Abstract
This study was conducted to determine whether quantitative trait loci (QTL) controlling traits of agronomic importance detected in recombinant inbred lines (RILs) are also expressed in testcross (TC) hybrids of rice. A genetic map was constructed using an RIL population derived from a cross between B5 and Minghui 63, a parent of the most widely grown hybrid rice cultivar in China. Four TC hybrid populations were produced by crossing the RILs with three maintaining lines for the widely used cytoplasmic male-sterile (CMS) lines and the genic male-sterile line Peiai64s. The mean values of the RILs for the seven traits investigated were significantly correlated to those of the F1 hybrids in the four TC populations. Twenty-seven main-effect QTL were identified in the RILs. Of these, the QTL that had the strongest effect on each of the seven traits in the RILs was detected in two or more of the TC populations, and six other QTL were detected in one TC population. Epistatic analysis revealed that the effect of epistatic QTL was relatively weak and cross combination specific. Searching publicly available QTL data in rice revealed the positional convergence of the QTL with the strongest effect in a wide range of populations and under different environments. Since the main-effect QTL is expressed across different testers, and in different genetic backgrounds and environments, it is a valuable target for gene manipulation and for further application in rice breeding. When a restorer line that expresses main-effect QTL is bred, it could be used in a number of cross combinations.
HETEROSIS has been very successfully exploited in diverse plants and animals. In agriculture, hybrid varieties contribute strongly worldwide to the production of many crop species, including the most important food crops, such as maize and rice (Stuber 1994; Yuan 1998; Khush 2001). Hybrid rice has a yield advantage of ∼15–20% over the best commercial rice varieties. The area planted to hybrid rice in China accounts for >50% of the total rice area of the country at present. The cultivation of hybrid rice has started on a large scale in many Asian countries.
Several hypotheses have been proposed to explain the genetic basis of heterosis. The dominance hypothesis (Bruce 1910) proposes that dominant factors from either parent mask deleterious recessive mutations from the other parent in the heterozygous F1 population. In contrast, the overdominance hypothesis (Shull 1908) holds that heterozygosity at single loci confers properties that are superior to either homozygote. The two hypotheses have been verified with molecular biology experiments (Stuber et al. 1992; Xiao et al. 1995). A third hypothesis suggests that heterosis may arise from epistasis between alleles at different loci (Yu et al. 1997; Goodnight 1999). More recently, further results have suggested that epistasis is the primary genetic basis of heterosis. It is suggested that separate efforts should be taken for breeding high-yielding inbred and hybrid cultivars in rice (Li et al. 2001; Luo et al. 2001).
In hybrid rice breeding programs in China, the breeders have made intense efforts to improve the traits of inbred lines and have obtained a number of elite lines, for example, Minghui 63, a restorer line of the most popular hybrid rice variety Shanyou 63, and 9311, a restorer line of the first super hybrid rice. The characteristics of the parental lines have a profound effect on those of the F1 offspring. Once an elite restorer line or male-sterile line has been developed, it is used to breed a series of hybrid varieties with strong heterosis that can be applied in rice production. The fact that the superior parental lines favorably enhance the performance of hybrid rice derived from many combinations in practice suggests that some common quantitative trait loci (QTL) may affect the performance of both the parental lines and the hybrids. Therefore, the detection of QTL controlling traits of the inbred lines and that of their hybrids is needed to understand the underlying genetic basis of the hybrid performance and to facilitate marker-aided breeding of hybrid rice.
The development of molecular markers in quantitative genetics greatly facilitates the study of quantitatively inherited complex traits related to F1 heterosis and has made it possible to dissect the polygenes associated with such traits into individual Mendelian factors (Paterson et al. 1988; Stuber et al. 1992). Molecular linkage genetic maps and QTL mapping techniques have been used to investigate the relationships between inbred lines and their hybrids and to identify QTL controlling agronomic traits and crop yields. Stuber et al. (1992) mapped QTL contributing to grain yield in two maize backcross (BC) populations derived from crosses between the F3 progeny from a B73 × Mo17 cross and their parental lines. The BC to B73 showed at least six QTL and the BC to Mo17 showed at least eight QTL for grain yield, and three of the QTL were detected in both of these BC populations. Xiao et al. (1995) investigated QTL controlling grain yield components in two rice BCF1 populations between 198 F7 recombinant inbred lines (RILs) and their parents. In all, 37 QTL were detected in the two populations: 27 QTL were detected in only one BCF1 population and the other 10 were detected in two BCF1 populations. Li et al. (2001) constructed five related rice mapping populations, including one RIL, two BC populations, and two testcross (TC) populations, which they analyzed in an attempt to detect QTL and found no correlation between the F1 populations and their maternal RILs in terms of biomass yield and grain yield. Using data from the same experiment, Luo et al. (2001) found no correlations between the F1 populations and their maternal RILs for the grain yield components PP (panicles per plant) and GP (grains per panicles). However, there was a significant correlation for GW (1000-grain weight). In current hybrid rice breeding, the male-sterile lines are crossed with elite restorer lines that are unrelated in pedigree, rather than backcrossed. The genetic relationships between the parental lines and F1 hybrids remain to be elucidated.
In the study reported in this article, we introduced an experimental design that produced TC populations by mating the RILs with maintaining lines for currently popular improved male-sterile lines with different types of cytoplasmic male sterilities and a genic male-sterile line, which are unrelated by pedigree to the RILs. The RILs were derived from a cross between Minghui 63, a parent of the most widely grown hybrid variety, Shanyou 63, and B5, a breeding line with superior resistance developed at Wuhan University. The RILs and their corresponding TC hybrids were evaluated for seven traits of agronomic importance. The objectives of this study were to detect and evaluate the QTL controlling the agronomic traits in RILs and the performance of TC hybrids and to understand the genetic relationship between the inbred lines and their F1 hybrids of improved modern rice.
MATERIALS AND METHODS
Plant materials:
Five related mapping populations were used in this study. One was an RIL population composed of 187 F8 lines derived by single-seed descent from a cross between Minghui 63 and B5 (Minghui 63/B5). Minghui 63 is a restorer line for many hybrid rice varieties. Three maintaining lines (Zhenshan 97B, II-32B, and YuetaiB) were selected to cross with the RILs because they have the same genomic composition as the corresponding cytoplasmic male-sterile (CMS) lines that are currently in common use. These lines are the maintaining lines for Zhenshan 97A, II-32A, and YuetaiA, which have WA-, IA-, and HL-type cytoplasmic male sterility, respectively. The other selected line was Peiai64s, a photo-thermo-sensitive genic male-sterile (PTGMS) line in which sterility is controlled by recessive nucleic genes. Four TC populations were developed, consisting of 160 Zhenshan 97B F1 hybrids (Zhenshan97B/RILs, TCP1), 181 II-32B F1 hybrids (II-32B/RILs, TCP2), 187 YuetaiB F1 hybrids (YuetaiB/RILs, TCP3), and 187 Peiai64s F1 hybrids (Peiai64s/RILs, TCP4).
Phenotypic evaluation:
The 902 F1 TC lines and 187 F8 RILs were laid out in a field in a randomized complete block design with two replications (plots) for phenotypic evaluation in the summer of 2002 at the experimental farm of the Hubei Academy of Agricultural Sciences (Wuhan, China). Each plot consisted of three rows, each with 10 hills. Seedlings, 30 days old, of all experimental materials were transplanted in the field with a spacing of 16.7 cm between plants within each row and 26.7 cm between the rows. The middle six plants in the central row of each plot were sampled for analysis. The seven quantitative traits investigated were: heading date (HD; in days), plant height (PH; in centimeters), panicles per plant (PPP), spikelets per panicle (SPP), grains per panicle (GPP), GW (in grams), and grain yield per plant (GYPP; in grams). For SPP, GPP, and GYPP, all panicles in a plant were counted. Means over replications, for each trait and for each population, were used for QTL and other analyses.
Molecular markers and linkage maps:
Preparation of genomic DNA from the parents and RILs followed the CTAB method as described by Murray and Thompson (1980). Two types of markers, RFLPs and SSRs, were used to survey DNA polymorphisms in the RILs. RFLP analyses, including restriction digestion, Southern blotting, and hybridization, were essentially as described by Huang et al. (2001). Six restriction enzymes (Apa, BamHI, HindIII, EcoRI, EcoRV, and DraI) were used for surveying RFLPs. The RFLP probes were kindly provided by the Japanese Rice Genome Research Project and S. D. Tanksley and S. McCouch, Cornell University. In addition, 300 primer pairs from published data were used to survey SSR polymorphisms between the parents. The analysis, including PCR reactions and detection, essentially followed the methods of Wu and Tanksley (1993). The DNA markers that detected polymorphisms between the parents were used to assay the entire population of 187 RILs. Molecular marker linkage maps were constructed using MAPMAKER/EXP version 3.0 (Lincoln et al. 1992).
