore, these changes in chromatin state coincide with the upregulation of patatin gene expression.
Figure 3.
Histone H4 lysine acetylation patterns of patatin genes at two different development stages (stolons vs. tubers). The letters in parentheses represent the PCR primer positions in BAC 14K07 (Figure 1C). All data were standardized to the reference primer set for the primer pair (a). Error bars indicate the standard deviation of two experimental replicates from the same ChIP DNA samples. The horizontal dashed line represents a relative H4 lysine acetylation enrichment of 1, which is the baseline enrichment of the non-patatin (a) region. All values >1 indicate a relative fold increase of histone H4 lysine acetylation compared to the non-patatin (a) region (see Table 2 for real-time PCR primer information and supplemental Figure 1 at http://www.genetics.org/supplemental/ for PCR primer pair quality assessment).
The different patatin gene regions exhibited variable levels of acetylation (Figure 3). Patatin exons 1 and 5 are highly enriched in the ChIPed DNA. Interestingly, the 3′-UTR region displayed the greatest enrichment in both the stolons and tubers. The only patatin gene region that showed no obvious histone H4 lysine acetylation enrichment was the third intron. It has been well demonstrated in both plant and animal species that there may be significant variation of histone modification throughout different regions within a single gene (He et al. 2004; Schneider et al. 2004). However, in the present study the regional variation in relative enrichment within individual gene copies needs to be interpreted with caution because the number of loci actually represented by each primer pair is unknown. For instance, exons tend to be more conserved than introns throughout the patatin gene family; thus multiple exon loci may be contributing to the PCR products in both the exon 1 and the 5 primer pairs; whereas the intron 3 primer pair is more likely to be specific to one or a few loci. Therefore, each primer pair may represent some range of patatin gene loci, and the relative enrichment observed may reflect the chromatin alteration within that range of loci.
The 3′-UTR structure of patatin genes is correlated with differential expression during tuber development:
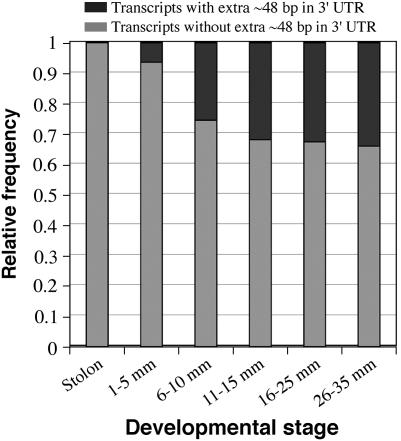
We used 3′ RACE to profile the expression of patatin genes preceding and throughout tuber development (see materials and methods). A universal patatin gene sense primer was developed for the 3′ RACE analysis (supplemental Figure 2 at http://www.genetics.org/supplemental/). We sequenced a total of 278 patatin 3′ RACE amplicons, with a minimum of 43 sequences from each of six developmental stages. The amplicons can be broadly grouped into two categories on the basis of the presence or absence of a 48-bp region located in the 3′-UTR (Figure 4). The patatin genes reported in the literature and found in BAC 14K07 display either form of this 3′-UTR (Table 3). Therefore, the presence/absence of this 48 bp appears to be a consequence of variation in the DNA template rather than of differential splicing in the 3′-UTR. The stolon amplicons consisted exclusively of transcripts lacking the 48-bp region. A gradual increase in transcripts with the 48-bp region was observed from early through later tuber growth (Figure 5).
Figure 4.
Structural diversity of patatin genes in the 3′-UTR. Shown here are eight examples of 3′ RACE PCR amplicon sequences amplified with the patatin universal sense primer. These amplicons were all sampled from the 1- to 5-mm developmental stage. Polymorphic nucleotides are indicated with shading. The terminal nucleotide of the stop codon is marked with an arrow. The 48-bp 3′-UTR insertion/deletion region is also marked by arrows. “PCR” notation indicates that the amplicon was derived from a colony PCR template. “Plas” notation indicates that the amplicon was derived from prepped plasmid template. However, the template source should not affect the results (GenBank accession numbers for the displayed sequences: DQ274226, DQ274227, DQ274232, DQ274237, DQ274244, DQ274252, DQ274260, and DQ274266).
TABLE 3.
