Abstract
Wheat–barley chromosome addition lines are useful genetic resources for a variety of studies. In this study, transcript accumulation patterns in Betzes barley, Chinese Spring wheat, and Chinese Spring–Betzes chromosome addition lines were examined with the Barley1 Affymetrix GeneChip probe array. Of the 4014 transcripts detected in Betzes but not in Chinese Spring, 365, 271, 265, 323, 194, and 369 were detected in wheat–barley disomic chromosome addition lines 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), and 1(7H), respectively. Thus, 1787 barley transcripts were detected in a wheat genetic background and, by virtue of the addition line in which they were detected, were physically mapped to barley chromosomes. We validated and extended our approach to physically map barley genes to the long and short arms of chromosome 6(6H). Our physical map data exhibited a high level of synteny with homologous sequences on the wheat and/or rice syntenous chromosomes, indicating that our barley physical maps are robust. Our results show that barley transcript detection in wheat–barley chromosome addition lines is an efficient approach for large-scale physical mapping of genes.
ALIEN chromosome addition lines have been developed for a variety of plant species and have been used for many purposes such as introducing valuable traits to the recipient species, mapping genes and markers on introgressed alien chromosomes, examining alien gene regulation, understanding meiotic pairing behavior and chromosome structure, and isolating individual chromosomes and genes of interest (e.g., Melzer et al. 1988; Islam and Shepherd 1990; Ananiev et al. 1997; Bass et al. 2000; Muehlbauer et al. 2000; Jin et al. 2004). Wheat (Triticum aestivum L.)–barley (Hordeum vulgare L.) disomic chromosome addition lines have been developed through wide hybridization between the hexaploid (2n = 6x = 42) wheat cultivar Chinese Spring (CS) and the diploid (2n = 2x = 14) barley cultivar Betzes (Islam et al. 1975). Each addition line contains the full complement of wheat chromosomes and a single homeologous chromosome pair from barley. Wheat–barley disomic addition lines for six of the seven barley chromosomes including 1(7H), 2(2H), 3(3H), 4(4H), 6(6H), and 7(5H) and ditelosomic addition lines harboring 13 of the 14 barley chromosome arms have been generated (Islam et al. 1981; Islam 1983; Islam and Shepherd 1990, 2000). A stable and self-maintaining wheat–barley disomic addition line for chromosome 5(1H) is not available due to one or more genes on 5L(1HL) causing sterility when present in wheat (Islam et al. 1981; Islam and Shepherd 1990).
Chromosome addition lines have been used often to map genes to donor chromosomes on the basis of the presence/absence of the genes on the chromosomes added to the recipient genome. Using wheat–barley chromosome addition lines, isozymes and DNA markers have been physically mapped to chromosomes and chromosome arms (e.g., Islam and Shepherd 1990; Garvin et al. 1998). Similar studies have been conducted in a set of oat–maize chromosome addition lines and their radiation hybrids (Okagaki et al. 2001; Kynast et al. 2004). Although the use of addition lines for physical mapping in both systems has been successful, the use of these lines as high-throughput mapping resources has been restricted by the lack of technologies to allocate markers to specific chromosomes.
Little is known about the regulation of alien gene expression after introgression into a foreign genetic background. Chromosome addition lines have made it possible to examine gene expression from the donor chromosome in the recipient genetic background. Isozyme mapping studies in the wheat–barley addition lines confirm that barley genes are expressed in wheat (Hart et al. 1980; Islam and Shepherd 1990). However, only a limited number of barley genes encoding isozymes are available for analysis. Similarly, examination of an oat–maize chromosome 3 addition line revealed expression of the maize liguleless3 gene, resulting in a liguleless phenotype (Muehlbauer et al. 2000). Although such studies show that introgressed alien genes can be expressed in a recipient genetic background, the number of genes examined has been small, and the extent of donor gene expression has not been quantified on a large scale.
Microarray analysis provides the opportunity to quantify the abundance of transcripts derived from thousands of genes simultaneously. The Barley1 Affymetrix GeneChip probe array (Affymetrix, Santa Clara, CA) represents 22,792 barley genes and was the first Affymetrix chip for a large-genome plant (Close et al. 2004). The probe sets on this GeneChip are highly specific for barley transcripts, with the number of detectable transcripts in the other grasses such as wheat, rice, and maize significantly lower than that in barley (Close et al. 2004). We sought to exploit this result by using GeneChip technology to examine barley transcript accumulation in wheat–barley addition lines. Our hypothesis was that the transcripts detected in Betzes and an addition line but not in CS were derived from Betzes genes on the particular donor barley chromosome present in the addition line.
To test this hypothesis, we examined transcript profiles of Betzes, CS, and the CS–Betzes chromosome addition lines harboring barley chromosome 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), 6S(6HS), 6L(6HL), or 1(7H) at a seedling stage, using the Barley1 Affymetrix GeneChip. Our data provided the opportunity to examine the extent of barley gene transcription in a wheat genetic background and enabled large-scale physical mapping of barley genes to chromosomes and chromosome arms.
MATERIALS AND METHODS
Genetic stocks:
Wheat cv. CS (barley chromosome recipient); the barley cv. Betzes (chromosome donor); the six different fertile and self-maintaining CS–Betzes chromosome disomic addition lines harboring barley chromosomes 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), or 1(7H); and ditelosomic addition lines for the long and short arms of Betzes chromosome 6(6H) were used (Islam et al. 1981). Barley chromosome nomenclature is abbreviated to 2H, 3H, 4H, 5H, 6H, 6HL, 6HS, and 7H in the rest of this article.
