Abstract
The evolution and maintenance of the phenomenon of postsegregational host killing or genetic addiction are paradoxical. In this phenomenon, a gene complex, once established in a genome, programs death of a host cell that has eliminated it. The intact form of the gene complex would survive in other members of the host population. It is controversial as to why these genetic elements are maintained, due to the lethal effects of host killing, or perhaps some other properties are beneficial to the host. We analyzed their population dynamics by analytical methods and computer simulations. Genetic addiction turned out to be advantageous to the gene complex in the presence of a competitor genetic element. The advantage is, however, limited in a population without spatial structure, such as that in a well-mixed liquid culture. In contrast, in a structured habitat, such as the surface of a solid medium, the addiction gene complex can increase in frequency, irrespective of its initial density. Our demonstration that genomes can evolve through acquisition of addiction genes has implications for the general question of how a genome can evolve as a community of potentially selfish genes.
ONE of the impressive features of genomes, which became evident through decoding, is their fluidity within the evolutionary timescale. By fluidity, we mean acquisition of genes and genetic elements from outside the cell (horizontal transfer); internal genome rearrangements; or continuous loss, duplication, and allelic substitutions. The genomes are full of mobile, symbiotic, or parasitic genetic elements. Genome comparison and evolutionary analysis have revealed extensive horizontal transfer of genes between organisms, especially in bacterial and archaeal worlds (Faguy and Doolittle 2000). Rather than being a well-designed blueprint, a genome appears to be a temporary community of potentially mobile genes that essentially act selfishly. Given this feature of the genome, how are symbiosis and a cohesive social order, within a genome, ever achieved?
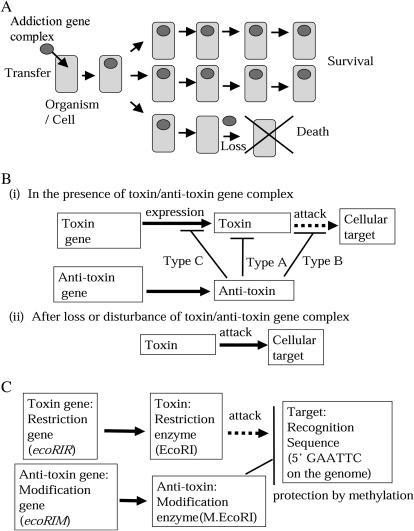
At the first glimpse inside the genomes, there are many genes, whose immediate advantage to the organism carrying them is unclear. An extreme and, therefore interesting, case is provided by the phenomenon of postsegregational killing or genetic addiction (Kobayashi 2004). In this phenomenon, the removal of a particular genetic element from the genome of an organism causes the product to induce death of the organism (Figure 1A), although the organism lived normally before acquisition of this genetic element. An intact form of the genetic element would survive in other members of the host population.
Figure 1.
Postsegregational host killing or genetic addiction. (A) Principle. Once transferred and established in a cell (or organism, in general), the addiction gene complex is difficult to eliminate because its loss, or some sort of threat to its persistence, leads to host-cell death. Intact copies of the gene complex survive in the other cells. (B) Simplest mechanisms. The addiction gene complexes consist of a set of closely linked genes that encodes a toxin (or killer) and an antitoxin (or antidote or anti-killer). The toxin's attack on a specific cellular target is inhibited by the antitoxin. After loss of the gene pair (or some sort of disturbance in the balance between the two gene products), the antitoxin becomes ineffective, thereby permitting the toxin to attack its target. Addiction systems can be classified into three types depending on which step leading to the toxin action is inhibited by the antitoxin. (C) Restriction–modification systems as addiction gene complexes. Several restriction–modification systems represent the only known example of the type B system (target protection by the antitoxin). Here, the toxin is a restriction enzyme (such as EcoRI) that attacks specific recognition sequences on the genome, while the antitoxin is a modification enzyme that protects these sequences by methylating them.
As shown in Figure 1B, a gene complex responsible for postsegregational killing (called an addiction gene complex here) consists of a set of closely linked genes that encodes a toxin and an antitoxin. The toxin's attack on a specific cellular target is blocked by the antitoxin at one of various steps. After loss of the gene complex (or some sort of disturbance in the balance between the two gene products), the antitoxin becomes ineffective, thereby permitting the toxin to attack its target.
Several type II restriction–modification systems (Figure 1C) have been experimentally proven to represent simple examples of postsegregational killing (Naito et al. 1995). Here, the toxin is a restriction enzyme that attacks specific recognition sequences on the chromosome, while the antitoxin is a modification enzyme that protects these sequences by methylating them. The loss of the gene complex, followed by cell division, eventually dilutes the antitoxin level, causing the target sites on the newly replicated chromosomes to be exposed to lethal attack by the toxin (Naito et al. 1995; Kobayashi 2004). In the second type of postsegregational killing system, called the classical proteic killer system or the toxin–antitoxin system, and labeled type A in Figure 1B, the antitoxin counteracts the toxin action through direct interaction. When cells fail to retain the gene complex, degradation of the antitoxin by a specific protease leads to release of the toxin. Some of these toxins turned out to be sequence-specific mRNA endonucleases (Gerdes et al. 2005). In the third type of postsegregational killing system, labeled type C in Figure 1B, expression of the toxin gene is inhibited by its own antisense RNA, which is encoded by the same locus and serves as the antitoxin. However, after the gene is lost, the antisense RNA decays faster due to digestion by an RNase. This allows the toxin to become expressed (Gerdes et al. 1997). Homologs of the postsegregational killing genes are found on mobile genetic elements and on the chromosomes and are quite abundant in some of the sequenced eubacterial and archaeal genomes (Pandey and Gerdes 2005). They are believed to have a role other than maintenance of plasmids (Gerdes et al. 2005).
The evolution and maintenance of the postsegregational killing process appear paradoxical. It is maintained in spite of its toxic effect of host killing (see Cooper and Heinemann 2000 for a discussion of conditions for plasmid maintenance). Bacterial cells sometimes lose their plasmid that carries an addiction gene complex upon, for example, cell replication resulting in postsegregational host killing. The killing decreases the numbers of both the responsible gene complex and its host, which seems to be detrimental for both of them. In fact, plasmids carrying a postsegregational killing gene complex did not outcompete in the liquid media, where equal numbers of cells, containing either a plasmid with or another plasmid without a postsegregational killing gene complex, were placed in direct competition (Cooper and Heinemann 2000). Thus, if plasmid maintenance depends on postsegregational host killing, selective pressure would eventually lead to a population that is free of the addiction gene complex and the plasmid. Indeed, a bacterial line carrying an addiction gene complex on its plasmid will eventually lose this gene complex after prolonged growth (Naito et al. 1995).
There have been repeated arguments that these addiction gene complexes are maintained because of properties beneficial to the host, despite their toxic effect of host killing (see, for example, Gerdes et al. 2005). An advantage of postsegregational killing, however, can be seen in the presence of a competitor genetic element. A plasmid carrying an addiction gene complex is difficult to replace by an incompatible plasmid because its loss leads to host death (Naito et al. 1995). A chromosomally located addiction gene complex likewise resists replacement by an allelic gene (Handa et al. 2001; Sadykov et al. 2003). Plasmids carrying a postsegregational killing gene are advantageous only when cell death is accompanied by the “death” of a competing plasmid (Cooper and Heinemann 2000). If this negative impact on the competing genes is great enough to overcome the cost of postsegregational host killing, the process will be maintained. Thus, addiction gene complexes would gain an advantage, not because they spitefully kill their host, but because they eliminate competing plasmids or genes.