The 5′- and 3′-UTR structural features of sequenced patatin genes
| Accession | 5′-UTR structure | 3′-UTR structure | Source | Reference |
|---|---|---|---|---|
| X01125 | Class I | 48 bp present | cDNA | Mignery et al. (1984) |
| U09331 | Class I | 48 bp present | cDNA | Banfalvi et al. (1994) |
| AF498099 | Class I | 48 bp present | cDNA | GenBank accession |
| X13179 | Class I | 48 bp present | cDNA | Stiekema et al. (1988) |
| M21879 | Class I | 48 bp present | cDNA | Stiekema et al. (1988) |
| X13178 | Class I | 48 bp present | cDNA | Stiekema et al. (1988) |
| Z27221 | Class I | 48 bp absent | cDNA | GenBank accession |
| StPat14K07.03 | Class I | 48 bp absent | Genomic gene | This study |
| M18880 | Class I | 48 bp present | Genomic gene | Mignery et al. (1988) |
| X03956 | Class I | 48 bp present | Genomic gene | Bevan et al. (1986) |
| X03932 | Class II | 48 bp absent | Genomic gene | Rosahl et al. (1986) |
| StPat14K07.08 | Class II | 48 bp present | Genomic pseudogene | This study |
| X04077 | Class II | 48 bp present | Genomic pseudogene | Pikaard et al. (1986) |
| X04078 | Class II | ∼20 bp present | Genomic pseudogene | Pikaard et al. (1986) |
Figure 5.
Relative abundance of patatin gene transcripts with the additional 48 bp in the 3′-UTR throughout tuber development. The proportion of 3′-UTR types is based on relative abundance per developmental stage.
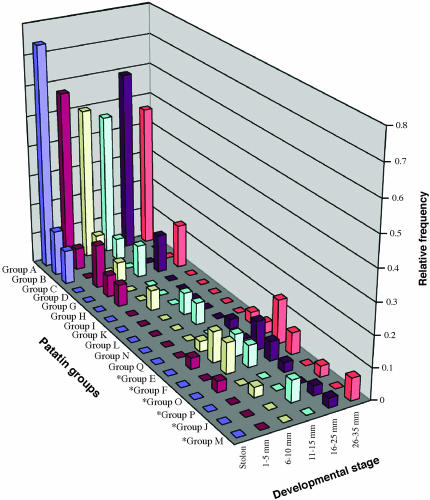
The amplicon sequences were further subdivided into groups on the basis of sequence identity within the final 401 bp of the 3′ coding region. The 3′-UTR sequences were not included as criteria in these groupings due to the occasional occurrence of ambiguous sequence reads in these regions, in particular around the polyadenylated sequences. A total of 180 amplicons were grouped into 17 different groups that are designated patatin gene groups A–Q (Figure 6). Another 98 amplicons did not have any identical sequence matches. The predicted amino acid sequences of the 17 groups were aligned and analyzed phylogenetically (supplemental Figure 3A at http://www.genetics.org/supplemental/). There was a correlation between group sequence similarity and the presence/absence of the 48 bp in the 3′-UTR (supplemental Figure 3B at http://www.genetics.org/supplemental/). Therefore, the increased frequency of transcripts with the 48 bp in the later tuber developmental stages suggests that there is an associated shift in the relative abundance of certain patatin protein forms.
Figure 6.
Relative abundance of different patatin gene groups throughout tuber development. Relative frequency of the 17 patatin groups is displayed per developmental stage. Groups that have an additional 48 bp in the 3′-UTR (see Figures 5 and 6) are denoted by asterisks (*); these transcripts become more abundant in the later developmental stages. The 98 “singleton” amplicons (described in results and discussion) were not included in the relative frequency calculation.
The cause and rationale for such a shift in different developmental stages remain unknown. Patatin genes show tissue- and cell-specific transcriptional regulation. Pikaard et al. (1987) divided patatin genes into two classes on the basis of the presence (class II) or absence (class I) of 22 nucleotides in the 5′-UTR and determined that the two classes are correlated with the tissue specificity of patatin gene transcription. It was subsequently demonstrated that patatin gene promoters determine the cell-specific transcription of some patatin genes (Koster-Topfer et al. 1989; Rocha-Sosa et al. 1989; Wenzler et al. 1989b). For example, a class I promoter showed transcriptional activity throughout tuber parenchyma cells (Rocha-Sosa et al. 1989), but expression of a class II promoter was limited to fewer and more specific cell types (Koster-Topfer et al. 1989). The relative proportion of certain cell types will change throughout stolon and early tuber development (Artschwager 1924; Reeve et al. 1969). Therefore, the transcriptional abundance of cell-type-specific patatin genes is probably, in part, determined by the relative abundance of cell types present at a given developmental stage. In the present study, this may explain the shift in later tuber development toward transcripts with the additional 48 bp in the 3′-UTR.