Plant growth and experimental design:
Seeds of each genotype were placed in sterile plastic petri dishes containing three layers of filter paper hydrated with 8 ml of distilled H2O. To achieve uniform seed germination and seedling development for all genetic stocks, seeds were cold treated in a 4° cooler for 96 hr in the dark. The petri dishes were then transferred to a growth chamber set at 22° with 16 hr light/8 hr dark periods at a light intensity of 360 μE m−1 sec−1. The petri dishes were arranged in a randomized complete block design with three replications. A total of 1.5 ml of sterile H2O was added to each petri dish after 24 and 48 hr. After 72 hr, seedlings at the growth stage “first leaf just emerging through the coleoptile” (GRO:0007059; see supplemental Figure S1 at http://www.genetics.org/supplemental/) were sampled by detaching the seedling from the seed and immediately freezing in liquid N2. At least 10 seedlings of each genotype were sampled from each replicate.
Barley1 Affymetrix GeneChip probe array:
The Barley1 GeneChip probe array (Affymetrix) consists of 22,792 probe sets designed from an exemplar set of barley sequences derived from expressed sequence tag (EST) contigs (Close et al. 2004). In general, probe sets were designed from the last 600 bp of each exemplar sequence. In some cases, probe sets were designed to paralogs and in other cases probe sets were designed to multiple alleles, indicating that there are multiple probe sets that may identify the same transcript.
RNA extraction and GeneChip hybridizations:
Total RNA was extracted from 1 g of tissue from each replicate using the Trizol (Invitrogen, Carlsbad, CA) protocol. Samples were treated with RNase-free DNase, and the RNA was purified on RNeasy columns (Promega, Madison, WI). Prior to labeling, total RNA quality was examined on an Agilent (Palo Alto, CA) 2100 bioanalyzer. The cRNA probes were prepared using GeneChip one-cycle target labeling and control reagents (Affymetrix), following the recommended protocol. Twenty micrograms of labeled cRNA was used for each hybridization reaction. The Barley1 GeneChip hybridizations and data acquisition were conducted in the Biomedical Image Processing Facility at the University of Minnesota following standard Affymetrix procedures (http://www.bipl.ahc.umn.edu/affymetrix.html).
Analysis of GeneChip data:
GeneChip data analysis was conducted using Genedata Expressionist Pro version 1.0 software (Genedata, San Francisco). Expressionist has two primary analysis tools called Refiner and Analyst. The Refiner tool condenses and normalizes the raw signal. The Analyst tool provides statistical analysis and data visualization capability. GeneChip and control gene statistics were calculated to check masked and outlier areas (threshold level: 0.2%), corner noise (threshold level: 0.015%) and 3′/5′ mean (threshold level: 2.5%), using the Refiner tool. Within the Refiner tool, detection signal condensation and normalization were conducted using Robust Multichip Analysis (RMA) (Irizarry et al. 2003). Further analysis of transcript abundance was conducted using the Analyst tool. The correlation of expression signals between replications for each genotype was ∼0.99 across all probe sets tested on the GeneChip.
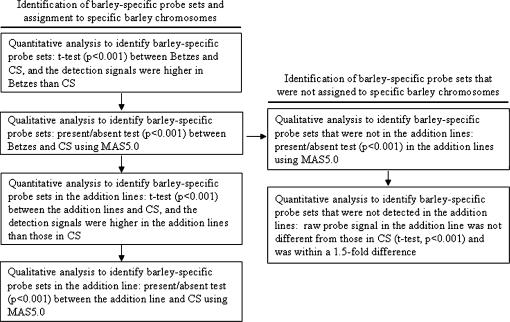
Each Barley1 GeneChip probe set was first tested for barley transcript specificity on the basis of quantitative and qualitative transcript abundance differences between Betzes and CS. The work flow for transcript detection is shown in Figure 1. Because of nonspecific signals of the Barley1 GeneChip probe sets hybridizing to wheat transcripts, only the transcripts showing significantly higher signals in Betzes compared to those in CS by t-tests at a P-value <0.001 were selected. Second, qualitative analysis of barley-specific transcript detection between Betzes and CS was conducted by a presence/absence test (detection P-value <0.001), using microarray analysis suite 5.0 (MAS 5.0) in the Analyst tool. A transcript was scored as present only if it was detected in at least two of three replications at a detection P-value <0.001, while it was scored as absent only if it did not satisfy the qualitative detection threshold level in all three replications. Therefore, only the transcripts showing significantly different abundance between Betzes and CS (t-test at a P-value <0.001) on the basis of RMA-normalized signals and scored as present in Betzes and absent in CS (MAS 5.0 detection P-value <0.001) were subjected to further analysis to identify barley transcripts in the wheat–barley chromosome addition lines.
Figure 1.
Work flow to identify barley-specific transcripts in Betzes and the wheat–barley chromosome addition lines. MAS 5.0, Microarray Analysis Suite 5.0.