Mongold (1992), in a study employing ordinary differential equations, theoretically analyzed this advantage and demonstrated that postsegregational killing might provide an advantage in competitive exclusion between incompatible plasmids. An increase in the postsegregational killing plasmid occurred when host killing was coupled with horizontal transfer of the plasmid, although conditions were restrictive, especially in that the plasmid had to have a high initial density. If the initial density was low, the plasmid-bearing cells were unlikely to invade any other population. In contrast, if the initial density was high and the plasmid was already established in a bacterial population, it was resistant to invasion by any other cell and the effect became greater when invading plasmids were incompatible with the postsegregational killing plasmid. According to this theoretical work, evolution of addiction gene complexes at the early stage (from a low density) remains unclear.
In the above work (Mongold 1992), the model did not include a spatial structure of the population, although the author mentioned its importance. In a spatially structured population, population density, measured in a local area, can be much higher than the average density and might provide an advantage for invasion of the minority type. In this work, we demonstrate that spatial structure indeed allows invasion of an addiction gene complex in the very early stage of evolutionary dynamics. Our population dynamics analysis employs a lattice model for spatial structure, through both mathematical analysis with pair approximation and computer simulation.
MODELS
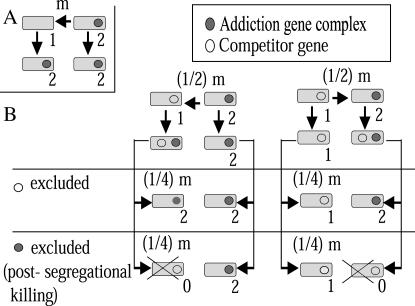
We consider two cases in which the addiction gene complex does not have a competitor (Figure 2A) and where it does have a competitor (Figure 2B). For each case, we consider a model without and with a spatial structure. Each of these four models is studied, both by mathematical analysis and by numerical (computer) simulation, within one of the four sections below.
Figure 2.
Diagrams of gene transfer and postsegregational host killing. (A) In the absence of a competitor. The addiction gene complex is transferred to a host that does not carry its competitor. (B) In the presence of a competitor. There are four possible outcomes. The addiction gene complex may be transferred to a cell carrying its competitor. Alternatively, the competitor genetic element may be transferred to a cell carrying the addiction gene complex at the same rate ((1/2)m). The two incompatible gene complexes may stay together for a while, but one of them will be eventually excluded. Loss of the addiction gene complex leads to cell death through the process of postsegregational host killing.
We consider two types of bacterial population, arranged on a two-dimensional regular square lattice, as applied in other spatially structured models of bacterial population dynamics (Frank 1994; Durrett and Levin 1997; Nakamaru et al. 1997; Iwasa et al. 1998). Each site is either occupied by a bacterial cell or vacant. One type of cell does not possess an addiction gene complex (type 1), while the other type does (type 2), as shown in Figure 2. The addiction gene complex resides on a plasmid, although it can also reside on the chromosome.
There are two models according to the states of type 1 bacteria. In the “single-plasmid model” (Figure 2A), type 1 bacteria have no competitor genetic element. In the “plasmid competition model” (Figure 2B), type 1 bacteria have a competitor genetic element, which is in a relationship of competitive exclusion with the addiction gene complex of type 2 bacteria. When the addiction gene complex is on a plasmid, an incompatible plasmid in type 1 is in this type of competitive exclusion. Due to conflicts in replication machinery, two incompatible plasmids cannot be stably maintained in the same host over a long period of time. When the addiction gene complex is on the chromosome, an allelic gene is regarded as such a competitor. The allelic genes will compete for the same locus during replacement through homologous recombination following transformation, general transduction, or any other forms of gene transfer.
Let 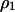 and
and 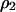 be the fraction of sites occupied by bacteria that do not carry an addiction gene complex (type 1) and that carry an addiction gene complex (type 2), respectively. Let
be the fraction of sites occupied by bacteria that do not carry an addiction gene complex (type 1) and that carry an addiction gene complex (type 2), respectively. Let 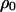 be the fraction of vacant sites. These satisfy
be the fraction of vacant sites. These satisfy 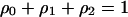 and are called global densities (Matsuda et al. 1992). For brevity, we call a site occupied by a type i cell an i-site (i = 1, 2) and a vacant site a 0-site from now on.
and are called global densities (Matsuda et al. 1992). For brevity, we call a site occupied by a type i cell an i-site (i = 1, 2) and a vacant site a 0-site from now on.
Let z be the number of neighbors for each site in the lattice (z = 4 in this study). The symbol b is the intrinsic birth rate for a neighboring pair of a vacant site and an occupied site. Bacterial reproduction to the 0-site occurs at rate b/z. We assume the same birth rate between type 1 and type 2 bacteria. Let d be the intrinsic death rate of bacteria type 1. The death rate of the cell type 2 is assumed to be d + c, which is larger than that of type 1 because the type 2 cell would die when it loses its addiction gene complex. We call the difference c in death rate between type 1 and type 2 “cost of host killing,” which is caused by occasional natural segregation of the addiction gene complex resulting in postsegregational killing. Growth cost is ignored here because it should be much smaller than cell death. Note that the effect of host killing caused by competitive exclusion of the plasmid carrying the addiction gene complex is not incorporated into c. The effect is, instead, incorporated into the ensuing transition events and changes in bacterial density, as is explained later. Therefore, the value of c is assumed to be very small.
We assume that neighboring bacteria will be involved in gene transfer at a constant rate, m. In the case of plasmids, this represents the rate of plasmid transfer between neighboring cells through conjugation. In the case of chromosomal genes, it represents the rate of successful establishment of an addiction gene complex by transformation. Because gene transfer between bacteria of the same type results in no recognizable change, only transfers between type 1 and type 2 bacteria were analyzed. The transfer may change the type of either or both bacteria. The effect of gene transfer is different between the two models, i.e., the single-plasmid model (model A, without competitor) and the plasmid competition model (model B, with competitor). Definitions of variables, parameters, and terms are summarized in Table 1.
TABLE 1.
Definitions
| Symbol | Definition | Comment |
|---|---|---|
| Model A | Single-plasmid model | Without competitor, Figure 2A |
| Model B | Plasmid competition model | With competitor, Figure 2B |
| Type 1 | A bacterial cell not carrying an addiction gene complex | It carries a competitor gene in the case with competitor |
| Type 2 | A bacterial cell carrying an addiction gene complex | |
| 0-Site | A vacant site | |
| 1-Site | A site occupied by a type 1 cell | |
| 2-Site | A site occupied by a type 2 cell | |
 |
Fraction of 0-sites | Global (or unconditional) density (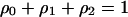 ) ) |
 |
Fraction of 1-sites | Global (or unconditional) density |
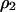 |
Fraction of 2-sites | Global (or unconditional) density |
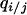 |
Conditional probability that a neighbor of a randomly chosen j-site is an i-site | Local (or conditional) density of i-sites among neighbors of a j-site |
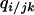 |
Conditional probability that a neighbor of a j-site of a randomly chosen jk-pair is a type i cell | Local (or conditional) density of i-sites among neighbors of a j-site of a randomly chosen jk-pair |
| z | No. of neighbors for each site in the lattice | z = 4 in this study |
| b | Intrinsic birth rate of each bacterium | Reproduction to a 0-site occurs at rate b/z |
| d | Intrinsic death rate of type 1 cell | Death rate of a type 2 cell is d + c |
| c | Cost of host killing | The difference in death rate between type 1 and type 2 by natural segregation of the addiction gene complex |
| m | Rate of gene transfer between neighboring cells | Rate of plasmid transfer between neighboring cells through conjugation in the case of plasmids; rate of successful establishment of an addiction gene complex by transformation in the case of chromosomal genes; we analyze only transfer between type 1 and type 2 bacteria |
Model A as a single-plasmid (without competitor) model:
In this model (Figure 2A), the addiction gene complex is always transferred from type 2 into type 1 bacteria, because type 1 bacteria do not carry any competitor genetic element.