The presence/absence of the 48 bp in the 3′-UTR and the different 5′-UTR classes characterized by Pikaard et al. (1987) appear to associate with gene expression in different ways. Previous studies showed that tubers express class I patatin genes 50- to 100-fold more frequently than class II forms, implying that tubers may silence certain patatin gene forms and selectively express forms with specific 5′-UTR structures (Pikaard et al. 1987; Mignery et al. 1988). However, in the present study tubers expressed transcripts both with and without the 48 bp in the 3′-UTR, while stolons expressed only the forms lacking the 48-bp sequence. This suggests that tuberization is correlated with activation of patatin genes with variable 3′-UTR structures that appear to be silenced in stolons. Interestingly, on the basis of 5′-UTR analyses Pikaard et al. (1987) suggested that a specific subset of patatin genes is predominant in tuber patatin expression, but our data suggest that tuber patatin forms become more diverse following tuber initiation and growth (also see Table 4).
TABLE 4.
Patatin gene groups detected in each developmental stage
| Developmental stage | Groups detected |
|---|---|
| Stolon (mm) | A, B, C |
| 1–5 | A, B, D, Ea, G, H, Oa |
| 6–10 | A, B, D, Ea, Fa, I, Pa, Q |
| 11–15 | A, B, D, Ea, Fa, Ja, K, L |
| 16–25 | A, D, Ea, Fa, Ja, Ma, N, Oa |
| 26–35 | A, D, Ea, Fa, Ma, N, Pa, Q |
Groups are associated with the additional 48 bp in the 3′-UTR.
It is unlikely that the 5′-UTR class structure is associated with the presence/absence of the 48 bp in the 3′-UTR. As stated above, class I transcripts are ∼50- to 100-fold more prevalent than class II transcripts in tubers (Mignery et al. 1988). Therefore, we assume that the vast majority of our sampled 3′ RACE amplicons are class I forms, including those with the extra 48 bp in the 3′-UTR, which is approximately one-third of the transcripts in the later tuber developmental stages (Figure 5). Additionally, structural analysis of genomic clones and full-length cDNAs indicated that the 5′-UTR class structure is not related to the presence/absence of the 48 bp in the 3′-UTR (Table 3). Patatin gene structures have been identified for all four possible combinations: class I 5′-UTR with the 48-bp 3′-UTR sequence, class I 5′-UTR without the 48-bp 3′-UTR sequence, class II 5′-UTR with the 48-bp 3′-UTR sequence, and class II 5′-UTR without the 48-bp 3′-UTR sequence (Table 3).
The 98 patatin 3′ RACE sequences that do not have any identical matches within the data set were designated as “singletons.” Five such singleton sequences (GenBank accessions DQ274223, DQ274236, DQ274254, DQ274326, and DQ274337) have frameshift mutations within the coding region, which may represent degenerated coding regions of transcribed pseudogenes or may result from experimental errors. Some singletons appear to be unique patatin gene transcripts, with several unique base substitutions and no codon frameshifts (for example, the sequence for clone “PCR30” in Figure 4 appears to be a very unique functional singleton). However, it is unlikely that all the singletons represent unique patatin genes. Sequence identity comparisons were made between the remaining 93 singleton sequences and the 17 patatin gene groups from this study. It was determined that 51 of the singletons (∼55%) had only one nucleotide mismatch from being identical to one of the 17 groups. This indicates that many of the singletons may represent true group matches; however, they exhibit nucleotide polymorphism as a consequence of sequencing errors or PCR-induced mutations.