To detect barley transcripts in the addition lines, differences in the abundance of barley transcript signals in the addition lines were tested against the background signal in CS by t-tests (P-value <0.001). Only those barley transcripts showing significantly higher transcript levels both in Betzes and in the addition lines compared to those in CS were subjected to qualitative analysis to detect barley-specific transcripts in the addition lines (presence/absence test, P-value <0.001 using MAS 5.0). For detection of chromosome-arm-specific transcripts on the long and short arms of 6H, probe sets targeting transcripts present both in Betzes and in the 6H disomic and ditelosomic addition lines and absent in CS were partitioned to the appropriate chromosome arm by hierarchical clustering.
All data from the Affymetrix scanner have been deposited at BarleyBase (http://www.barleybase.org/) in the form of DAT, CEL, EXP, and CHP files. The accession number for the experiment is BB8.
PCR validation of GeneChip transcript accumulation patterns and physical map locations:
To validate the transcript accumulation data obtained from the Barley1 GeneChip, we conducted RT-PCR on a subset of barley genes identified in Betzes and the chromosome addition lines. RT-PCR was carried out from first-strand cDNA synthesized from total RNA samples pooled from the three replications of the experiment using the ThermoScript RT-PCR system (Invitrogen). Barley-specific primers (see supplemental Tables S1 and S2 at http://www.genetics.org/supplemental/) were designed from the exemplar sequences corresponding to the probe sets on the Barley1 GeneChip (http://www.barleybase.org/probesetlist.php?start=1&end=200&chipdesign=Barley1), using the Primer3 program (Rozen and Skaletsky 2000).
The physical presence/absence of barley genes corresponding to transcripts detected in the addition lines was tested by genomic PCR on a subset of genes (for the lists of tested probe sets and primer sequences, see supplemental Tables S1–S5 at http://www.genetics.org/supplemental/). Separation of PCR products was conducted either by agarose (1.5%) gel electrophoresis or by single-stranded conformation polymorphism analysis on 0.5× mutation detection enhancement (MDE) gels (Cambrex Bioscience, Rockland, ME).
In silico comparative mapping to the rice genome and mapped wheat ESTs:
Genomic locations of wheat and rice sequences homologous to barley genes were allocated to chromosomes on the basis of addition line transcript accumulation patterns determined by BLASTN searches. Target barley sequences from the Barley1 GeneChip probe sets used for the searches are available in fasta format at the Barley1 Affymetrix GeneChip technical support web page (http://www.affymetrix.com/support/technical/byproduct.affx?product=barley1). The wheat EST map database (Graingenes, a database for Triticeae and Avena: www.graingenes.org/cgi-bin/ace/custom/goBlast/graingenes) and the whole rice genome database (National Institute of Agrobiological Sciences DNA bank, Japan: http://riceblast.dna.affrc.go.jp/cgi-bin/robo-blast/blast2.cgi?dbname=all) were used to conduct the BLASTN searches (E < e−10). Multiple hits to wheat and/or rice sequences were counted as many times as they were found when the hits were to different chromosomes. In contrast, multiple hits to wheat and/or rice sequences were counted as one hit when they were found on the same chromosomal location.
RESULTS
Barley gene transcription in wheat–barley chromosome addition lines:
Transcript abundance patterns in seedling tissues of Betzes barley, CS wheat, and six wheat–barley chromosome addition lines harboring barley chromosome 2H, 3H, 4H, 5H, 6H, or 7H were examined with the Barley1 GeneChip. The work flow to determine the presence of barley-specific transcripts in the addition lines is shown in Figure 1. Among 22,792 probe sets on this GeneChip, 7828 showed significantly higher detection signals in Betzes compared to CS (t-test, P < 0.001). Of these probe sets, 4097 were subsequently classified as present in Betzes but not in CS by a presence/absence test using MAS 5.0 (detection P-value <0.001). The Barley1 GeneChip platform has some probe sets targeting paralogs or different alleles of genes (Close et al. 2004). For the purposes of this study, we removed all “redundant” probe sets targeting paralogs or different alleles to remove any bias from counting a single transcript two or more times due to its identification by multiple probe sets. Subtraction of these redundant probe sets resulted in 4014 probe sets (transcripts) detected in Betzes but not in CS.
To detect Betzes transcripts in the addition lines, we compared the transcript abundance of these 4014 probe sets in the addition lines to CS both by t-tests (P < 0.001) and by a presence/absence test (detection P-value <0.001 using MAS 5.0). This resulted in the detection of 365, 271, 265, 323, 194, and 369 transcripts (1787 in total) showing higher detection signals in the addition lines carrying barley chromosome 2H, 3H, 4H, 5H, 6H, and 7H, respectively (Table 1 and see supplemental Figure S2 at http://www.genetics.org/supplemental/). These 1787 transcripts were then examined for barley chromosome specificity. We found 344, 243, 240, 301, 187, and 342 (1657 in total) transcripts that were specific to single addition lines carrying barley chromosomes 2H, 3H, 4H, 5H, 6H, and 7H, respectively. These transcripts were termed “single copy.” The remaining 130 transcripts were detected in two or more chromosome addition lines (Table 1 and see supplemental Figure S2 at http://www.genetics.org/supplemental/). These transcripts (21, 28, 25, 22, 7, and 27 transcripts detected in addition lines 2H, 3H, 4H, 5H, 6H, and 7H, respectively) were identified by 55 unique probe sets. Because these transcripts were each detected in more than one wheat–barley chromosome addition line by identical probe sets, we considered them to be products of multiple genes belonging to 55 different barley gene families.
TABLE 1.