In Equation 1a, each number refers to each type of neighboring bacterial cell. The number attached to the arrow indicates the rate of transition of bacterial types triggered by gene transfer. We assume z = 4 from now on. The type 1 bacterial cell, thus, becomes a type 2 bacterial cell when gene transfer between it and a neighboring type 2 cell takes place:
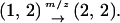 |
(1a) |
The pair of numbers in parentheses shows the doublet state of the nearest neighboring pair. This model should apply to any mode of gene transfer and any status of the addiction gene complex. These include plasmid transfer by conjugation and gene integration into the chromosome by any process, i.e., homologous recombination, transposition, or any form of nonhomologous recombination.
Model B as plasmid competition (with competitor) model:
In this model (Figure 2B), type 1 bacteria carry a competitor genetic element that is incompatible with the addiction gene complex. This model applies to two plasmids of the same incompatibility group, one of which carries an addiction gene complex (Naito et al. 1995). Another case involves an addiction gene complex on the chromosome and its allele. They would compete for the same locus in homologous replacement during natural transformation (Handa et al. 2001; Sadykov et al. 2003) and during other forms of gene transfer.
When a type 1 and a type 2 bacterial cell are in contact, the competitor gene complex is assumed to be transferred from a type 1 into a type 2 cell with the probability of (1/2)m/z. On the other hand, the addiction gene complex is transferred in the opposite direction with the probability of (1/2)m/z (Figure 2B). In each case, the recipient bacterial cell temporarily possesses both the addiction gene complex and its competitor. However, one of them will soon be lost from the bacterial cell. If the addiction gene complex is lost, the host bacterial cell will die through the process of postsegregational host killing. If the competitor is removed, the host bacterial cell changes into type 2. It is assumed that this process takes place over a short time period. Then, the transition of bacterial types triggered by gene transfer in this model (Figure 2B) is summarized as follows:
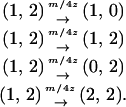 |
(1b) |
Equation 1b is similar to Equation 1a, but differs in that 0 means a vacant site created after cell death and each transition rate is divided by 4 because type 1 or type 2 bacterial cells, temporarily carrying a pair of incompatible plasmids, are assumed to lose one of them with an equal probability. The sum of these transition rates is m/z.
RESULTS
Equations:
The basic equations used here are based on those employed by Mongold (1992), while some parts are simplified to enable mathematical analysis with the inclusion of spatial structure. In computing temporal changes in global densities, we have to consider the correlation between neighbors. Due to the spatial pattern formed automatically by the demographic processes of reproduction, mortality, and interaction occurring between neighbors, the temporal change in global density cannot be expressed in terms of global densities only. The dynamics of 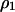 and
and 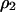 in model A are
in model A are
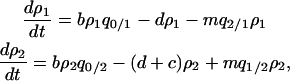 |
(2a) |
where 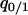 is the conditional probability that a neighbor of a randomly chosen 1-site is vacant (0-site). It is called local (or conditional) density of 0-sites among neighbors of a 1-site (Matsuda et al. 1992; Sato et al. 1994). In general, it is different from
is the conditional probability that a neighbor of a randomly chosen 1-site is vacant (0-site). It is called local (or conditional) density of 0-sites among neighbors of a 1-site (Matsuda et al. 1992; Sato et al. 1994). In general, it is different from 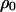 , the global (or unconditional) density of 0-sites. In the last term of the first equation,
, the global (or unconditional) density of 0-sites. In the last term of the first equation, 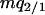 indicates the rate of loss of type-1 bacteria by gene transfer. The variable
indicates the rate of loss of type-1 bacteria by gene transfer. The variable 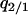 is the local density of 2-sites among neighbors of a 1-site. The variables
is the local density of 2-sites among neighbors of a 1-site. The variables 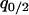 and
and 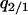 are defined similarly.
are defined similarly.
Similarly, the dynamics of 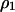 and
and 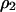 in model B are
in model B are
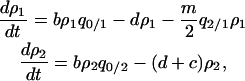 |
(2b) |
where the term 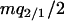 of the former equation indicates the loss of type 1 following gene transfer, which is the sum of the third and fourth transitions from type 1 in expression (1b). On the other hand, the effect of gene transfer is canceled in the dynamics of type 2, as shown in the latter equation.
of the former equation indicates the loss of type 1 following gene transfer, which is the sum of the third and fourth transitions from type 1 in expression (1b). On the other hand, the effect of gene transfer is canceled in the dynamics of type 2, as shown in the latter equation.
Population dynamics in the absence of spatial structure
These models were analyzed at different levels of approximation. We first summarize the population dynamic behavior of the two types of bacterial cells in the absence of spatial structure. This can be calculated simply by replacing a local density (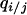 ) by a global density (
) by a global density (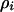 ) in Equations 2a and 2b. This is called the “mean-field approximation” of the lattice model (Iwasa et al. 1998). This approximation is valid if there is an additional process mixing the spatial configuration, as in a well-mixed liquid culture.
) in Equations 2a and 2b. This is called the “mean-field approximation” of the lattice model (Iwasa et al. 1998). This approximation is valid if there is an additional process mixing the spatial configuration, as in a well-mixed liquid culture.
Considering 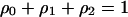 , dynamics (2a) and (2b) become:
, dynamics (2a) and (2b) become:
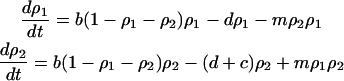 |
(3a) |
and
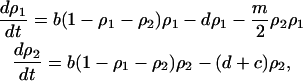 |
(3b) |
respectively.
Next, we consider the case in which the lattice population is close to equilibrium, in which only one type (type 1 or type 2) predominates, and we ask whether the other initially rare type (type 2 or type 1) can increase. In other words, we consider invasion conditions under which a rare type can invade an environment dominated by another type. This is our general definition of “invasion condition.” Using the assumption that the invading type is rare, we can simplify the dynamics.
Without competitor:
In the model without a competitor (3a), we first calculate the condition for invasion of type 2. If we consider an equilibrium population consisting only of type 1 bacteria, a rare type 2 carrying an addiction gene complex will invade and become established only if per capita growth rate 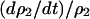 is positive. From Equation 3a, this condition is
is positive. From Equation 3a, this condition is
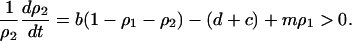 |
By neglecting the density of the rare invader (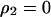 ), this condition becomes
), this condition becomes
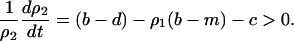 |
Since the dynamics satisfy 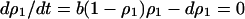 at equilibrium with type 1 bacteria alone, we calculate the density of type 1 bacteria at nontrivial equilibrium to
at equilibrium with type 1 bacteria alone, we calculate the density of type 1 bacteria at nontrivial equilibrium to 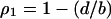 . We substitute this for the above inequality to arrive at the following condition:
. We substitute this for the above inequality to arrive at the following condition:
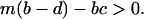 |
(4) |
Similarly, we can also calculate the condition for a type 1 invasion:
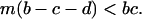 |
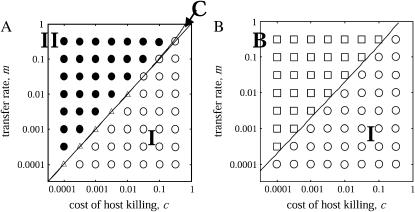
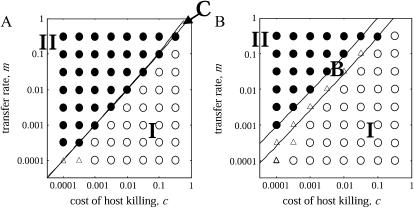
Under these conditions, we illustrate the parameter region on a m − c plane in Figure 3A, which is divided into three regions. The first region (labeled I) is a “type 2 extinct (type 1 win)” region, in which bacteria without the addiction gene complex (type 1) always win and drive those with the gene complex (type 2) to extinction. The second region (labeled II) is a “type 1 extinct (type 2 win)” region, in which type 2 always wins and drives type 1 to extinction. In the third narrow region (labeled C), both the conditions for type 1 and type 2 invasion are satisfied. Therefore, the two types can invade each other and coexist in the population to maintain equilibrium densities. Isocline diagrams are shown in Figure 4.