Data analysis:
For mapping main-effect and epistatic QTL, QTLMapper version 1.0 (Wang et al. 1999) was employed to identify loci affecting quantitative traits on the basis of composite interval analysis. Here, the main-effect QTL and epistatic QTL were defined as QTL with main effect and interaction between a pair of QTL, respectively (Li et al. 2001). A LOD score of 3.0 was selected as the threshold for the presence of a main-effect QTL on the basis of the total map distance and the average distance between markers; a LOD of 5.0 was used for declaring the existence of a putative pairs of epistatic QTL. With such a threshold, a false-positive QTL would be detected anywhere in the entire genome with a probability of ∼0.05 (Lynch and Walsh 1998). An independence test in which the initial scan suggested that two or more QTL were located on the same chromosome was performed, as described by Paterson et al. (1988) and Lander and Botstein (1989). The total phenotypic variation explained by all QTL was estimated by fitting a multiple regression model into the QTLMapper program.
RESULTS
Molecular marker linkage map:
A molecular marker linkage map was constructed on the basis of the RILs of Minghui 63/B5 with 187 lines that served as the base population for generating the four TC populations employed in this study. A total of 244 molecular markers, including 190 RFLP and 54 SSR loci, were mapped on 12 linkage groups, covering 1478 cM according to the Kosambi function with an average interval of 6.1 cM between adjacent markers. The markers distributed relatively evenly among the chromosomes, and marker orders on the maps were in good agreement with those on previously published maps (Causse et al. 1994; Harushima et al. 1998). Genotype segregation ratios of Minhui 63 and B5 followed the expected Mendelian ratio of 1:1 for most of the markers, except 12 markers (RM233, R2724, Y0193, R569, Y143D, S1520, R1679, RM242, R562, S1559B, R1506, and R2672) displayed distorted segregation ratios. This map is suitable for QTL analysis.
The performance of the populations:
The means, SDs, and heritability of seven quantitative traits measured in the RILs and the four TC progenies are listed in Table 1. The measurements of seven traits varied widely in the RILs and the four TC progenies. The values for all of the traits were approximately normally distributed (data not shown), indicating the feasibility of QTL mapping for all these traits in the RILs and the four TC populations. The mean values showed that the grain yield and its components, except for GW, of the TC populations were higher than the corresponding values of the RIL population. The TC plants were also taller than the RIL plants. The heritability was high for PH, HD, SPP, GPP, and GW and low for PPP and GYPP in the RIL and the four F1 populations.
TABLE 1.
Summary statistics for the seven quantitative traits measured in RILs and four TC populations
| Population | Items | PH (cm) | HD (days) | PPP | SPP | GPP | GW (g) | GYPP (g) |
|---|---|---|---|---|---|---|---|---|
| RILs | Mean ± SD | 99.7 ± 5.57 | 90.3 ± 2.97 | 12.0 ± 1.30 | 89.9 ± 13.68 | 69.9 ± 13.65 | 26.4 ± 1.88 | 20.8 ± 8.4 |
| Heritabilitya | 73.1 | 89.3 | 47.3 | 86.7 | 82.5 | 80.3 | 42.3 | |
| TCP1 | Mean ± SD | 105.5 ± 6.47 | 88.6 ± 3.34 | 14.5 ± 3.45 | 131.4 ± 16.21 | 108.5 ± 15.89 | 26.0 ± 1.26 | 34.2 ± 9.08 |
| Heritability | 67.2 | 87.5 | 38.6 | 84.9 | 65.7 | 68.8 | 45.8 | |
| TCP2 | Mean ± SD | 106.3 ± 5.90 | 92.6 ± 4.51 | 12.9 ± 3.61 | 140.1 ± 16.33 | 122.6 ± 16.78 | 25.9 ± 1.23 | 33.1 ± 11.13 |
| Heritability | 69.3 | 84.3 | 35.2 | 85.2 | 64.6 | 67.7 | 53.6 | |
| TCP3 | Mean ± SD | 109.2 ± 3.51 | 85.0 ± 1.42 | 14 ± 2.46 | 136.3 ± 14.80 | 101.6 ± 14.00 | 24.5 ± 1.11 | 32.4 ± 5.56 |
| Heritability | 58.1 | 79.6 | 37.4 | 75.9 | 67.4 | 72.7 | 51.7 | |
| TCP4 | Mean ± SD | 107.1 ± 3.04 | 86 ± 1.44 | 12.7 ± 2.42 | 142.4 ± 12.90 | 114.6 ± 12.44 | 23.5 ± 1.05 | 31.8 ± 4.37 |
| Heritability | 65.3 | 82.7 | 45.5 | 79.8 | 77.2 | 76.5 | 48.2 |
PH, HD, PPP, SPP, GPP, GW, and GYPP indicate plant height, heading date, panicles per plant, spikelets per panicle, grains per panicle, 1000-grain weight, and grain yield per plant, respectively.
TCP1, TCP2, TCP3, and TCP4 indicate the TC populations comprising the F1 offspring of Zhenshan 97B × RILs, II-32B × RILs, YuetaiB × RILs, and Peiai64s × RILs, respectively.
Heritability was broad-sense Heritability (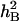 ),
), 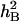 = VG/(VG + VE) × 100%.
= VG/(VG + VE) × 100%.
Relationships between the trait values of RILs and F1 populations:
Table 2 shows the phenotypic correlation coefficients between the values of individual F1 hybrids and the values of their paternal RILs for the seven traits investigated. For all traits evaluated, there was a significant correlation between the means of the RILs and their F1 performance in four TC populations, and the performance of TC hybrids was related to that of the RILs. The correlation coefficients were high for PH, HD, and GW; intermediate for PPP, SPP, and GPP; and low for GYPP. The presence of significant correlation between RILs and F1's for the investigated seven traits differs from the findings of Luo et al. (2001), possibly because of differences in the experimental materials and design between the two studies.
TABLE 2.
Phenotypic correlation (r) coefficients for traits of agronomic importance between the mean trait values of RILs and F1 performance
| Traits
|
|||||||
|---|---|---|---|---|---|---|---|
| Population | PH | HD | PPP | SPP | GPP | KGW | GYPP |
| TCP1 | 0.412** | 0.761** | 0.468** | 0.468** | 0.430** | 0.594** | 0.222* |
| TCP2 | 0.465** | 0.851** | 0.671** | 0.671** | 0.685** | 0.680** | 0.321** |
| TCP3 | 0.604** | 0.576** | 0.350** | 0.350** | 0.495** | 0.725** | 0.234* |
| TCP4 | 0.610** | 0.765** | 0.285** | 0.285** | 0.453** | 0.750** | 0.356** |
Significance levels of P < 0.05; **significance levels of P < 0.01.
The contributions of six component traits to GYPP:
The partial R2 of six component traits to the total variances of grain yield per plant in the five populations are listed in Table 3. Regression analyses indicated that the three main yield traits PPP, GPP, and GW had high contributions (partial R2) to the total variances of GYPP and that the contributions of the other traits were very low in the five populations. The partial R2 in the testcross populations (TCPs) TCP1, TCP2, TCP3, and TCP4 were 65.4, 42.3, 38.0, and 65.6% for PPP; 23.6, 8.8, 84.4, and 77.0% for GPP; and 1.6, 3.8, 7.5, and 9.7% for GW, respectively. For the RILs, the partial R2 was 24.5, 72.3, and 9.8% for PPP, GPP, and GW. For the three traits, the observed levels of the partial R2 were higher for PPP and GPP than for GW.
TABLE 3.
The partial R2 of six traits to the total variances of grain yield per plant in the five populations
| Traits
|
||||||
|---|---|---|---|---|---|---|
| Population | PH | HD | PPP | SPP | GPP | GW |
| RILs | 6.27 E-04 | 1.87E-05 | 0.245** | 4.52E-05 | 0.723** | 0.098** |
| TCP1 | 2.72 E-03** | 1.55 E-04 | 0.654** | 6.19 E-03** | 0.236** | 0.016** |
| TCP2 | 0.034** | 1.96 E-03 | 0.423** | 0.097** | 0.088** | 0.038** |
| TCP3 | 1.95 E-03 | 1.24 E-03 | 0.380** | 8.08 E-03** | 0.844** | 0.075** |
| TCP4 | 1.8E-05 | 6.62 E-03** | 0.656** | 7.14 E-03** | 0.770** | 0.097** |
Significance levels of P < 0.05; **significance levels of P < 0.01.