Detection patterns of barley-specific transcripts in Betzes, Chinese Spring, and Chinese Spring–Betzes chromosome addition lines harboring barley chromosome 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), or 1(7H)
| Genetic stock
|
No. of barley transcripts
|
No. of single-copy transcriptsa
|
No. of multiple-copy transcriptsa
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Betzes | CS | 2H | 3H | 4H | 5H | 6H | 7H | |||
| + | − | + | 365 | 344 | 21 | |||||
| + | − | + | 271 | 243 | 28 | |||||
| + | − | + | 265 | 240 | 25 | |||||
| + | − | + | 323 | 301 | 22 | |||||
| + | − | + | 194 | 187 | 7 | |||||
| + | − | + | 369 | 342 | 27 | |||||
Transcript accumulation patterns among Betzes, CS, and the wheat–barley disomic chromosome addition lines were determined on the basis of t-tests (P < 0.001) of transcript signals normalized by robust multichip analysis (RMA) followed by presence (+) or absence (−) test (detection P < 0.001 using microarray analysis suite 5.0) of 22,792 probe sets on the Barley1 Affymetrix GeneChip. See Figure 1 for details on the work flow. CS, Chinese Spring; 2H, 3H, 4H, 5H, 6H, and 7H are the wheat–barley chromosome addition lines carrying barley chromosomes 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), and 1(7H), respectively.
Determination of single- or multiple-copy transcripts was based on specificity of a given transcript to each barley chromosome added to wheat.
While 1787 of the 4014 unique Betzes transcripts were assigned to one or more addition lines, 2227 transcripts could not be assigned to an addition line on the basis of our statistical thresholds. To detect transcripts that met a statistical significance for not accumulating in the addition lines, we compared raw transcript levels of these 2227 transcripts in CS and in the disomic addition lines (t-tests, P < 0.001, and >1.5-fold difference). The work flow to determine absence of barley-specific transcripts in the addition lines is shown in Figure 1. Using these criteria, we identified 687 transcripts showing higher transcript abundance in Betzes compared to CS and the addition lines. These transcripts were designated as exclusive to Betzes and not found in CS or in the addition lines. The remaining 1540 (2267 − 687) were classified as “marginal transcripts” because the detection signals in the addition lines were different from CS at a relaxed threshold level. However, they did not satisfy our original statistical thresholds to be assigned to a particular addition line and were not considered in further analysis.
Chromosome arm-specific barley gene transcript patterns:
To validate the approach of assigning transcript accumulation on a chromosome-specific basis, we examined barley transcript accumulation in wheat–barley ditelosomic addition lines carrying either the short or the long arm of chromosome 6H. We postulated that transcripts identified in Betzes, the chromosome 6H disomic addition line, and either the 6HL or 6HS ditelosomic addition line but not in CS were derived from genes on the particular arm of barley chromosome 6H. We detected 194 transcripts in the disomic chromosome 6H addition line. Hierarchical clustering partitioned 111 and 81 transcripts to 6HL and 6HS, respectively (Table 2, transcript patterns 1 and 2, and see supplemental Figure S2 at http://www.genetics.org/supplemental/). The remaining two chromosome 6H transcripts, detected by the probe sets rbags22p06_s_at and HD12H12r_at, were not detected in either of the ditelosomic chromosome 6HL or 6HS addition lines (Table 2, transcript pattern 3). Genomic PCR of the target sequence of rbags22p06_s_at amplified the target sequence in Betzes, the 6H disomic addition line, and the 6HL ditelosomic addition line (see supplemental Table S1 at http://www.genetics.org/supplemental/), showing that the gene was present on chromosome 6HL. We could not amplify genomic sequence for the gene represented by probe set HD12H12r_at. Thus, 193 of 194 transcripts assigned to chromosome 6H were validated by the results obtained from analysis of the chromosome 6HS and 6HL ditelosomic addition lines.
TABLE 2.
Transcript detection patterns in Betzes, Chinese Spring, and Chinese Spring–Betzes chromosome 6(6H) addition lines
| Pattern | Betzes | CS | 6H | 6HL | 6HS | No. of transcripts |
|---|---|---|---|---|---|---|
| 1 | + | − | + | + | − | 111 |
| 2 | + | − | + | − | + | 81 |
| 3 | + | − | + | − | − | 2 |
Partitioning of 194 barley chromosome 6(6H)-specific transcripts to barley chromosome 6(6H) arms, 6L(6HL) and 6S(6HS) was based on hierarchical clustering analysis of accumulation patterns of these 194 transcripts in Betzes, CS, the wheat–barley disomic 6(6H) addition line, and the wheat–barley ditelosomic chromosome 6L(6HL) and 6S(6HS) addition lines. CS, Chinese Spring; 6H, the Chinese Spring–Betzes chromosome 6(6H) disomic addition line; 6HL and 6HS, the Chinese Spring–Betzes chromosome 6L(6HL) and 6S(6HS) ditelosomic addition lines, respectively.
Validation of barley transcript accumulation in wheat–barley chromosome addition lines:
Confirmation of barley chromosome-specific transcript detection patterns was sought by RT-PCR for a random group of transcripts detected by 12 probe sets in the chromosome 6H addition lines. Four of these transcripts showed a 6HL-specific detection pattern and 8 showed a 6HS-specific detection pattern (see supplemental Figure S3 and supplemental Table S1 at http://www.genetics.org/supplemental/). The RT-PCR results were concordant with the GeneChip transcript patterns for all 12 transcripts. In addition, 8 probe sets detecting barley transcripts when a relaxed presence/absence detection significance level (P < 0.04) was used were similarly evaluated and found to confirm the GeneChip data, although they are not included in the final transcript list (see supplemental Table S2 at http://www.genetics.org/supplemental/). These results show that the quantitative and qualitative analysis of differential transcript abundance in this study was both robust and stringent.