Figure 3.
Phase diagram in the absence of spatial structure (complete mixing). An overlay of the results of analytical solutions and numerical simulations is shown. The lines show the boundaries where the analytically predicted dynamic behavior changes on the parameter space. The analytically predicted behavior in each region is: type 1 drives type 2 to extinction (I); type 2 drives type 1 to extinction (II); the winner is either type depending on the initial density (bistable, B); and the two types of bacteria coexist (coexistence, C). The results of computer simulations are shown by each symbol: (○) type 1 drives type 2 to extinction ≥90%; (•) type 2 drives type 1 to extinction ≥90%;(□) either type extinct ≥90% depending on the initial density; and (△) extinction of either type is <90%. Division rate of bacteria b is 2.0 times per generation. The intrinsic death rate d is assumed to be 0.001 per generation. The gene transfer rate m and cost of host killing c varied from 0.0001 to 1.0 per generation. (A) In the absence of a competitor. Either type 1 or type 2 drives the other type of bacteria to extinction. (B) In the presence of a competitor. The result is either bistable or dominance of type 1. Even when the transfer rate is very high, type 2 bacteria cannot invade the initial population.
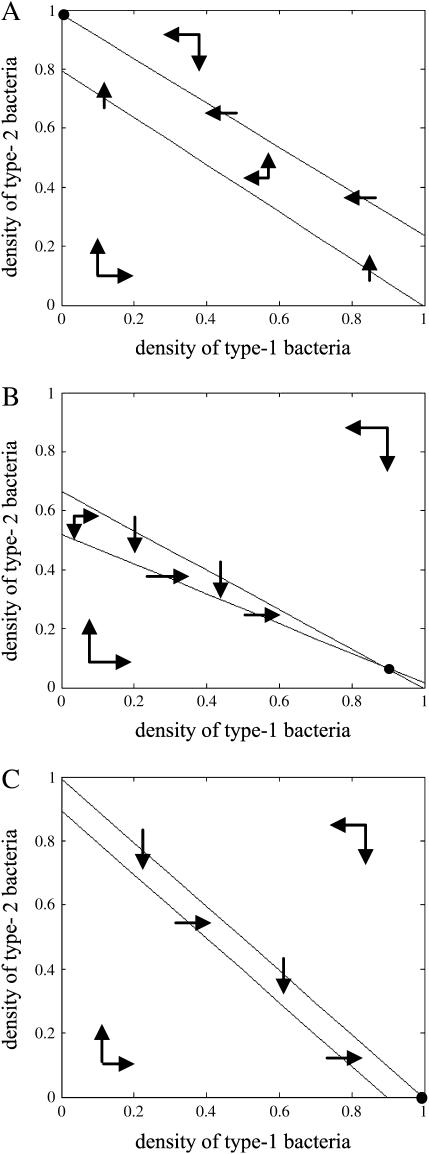
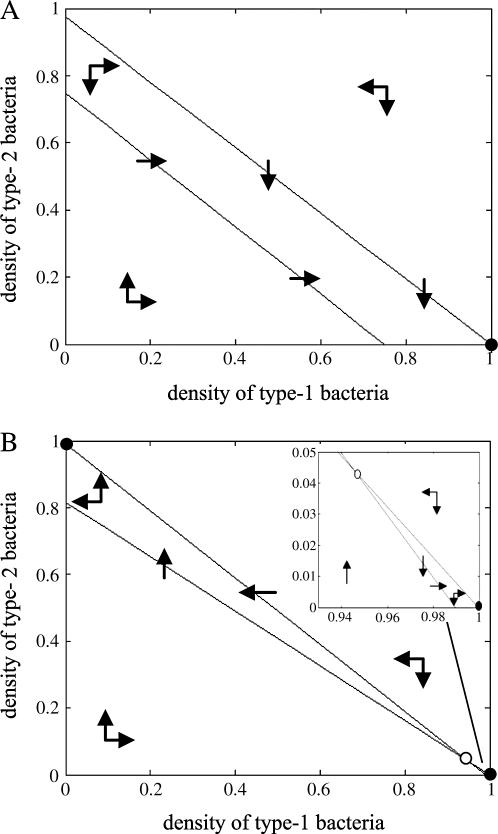
Figure 4.
Isocline diagram without competitor in the absence of spatial structure. Population dynamics of both bacteria are shown by arrows, and the resulting equilibrium is shown by dots. The solid circle (•) represents a stable equilibrium. (A) Type 1 extinction (type 2 win). Type 2 bacteria always win and the equilibrium is globally stable. The parameter values are m = 0.5, c = 0.02. (B) Coexistence. Both bacteria always coexist and the equilibrium is globally stable. The parameter values are m = 0.99, c = 0.95. (C) Type 2 extinction (type 1 win). Type 1 bacteria always win and the equilibrium is globally stable. The parameter values are m = 0.001, c = 0.2.
We also carried out computer simulations (see appendix a for details). The results for different m- and c-values are shown, together with the analytical results, in Figure 3A. In the single-plasmid model (model A), the predicted behaviors (type 1 extinction, type 2 extinction) for the different values of m and c are observed under the predicted conditions.
These results mean that, even when there is no competitor, the addiction gene complex can evolve to increase in frequency within the bacterial population and maintain itself under the condition that the transfer rate is higher than the cost of host killing. However, Equation 4 and Figure 3A indicate that host killing by the addiction gene complex (represented as c, cost of host killing) does not help its increase. The addiction gene complex increases its frequency only when the above condition is satisfied, although it is disadvantageous to the host. Therefore, this model does not provide an answer to the question as to why genetic addiction systems persist.
With competitor:
On the other hand, analysis of the plasmid competition model (3b) reveals that there is no condition for invasion of the type 2 bacteria in the absence of spatial structure. Instead, the dynamics show type 2 extinction or bistability, depending on parameter values (Figure 3B). Bistability is defined as the ability of the system to exist in two stable equilibrium configurations; selection of a particular equilibrium depends on the initial state.
The condition for invasion of type 2 into a population of type 1 dominant equilibrium is
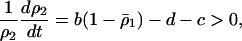 |
where 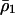 is the density of type 1 at equilibrium. By substituting
is the density of type 1 at equilibrium. By substituting 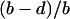 for
for 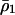 in the above inequality, the invasion condition for type 2 is
in the above inequality, the invasion condition for type 2 is 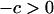 , which is never satisfied. Similarly, the condition for invasion of type 1 into type 2 dominant equilibrium is
, which is never satisfied. Similarly, the condition for invasion of type 1 into type 2 dominant equilibrium is
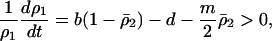 |
where 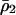 is the density of type 2 at equilibrium. By substituting
is the density of type 2 at equilibrium. By substituting 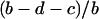 for
for 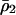 in the above inequality, the invasion condition for type 1 is
in the above inequality, the invasion condition for type 1 is 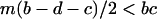 .
.
That is, type 1 always wins, if
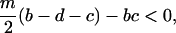 |
(5a) |
and bistability takes place, if
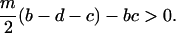 |
(5b) |
The parameter space in Figure 3B is, thus, divided into two regions. In the first region (labeled I), type 1 always wins and drives type 2, carrying the addiction gene complex, to extinction. In the second region (labeled B), the final winner is type 1 or type 2, depending on the initial density of the two types. In this region, neither one of the invasion conditions for type 1 or type 2 is satisfied. Both bacteria resist invasion by the rare opponent type: the system is bistable. The addiction gene complex of type 2 can never invade the equilibrium population dominated by type 1 bacteria with its competitor. This result means that the addiction gene complex cannot increase in the early stage of evolution if there is any competitor gene complex. Isocline diagrams are shown in Figure 5.