Main-effect QTL across RILs and TC progeny:
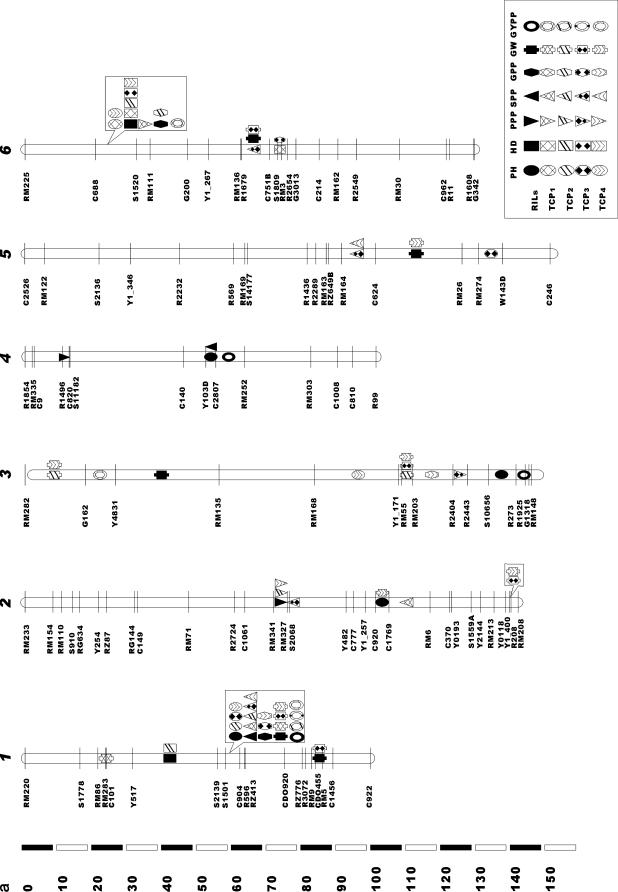
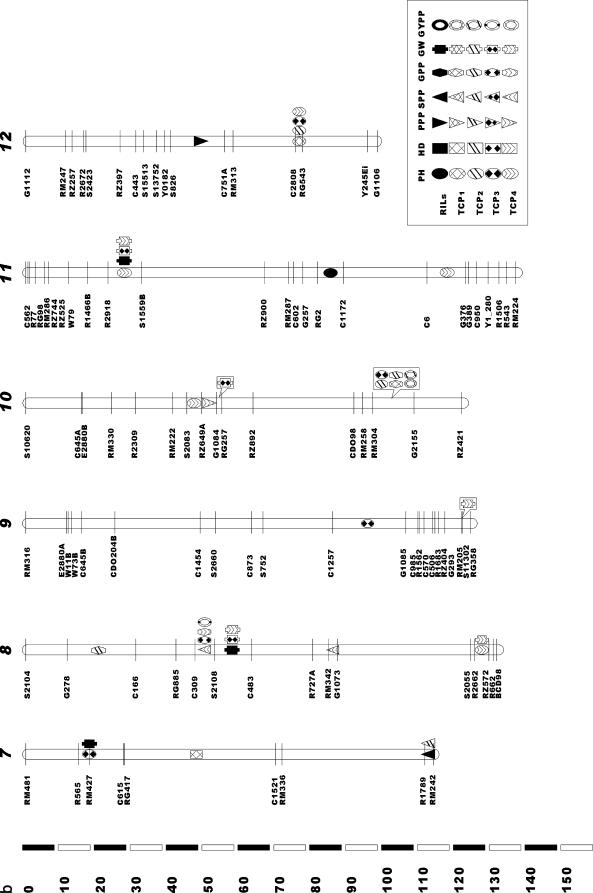
A total of 67 main-effect QTL affecting the seven traits in the RILs and the four TC populations were identified (Table 4, Figure 1). Seventeen main-effect QTL, dispersed among all 12 chromosomes, were mapped for PH, which explained 50.1, 37.1, 53.0, 40.2, and 55.7% of the variance of this trait in the RILs, TCP1, TCP2, TCP3, and TCP4, respectively. Five of these main-effect QTL were detected in the RILs. The main-effect QTL ph1, which is bordered by markers S1501-C904 on chromosome 1 and had the strongest effect in the RILs, was detected in three TC populations (TCP2, TCP3, and TCP4). The other 5 main-effect QTL detected in the RILs were not detected in any TC populations. In all, 12 main-effect QTL were detected in TC populations but not in RILs: ph12b (C2808-RG543) was detected in all four TC populations, ph6 (C688-S1520) and ph10a (RM304-G2155) were detected in two TC populations [(TCP1 and TCP4) and (TCP1 and TCP2,) respectively], while each of the other 9 main-effect QTL was detected in only one TC population. For the common main-effect QTL, for example, ph1, which was detected in the RILs and three TC progenies, the direction of the parental (B5) contribution was the same. However, the magnitude of ph1's effect was not consistent across the RILs and the three TC progenies (4.2, 3.0, 2.1, and 3.0 in the RILs, TCP2, TCP3, and TCP4, respectively; see Table 4); the difference in main effect was significant between RILs and TCP3, and not among other populations. The same tendencies were observed for the other common main-effect QTL.
TABLE 4.
Main-effect QTL affecting traits of agronomic importance detected in RILs and four TC populations
| Main-effect QTLa
|
Marker interval
|
RILs
|
TCP1
|
TCP2
|
TCP3
|
TCP4
|
Significant teste
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | LODb | Effectc | R2%d | LOD | Effect | R2% | LOD | Effect | R2% | LOD | Effect | R2% | LOD | Effect | R2% | |||
| ph1 | 1 | S1501-C904 | 15.67 | −4.2 | 23.4 | 9.63 | −3.0 | 15.0 | 6.04 | −2.1 | 9.3 | 12.89 | −3.0 | 14.2 | NS, ** | |||
| ph2 | 2 | C920-C1769 | 4.12 | −2.6 | 5.6 | |||||||||||||
| ph3a | 3 | S10656-r273 | 6.26 | 3.0 | 9.3 | |||||||||||||
| ph3b | 3 | RM168-yl-171 | 5.42 | 1.9 | 5.7 | |||||||||||||
| ph4 | 4 | y103D-C2807 | 3.60 | −2.4 | 4.7 | |||||||||||||
| ph5 | 5 | RM274-w143D | 3.83 | −1.9 | 5.4 | |||||||||||||
| ph6 | 6 | C688-S1520 | 4.95 | −3.6 | 14.1 | 6.63 | −1.8 | 4.9 | ** | |||||||||
| ph7 | 7 | C615-R565 | 4.89 | 2.0 | 7.5 | |||||||||||||
| ph8 | 8 | R2662-RZ572 | 8.79 | −2.0 | 3.8 | |||||||||||||
| ph9a | 9 | C1257-G1085 | 4.17 | 2.0 | 6.6 | |||||||||||||
| ph9b | 9 | R1562-C570 | 11.63 | 2.1 | 8.5 | |||||||||||||
| ph10a | 10 | RM304-G2155 | 7.74 | −3.8 | 16.8 | 15.63 | −3.8 | 30.0 | ||||||||||
| ph10b | 10 | S2083-RZ649A | 3.47 | −1.6 | 3.0 | |||||||||||||
| ph11a | 11 | RG2-C1172 | 4.99 | 2.8 | 7.1 | |||||||||||||
| ph11b | 11 | R2918-S1559B | 4.16 | 1.7 | 3.8 | |||||||||||||
| ph11c | 11 | C6-G376 | 7.37 | 2.0 | 7.4 | |||||||||||||
| ph12 | 12 | C2808-RG543 | 3.56 | −2.7 | 6.2 | 4.62 | −2.5 | 8.0 | 7.07 | −2.3 | 11.4 | 5.87 | −1.8 | 4.4 | NS | |||
| hd1 | 1 | y517-S2139 | 6.71 | −2.2 | 7.2 | 3.37 | −1.9 | 2.6 | NS | |||||||||
| hd6a | 6 | C688-S1520 | 12.52 | 4.3 | 54.7 | 14.27 | 3.9 | 57.9 | 15.09 | 5.7 | 69.0 | 4.99 | 1.7 | 21.5 | 6.63 | 1.9 | 36.5 | NS, ** |
| hd6b | 6 | S1809-RM3 | 3.28 | 1.7 | 3.3 | |||||||||||||
| hd7 | 7 | R565-C1521 | 5.82 | 1.9 | 5.9 | |||||||||||||
| ppp2 | 2 | RM341-RM327 | 5.99 | 0.6 | 13.2 | 4.78 | 0.8 | 11.6 | 8.92 | 0.6 | 22.6 | NS | ||||||
| ppp3 | 3 | R2404-R2443 | 4.26 | −0.6 | 10.3 | |||||||||||||
| ppp4 | 4 | R1496-C820 | 3.88 | 0.4 | 7.7 | |||||||||||||
| ppp6 | 6 | C688-S1520 | 3.75 | −1.2 | 14.1 | |||||||||||||
| ppp10 | 10 | RZ649A-G1084 | 3.76 | 0.4 | 8.3 | |||||||||||||
| ppp12 | 12 | S826-C751A | 3.83 | −0.4 | 7.7 | |||||||||||||
| spp1 | 1 | S1501-C904 | 9.64 | −15.1 | 26.1 | 13.52 | −8.5 | 32.8 | 6.02 | −8.4 | 22.1 | 10.85 | −5.3 | 18.4 | 7.25 | −4.5 | 11.9 | NS, *, ** |
| spp2a | 2 | C1769-RM6 | 3.22 | −3.9 | 7.1 | |||||||||||||
| spp2b | 2 | RM327-S2068 | 8.52 | −4.5 | 13.0 | |||||||||||||
| spp4 | 4 | y103D-C2807 | 4.27 | −5.4 | 8.5 | |||||||||||||
| spp5 | 5 | RM164-C624 | 6.38 | −4.1 | 10.9 | 4.57 | −3.5 | 7.2 | NS | |||||||||
| spp6 | 6 | R1679-C751B | 8.20 | 4.2 | 11.7 | |||||||||||||
| spp7 | 7 | R1789-RM242 | 4.29 | 7.0 | 12.9 | 9.36 | 7.0 | 8.4 | ||||||||||
| spp8a | 8 | C309-S2108 | 6.89 | −4.5 | 11.8 | |||||||||||||
| spp8b | 8 | RM342-G1073 | 4.91 | −4.1 | 9.8 | |||||||||||||
| gpp1 | 1 | S1501-C904 | 10.40 | −18.2 | 27.6 | 13.27 | −7.2 | 25.0 | 15.74 | −6.0 | 23.3 | NS, ** | ||||||
| gpp3 | 3 | RM203-R2404 | 10.07 | 4.6 | 13.4 | |||||||||||||
| gpp6a | 6 | C688-S1520 | 4.29 | −13.0 | 12.9 | 3.23 | −3.0 | 4.4 | ** | |||||||||
| gpp6b | 6 | RM3-R2654 | 3.21 | 3.4 | 5.6 | |||||||||||||