The most abundant patatin gene group remains dominant throughout tuber development:
Despite the shift in transcript frequency correlated with the 3′-UTR structure, patatin gene group A (GenBank accession DQ274461), which lacks the 48-bp 3′-UTR insertion, is the most abundant patatin gene group in all stages of tuber development (Figure 6). We found that 99 of the 278 total amplicons are 100% identical to the patatin gene group A sequence throughout the terminal 401-bp coding region. However, it is unclear whether these transcripts are derived from a single locus or from multiple loci with identical sequences in the final 401 bp of their coding region. Patatin gene group A has no identical sequence matches in GenBank. The most similar sequence match is a class I patatin cDNA from a Korean potato cultivar (GenBank accession Z27221). This sequence is identical to 400 of the 401 coding bases in patatin gene group A and has identical sequence matches for almost the entire 3′-UTR. The next most similar patatin gene match in GenBank is only ∼94% identical to the coding region of group A.
We used PCR to amplify and sequence the entire coding regions of the patatin gene group A transcript. We designed a primer matching the 3′-UTR of several patatin gene groups, including group A (ATTTACAACTACAACCCGAGACCTTGAAT), and a near-universal class I patatin 5′-UTR primer (CMAACAAAATTTAAAAACAMWTTGAACATTTGC, where M = C and A and W = A and T) to amplify the entire coding regions of 1- to 5-mm and 26- to 35-mm tuber cDNAs. A total of 34 amplicons were cloned and sequenced. The entire coding regions were determined for these 34 clones; thus we refer to these data as patatin coding sequences (CDS).
Several different patatin CDSs were identified by this approach, including transcripts that match the terminal 401 coding nucleotides from patatin gene groups A, D, M, and J (Table 5, GenBank accessions DQ274478–DQ274487). Additionally, 15 unique CDSs were identified (GenBank accessions DQ274488–DQ274502); these CDSs do not match any of the patatin gene groups identified by 3′ RACE. All 34 CDSs, however, display perfect conservation of the patatin gene universal sense 27-bp priming region used for generating the 3′ RACE libraries, further confirming the unbiased nature of the RACE profiling approach used in this study.
TABLE 5.
List of fully sequenced patatin coding sequences (CDS)
| CDS name | No. of clonesa | Notes |
|---|---|---|
| Patatin A-1 | 7 | |
| Patatin A-2 | 1 | 1 nt different from A-1 |
| Patatin A-3 | 1 | 1 nt different from A-1 |
| Patatin D-1 | 2 | |
| Patatin D-2 | 1 | 1 nt different from D-1 |
| Patatin D-3 | 1 | 1 nt different from D-1 |
| Patatin M-1 | 3 | |
| Patatin M-2 | 1 | 1 nt different from M-1 |
| Patatin M-3 | 1 | 1 nt different from M-1 |
| Patatin J-1 | 1 | |
| Unique CDSsb | 15 |
3′-UTR primer, ATTTACAACTACAACCCGAGACCTTGAAT; 5′-UTR primer, CMAACAAAATTTAAAAACAMWTTGAACATTTGC. There is differential homology of these primers to potential cDNA templates (see text). Therefore these data should not be interpreted as an unbiased estimate of the relative frequency of each CDS.
Unique CDSs have no identical matches to the patatin groups defined by the terminal 401 nucleotides or to any other CDS in this study.
Nine CDSs matched the terminal 401-bp coding region of patatin gene group A (Table 5). Seven of these nine CDSs were identical throughout the entire CDS and were thus designated “patatin gene group A-1” (GenBank accession DQ274478). Interestingly, the patatin gene group A-1 CDS is identical to the predicted coding sequence of gene StPat14K07.03 from BAC 14K07. The remaining two group A sequences each had one nonsynonymous nucleotide difference relative to the patatin gene group A-1 sequence and were thus designated A-2 and A-3, respectively (GenBank accessions DQ274479 and DQ274480). It is unclear whether these polymorphic bases represent true nucleotide variation or variation caused by technical error. Nevertheless, it appears that the patatin gene group A-1 sequence (supplemental Figure 4 at http://www.genetics.org/supplemental/) at minimum represents the majority of the group A transcripts. Thus, we propose that the patatin gene group A-1 represents the most abundant form of patatin found throughout tuber development in potato cultivar Kennebec.
It would be interesting to know why patatin gene group A is so abundant throughout early tuber development. Patatin gene coding regions are highly conserved; however, patatin gene promoters are highly variable in terms of sequence motifs and structure. Different patatin gene promoters show variable expression abundance and localization in promoter-GUS fusion assays (Koster-Topfer et al. 1989; Rocha-Sosa et al. 1989; Wenzler et al. 1989b). Therefore, it appears that the variations in patatin gene promoter sequence structure ultimately determine the activity and function of any given patatin gene. The patatin group A promoter may have features that confer the high activity of this gene(s).