Physical mapping of barley gene transcripts detected in the addition lines:
To test the feasibility of direct physical mapping of barley genes based on their expression patterns in the addition lines, we conducted genomic PCR for a subset of 19 barley genes detected in our study (2 genes on 2H, 1 gene on 3H, 3 genes on 5H, 11 genes on 6H, and 2 genes on 7H) (see supplemental Tables S1 and S3 at http://www.genetics.org/supplemental/). Of these 19 genes, 17 were confirmed to reside on the predicted barley chromosomes in a manner consistent with their transcript accumulation patterns. One 6H-specific gene (Contig3307_at) and 1 5H-specific gene (Contig3898_at) were not amplified in any of the addition lines. We also tested the physical locations of 9 genes exhibiting chromosome arm 6HL- and 6HS-specific transcript patterns by genomic PCR. For each of these genes, their physical locations were congruent with their transcript accumulation patterns (see supplemental Figure S3 and Table S1 at http://www.genetics.org/supplemental/). In addition to confirming the chromosome locations of barley genes whose transcripts were identified with highly stringent P-values, we also tested the locations of an additional 18 genes that exhibited barley chromosome-specific detection patterns in the addition lines when a relaxed statistical stringency (P-value <0.04) was used (see supplemental Tables S2 and S4 at http://www.genetics.org/supplemental/). All 18 of these genes were found to be located on the predicted barley chromosomes. Thus, of the 37 genes tested for their physical location on barley chromosomes (19 genes detected at a P-value <0.001 and 18 genes detected at a P-value <0.04), 35 were found to be located on the same barley chromosomes predicted on the basis of their barley chromosome-specific transcript patterns.
To determine if barley-specific sequences for the 687 transcripts detected in Betzes but not in CS or the addition lines were present in the addition lines, genomic PCR was conducted for 16 randomly selected genes. Only 1 of 16 genes (Contig3198_s_at) was amplified in an addition line (see supplemental Table S5 at http://www.genetics.org/supplemental/). These PCR results indicate that our ability to identify gene transcripts that are not present in the addition lines is robust.
In silico comparative mapping of the barley transcriptome:
We assigned 365, 271, 265, 323, 194, and 369 barley gene transcripts to barley chromosomes 2H, 3H, 4H, 5H, 6H, and 7H, respectively, on the basis of chromosome-specific transcript patterns observed in the wheat–barley chromosome addition lines. Additional evidence for the chromosome assignment of barley genes encoding these transcripts was obtained by determining the locations of wheat and rice homologs in their respective genomes. Genomic synteny among wheat, rice, and barley has been established through a variety of studies (e.g., Van Deynze et al. 1995; Gale and Devos 1998; Sorrells et al. 2003). Barley chromosomes 2H, 3H, 4H, 5H, 6H, and 7H are syntenic to wheat chromosome groups 2, 3, 4, 5, 6, and 7, respectively. Barley chromosomes exhibit the following syntenic relationships with rice chromosomes: 2H–rice 4 and 7; 3H–rice 1; 4H–rice 3 and 11; 5H–rice 3, 9, and 12; 6H–rice 2; and 7H–rice 6 and 8. Thus, if wheat and rice sequences homologous to our tentatively mapped barley genes are found in syntenous regions within their respective genomes, this would provide additional support for our barley gene chromosome assignments.
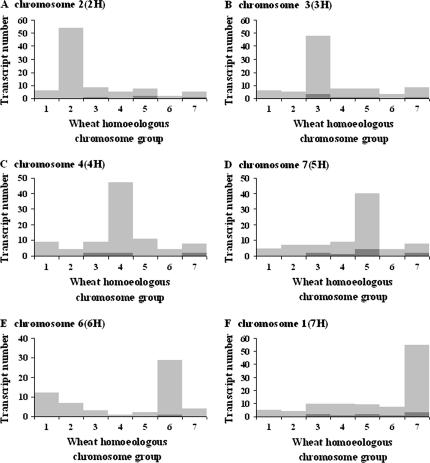
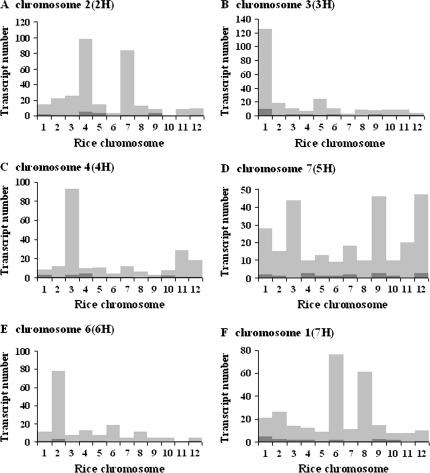
To test this, barley target sequences from the Barley1 GeneChip probe sets were used for BLASTN searches against wheat ESTs and rice genomic sequences. These extracted 371 wheat ESTs and 979 rice sequences with similarity to the 1787 physically mapped barley target sequences. Detection of homologous wheat ESTs on syntenous wheat chromosomes ranged from 69% (5H addition line) to 78% (2H addition line), while for rice this ranged from 72% (7H addition line) to 89% (2H addition line) (Figures 2 and 3 and see supplemental Tables S6 and S7 at http://www.genetics.org/supplemental/). Across all chromosomes, ∼74 and 79% of our physically mapped barley genes exhibited genomic synteny to wheat and rice, respectively. These comparative mapping results provide compelling support for the accuracy of physical mapping of barley genes based on their transcript accumulation patterns in the addition lines. For a complete description of all physically mapped genes and comparative mapping data see supplemental Tables S8–S13 at http://www.genetics.org/supplemental/.