| gpp7 | 7 | R1789-RM242 | 4.27 | 12.4 | 8.5 | |||||||||||||
| gpp8a | 8 | G278-C166 | 3.51 | −8.2 | 4.0 | |||||||||||||
| gpp8b | 8 | C309-S2108 | 4.52 | −3.9 | 7.6 | 8.55 | −4.4 | 12.3 | NS | |||||||||
| gpp10 | 10 | RM304-G2155 | 7.89 | −9.7 | 21.5 | 6.96 | −10.8 | 20.8 | NS | |||||||||
| gw1a | 1 | C904-R596 | 10.6 | −1.1 | 16.2 | 5.51 | −0.5 | 13.1 | 9.38 | −0.4 | 6.3 | 5.38 | −0.3 | 4.5 | NS, ** | |||
| gw1b | 1 | CDO455-RM5 | 4.18 | 0.6 | 5.6 | 4.66 | 0.3 | 4.4 | NS | |||||||||
| gw1c | 1 | RM283-C101 | 3.46 | 0.4 | 7.1 | |||||||||||||
| gw2a | 2 | Y1-400-R208 | 10.2 | 0.5 | 8.5 | 14.00 | 0.6 | 8.3 | NS | |||||||||
| gw2b | 2 | C920-C1769 | 3.33 | −0.3 | 2.7 | |||||||||||||
| gw3a | 3 | y4831-RM135 | 5.27 | −0.8 | 9.7 | |||||||||||||
| gw3b | 3 | RM55-RM203 | 3.02 | −0.4 | 7.3 | 5.44 | −0.3 | 5.2 | 4.07 | −0.3 | 4.5 | NS | ||||||
| gw3c | 3 | RM282-G162 | 3.59 | −0.4 | 7.9 | 3.13 | −0.3 | 4.3 | NS | |||||||||
| gw5 | 5 | C624-RM26 | 3.14 | −0.6 | 5.5 | 4.55 | −0.3 | 3.1 | NS | |||||||||
| gw6 | 6 | R1679-C751B | 5.40 | −0.7 | 6.8 | 3.16 | −0.3 | 4.3 | * | |||||||||
| gw7 | 7 | C615-R565 | 8.13 | 0.9 | 13.1 | |||||||||||||
| gw8a | 8 | S2108-C483 | 6.08 | 0.7 | 6.3 | 7.08 | 0.3 | 5.5 | 3.62 | 0.3 | 3.6 | NS, * | ||||||
| gw8b | 8 | R2662-RZ572 | 8.04 | −0.5 | 8.2 | |||||||||||||
| gw9 | 9 | RM205-S11302 | 5.33 | 0.4 | 3.0 | |||||||||||||
| gw10 | 10 | G1084-RG257 | 4.49 | −0.3 | 3.5 | |||||||||||||
| gw11 | 11 | R2918-S1559B | 4.24 | −0.7 | 6.9 | 17.66 | −0.9 | 21.4 | 8.04 | −0.5 | 6.2 | NS, * | ||||||
| gypp1 | 1 | S1501-C904 | 18.08 | −3.4 | 31.3 | 6.08 | −2.6 | 13.3 | 4.23 | −1.6 | 10.8 | 6.93 | −1.5 | 15.4 | NS, *, ** | |||
| gypp3a | 3 | R1925-G1318 | 4.53 | 1.4 | 6.6 | |||||||||||||
| gypp3b | 3 | G162-y4831 | 3.13 | 1.1 | 7.8 | |||||||||||||
| gypp4 | 4 | C2807-RM252 | 3.06 | −1.2 | 4.6 | |||||||||||||
| gypp6 | 6 | C688-S1520 | 3.37 | −5.2 | 10.5 | 2.95 | −4.3 | 7.7 | NS | |||||||||
| gypp8 | 8 | C309-S2108 | 6.89 | −4.5 | 11.8 | |||||||||||||
| gypp10 | 10 | RM304-G2155 | 5.68 | −6.6 | 17.1 | 8.08 | −2.6 | 16.4 | ** | |||||||||
NS, no significance; *significance levels of P < 0.05; **significance levels of P < 0.01.
QTLs are named by trait abbreviations plus chromosomal number.
A LOD score of 3.0 was used for declaring the existence of a putative QTL, according to the method of Lynch and Walsh (1998).
In the RILs, QTL effect is the additive effect, i.e., the average substitution effect of the B5 allele by the Minhui allele. In the TC populations, QTL effects were estimated by the difference between the heterozygote tester/Minhui and the heterozygote tester/B5. The heterozygosity was 17.22, 16.67, 14.44, and 17.78% for Zhenshan97B/B5, II-32B/B5, YuetaiB/B5, and Peiai64s/B5, respectively, and 19.94, 20.00, 19.44, and 18.89% for Zhenshan 97B/Minhui, II-32B/Minhui, YuetaiB/Minhui, and Peiai64s/Minhui, respectively. The genetic expectation of the QTL effect is the additive gene effect (α) when estimated from the RILs, the additive effects, and the dominance effects (α + d) from the F1 mean values.
Variation explained by each QTL.
For testing the difference of QTL effect estimated among different populations.
Figure 1.
Genomic locations of main-effect QTL for the seven investigated traits of agronomic importance in the Minghui63/B5 RILs and four testcross F1 populations.
Only four main-effect QTL were detected for HD, accounting for 61.9, 67.1, 71.6, 21.5, and 36.5% of the total variance of the trait in the RILs, TCP1, TCP2, TCP3, and TCP4, respectively. Two main-effect QTL were detected in the RILs. Hd6a, which is bordered by markers C688-S1520 on chromosome 6 and has the largest effect in the RILs, was also identified in four TC populations. This allele contributed a considerable portion to the total heading date variation in the TCPs (47.9% on average, ranging from 21.5 to 69.0%). Another main-effect QTL detected in RILs, hd1, was also identified in TCP2. The other two main-effect QTL (hd6b and hd7) were identified in only one TC population. For the common main-effect QTL (hd6a), detected across the RILs and four TC progenies, the source of parental contribution was the same (Minghui 63).
Six main-effect QTL affecting the number of PPP were revealed, explaining 28.6, 14.1, 11.6, 10.3, and 30.9% of the total variance of this trait in the RILs, TCP1, TCP2, TCP3, and TCP4, respectively. Three of these main-effect QTL were detected in the RILs. The QTL ppp2, which is bordered by markers RM341-RM327 on chromosome 2 and has the largest effect in the RILs, was detected in three TC populations (TCP2, TCP3, and TCP4). The other two main-effect QTL (ppp4 and ppp12) found in the RILs were not detected in any TC populations. Four main-effect QTL (ppp3, ppp6a, ppp6b, and ppp10) were identified in TC populations, but not in the RILs. The direction of parental contribution and the magnitude of the effect for the common QTL on the number of panicles per plant displayed the same trends as those affecting PH and HD.