Figure 5.
Isocline diagram with competitor in the absence of spatial structure. Symbols are the same as those in Figure 4, except that the open circle (○) represents an unstable equilibrium. (A) Type 2 extinction (type 1 win). Type 1 bacteria always win and the equilibrium is globally stable. The parameter values are m = 0.1, c = 0.9. (B) Bistability. The final state of the dynamics is either extinction of type 1 or extinction of type 2, depending on their initial density. Either equilibrium is locally stable. The parameter values are m = 0.5, c = 0.5.
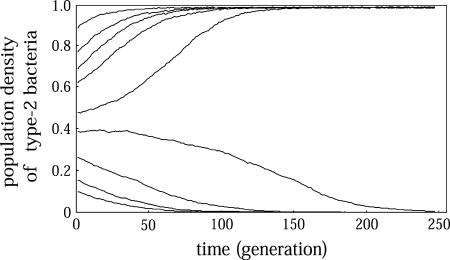
Computer simulation results for different m and c are shown, together with the analytical results, in Figure 3B. The simulation results follow very well the prediction of mean-field analysis in the absence of spatial structure. We also show the dynamical change in the density of type 2 in Figure 6. When the initial density of type 2 bacteria is small, they cannot invade the type 1 bacterial population. This result confirms the result of Mongold (1992). Her study was the first to claim that a postsegregational killing plasmid cannot increase against competition from an incompatible plasmid in the early stage of evolution.
Figure 6.
Bistability in population density in the absence of spatial structure and in the presence of a competitor. The final state of the dynamics is either extinction of type 1 or that of type 2, depending on the density in the initial state. The parameter values are m = 0.1, c = 0.02. Any other parameter set in the B region of Figures 3B and 9B should produce the same dynamics.
Population dynamics in the presence of spatial structure
Under these two conditions corresponding to the absence of spatial structure, we were unable to identify a definitive evolutionary advantage of genetic addiction. However, the results change dramatically when we include spatial structure in our model. To analyze the effects of the spatial localization of interactions, we focused on the dynamics of conditional local densities in addition to those of global densities 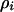 . The “pair approximation” is a method of explicitly constructing a dynamical system, including changes in pair-state densities of neighboring sites and ignoring the dynamics of triplet, quadruplet, or larger states (Matsuda et al. 1992; Harada and Iwasa 1994; Iwasa et al. 1998). In this method, long-range spatial correlations are replaced by short-range correlations (conditional local density
. The “pair approximation” is a method of explicitly constructing a dynamical system, including changes in pair-state densities of neighboring sites and ignoring the dynamics of triplet, quadruplet, or larger states (Matsuda et al. 1992; Harada and Iwasa 1994; Iwasa et al. 1998). In this method, long-range spatial correlations are replaced by short-range correlations (conditional local density 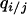 ), and it can predict population dynamic behavior with spatial structure very well (Harada and Iwasa 1994; Iwasa et al. 1998), although we could not construct isocline diagrams as in the case with the absence of spatial structure. Here, we follow the method of Iwasa et al. (1998).
), and it can predict population dynamic behavior with spatial structure very well (Harada and Iwasa 1994; Iwasa et al. 1998), although we could not construct isocline diagrams as in the case with the absence of spatial structure. Here, we follow the method of Iwasa et al. (1998).
Without competitor:
We consider invasion conditions from the dynamics of Equation 2a similarly to analysis in the absence of spatial structure. This time, we do not approximate conditional density 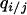 by local density
by local density 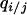 . Instead, we consider
. Instead, we consider 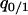 ,
, 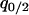 , and
, and 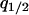 as dynamical variables and describe the changes by differential equations, as shown in appendix b. Note that
as dynamical variables and describe the changes by differential equations, as shown in appendix b. Note that 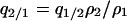 , and we do not have to consider the dynamics of either
, and we do not have to consider the dynamics of either 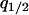 or
or 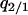 . The dynamical system considered here includes five independent variables:
. The dynamical system considered here includes five independent variables:  , and
, and 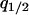 . The condition for type 2 invasion into the type 1 dominant population in this system is calculated by the following procedure. First, we consider the equilibrium population,
. The condition for type 2 invasion into the type 1 dominant population in this system is calculated by the following procedure. First, we consider the equilibrium population, 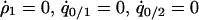 , and
, and 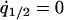 under the condition of
under the condition of 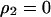 . Second, we substitute the equilibrium values of
. Second, we substitute the equilibrium values of 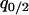 and
and 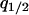 into the per capita growth rate of the type 2 cell derived from Equation 2a. Then, type 2 can invade into a population dominated by type 1, if
into the per capita growth rate of the type 2 cell derived from Equation 2a. Then, type 2 can invade into a population dominated by type 1, if
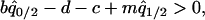 |
(6a) |
where 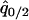 and
and 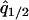 are the equilibrium values of
are the equilibrium values of 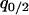 and
and 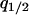 in a type 1 dominant population. We cannot determine the analytical form of the equilibrium
in a type 1 dominant population. We cannot determine the analytical form of the equilibrium 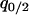 and that of
and that of 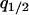 due to the nonlinearity of the dynamics. We calculated the left-hand side of Equation 6a by numerically changing
due to the nonlinearity of the dynamics. We calculated the left-hand side of Equation 6a by numerically changing 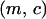 and determined the boundary line that satisfies the left-hand side of Equation 6a equal to 0. We show the lines on the
and determined the boundary line that satisfies the left-hand side of Equation 6a equal to 0. We show the lines on the 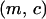 plane in Figure 7. Details of the analysis are shown in appendix b.
plane in Figure 7. Details of the analysis are shown in appendix b.
Figure 7.
Phase diagram in the presence of spatial structure. Methods and parameters are the same as those in Figure 3. (A) In the absence of a competitor. Either type 1 or type 2 drives the other type of bacteria to extinction. (B) In the presence of a competitor. The type 2 bacteria can invade into the population when the transfer rate m is higher than the cost of host killing c.
The invasion condition of type 1 into a population dominated by type 2 was determined similarly. We calculated equilibrium values of 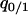 and
and 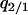 by solving
by solving 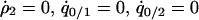 , and
, and 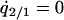 under the condition that
under the condition that 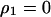 and substituted them into the per capita growth rate of type 1. Then, type 1 can invade into a population dominated by type 2, if
and substituted them into the per capita growth rate of type 1. Then, type 1 can invade into a population dominated by type 2, if
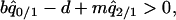 |
(6b) |
where 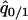 and
and 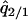 are the equilibrium values of
are the equilibrium values of 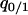 and
and 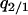 in a type 2 dominant population. Figure 7A shows the result. In this single-plasmid model (model A, in the absence of a competitor), the phase diagram is almost the same as that derived from the mean-field analysis (Figure 3A). The dynamical behavior is labeled as type 1 extinction, type 2 extinction, or “coexistence,” and the condition for each behavior is almost the same as that in the mean-field analysis. In this model of no competitor, the effect of spatial structure appears small, if existent.
in a type 2 dominant population. Figure 7A shows the result. In this single-plasmid model (model A, in the absence of a competitor), the phase diagram is almost the same as that derived from the mean-field analysis (Figure 3A). The dynamical behavior is labeled as type 1 extinction, type 2 extinction, or “coexistence,” and the condition for each behavior is almost the same as that in the mean-field analysis. In this model of no competitor, the effect of spatial structure appears small, if existent.
We also carried out computer simulations of the dynamical model. The results for different m- and c-values are shown, together with the analytical result, in Figure 7A. In this single-plasmid model (model A), the results are almost the same as the analytical prediction by pair approximation. The predicted behaviors (type 1 extinction, type 2 extinction, or coexistence) for the values of m and c are observed by computer simulations under the predicted conditions.