Figure 2.
In silico comparative mapping of barley genes to mapped wheat ESTs. (A–F) Barley chromosome 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), and 1(7H), respectively. Bars with light and dark shading represent single- and multiple-copy transcripts, respectively. Barley chromosomes 2(2H), 3(3H), 4(4H), 7(5H), 6(6H), and 1(7H) are syntenic to wheat chromosome groups 2, 3, 4, 5, 6, and 7, respectively (Van Deynze et al. 1995; Gale and Devos 1998). The transcript number shown in each wheat homeologous chromosome group represents the total number of wheat genes found to be homologous to barley sequences by BLASTN (E < e−10) search using the wheat EST map database (www.graingenes.org/cgi-bin/ace/custom/goBlast/graingenes).
Figure 3.
In silico comparative mapping of barley genes to the rice genome sequence. (A–F) Barley chromosomes 2H, 3H, 4H, 5H, 6H, and 7H, respectively. Bars with light and dark shading represent single- and multiple-copy transcripts, respectively. Genomic synteny of barley chromosomes to rice chromosomes is as follows: barley 2H to rice 4 and 7; barley 3H to rice 1; barley 4H to rice 3 and 11; barley 5H to rice 3, 9, and 12; barley 6H to rice 2; and barley 7H to rice 6 and 8 (Van Deynze et al. 1995; Gale and Devos 1998). The transcript number shown in each rice chromosome represents the total number of rice genes found to be homologous to barley sequences by BLASTN (E < e−10) search using the rice genome database (http://riceblast.dna.affrc.go.jp/cgi-bin/robo-blast/blast2.cgi?dbname=all).
DISCUSSION
Barley gene transcript accumulation in wheat–barley chromosome addition lines:
The Barley1 GeneChip was used to detect barley gene transcripts in seedling tissues of wheat–barley chromosome addition lines. This is the first large-scale examination of the extent of alien gene expression in chromosome addition lines. In the six wheat–barley disomic addition lines used here, we observed the accumulation of 1787 barley transcripts expressed at a single developmental stage, indicating that a substantial number of barley genes are actively transcribed in a wheat genetic background. Examining the transcript abundance at different developmental stages or in different tissue types would likely increase the number of barley gene transcripts observed. In addition, selection of statistical stringency for transcript detection dramatically alters the number of transcripts detected. Our empirical studies with this same data set indicate that at lower stringencies ∼4000–5000 barley transcripts can be detected in the wheat genetic background. However, for this report we used highly conservative conditions to detect the number of barley genes expressed in wheat to ensure a high degree of confidence in the results. Further examination of the data set will undoubtedly uncover many more barley transcripts in the wheat genetic background than described here. As the data set is publicly available for data mining by interested researchers, statistical stringencies can be selected as desired. For instance, if one is interested in obtaining as many genes as possible in a given chromosome, a lower stringency could be used and the possibility of a misread may be acceptable relative to the identification of larger numbers of genes.
For the individual barley chromosomes, the transcript numbers detected were 365, 271, 265, 323, 194, and 369 for chromosomes 2H, 3H, 4H, 5H, 6H, and 7H, respectively. The estimated physical sizes of chromosomes 2H, 3H, 4H, 6H, and 7H are approximately equal (Brown et al. 1999). Interestingly, the number of detected transcripts between chromosome 6H with the smallest number of transcribed genes and chromosome 7H, with the largest number of detected transcripts, differed nearly by a factor of two. Taken together, our results demonstrate that the extent of barley gene expression from individual chromosomes introgressed into wheat is substantial and that the physical size of the barley chromosomes is not necessarily associated with the number of expressed genes that were detected in this study.
The Barley1 GeneChip is an efficient platform for physical mapping of barley genes:
As of June 2005, 394,937 ESTs of H. vulgare subsp. vulgare are publicly available in dbEST (www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html). However, of the ∼4000 markers mapped in barley, only ∼700 are expressed sequences (http://barleygenomics.wsu.edu/xl061201.xls; Thiel et al. 2003). To accelerate EST mapping in barley, we used the Barley1 GeneChip in combination with the wheat–barley addition lines to assign genes to chromosomes. We did so by considering the presence of a specific barley transcript in a chromosome addition line to be the signature of the presence of the barley gene encoding the transcript. In this study, we successfully detected barley transcripts in six wheat–barley chromosome addition lines, and 1787 barley genes were physically assigned to six barley chromosomes. This represents a substantial increase in the number of barley genes mapped to barley chromosomes. For a listing of all physically mapped genes, see supplemental Tables S8–S13 at http://www.genetics.org/supplemental/.