Comparison of patatin and maize zein gene families:
The zein gene family of maize is one of the best-characterized storage protein gene families. Extensive BAC-based sequence analysis has been conducted on the zein gene clusters (Song et al. 2001; Song and Messing 2002, 2003). Sequence analysis of BAC 14K07 indicates that the patatin gene clusters have similar structural features to the zein gene clusters. Similar to the patatin gene family, functional zein genes are interspersed with inactive zein genes (Song et al. 2001; Song and Messing 2002, 2003). These inactive forms have features similar to the patatin pseudogenes described above, including gene truncation, internal deletion, and premature stop codons (Song et al. 2001; Song and Messing 2002). One extensively studied gene cluster, the 22-kDa α-zein cluster, is 346 kb in length and includes six active genes and 16 copies inactive due to in-frame stop codons (Song et al. 2001). The zein gene density in this cluster is ∼1 copy per 15.7 kb. The BAC 14K07 sequence indicates that the patatin locus may have a similar patatin gene density and a similar ratio of active to inactive copies. Additionally, all active and inactive copies of the 22-kDa α-zein cluster are orientated with the same directional polarity (Song et al. 2001; Song and Messing 2003), which is also true of the BAC 14K07 patatin gene copies.
The functional and structural similarities of patatin and zein gene families suggest that they may have experienced similar evolutionary pathways. As discussed above, patatin proteins likely have secondary functions, and evolutionary selection for these functions may explain the current patatin gene structure. The zein protein clusters have similar structural features to patatin, but have not been shown to have a secondary function beyond seed protein storage. This suggests that storage protein gene families may be under selective pressure directly for the protein storage trait. Differential gene expression of zein gene copies in different genetic backgrounds (Song and Messing 2003) and of patatin gene copies throughout tuber development supports the hypothesis that each protein storage gene copy may differentially contribute to the plant phenotype and is thereby subject to selection for protein storage function.
Sequencing of 3′ RACE amplicons as a tool for transcript profiling of multicopy gene families:
The analysis of transcriptional regulation of multicopy gene families is problematic in part because it is difficult to distinguish between gene copies with similar structure and sequence. Hybridization-based techniques, such as RNA gel blot or microarray analysis, cannot distinguish between the various transcripts from multiple loci. Quantitative real-time PCR is also of limited value for similar reasons; PCR primers may be difficult to design for individual gene copies due to the high sequence similarity among the different gene members. Song and Messing (2003) were able to profile the expression of the zein gene family in maize by cloning and sequencing PCR amplicons from zein gene transcripts. Using cDNA templates, they performed PCR reactions with primers that are conserved among all zein genes. Their PCR amplicons had sufficient internal polymorphism that allowed different gene copies to be distinguished by sequence analysis.
In this study, we were able to design one conserved patatin sense primer toward the 3′ end of the transcript. However, we were unable to design a compatible antisense primer because of sequence divergence among known patatin gene transcripts in the compatible region. Therefore, the conserved sense primer was used to generate 3′ RACE amplicons, essentially exploiting the polyadenylation tail as the antisense primer in the PCR reaction. The cloning and sequencing of these amplicons generated a data set from which transcripts could be analyzed and categorized on the basis of sequence identity and polymorphism. We were then able to utilize priming sites from the 3′-UTR sequence of abundant transcripts and from a near-universal 5′-UTR region to PCR amplify and sequence the complete CDS of the most transcribed patatin genes. Our results show that this strategy was effective in profiling the expression of the potato patatin gene family and can be potentially used to analyze other multicopy gene families.
Acknowledgments
We thank Bala Pudota for assisting in the growth of plant materials. We are grateful to Robin Buell, Jia Liu, and Bill Belknap for their contributions in sequence annotation of BAC 14K07 and to Robin Buell for critical reading of this manuscript. This work was supported by National Science Foundation Plant Genome Research Program grants DBI-9975866 and DBI-0218166.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under the following accession nos.: BAC 14K07, accession no. DQ274179; the 3′ RACE sequence data, accession nos. DQ274180–DQ274460; the 401 bp of the 3′ coding regions for patatin gene groups A–Q, accession nos. DQ274461–DQ274477; and the patatin complete CDS sequence data, accession nos. DQ274478–DQ274502.