With competitor:
Similarly, we calculated the dynamical results of model B (in the presence of competitor) in the presence of spatial structure by the pair-approximation method. The invasion condition of type 2 into a population dominated by type 1 is as follows:
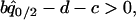 |
(7a) |
where 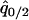 was calculated by solving
was calculated by solving 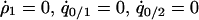 , and
, and 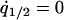 under the condition that
under the condition that 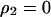 . The invasion condition of type 1 into a population dominated by type 2 is
. The invasion condition of type 1 into a population dominated by type 2 is
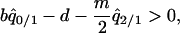 |
(7b) |
where 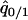 and
and 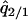 were calculated by solving
were calculated by solving 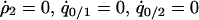 , and
, and 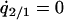 under the condition that
under the condition that 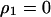 . The lines in Figure 7B show the boundaries where the left-hand sides of (7a) and (7b) equal 0, respectively. The lines are determined numerically, as in model A. The result for the plasmid competition model (model B in the presence of a competitor) in the presence of spatial structure, as analyzed by the pair-approximation method (Figure 7B), is dramatically different from that obtained by mean-field approximation (Figure 3).
. The lines in Figure 7B show the boundaries where the left-hand sides of (7a) and (7b) equal 0, respectively. The lines are determined numerically, as in model A. The result for the plasmid competition model (model B in the presence of a competitor) in the presence of spatial structure, as analyzed by the pair-approximation method (Figure 7B), is dramatically different from that obtained by mean-field approximation (Figure 3).
In the pair-approximation analysis, type 2 bacteria carrying the addiction gene complex can invade, irrespective of its initial density, when transfer rate m is sufficiently larger than the cost of host killing c. Under such conditions, the bacteria carrying the addiction gene complex (type 2) always win and drive the other bacteria with the competitor (type 1) to extinction. The m–c plane is divided into three regions, type 1 extinction, type 2 extinction, or “bistable” as shown in Figure 7B; only the latter two are observed in the mean-field analysis.
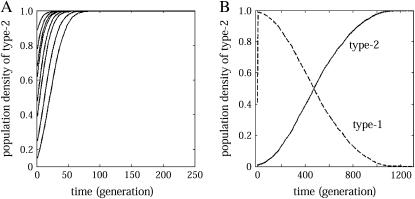
The predicted behaviors (type 1 extinction, type 2 extinction, or bistable) are observed also by computer simulation, when m and c are varied. Figure 7B shows the good match of analysis and simulation. We also illustrate the dynamical change of density of type 1 and type 2 bacteria under the condition of type 1 extinction (Figure 8).
Figure 8.
Increase of cells carrying the addiction gene complex in the presence of spatial structure and of a competitor. (A) The type 2 bacteria always drive type 1 to extinction, irrespective of their initial density. (B) The density of type 1 bacteria is included as a dashed line, which results in its extinction. The parameter values are m = 0.1, c = 0.001. Any other parameter in the II region of Figures 7B and 9D should produce the same dynamics.
From the above results, we can say that the addiction gene complex can invade into the bacterial population, increase in frequency, and maintain itself under the condition where the ratio of m (the rate of gene transfer) to c (the cost of host killing) is large enough. By considering the spatial structure, the early evolution of the addiction gene complex is possible when there are any competitor genetic elements.
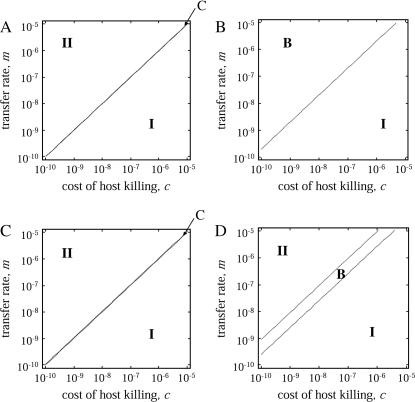
Transfer and postsegregational host killing in the natural environments could be very rare. Figure 9 shows analytical results obtained by the pair approximation when the transfer rate m and the cost of host killing c are as small as 10−5–10−10 and 10−5–10−10 per generation, respectively. Even with these parameters being much smaller than those in the previous analysis, we obtained qualitatively the same results as in Figures 3 and 7.
Figure 9.
Phase diagram with lower parameter values. Analytically predicted dynamic behaviors are shown in the regions on the parameter space as in Figures 3 and 7. Methods and parameters are the same as those in previous analyses, except that the values of transfer rate m and the cost of host killing c are much smaller. The four diagrams are those without (A and C) and with (B and D) competitor and in the absence (A and B) and presence (C and D) of spatial structure, respectively.
DISCUSSION
We studied evolution of genetic addiction in the presence and absence of spatial structure using mathematical models and computer simulations of population dynamics. This is the first study analyzing the contribution of spatial structure in postsegregational killing or genetic addiction systems, although the method of pair approximation for spatial structure (Nakamaru et al. 1997; Iwasa et al. 1998) and a basic model analyzing evolution of postsegregational killing in the absence of a spatial structure (Mongold 1992) were already developed. We demonstrated that the property of genetic addiction itself allows the responsible gene complex to increase in frequency in a bacterial population irrespective of its initial density. If we consider the spatial structure by the pair approximation and computer simulation of a spatially structured model, no matter how low the initial density is, an addiction gene complex is able to invade into a bacterial cell population with its competitor and maintain itself under dynamical conditions, where the rate of gene transfer is higher than the cost of host killing.
The intuitive interpretation of this result is as follows. Cell death caused by horizontal transfer of the addiction gene complex and competitive exclusion makes an occupied site temporarily vacant. Whether type 2 can spread to this site or not is most important for evolution. In the presence of a spatial structure, the spatial pattern, in which the same types of bacterial cells are present close together, is formed automatically. This pattern produces a higher probability that the bacterial cells neighboring the dead cell are of type 2, which enables them and the addiction gene complexes to spread to the site.
In a preceding theoretical study (Mongold 1992), the plasmid carrying a postsegregational killing gene was able to evolve due to an advantage in competitive exclusion between incompatible plasmids. The conditions are that it has a high initial density, a high transfer rate into cells carrying the competing plasmid, and strong incompatibility resulting in a high segregation rate in the cells carrying both the plasmids.
Our models are based on Mongold's (1992), while some parts are simplified to enable mathematical analysis of spatial structure. For example, she considered both growth and segregational effects for the cost of a plasmid carrying a postsegregational killing gene. She also considered a segregation rate for each plasmid separately, quoting that the value could be no more than 0.5 (Simonsen 1990) and assuming that the rate of plasmid transfer to other cells carrying the other plasmid became lower because of surface exclusion. In contrast, our model neglects the effect of surface exclusion and growth cost of the plasmid to its host. In addition, our model assumes that the segregation rate in cells carrying two competing plasmids is 1.0, although bacterial cells carrying pairs of incompatible plasmids indeed may carry both plasmids for a few generations before one of them is lost. However, her model did not include spatial structure at all. Our model instead includes spatial structure explicitly.
These simplifications in our model seem to make the evolution of type 2 and the addiction gene complex easier. However, our model, in the absence of a spatial structure and with a competitor, which corresponds to her model, produced essentially the same result as her model. That is, addiction gene complexes cannot evolve without their high initial density. Only after taking the effect of spatial structure into account is evolution from their low initial density made possible in our simplified model. Therefore, it is revealed mathematically that the most important factor enabling the addiction gene complex to evolve in the very early stage of evolutionary dynamics is the spatial structure. The spatial structure causes the major difference in the invasion conditions between our model and Mongold's.