Chromosome addition lines and chromosome deletion lines have been used for large-scale physical mapping. The oat–maize chromosome addition lines were used to assign 300 EST and 50 STS markers to maize chromosomes (Okagaki et al. 2001). In wheat, using 101 wheat chromosome deletion lines, 24 ditelosomic lines, and 21 nullisomic-tetrasomic lines, 7873 unique ESTs were mapped to 21 chromosomes in three genomes (Akhunov et al. 2003; http://wheat.pw.usda.gov/NSF/progress_mapping.html). However, in spite of the accuracy of these approaches, substantial commitments of personnel and physical resources were required to carry out these projects due to the techniques used. Our novel GeneChip-based method of mapping expressed sequences to specific chromosomes or chromosome arms has proven to be an efficient alternative strategy to physically map a large number of genes in a short period of time. Additional genes can be physically mapped in barley using this same approach by examining transcript accumulation in different tissues or by using different bioinformatics stringencies. Our approach can also be applied to other chromosome addition line stocks for which appropriate microarray resources exist.
Genomic synteny of barley to other grass species:
Our results revealed that ∼74 and 79% of the 1787 barley genes physically mapped in our study exhibited synteny with wheat and rice, respectively. Previous comparative genomics studies in grass species also showed significant genomic synteny between barley and rice (Kilian et al. 1995; Saghai Maroof et al. 1996; Smilde et al. 2001; Goff et al. 2002; Gottwald et al. 2004; Perovic et al. 2004), between barley and wheat (Hohmann et al. 1995; Foote et al. 1997; Feuillet and Keller 1999), and between rice and wheat (Goff et al. 2002; Sorrells et al. 2003). Large-scale comparative mapping between wheat and rice showed that the degree of synteny ranged from 58% between wheat chromosome group 6 and rice chromosome 2 to 86% between wheat chromosome group 5 and rice chromosomes 3, 9, and 12 (Sorrells et al. 2003; Conley et al. 2004; Hossain et al. 2004; Linkiewicz et al. 2004; Miftahudin et al. 2004; Munkvold et al. 2004; Peng et al. 2004; Randhawa et al. 2004). The results obtained from our study using microarray technology applied to wheat–barley chromosome addition lines are consistent with these prior results and further extend our knowledge of the syntenic relationships between members of the grass family.
Acknowledgments
We acknowledge Zheng Jin Tu at the Computational Genetics Laboratory, University of Minnesota and Joel Mason at the U.S. Department of Agriculture (USDA)-Agricultural Research Service, Plant Science Research Unit for technical support and James Anderson and Howard Rines for manuscript review. This work was funded by grants from the North American Barley Genome Project to G.J.M. and D.F.G. and by the USDA-Initiative for Future Agriculture and Food Systems program to G.J.M.
References
- Akhunov, E. D., J. A. Goodyear, S. Geng, L.-L. Qi, B. Echalier et al., 2003. The organization and rate of evolution of wheat genomes are correlated with recombination rates along chromosome arms. Genome Res. 13: 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E. V., O. Riera-Lizarazu, H. W. Rines and R. L. Phillips, 1997. Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 94: 3524–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, H. W., O. Riera-Lizarazu, E. V. Ananiev, S. Bordoli, H. W. Rines et al., 2000. Evidence of the coincident initiation of homolog pairing and synapsis during telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Brown, S. E., J. L. Stephens, N. L. V. Lapitan and D. L. Knudson, 1999. FISH landmarks for barley chromosomes (Hordeum vulgare L.). Genome 42: 274–281. [PubMed] [Google Scholar]
- Close, T. J., S. I. Wanamaker, R. A. Caldo, S. M. Turner, D. A. Ashlock et al., 2004. A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol. 134: 960–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley, E. J., V. Nduati, J. L. Gonzalez-Hernandez, A. Mesfin, M. Trudeau-Spanjers et al., 2004. A 2600-locus chromosome bin map of wheat homoeologous group 2 reveals interstitial gene-rich islands and colinearity with rice. Genetics 168: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 1999. High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96: 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote, T., M. Roberts, N. Kurata, T. Sasaki and G. Moore, 1997. Detailed comparative mapping of cereal chromosome regions corresponding to the Ph1 locus in wheat. Genetics 147: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95: 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin, D. F., J. E. Miller-Garvin, E. A. Viccars, J. V. Jacobsen and A. H. D. Brown, 1998. Identification of molecular markers linked to ant28, a mutation that eliminates proanthocyanidin in barley seeds. Crop Sci. 38: 1250–1255. [Google Scholar]
- Goff, S. A., D. Ricke, T.-H. Lan, G. Presting, R. Wang et al., 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Gottwald, S., N. Stein, A. Börner, T. Sasaki and A. Graner, 2004. The gibberellic-acid insensitive dwarfing gene sdw3 of barley is located on chromosome 2HS in a region that shows high colinearity with rice chromosome 7L. Mol. Gen. Genomics 271: 426–436. [DOI] [PubMed] [Google Scholar]
- Hart, G. E., A. K. M. R. Islam and K. W. Sheperd, 1980. Use of isozymes as chromosome markers in the isolation and characterization of wheat-barley chromosome addition lines. Genet. Res. 36: 311–326. [Google Scholar]
- Hohmann, U., A. Graner, T. R. Endo, B. S. Gill and R. G. Herrmann, 1995. Comparison of wheat physical maps with barley linkage maps for group 7 chromosomes. Theor. Appl. Genet. 91: 618–626. [DOI] [PubMed] [Google Scholar]
- Hossain, K. G., V. Kalavacharla, G. R. Lazo, J. Hegsted, M. J. Wentz et al., 2004. A chromosome bin map of 2148 expressed sequence tag loci of wheat homoeologous group 7. Genetics 168: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis et al., 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Islam, A. K. M. R., 1983. Ditelosomic additions of barley chromosomes to wheat, pp. 233–238 in Proceedings of the 6th International Wheat Genetics Symposium, edited by S. Sakamato. Maruzen, Kyoto, Japan.