References
- Artschwager, E., 1924. Studies on the potato tuber. J. Agric. Res. 27: 809–835. [Google Scholar]
- Bachem, C., R. van Der Hoeven, J. Lucker, R. Oomen, E. Casarini et al., 2000. Functional genomic analysis of potato tuber life-cycle. Potato Res. 43: 297–312. [Google Scholar]
- Banfalvi, Z., Z. Kostyal and E. Barta, 1994. Solanum brevidens possesses a non-sucrose-inducible patatin gene. Mol. Gen. Genet. 245: 517–522. [DOI] [PubMed] [Google Scholar]
- Bevan, M., R. Barker, A. Goldsbrough, M. Jarvis, T. Kavanagh et al., 1986. The structure and transcription start site of a major potato tuber protein gene. Nucleic Acids Res. 14: 4625–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresee, K. A., 2002. Molecular characterization of the major storage protein family in potato. M.S. Thesis, University of Wisconsin, Madison, WI.
- Burge, C., and S. Karlin, 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Feng, Q., Y. J. Zhang, P. Hao, S. Y. Wang, G. Fu et al., 2002. Sequence and analysis of rice chromosome 4. Nature 420: 316–320. [DOI] [PubMed] [Google Scholar]
- Ganal, M. W., M. W. Bonierbale, M. S. Roeder, W. D. Park and S. D. Tanksley, 1991. Genetic and physical mapping of the patatin genes in potato and tomato. Mol. Gen. Genet. 225: 501–509. [DOI] [PubMed] [Google Scholar]
- Grierson, C., J. S. Du, M. de Torres Zabala, K. Beggs, C. Smith et al., 1994. Separate cis sequences and trans factors direct metabolic and developmental regulation of a potato tuber storage protein gene. Plant J. 5: 815–826. [DOI] [PubMed] [Google Scholar]
- He, Y., M. R. Doyle and R. M. Amasino, 2004. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 18: 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks, T., D. Vreugdenhil and W. J. Stiekema, 1991. Patatin and four serine proteinase inhibitor genes are differentially expressed during potato tuber development. Plant Mol. Biol. 17: 385–394. [DOI] [PubMed] [Google Scholar]
- Jiang, J., B. S. Gill, G. L. Wang, P. C. Ronald and D. C. Ward, 1995. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92: 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster-Topfer, M., W. B. Frommer, M. Rocha-Sosa, S. Rosahl, J. Schell et al., 1989. A class II patatin promoter is under developmental control in both transgenic potato and tobacco plants. Mol. Gen. Genet. 219: 390–396. [DOI] [PubMed] [Google Scholar]
- Liu, Y. W., C. H. Han, M. H. Lee, F. L. Hsu and W. C. Hou, 2003. Patatin, the tuber storage protein of potato (Solanum tuberosum L.), exhibits antioxidant activity in vitro. J. Agric Food Chem. 51: 4389–4393. [DOI] [PubMed] [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lukashin, A. V., and M. Borodovsky, 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Mignery, G. A., C. S. Pikaard, D. J. Hannapel and W. D. Park, 1984. Isolation and sequence analysis of cDNAs for the major potato tuber protein, patatin. Nucleic Acids Res. 12: 7987–8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignery, G. A., C. S. Pikaard and W. D. Park, 1988. Molecular characterization of the patatin multigene family of potato. Gene 62: 27–44. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., P. B. Talbert, C. X. Zhong, R. K. Dawe, S. Henikoff et al., 2003. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163: 1221–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W. D., C. Blackwood, G. A. Mignery, M. A. Hermodson and R. M. Lister, 1983. Analysis of the heterogeneity of the 40,000 molecular weight tuber glycoprotein of potatoes by immunological methods and by NH2-terminal sequence analysis. Plant Physiol. 71: 156–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard, C. S., G. A. Mignery, D. P. Ma, V. J. Stark and W. D. Park, 1986. Sequence of two apparent pseudogenes of the major potato tuber protein, patatin. Nucleic Acids Res. 14: 5564–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard, C. S., J. S. Brusca, D. J. Hannapel and W. D. Park, 1987. The two classes of genes for the major potato tuber protein, patatin, are differentially expressed in tubers and roots. Nucleic Acids Res. 15: 1979–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pots, A. M., H. Gruppen, M. Hessing, M. A. J. S. van Boekel and A. G. J. Voragen, 1999. Isolation and characterization of patatin isoforms. J. Agric. Food Chem. 47: 4587–4592. [DOI] [PubMed] [Google Scholar]