We identified nine main-effect QTL affecting SPP, which explained 47.5, 39.9, 30.5, 54.0, and 40.7%, respectively, of the total variance of this trait in the five related populations. Three of these QTL were detected in the RILs. The QTL spp1, which is flanked by markers S1501-C904 on chromosome 1 and has the largest effect in the RILs, was also detected in four TC populations. Main-effect QTL spp7 was identified in only one TC population (TCP2) and spp4 was not identified in any TC populations. Six main-effect QTL were revealed in TC populations, but not in RILs. Of these, spp5 (RM164-C624) was identified in two TC populations (TCP3 and TCP4), and each of the other five was detected in only one of the four TC populations.
Eight main-effect QTL related to the number of GPP were found, which explained 49.0, 21.5, 29.2, 38.2, and 49.0% of the total variance of this trait in the five related populations, respectively. Three main-effect QTL were detected in the RILs: gpp1 (which is flanked by markers S1501-C904 on chromosome 1 and has the largest effect in RILs) was detected in two TC populations (TCP3 and TCP4); the QTL gpp6a (C688-S1520) was identified in one TC population (TCP2); and gpp7 (R1789-RM242) was not detected in any TC populations. Five main-effect QTL were revealed in TC populations, but not in RILs: gpp8b and gpp10 were identified in two of the four TC populations [(TCP3 and TCP4) and (TCP1 and TCP2)], respectively, while each of the other three was detected in only one of the four TC populations.
As many as 16 main-effect QTL affecting GW were detected, which explained 70.1, 20.2, 15.2, 59.1, and 48.4% of the total variance of this trait in the five related populations, respectively. Eight main-effect QTL were detected in the RILs: gw1a, which mapped between markers C904 and R596 on chromosome 1, had the largest effect in RILs and was also detected in three TC populations (TCP1, TCP3, and TCP4); gw8a and gw11 were also identified in TCP3 and TCP4; gw1b, gw5, and gw6 were each identified in just one TC population. Two main-effect QTL (gw3a and gw7) detected in the RILs were not identified in any TC populations. Eight main-effect QTL were found in TC populations but not in the RILs. Of these, gw3b was detected in three TC populations, and both gw2a and gw3c were detected in two TC populations. The other 5 main-effect QTL were each detected in only one of the four TC populations.
Seven main-effect QTL affecting GYPP were identified, which explained 42.5, 27.6, 37.4, 22.6, and 23.2% of the total variance of this trait in the five populations, respectively. Three of these main-effect QTL were detected in the RIL population; gypp1, which is located between S1501 and C904 on chromosome 1 and has the largest effect in RILs, was also detected in three TC populations, while the other two (gypp3a and gypp4) were not identified in any TC populations. Four main-effect QTL that were not detected in the RIL population were found in TC populations: gypp6 and gypp10 were both identified in two TC populations, while each of the other two (gypp3b and gypp8) were found in only one TC population.
Epistatic QTL detected in the RILs and TC progenies:
Table 5 shows 29 digenic epistatic QTL pairs identified in the RIL and four TC populations. No common digenic epistatic QTL pairs were detected in the RILs and all of the TC populations, and no epistatic QTL were identified for HD in the five populations.
TABLE 5.
Epistaic QTL affecting traits of agronomic importance detected in RILs and TC progenies
| Variance (%)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | Population | Chromosome | Interval i | Chromosome | Interval j | LODa | Locusb | Traitc | Phenotypic effectd |
| PH | RILs | 6 | RM30-C962 | 11 | C602-G257 | 5.25 | 6.1 | 6.1 | −2.8 |
| TCP2 | 2 | Yl257-C920 | 6 | R11-R1608 | 5.48 | 4.9 | −2.3 | ||
| 2 | Y0193-S1559A | 10 | RM304-G2155 | 18.2 | 4.0 | 8.9 | −2.2 | ||
| TCP4 | 4 | R1854-RM335 | 10 | S2083-RZ649A | 5.23 | 2.8 | 2.8 | −1.6 | |
| PPP | RILs | 3 | yl-171-RM55 | 9 | C985-R1562 | 5.58 | 5.9 | 5.9 | −0.4 |
| TCP1 | 8 | RM337-G278 | 10 | E2880B-RM330 | 5.21 | 11.0 | 11.0 | −1.2 | |
| TCP3 | 2 | y254-RZ87 | 12 | RM313-C2808 | 5.47 | 13.5 | 13.5 | 0.5 | |
| TCP4 | 2 | y2144-RM213 | 4 | RM303-C1008 | 5.00 | 6.2 | −0.4 | ||
| 4 | C140-y103D | 9 | G293-RM205 | 5.03 | 5.6 | 0.4 | |||
| 5 | R2289-RM163 | 6 | R11-R1608 | 5.06 | 6.7 | 18.5 | 0.4 | ||
| SPP | RILs | 4 | R1854-RM335 | 9 | C1257-G1085 | 5.88 | 5.9 | 3.9 | |
| 11 | R2918-S1559B | 12 | S13752-Y0182 | 6.00 | 6.2 | 12.1 | 4.0 | ||
| TCP1 | 2 | C1769-RM6 | 3 | R2404-R2443 | 6.09 | 3.7 | 3.7 | 3.6 | |
| TCP4 | 2 | RG144-C149 | 8 | C309-S2108 | 10.2 | 3.3 | −2.7 | ||
| 8 | RM342-G1073 | 11 | R1466B-R2918 | 6.77 | 7.8 | 11.1 | 4.1 | ||
| GPP | TCP1 | 6 | C688-S1520 | 10 | RM304-G2155 | 17.9 | 14.4 | 14.4 | −16.2 |
| TCP4 | 5 | RZ649B-RM164 | 10 | RM330-R2309 | 6.60 | 6.9 | 6.9 | 3.5 | |
| GW | TCP1 | 4 | RM303-C1008 | 10 | E2880B-RM330 | 5.00 | 10.1 | 10.1 | −0.4 |
| TCP3 | 1 | R596-RZ413 | 2 | S910-RG634 | 9.38 | 3.2 | 0.2 | ||
| 3 | RM168-yl-171 | 7 | C615-R565 | 6.20 | 6.7 | 0.3 | |||
| 8 | S2108-C483 | 11 | RG543-y245EI | 7.08 | 2.8 | 12.7 | −0.2 | ||
| TCP4 | 2 | C370-Y0193 | 11 | G257-RG2 | 5.46 | 1.9 | −0.2 | ||
| 5 | C624-RM26 | 9 | S752-C1257 | 7.30 | 3.8 | 5.7 | −0.3 | ||
| GYPP | TCP2 | 2 | RG634-y254 | 4 | y103D-C2807 | 5.40 | 5.9 | 3.6 | |
| 8 | RM337-G278 | 10 | RM304-G2155 | 8.73 | 5.8 | 10.7 | −3.5 | ||
| TCP3 | 4 | y103D-C2807 | 5 | R2232-R568 | 5.00 | 5.3 | 5.3 | −1.4 | |
| TCP4 | 1 | y517-S2139 | 1 | C904-R596 | 9.81 | 9.2 | 1.8 | ||
| 2 | RM233-RM154 | 6 | R1679-C751B | 5.26 | 3.6 | −1.1 | |||
| 8 | G1073-S2055 | 9 | C873-S752 | 5.07 | 6.2 | 19.0 | −1.5 | ||
A LOD score of 5.0 was used for declaring the existence of putative pairs of epistatic QTL, according to the method of Lynch and Walsh (1998).
Variance (%) for locus is the proportion of variance explained by the component epistasis.
Variance (%) for trait is the total variance of this trait explained by all epistatic QTL detected in a population.
Phenotypic effect is the effect arising from interactions between alleles at the loci i and j, as defined by Mather and Jinks (1982).
For plant height, four pairs of epistatic QTL (one in both the RILs and TCP4, two in TCP2) were identified, which explained 6.1, 8.9, and 2.8% of the total variation of this trait in the RILs and the two TC F1 populations, respectively. For panicles per plant, six pairs of epistatic QTL (one in each of the RILs, TCP1, and TCP3, and three in TCP4) were detected, explaining 5.9, 11.0, 13.5, and 18.5% of the total variation of this trait in the RILs and three TC populations, respectively. For spikelets per panicle, five pairs of epistatic QTL (one in TCP1, two in both the RILs and TCP4) were found, accounting for 12.1, 3.7, and 11.1% of the total variation of this trait in the RILs and two TC F1 hybrids, respectively. Only two pairs of epistatic QTL (one in both TCP1 and TCP4) were revealed for grain number per panicle, explaining 14.4 and 6.9% of total phenotypic variation in two TC F1 hybrids, respectively. For 1000-grain weight, six pairs of epistatic QTL (one in TCP1, two in TCP4, three in TCP3) were detected, which explained 10.1, 12.7, and 5.7% of total phenotypic variation in three TC F1 hybrids, respectively. For grain yield per plant, six pairs of epistatic QTL (one in TCP3, two in TCP2, and three in TCP4) were identified, explaining 10.7, 5.3, and 19.0% of total phenotypic variation in the three TC F1 populations, respectively. In summary, only a small number of epistatic QTL were detected and the degrees of variation that they explained were relatively small compared to the main-effect QTL.