In fact, in an experimental population of bacteria with and without a bacteriocin, the dynamics of the bacterial population were shown to be affected by spatial structure (Chao and Levin 1981). In an experimental fluid culture, the result of the dynamics was bistable and depended on the initial ratio of the bacteria. The colicinogenic cells could not increase from a low initial frequency. On the other hand, if bacteria were cultured on a solid surface, the result of the dynamics did not depend on the bacterial ratio. The colicinogenic cells were able to increase from a very low frequency. This experimental result is analogous to our modeling, although it does not completely correspond to our model in that the two bacterial populations are competing as colonies, not as individual cells.
Our lattice model was never meant to be a physically realistic and an accurate analog of bacteria in surface culture. The purpose of this type of model is not to reflect the reality but to examine the qualitative difference caused by an important novel factor. Our model indeed revealed that spatial structure, in which the same types of bacteria naturally get close together, causes the dramatic change in invasion condition for addiction gene complexes. Simplifying the model made mathematical analysis much easier and demonstrated the effect of spatial structure more clearly.
In fact, in surface culture or other physically structured habitats, bacteria exist as colonies or microcolonies. On the other hand, our model assumes that each lattice site is occupied by a bacterial cell. This situation might seem to be unrealistic, and one can interpret a cell in our model as a colony by replacing values m, c, b, and d with those concerning colony-level events. However, more unrealistic assumptions are needed for this interpretation because it requires that all bacteria in each colony change (experience gene transfer or postsegregational killing) all at once. In reality, plasmid (and gene) transfer occurs first on the interface between two colonies (Molin and Tolker-Nielsen 2003), which better corresponds to our model, in which most sites are occupied by either type and the same types of bacteria naturally get closer together.
We analyzed our model using a wide range of parameter values of m and c, as small as 10−5–10−10 per generation. The corresponding region in Figure 9, B and D, indeed shows the most important result of this research, i.e., the major difference in the invasion conditions in the presence of a competitor genetic element. Without spatial structure, addiction gene complexes cannot evolve without a high initial density (bistable region, B, in Figure 9B). With spatial structure, their evolution is possible, irrespective of their initial density (type-2 winning region, II, in Figure 9D).
Intuitively, evolution of the addiction gene complex gets easier if the transfer rate is higher and the cost of host killing is lower. The rate of plasmid transfer in the natural environments could be much higher than earlier estimates. According to a recent compilation of the experiments in aquatic microcosms (Van Elas et al. 2000), the rates can be as high as 5 × 10−2 and 4 × 10−2 transconjugants/recipient. The cost of host killing by natural segregation in the absence of competitor genetic elements could be very rare. When an addiction gene complex establishes itself in a new host cell, it attempts to avoid cell killing by expressing its antitoxin first and then its toxin (Nakayama and Kobayashi 1998; Kobayashi 2004). In the maintenance phase, the antitoxin, together with the toxin, has tight autoregulation of the gene complex, so that they can suppress toxin activity until they receive a signal to attack the host (de Feyter et al. 1989; Kobayashi 2004). In addiction, many of the plasmids carrying addiction gene complexes often have other means of stable maintenance to minimize host killing. For example, the F plasmid carries machinery (Sop) for active portioning of plasmid molecules to daughter cells and a site-specific recombination system to resolve a dimeric plasmid molecule into monomers, in addition to two addiction gene complexes (Ccd and SrnB) (Funnell and Phillips 2004). Therefore, the parameter values for the evolution of addiction genes may well be present in the natural environments.
A testable prediction of our theoretical results would be that an addiction gene complex should be able to increase in frequency from near zero in a mixed culture of its carrier and noncarrier on a solid surface. The principle of postsegregational killing or genetic addiction was first identified by bacteriologists as a mechanism used by several bacterial extrachromosomal genetic elements (plasmids) to stably maintain themselves within their bacterial host. Yarmolinsky called this phenomenon plasmid addiction (Lehnherr et al. 1993). However, similar genetic elements and phenomena have also been identified, in addition to plasmid-related ones. Consequently, programmed cell death in bacteria was proposed to be often mediated through these “addiction modules” (Engelberg-Kulka and Glaser 1999; Zhang et al. 2003). The term genetic addiction, which we propose (Kobayashi 2004), is based on preceding terms, but more generalized. It is possible that, rather than being a curious exception to the general rules governing symbiosis of genes in a genome, genetic addiction may represent a general principle in itself. When knockout of a gene leads to death of an organism, the gene is called essential. However, these genes may have evolved to be good at causing host death at their loss after their appearance in the genome. Therefore, this phenomenon may have implications for the general issue of genome evolution.
Several theoretical works have shown that otherwise costly behaviors can evolve in spatially structured populations (Nakamaru et al. 1997; Iwasa et al. 1998; Keller 1999). However, postsegregational host killing does not represent costly and spiteful behavior by an organism. Instead, it represents a strategy of a gene complex, as analyzed here. Postsegregational killing may represent a “green beard effect,” which is a form of genetic self and nonself recognition, resulting in directing benefits to individuals with the relevant (self) gene or rejecting individuals that do not possess the (self) gene (Haig 1997).
Although our results demonstrate that genetic addiction is sufficient only for evolution of addiction gene complexes, they do not exclude the possibility that these addiction gene complexes may also have beneficial traits for their hosts. For example, some restriction–modification gene complexes may serve as a defense against bacteriophages. In other words, genome evolution, through acquisition of beneficial genes, may be assisted by genetic addiction.
Acknowledgments
The computer simulation was run on a UNIX workstation of the National Institute of Basic Biology and Human Genome Center at the Institute of Medical Science, the University of Tokyo. This work was supported by a grant from 21st Century Center of Excellence program (genome and language) to Ichizo Kobayashi; by grants-in-aid for scientific research by the Ministry of Education, Science, Sports, and Culture, Japan (genome biology, genome homeostasis) to Ichizo Kobayashi; and by a grant from the Foundation for the Fusion of Science and Technology to Koji Yahara.
APPENDIX A
Details of the simulation are as follows (Durrett and Levin 1997; Nakamaru et al. 1997). We have chosen a lattice of size 100 × 100 with a periodic boundary condition so that the lattice is mapped onto a torus by identifying its opposite edges. In the case without spatial structure, each bacterium is on a lattice point but can interact with another bacterium wherever it is on the lattice. In each computation step, two sites of the lattice are randomly selected. If one of these two sites is vacant and the other is occupied by a bacterial cell, then the cell reproduces at the rate b/z, and the new bacterial cell occupies the vacant site. In the same way, randomly selected type 1 and type 2 bacterial cells will be engaged in gene transfer at rate 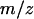 . The bacterial death occurs with a given rate of d. The initial density of both types of bacteria
. The bacterial death occurs with a given rate of d. The initial density of both types of bacteria 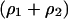 was 0.05. The initial distribution pattern was randomly selected and the local densities were the same as the corresponding global ones. The cases in which
was 0.05. The initial distribution pattern was randomly selected and the local densities were the same as the corresponding global ones. The cases in which 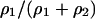 is 0.01, 0.02, 0.05, 0.1, 0.5, 0.9, 0.95, 0.98, and 0.99 were examined. (We tried the extreme fraction of 0.9998 in some cases.) We stopped the computation when the time reached 1,000,000 steps or if either bacterium became extinct. We iterated the calculation 10 times for each initial pattern.
is 0.01, 0.02, 0.05, 0.1, 0.5, 0.9, 0.95, 0.98, and 0.99 were examined. (We tried the extreme fraction of 0.9998 in some cases.) We stopped the computation when the time reached 1,000,000 steps or if either bacterium became extinct. We iterated the calculation 10 times for each initial pattern.
If either bacterium drives the other to extinction ≥90% (9 or 10 times out of 10 times), we decide this type is the winner. If the final winner is type 1 or type 2 depending on the initial ratio 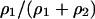 , we decide that the result is bistable. In bistable systems, both bacteria resist invasion by the rare opponent type.
, we decide that the result is bistable. In bistable systems, both bacteria resist invasion by the rare opponent type.