- Prat, S., W. B. Frommer, R. Hofgen, M. Keil, J. Kossmann et al., 1990. Gene expression during tuber development in potato plants. FEBS Lett. 268: 334–338. [DOI] [PubMed] [Google Scholar]
- Reeve, R. M., E. Hautala and M. L. Weaver, 1969. Anatomy and compositional variation of potatoes. I. Developmental histology of the tuber. Am. Potato J. 46: 361–373. [Google Scholar]
- Rocha-Sosa, M., U. Sonnewald, W. Frommer, M. Stratmann, J. Schell et al., 1989. Both developmental and metabolic signals activate the promoter of a class I patatin gene. EMBO J. 8: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosahl, S., R. Schmidt, J. Schell and L. Willmitzer, 1986. Isolation and characterization of a gene from Solanum tuberosum encoding patatin, the major storage protein of potato tubers. Mol. Gen. Genet. 203: 214–220. [Google Scholar]
- Rydel, T. J., J. M. Williams, E. Krieger, F. Moshiri, W. C. Stallings et al., 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42: 6696–6708. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson et al., 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6: 73–77. [DOI] [PubMed] [Google Scholar]
- Sharma, N., H. A. Gruszewski, S. W. Park, D. G. Holm and J. M. Vivanco, 2004. Purification of an isoform of patatin with antimicrobial activity against Phytophthora infestans. Plant Physiol. Biochem. 42: 647–655. [DOI] [PubMed] [Google Scholar]
- Shewry, P. R., 2003. Tuber storage proteins. Ann. Bot. 91: 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., F. Dong and J. Jiang, 2000. Construction of a bacterial artificial chromosome (BAC) library for potato molecular cytogenetics research. Genome 43: 199–204. [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2002. Contiguous genomic DNA sequence comprising the 19-kD zein gene family from maize. Plant Physiol. 130: 1626–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., and J. Messing, 2003. Gene expression of a gene family in maize based on noncollinear haplotypes. Proc. Natl. Acad. Sci. USA 100: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, R., V. Llaca, E. Linton and J. Messing, 2001. Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res. 11: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiekema, W. J., F. Heidekamp, W. G. Dirkse, J. Vanbeckum, P. Dehaan et al., 1988. Molecular cloning and analysis of 4 potato tuber messenger RNAs. Plant Mol. Biol. 11: 255–269. [DOI] [PubMed] [Google Scholar]
- Strahl, B. D., and C. D. Allis, 2000. The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Strickland, J. A., G. L. Orr and T. A. Walsh, 1995. Inhibition of diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiol. 109: 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Q., Y. L. Chang, J. Wang, H. Chen, M. N. Islam-Faridi et al., 2001. Bacterial artificial chromosome-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics 158: 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., and G. Ooms, 1988. Structural diversity of the patatin gene family in potato cv. Desiree. Mol. Gen. Genet. 212: 325–336. [DOI] [PubMed] [Google Scholar]
- Wenzler, H., G. Mignery, L. Fisher and W. Park, 1989. a Sucrose-regulated expression of a chimeric potato tuber gene in leaves of transgenic tobacco plants. Plant Mol. Biol. 13: 347–354. [DOI] [PubMed] [Google Scholar]
- Wenzler, H. C., G. A. Mignery, L. M. Fisher and W. D. Park, 1989. b Analysis of a chimeric class-I patatin-GUS gene in transgenic potato plants: high-level expression in tubers and sucrose-inducible expression in cultured leaf and stem explants. Plant Mol. Biol. 12: 41–50. [DOI] [PubMed] [Google Scholar]
- Yuan, Q., J. Hill, J. Hsiao, K. Moffat, S. Ouyang et al., 2002. Genome sequencing of a 239 Kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast insertion. Mol. Genet. Genomics 267: 713–720. [DOI] [PubMed] [Google Scholar]
- Zourelidou, M., M. de Torres-Zabala, C. Smith and M. W. Bevan, 2002. Storekeeper defines a new class of plant-specific DNA-binding proteins and is a putative regulator of patatin expression. Plant J. 30: 489–497. [DOI] [PubMed] [Google Scholar]