DISCUSSION
Selection of TC populations and improved modern rice as experiment materials:
It is essential to use appropriate experimental designs and materials for QTL mapping, and strenuous efforts have been made to construct experimental populations for detecting and analyzing QTL in the last decade. Plant populations with various genetic structures have been developed for the purpose, mainly consisting of F2/F3, BC, double haploids (DHs), RILs, and backcross inbred lines (BILs). In rice, permanent populations, such as DHs and RILs, are used most often, because of their inherent advantages of providing a permanent DNA supply and phenotyping opportunities for many different studies. Allelic differences are limited in these populations, since only two alleles segregate at each polymorphic locus. In rare cases, TC progenies and immortalized F2 populations have been used (Li et al. 2001; Hua et al. 2002). In studies seeking common QTL in inbred lines and their hybrid progenies, backcross populations are usually employed. In the study reported in this article, we introduced a variant of design III (Comstock and Robison 1948), which produced four TC progenies by mating three maintaining lines for different types of cytoplasmic male sterility and one PTGMS line with the RILs. With such populations, we could compare the QTL across the RILs and the four TC populations with a set of common male parents (RILs). Our design was expected to increase the scope to identify QTL, and even multiple alleles at QTL loci, because of the inclusion of four testers as parents of the mapping populations. This was particularly true since the four testers were genetically divergent and unrelated in pedigree to the RILs. As shown by the data presented here, we were able to detect a total of 67 main-effect QTL (or 9.6 QTL/trait) and 29 epistatic QTL pairs (or 4.1 pair/trait) in the populations.
Another feature of this experiment is the selection of improved modern rice lines as the testers for TCs and the parents for the RILs. Minghui63, a parental variety of the RILs, is a restorer line of the most popular hybrid rice variety, Shanyou 63. The total planting area of Shanyou63 has exceeded 67 million hectares (ha) in China and other Asian countries. Minghui63 is also a restorer line for many other hybrid rice varieties, with a combined total planting area of 130 million ha. The high general combining ability and the genetic basis of Minghui63 are attractive to rice breeders. Three maintaining lines used in this experiment are among the best in China. Zhenshan97B is the maintainer of Zhenshan97A, which is the female parent of Shanyou63 and a number of other hybrid varieties with a total planting area of 108 million ha. In addition, the respective CMS lines of II-32B and YuetaiB, which have different types of cytoplasmic male sterility, are widely employed in rice breeding programs and the areas planted with them are rapidly growing. Another tester, Peiai64S, is a PTGMS rice line that is the parental line of the first super hybrid rice in China. Utilizing Peiai64s, many hybrid rice combinations have been released for rice production. For field trials, we used TC progenies derived directly from crossing the breeding lines (RILs) with maintaining lines for different types of CMS and PTGMS lines, closely resembling practices in modern rice breeding strategies. Data on QTL and the genetic features associated with them, detected using improved modern rice varieties, are expected to be more attractive to breeders than data obtained using other types of material since such results are likely to be highly applicable to contemporary rice breeding programs.
Comparison of QTL mapped in RILs and their TC populations in rice:
In this study, F1 performance was related to the performance of the parental lines according to both the phenotypic correlation analysis (Table 2) and the QTL mapping (Table 4). In all, 67 distinct main-effect QTL were identified for seven traits in the RILs and four TC populations. Of these, 27 were identified in the RIL population. For each trait, the QTL that had the strongest effect in the RILs (giving a total of 9 such QTL) were also detected in two, three, or four TC populations. Six main-effect QTL in RILs were identified in one TC populations. Another 12 main-effect QTL detected in RILs were not identified in any TC populations. In all instances of common main-effect QTL across the RILs and multiple TC progenies, the direction of parental contribution was the same. Several features of the distribution of main-effect QTL in the RILs and TC progenies for the seven traits under study can be noted. First, only the main-effect QTL with the largest effects in the RILs for each trait were detected in two or more TC populations, and except for GW only one such QTL was identified for the other six traits considered. Second, some main-effect QTL that were detected in RILs were identified in one TC population, and more than half of the main-effect QTL detected in RILs were identified in TC populations. Third, effects of the common main-effect QTL among the RILs and four TC progenies may vary in the magnitude of their substitution effects, but do not change in parental contribution. Fourth, both Minghui 63 and B5 contributed to increased trait values across the RILs and four TC populations for the seven traits evaluated.
A number of main-effect QTL were detected either in TC populations or in RILs. When comparing QTL mapped in RILs and their TC populations, we have to take into account the fact that TC progenies are likely to show only half of the difference in performance attachable to any specific marker compared to the difference between the testers and the RILs. In RILs, a QTL is identified when the additive effect between lines homozygous for the allele from the parents is significant. TC progenies carry only one allele from RILs in combination with another allele from the tester. A QTL is detected when the substitution effect of replacing an allele from RILs with the allele from the tester is significant. Possible interactions between the parental allele with the tester allele also have to be considered when comparing different types of progeny. The discrepancies between QTL mapped in RILs and their TC populations could be caused by genetic effects and are easy to explain by considering the genetic components of the RILs and TC populations. Main-effect QTL effects detected in the TC populations represent the differential intralocus interactions between the parental alleles from the RILs and those from the respective testers—due to the segregation of alleles with additive, partial recessiveness, intermediate gene action, or dominance—or the difference between the two heterozygous loci (Minghui63/tester–B5/tester). If a tester carries an allele that is fully dominant over the alleles carried by RILs, the corresponding QTL will not be detected in RILs but may be detected in the TC progenies. Overdominance of the RIL alleles over tester alleles can also lead to divergent results among TC progenies unless the four testers carry the same allele. The parental (Minghui 63 and B5) alleles in QTL associated with a single tester presumably have specific dominance interactions with the respective testers that do not occur with the other testers. Another possible explanation for the differences between QTL mapped in RILs and TC populations is the presence of epistatic effects. Twenty-nine pairs of distinct epistatic QTL were identified for six traits, and no common epistatic QTL were identified for the same trait across the RILs and the four TC progenies in this study. Hence, epistasis is very likely a major cause for the inconsistencies of QTL detection across RILs and their TC populations and is specific to the cross combination.
Cluster distribution of the main-effect QTL:
In this study, 67 distinct main-effect QTL distributed among 12 chromosomes were identified for seven traits across the RILs and four TC populations. A very interesting feature is the highly concentrated distribution of the QTL in a few chromosomal regions and the existence of QTL hot spots (Figure 1). This is particularly true for the region around the S1501-C904 locus on chromosome 1 and the C688-S1520 locus on chromosome 6, where QTL for several traits were detected in the RILs and the four TC populations. Similar concentrated distributions of QTL have also been observed in previous studies (Xiong et al. 1999; Li et al. 2000). Particular attention should be given to such QTL hot spots in future studies of gene cloning and functional genomics.
Positional convergence of main-effect QTL in rice:
Traits of agronomic importance, including those in this study, are useful characters for QTL analysis in rice. A QTL associated with grain weight or length has been reported in the centromere region of rice chromosome 3 in at least 10 different inter- and intraspecific populations of independent studies, suggesting that a homologous gene determining seed weight or size may be associated with domestication and subsequent selection (Li et al. 2004). For the seven QTL with strongest main effect (i.e., ph1, ppp2, spp1, gpp1, gw1, gypp1, and hd6a), we explored the publicly available QTL database (http://www.gramene.org) to search their alignment QTL identified in the same chromosome region in previous studies. For ph1, the same QTL associated with plant height has been reported in six populations: CNHZAU Zh97/Ming63 RI (Cui et al. 2002), Cornell 9024/LH422 RI (Xiao et al. 1996), Cornell Jef/Oruf BC (Thomson et al. 2003), IRRI Mor/CO39 (Huang et al. 1996), IRRI IR64/Azu DH (Venuprasad et al. 2002), and IRRI Lem/Teq RI QTL (Mei et al. 2003). In the vicinity of ph1, two genes associated with plant height have been identified and isolated: one is the gibberellin biosynthetic gene OsGA3ox2 (Itoh et al. 2001) and the other is the brassinosteroid biosynthetic gene D2 (Hong et al. 2003). The same QTL of hd6a has been identified in four populations, including CNHZAU Zh97/Ming63 RI (Yu et al. 2002), IRRI IR64/Azu DH (Li et al. 2003), JRGP Nip/Kas F2 (Lin et al. 1998), and NIAS Kosh/Kas BIL (Yamamoto et al. 2001). One allele of the gene (Hd3a) has been cloned, which encodes a protein closely related to Arabidopsis FT (Kojima et al. 2002). For five yield-related QTL (i.e., ppp2, spp1, gpp1, gw1, and gypp1), the same QTL have been reported in the corresponding chromosome regions in 5, 9, 3, 3, and 4 populations, respectively, among total 13 populations: CNHZAU Zh97/Ming63 RI (Cui et al. 2002; Hua et al. 2002, 2003), CNHZAU Zhe97/Wuy2 (Jiang et al. 2004), CNRRI Tes/CB (Zhuang et al. 1997), CNRRI Zh97B/Mil46 RI (Zhuang et al. 2001, 2002), Cornell 9024/LH422 RI (Xiao et al. 1996), Cornell IR64/IRG105 (Septiningsih et al. 2003), Cornell Jef/Oruf BC (Thomson et al. 2003), HNAES MIL23/Aki RI (Yagi et al. 2001), IGCAS ZYQ8/JX17 F2 (Xu et al. 1995), IRRI Lem/Teq RI (Mei et al. 2003), IRRI Mil23/Aki RI (Nagata et al. 2002; Kobayashi et al. 2003), JRGP Nip/Kas F2 (Yamaya et al. 2002), and NIAS Kosh/Kas near isogenic lines (Obara et al. 2004). Recently, the gene Gn1a, which produces more grains per panicle near the QTL gpp1 and encodes a cytokinin oxidase, has been cloned (Ashikari et al. 2005). The results show that these main-effect QTL have been identified in a much wider range of populations and under different environments in independent studies in rice. Correspondence in the location of QTL in different taxa suggests that some of the underlying genes are identical (Paterson et al. 1995). On the basis of the positional convergence of QTL across different populations, the structure and function of the underlying genes might be conserved across different varieties in the rice gene pool.