- Islam, A. K. M. R., and K. W. Shepherd, 1990. Incorporation of barley chromosomes in wheat, pp. l28–151 in Wheat (Biotechnology in Agriculture and Forestry, Vol. 13), edited by Y. P. S. Bajaj. Springer-Verlag, Berlin.
- Islam, A. K. M. R., and K. W. Shepherd, 2000. Isolation of a fertile wheat-barley addition line carrying the entire barley chromosome 1H. Euphytica 111: 145–149. [Google Scholar]
- Islam, A. K. M. R., K. W. Shepherd and D. H. B. Sparrow, 1975. Addition of individual barley chromosomes to wheat, pp. 260–270 in Proceedings of the 3rd International Barley Genetics Symposium, edited by H. Gaul. Verlag Karl Thiemig, Munich, Germany.
- Islam, A. K. M. R., K. W. Shepherd and D. H. B. Sparrow, 1981. Isolation and characterization of euplasmic wheat-barley chromosome addition lines. Heredity 46: 16l–174. [Google Scholar]
- Jin, W. W., J. R. Melo, K. Nagaki, P. B. Talbert, S. Henikoff et al., 2004. Maize centromeres: organization and functional adaptation in the genetic background of oat. Plant Cell 16: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian, A., D. A. Kudrna, A. Kleinhofs, M. Yano, N. Kurata et al., 1995. Rice-barley synteny and its application to saturation mapping of the barley Rpg1 region. Nucleic Acids Res. 23: 2729–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast, R. G., R. J. Okagaki, M. W. Galatowitsch, S. R. Granath, M. S. Jacobs et al., 2004. Dissecting the maize genome by using chromosome addition and radiation hybrid lines. Proc. Natl. Acad. Sci. USA 101: 9921–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkiewicz, A. M., L. L. Qi, B. S. Gill, A. Ratnasiri, B. Echalier et al., 2004. A 2500-locus bin map of wheat homeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics 168: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer, J. M., A. Kleinhofs, D. A. Kudrna, R. L. Warner and T. K. Blake, 1988. Genetic mapping of the barley nitrate reductase deficient Nar1 and Nar2 loci. Theor. Appl. Genet. 75: 767–771. [Google Scholar]
- Miftahudin, K. Ross, X.-F. Ma, A. A. Mahmoud, J. Layton et al., 2004. Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer, G. J., O. Riera-Lizarazu, R. G. Kynast, D. Martin, R. L. Phillips et al., 2000. A maize-chromosome 3 addition line of oat exhibits expression of the maize homeobox gene liguleless3 and alterations of cell fates. Genome 43: 1055–1064. [PubMed] [Google Scholar]
- Munkvold, J. D., R. A. Greene, C. E. Bermudez-Kandianis, C. M. La Rota, H. Edwards et al., 2004. Group 3 chromosome bin maps of wheat and their relationship to rice chromosomes. Genetics 168: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki, R. J., R. G. Kynast, S. M. Livingston, C. D. Russell, H. W. Rines et al., 2001. Mapping maize sequences to chromosomes using oat-maize chromosome addition materials. Plant Physiol. 125: 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J. H., H. Zadeh, G. R. Lazo, J. P. Gustafson, S. Chao et al., 2004. Chromosome bin map of expressed sequence tags in homeologous group 1 of hexaploid wheat and homeology with rice and Arabidopsis. Genetics 168: 609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perovic, D., N. Stein, H. Zhang, A. Drescher, M. Prasad et al., 2004. An integrated approach for comparative mapping in rice and barley with special reference to the Rph16 resistance locus. Funct. Integr. Genomics 4: 74–83. [DOI] [PubMed] [Google Scholar]
- Randhawa, H. S., M. Dilbirligi, D. Sidhu, M. Erayman, D. Sandhu et al., 2004. Deletion mapping of homeologous group 6-specific wheat expressed sequence tags. Genetics 168: 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Saghai Maroof, M. A., G. P. Yang, R. M. Biyashev, P. J. Maughan and Q. Zhang, 1996. Analysis of the barley and rice genomes by comparative RFLP linkage mapping. Theor. Appl. Genet. 92: 541–551. [DOI] [PubMed] [Google Scholar]
- Smilde, W. D., J. Halukova, T. Sasaki and A. Graner, 2001. New evidence for the synteny of rice chromosome 1 and barley chromosome 3H from rice expressed sequence tags. Genome 44: 361–367. [PubMed] [Google Scholar]
- Sorrells, M. E., C. M. La Rota, C. E. Bermudez-Kandianis, R. A. Greene, R. Kantety et al., 2003. Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13: 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, T., W. Michalek, R. K. Varshney and A. Graner, 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422. [DOI] [PubMed] [Google Scholar]
- Van Deynze, A. E., J. C. Nelson, E. S. Yglesias, S. E. Harrington, D. P. Braga et al., 1995. Comparative mapping in grasses. Wheat relationships. Mol. Gen. Genet. 248: 744–754. [DOI] [PubMed] [Google Scholar]