The data listed in Table 4 and Table 6 indicate that the genes underlying the main-effect QTL, with large effects, might determine the phenotype of the traits studied in RILs and TC hybrid populations. The effect of these QTL is more evident in modern improved varieties than in traditional varieties, indicating that the loci have been the targets of selection associated with breeding practice. The main-effect QTL detected in RILs are expressed in heterozygous F1 as showed in this report, which is an important part of the genetic basis of heterosis. The QTL are expressed in different genetic backgrounds and environments, making them valuable targets for gene manipulation and also for application in rice breeding. By comparing QTL mapped in RILs and their hybrid populations in rice, and with a publicly available QTL data reservoir, specific candidate loci will be identified to address fundamental problems in rice improvement. Recently, a fine mapping of a grain-weight QTL has been constructed (Li et al. 2004). Such information should be valuable for positional cloning genes underlying QTL and for marker-aided selection of QTL in rice breeding programs. As correspondence in the location of QTL exists in different taxa of grasses (Paterson et al. 1995), main-effect QTL identified in rice will be useful in other cereal crops.
TABLE 6.
Phenotypic variance (%) for seven traits explained by main-effect QTL and epistaic QTL detected in RILs and TC progenies
| Trait | Loci type | RILs | TCP1 | TCP2 | TCP3 | TCP4 |
|---|---|---|---|---|---|---|
| PH | Main-effect QTL | 50.1 | 37.1 | 53.0 | 40.2 | 55.7 |
| Epistaic QTL | 6.1 | — | 8.9 | — | 2.8 | |
| Total | 56.2 | 37.1 | 61.9 | 40.2 | 58.5 | |
| HD | Main-effect QTL | 61.9 | 67.1 | 71.6 | 21.5 | 36.5 |
| Epistaic QTL | — | — | — | — | — | |
| PPP | Main-effect QTL | 28.6 | 14.1 | 11.6 | 10.3 | 30.9 |
| Epistaic QTL | 5.9 | 11.0 | — | 13.5 | 18.5 | |
| Total | 34.5 | 25.1 | 11.6 | 23.8 | 32.4 | |
| SPP | Main-effect QTL | 47.5 | 39.9 | 30.5 | 54.0 | 40.7 |
| Epistaic QTL | 12.1 | 3.7 | — | — | 11.1 | |
| Total | 59.6 | 43.6 | 30.5 | 54.0 | 51.8 | |
| GPP | Main-effect QTL | 49.0 | 21.5 | 29.2 | 38.2 | 49.0 |
| Epistaic QTL | — | 14.4 | — | — | 6.9 | |
| Total | 49.0 | 35.9 | 29.2 | 38.2 | 55.9 | |
| GW | Main-effect QTL | 70.1 | 20.2 | 15.2 | 59.1 | 48.4 |
| Epistaic QTL | — | 10.1 | — | 12.7 | 5.7 | |
| Total | 70.1 | 30.3 | 15.2 | 71.8 | 54.1 | |
| GYPP | Main-effect QTL | 42.5 | 27.6 | 37.4 | 22.6 | 23.2 |
| Epistaic QTL | — | — | 10.7 | 5.3 | 19.0 | |
| Total | 42.5 | 27.6 | 48.1 | 27.9 | 19.0 |
—, indicates that no epistaic QTL was identified in this population for the trait evaluated.
Implications for genetic improvement and marker-aided breeding of hybrid rice:
In our study, the detected main-effect QTL had distinct effects on both the RILs and the TC hybrids. The results indicate that the contributions of the inbred line on F1 performance were quite stable across unrelated testers. The common main-effect QTL showing effects on multiple TC progenies may be associated with the general combining ability in hybrid breeding. On the other hand, a number of epistatic QTL were detected. Compared with the main-effect QTL, the effects of the epistatic QTL were relatively weak and present in some, but not all, crosses (Table 6). These cross-specific epistatic QTL are likely to be related to the special combining ability in hybrid breeding. Our findings have several implications for contemporary hybrid rice breeding. First, the elite varieties can be selected for use as restorers, as long as they carry fertility-restoring genes, and may contain a relatively high number of main-effect QTL that give large contributions to yields. Second, when a restorer line that carries the common main-effect QTL is bred, it can be used in a number of cross combinations. The direction of the contribution from the main-effect QTL of the restorer will be consistent in the cross combinations, according to our results. Third, to further improve the hybrid performance, the selection of the genetic background of the restorer line and the maintaining line should be seriously considered in rice breeding, since the epistatic QTL are cross combination dependent.
Since most quantitative traits of interest to plant breeders are considered to have poor heritabilities, plant breeders are more interested in the phenotypic variability among progenies of crosses between the inbred lines than in the lines per se. Breeders of hybrid crops need to improve the performance of the traits in the inbred lines and evaluate the progenies of testcrosses with unrelated testers. A crucial question in hybrid rice breeding is whether the QTL in inbred lines are stable across different testers. The identification of QTL in RILs that affect the performance of F1 populations and the accurate estimation of their genetic effects, including epistasis, is essential for efficient hybrid breeding of crops. Common QTL exhibiting effect across RILs and the F1 population in this study have also been detected in the large number of previous studies in rice. With the development of dense linkage maps based on molecular markers, marker-aided transfer and selection of the common QTL to improve hybrid productivity is expected to be possible. The common main-effect QTL for each trait with the largest effects in both the RILs and different TC populations, and the chromosome regions harboring multiple main-effect QTL for different traits such as those between S1501 and C904 on chromosome 1 and between C688 and S1520 on chromosome 6, will be valuable targets in marker-aided selection for efficient hybrid rice breeding and functional genomic studies.
Acknowledgments
We thank S. D. Tanksley and S. McCouch of Cornell University and the Japanese Rice Genome Research Program for providing us with DNA probes. This research was supported by a grant from the National Program of High Technology Development of China.
References
- Ashikari, M., H. Sakakibara, S. Lin, T. Yamamoto, T. Takashi et al., 2005. Cytokinin oxidase regulates rice grain production. Science 309: 741–745. [DOI] [PubMed] [Google Scholar]
- Bruce, A. B., 1910. The Mendelian theory of heredity and the augmentation of vigor. Science 32: 627–628. [DOI] [PubMed] [Google Scholar]
- Causse, M., T. Fulton, Y. Cho, S. Ahn, J. Chunwongse et al., 1994. Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138: 1251–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock, R. E., and H. F. Robison, 1948. The components of genetic variance in populations of biparental progenies and their use in estimating the average degree of dominance. Biometrics 4: 254–266. [PubMed] [Google Scholar]
- Cui, K. H., S. Peng, Y. Xing, S. B. Yu and C. G. Xu, 2002. Molecular dissection of relationship between seedling characteristics and seed size in rice. Acta Bot. Sin. 44: 702–707. [Google Scholar]
- Goodnight, C. J., 1999. Epistasis and heterosis, pp. 59–67 in The Genetics and Exploitation of Heterosis in Crops, edited by J. G. Coors and S. Pandey. American Society of Agronomy, Crop Science Society of America and Soil Science Society of America, Madison, WI.
- Harushima, Y., M. Yano, A. Shormura, M. Sato, T. Shimano et al., 1998. A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148: 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Z., M. Ueguchi-Tanaka, K. Umemura, S. Uozu and M. Matsuoka, 2003. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15: 2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J. P., Y. Z. Xing, C. G. Xu, X. L. Sun, S. B. Yu et al., 2002. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genetics 162: 1885–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J. P., Y. Xing, W. Wu, C. G. Xu, X. Sun et al., 2003. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 100: 2574–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N., B. Courtois, G. S. Khush, H. X. Lin, G. L. Wang et al., 1996. Association of quantitative trait loci for plant height with major dwarfing genes in rice. Heredity 77: 130–137. [Google Scholar]
- Huang, Z., G. C. He, L. Shu, X. Li and Q. F. Zhang, 2001. Identification and mapping of two brown planthopper resistance genes in rice. Theor. Appl. Genet. 102: 929–934. [Google Scholar]
- Itoh, H., M. Ueguchi-Tanaka, N. Sentoku, H. Kitano, M. Matsuoka et al., 2001. Cloning and functional analysis of two gibberellin 3 beta-hydroxylase genes that are differently expressed during the growth of rice. Proc. Natl. Acad. Sci. USA 98: 8909–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, G. H., C. G. Xu, X. H. Li, Y. Q. He, 2004. Characterization of the genetic basis for yield and its component traits of rice revealed by doubled haploid population. Acta Genet. Sin. 31: 63–72. [PubMed] [Google Scholar]
- Khush, G. S., 2001. Green revolution: the way forward. Nat. Genet. Rev. 2: 815–822. [DOI] [PubMed] [Google Scholar]
- Kobayashi, S., Y. Fukuta, T. Sato, M. Osaki and G. S. Khush, 2003. Molecular marker dissection of rice (Oryza sativa L.) plant architecture under temperate and tropical climates. Theor. Appl. Genet. 107: 1350–1356. [DOI] [PubMed] [Google Scholar]
- Kojima, S., Y. Takahashi, Y. Kobayashi, L. Monna, T. Sasaki et al., 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., and D. Botstein, 1989. Mapping Menedelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. M., M. Thomson and S. R. McCouch, 2004. Fine mapping of a grain-weight quantitative trait locus in pericentromeric region of rice chromsome 3. Genetics 168: 2187–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. X., S. B. Yu, C. G. Xu, Y. F. Tan, Y. J. Gao et al., 2000. Analyzing quantitative trait loci for yield using a vegetatively replicated F2 population from a cross between the parents of an elite hybrid. Theor. Appl. Genet. 101: 248–254. [Google Scholar]
- Li, Z. K., L. J. Luo, H. W. Mei, D. L. Wang, Q. Y. Shu et al., 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. I. Biomass and grain yield. Genetics 158: 1737–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. K., S. B. Yu, H. R. Lafitte, N. Huang, B. Courtois et al., 2003. QTL × environment interactions in rice. I. Heading date and plant height. Theor. Appl. Genet. 108: 141–153. [DOI] [PubMed] [Google Scholar]
- Lin, S. Y., T. Sasaki and M. Yano, 1998. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred Theor. Appl. Genet. 96: 997–1003. [Google Scholar]
- Lincoln, S., M. Daly and E. Lander, 1992. Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report, Whitehead Institute, Cambridge, MA.
- Luo, L. J., Z. K. Li, H. W. Mei, Q. Y. Shu, R. Tabien et al., 2001. Overdominant epistatic loci are the primary genetic basis of inbreeding depression and heterosis in rice. II. Grain yield components. Genetics 158: 1755–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mather, K., and J. L. Jinks, 1982. Bimetrical Genetics, Ed. 3. Chapman & Hall, London/New York.
- Mei, H. W., L. J. Luo, C. S. Ying, Y. P. Wang, X. Q. Yu et al., 2003. Gene actions of QTLs affecting several agronomic traits resolved in a recombinant inbred rice population and two testcross populations. Theor. Appl. Genet. 107: 89–101. [DOI] [PubMed] [Google Scholar]
- Murray, M. G., and W. F. Thompson, 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, K., Y. Fukuta, H. Shimizu, T. Yagi and T. Terao, 2002. Quantitative trait loci for sink size and ripening traits in rice (Oryza sativa L.). Breed. Sci. 52: 259–273. [Google Scholar]
- Obara, M., T. Sato, S. Sasaki, K. Kashiba, A. Nagano et al., 2004. Identification and characterization of a QTL on chromosome 2 for cytosolic glutamine synthetase content and panicle number in rice. Theor. Appl. Genet. 110: 1–11. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., E. S. Lander, J. D. Had, S. Paterson, S. E. Lincoln et al., 1988. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature 335: 721–726. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., Y. R. Lin, Z. Li, K. F. Schertz, J. F. Doebley et al., 1995. Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1718. [DOI] [PubMed] [Google Scholar]
- Septiningsih, E. M., J. Prasetiyono, E. Lubis, T. H. Tai, T. Tjubaryat et al., 2003. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Shull, G., H., 1908. The composition of a field of maize. Ann. Breed. Assoc. 4: 296–301. [Google Scholar]
- Stuber, C. W., 1994. Heterosis in plant breeding. Plant Breed. Rev. 12: 227–251. [Google Scholar]
- Stuber, C. W., S. W. Lincoln, D. W. Wolff, T. Helentjaris and E. S. Lander, 1992. Identification of genetic factors contributing to heterosis in a hybrid from two elite maize inbred lines using molecular markers. Genetics 132: 823–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, M. J., T. H. Tai, A. M. McClung, H. X. Lai, M. E. Hinga et al., 2003. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Venuprasad, R., H. E. Shashidhar, S. Hittalmani and G. S. Hemamalini, 2002. Tagging quantitative trait loci associated with grain yield and root morphological traits in rice (Oryza sativa L.) under contrasting moisture regimes. Euphytica 128: 293–300. [Google Scholar]
- Wang, D. L., J. Zhu, Z. K. Li and A. H. Paterson, 1999. Mapping QTL with epistatic effects and QTL × environment interactions by mixed model approaches. Theor. Appl. Genet. 99: 1255–1264. [Google Scholar]
- Wu, K. S., and S. D. Tanksley, 1993. Abundance polymorphism and genetic mapping of microsatellites in rice. Mol. Gen. Genet. 241: 225–235. [DOI] [PubMed] [Google Scholar]
- Xiao, J. H., J. Li, L. P. Yuan and S. D. Tanksley, 1995. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. H., J. M. Li, L. P. Yuan and S. D. Tanksley, 1996. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 92: 230–244. [DOI] [PubMed] [Google Scholar]
- Xiong, L. Z., K. D. Liu, X. K. Dai, C. G. Xu and Q. F. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]
- Xu, Y. B., Z. Shen, J. Xu, H. Zhu, Y. Chen et al., 1995. Interval mapping of quantitative trait loci by molecular markers in rice (Oryza sativa L.). Sci. China Ser. C Life Sci. 38: 422–428. [Google Scholar]
- Yagi, T., K. Nagata, Y. Fukuta, K. Tamura, I. Ashikawa et al., 2001. QTL mapping of spikelet number in rice (Oryza sativa L.). Breed. Sci. 51: 53–56. [Google Scholar]
- Yamamoto, T., S. F. Taguchi, Y. Ukai, T. Sasaki and M. Yano, 2001. Mapping quantitative trait loci for days-to-heading, and culm, panicle and internode lengths in a BC1F3 population using an elite rice variety, Koshihikari, as the recurrent parent. Breed. Sci. 51: 63–71. [Google Scholar]
- Yamaya, T., M. Obara, H. Nakajima, S. Sasaki, T. Hayakawa et al., 2002. Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53: 917–925. [DOI] [PubMed] [Google Scholar]
- Yu, S. B., J. X. Li, C. G. Xu, Y. F. Tan, Y. J. Gao et al., 1997. Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc. Natl. Acad. Sci. USA 94: 9226–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S. B., J. X. Li, C. G. Xu, Y. F. Tan, X. H. Li et al., 2002. Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor. Appl. Genet. 104: 619–625. [DOI] [PubMed] [Google Scholar]
- Yuan, L. P., 1998. Hybrid rice breeding in China, pp. 27–33 in Advances in Hybrid Rice Technology, edited by S. S. Virmani, E. A. Siddiq and K. Muralidharan. International Rice Research Institute, Los Baños, Philippines.
- Zhuang, J. Y., H. X. Lin, J. Lu, H. R. Qian, S. Hittalmani et al., 1997. Analysis of QTL × environment interaction for yield components and plant height in rice. Theor. Appl. Genet. 95: 799–808. [Google Scholar]
- Zhuang, J. Y., Y. Y. Fan, J. L. Wu, Y. W. Xia, K. L. Zheng, 2001. Comparison of the detection of QTL for yield traits in different generations of a rice cross using two mapping approaches. Acta Genet. Sin. 28: 458–464. [PubMed] [Google Scholar]
- Zhuang, J. Y., Y. Y. Fan, Z. M. Rao, J. L. Wu, Y. W. Xia et al., 2002. Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor. Appl. Genet. 105: 1137–1145. [DOI] [PubMed] [Google Scholar]