In the case with spatial structure, each bacterium can interact only with the nearest neighbor lattice site. In each computational step, a randomly chosen bacterial cell can reproduce only when there is a vacant site among the z nearest neighbors. The newly born bacterial cell would be set on the vacant site. In the same way, a randomly selected type 1 bacterial cell can be engaged in gene transfer with a nearest neighbor type 2 cell when there is one. Other methods are the same as in the case without spatial structure.
APPENDIX B
To analyze the effects of the spatial localization of interactions, we introduce the dynamics of conditional local densities in addition to those of the global densities 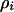 (Matsuda et al. 1992; Harada and Iwasa 1994; Iwasa et al. 1998). There are nine different local densities (
(Matsuda et al. 1992; Harada and Iwasa 1994; Iwasa et al. 1998). There are nine different local densities (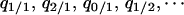 ), although not all of them are independent. The number of variables can be reduced, leaving five independent variables without loss of generality. When we analyze the condition for type-2 invasion into a population dominated by type 1, we choose
), although not all of them are independent. The number of variables can be reduced, leaving five independent variables without loss of generality. When we analyze the condition for type-2 invasion into a population dominated by type 1, we choose 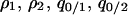 , and
, and 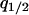 as the five independent variables and express all the other variables in terms of these five.
as the five independent variables and express all the other variables in terms of these five.
Each conditional density may be expressed as a “pair” probability (doublet density) (Matsuda et al. 1992) divided by a global density. For example, the local density of a vacant site (0-site) in the neighborhood of a randomly chosen type 1 cell is 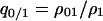 , i.e., the ratio of
, i.e., the ratio of 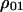 , the probability for a randomly chosen nearest neighbor pair to be type 1, divided by the global density
, the probability for a randomly chosen nearest neighbor pair to be type 1, divided by the global density 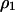 . The higher-order conditional probability
. The higher-order conditional probability 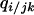 is defined as the probability that a neighbor of the j-site of a randomly chosen jk-pair is a type i cell.
is defined as the probability that a neighbor of the j-site of a randomly chosen jk-pair is a type i cell.
Let us consider the dynamics of 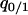 in the single-plasmid model (without competitor). We first calculate the dynamics of summation of “01” pairs and “10” pairs:
in the single-plasmid model (without competitor). We first calculate the dynamics of summation of “01” pairs and “10” pairs:
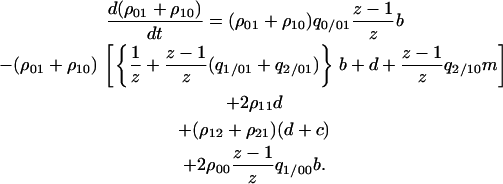 |
(B1) |
The first two lines of the right-hand side indicate the transition from 01 or 10 pairs: increase and decrease of the pairs by the birth-and-death process. The third line indicates increase of 01 or 10 pairs by the death of either cell of the 11 pair. The fourth line indicates increase of 01 or 10 pairs by the death of the type 2 cell of 12 or 21 pairs. The fifth line indicates transition from the 00 pair to the 01 pair by the reproduction of possible neighboring type 1 cells that may exist in the 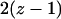 sites neighboring the 0-site.
sites neighboring the 0-site.
To calculate the rate of the latter event, we need a higher-order conditional probability, such as 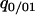 or
or 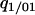 . Pair approximation assumes that
. Pair approximation assumes that 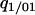 can be replaced by
can be replaced by 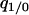 , on the assumption that the indirect dependence is weak (Matsuda et al. 1992). This approximation is more accurate than the mean-field version. Under this simplification, we can calculate the dynamics of the doublet density in terms of global and local densities:
, on the assumption that the indirect dependence is weak (Matsuda et al. 1992). This approximation is more accurate than the mean-field version. Under this simplification, we can calculate the dynamics of the doublet density in terms of global and local densities:
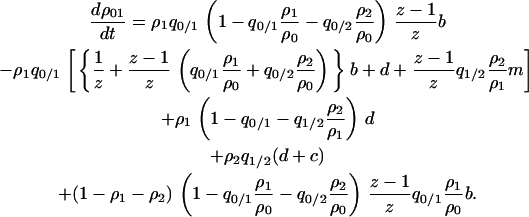 |
(B2) |
Here, we used the formulas 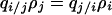 and
and 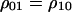 to derive (B2) from (B1) and removed the coefficient 2. By differentiating
to derive (B2) from (B1) and removed the coefficient 2. By differentiating 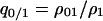 with respect to time t, we have
with respect to time t, we have
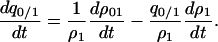 |
Using (2a) and (B2), we have the dynamics of the local density as
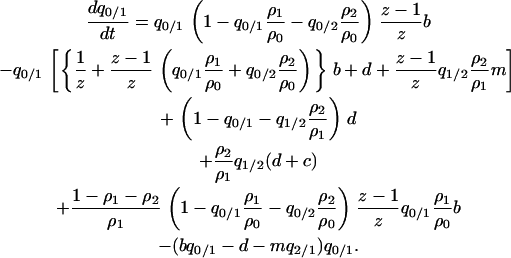 |
(B3) |
In a similar way, we can calculate the dynamics of the other doublet densities, such as 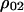 and
and 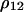 , from which we derive the dynamics of local densities, such as
, from which we derive the dynamics of local densities, such as 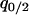 and
and 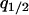 . The right-hand side of all the differential equations can be written in functions of
. The right-hand side of all the differential equations can be written in functions of 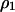 ,
, 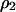 ,
, 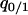 ,
, 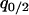 , and
, and 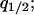 i.e., it is a closed system.
i.e., it is a closed system.
Pair approximation dynamics allow us to analytically derive conditions for successful invasion of one type of cell into a population dominated by the other type. Here, we consider the case in which the lattice population is near the equilibrium in which only type 1 cells exist, and we ask whether type 2 cells (which are initially rare) can increase in the system. The values of the densities 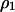 ,
, 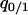 ,
, 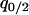 , and
, and 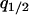 can be calculated at the type 1 dominant equilibrium by solving
can be calculated at the type 1 dominant equilibrium by solving 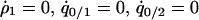 , and
, and 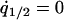 under the condition that
under the condition that 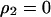 , although we cannot determine the equilibrium formulas explicitly. We determined them numerically. It is important to note that the invader typically develops a clumped spatial distribution and, therefore, that the density of the invader in the neighborhood of another invader (i.e., local density
, although we cannot determine the equilibrium formulas explicitly. We determined them numerically. It is important to note that the invader typically develops a clumped spatial distribution and, therefore, that the density of the invader in the neighborhood of another invader (i.e., local density 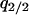 ) is not negligible.
) is not negligible.
Invasion condition is determined by the sign of its per capita growth rate. From (2a), we have that 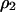
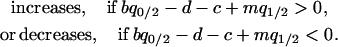 |
Note that it depends on the equilibrium values of local densities 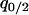 and
and 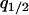 only. As we do not have the explicit solutions of these equilibria, we calculated numerically the left-hand side of the above inequality for each pair of
only. As we do not have the explicit solutions of these equilibria, we calculated numerically the left-hand side of the above inequality for each pair of 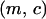 .
.
The invasion condition for the type 1 cell is also determined. The procedure is just the same as that for the type 2 invasion, except that we use 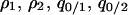 , and
, and 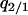 (instead of
(instead of 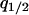 ) as the independent variables.
) as the independent variables.
References
- Chao, L., and B. R. Levin, 1981. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc. Natl. Acad. Sci. USA 78: 6324–6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T. F., and J. A. Heinemann, 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. USA 97: 12643–12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Feyter, R., C. Wallace and D. Lane, 1989. Autoregulation of the ccd operon in the F plasmid. Mol. Gen. Genet. 218: 481–486. [DOI] [PubMed] [Google Scholar]
- Durrett, R., and S. Levin, 1997. Allelopathy in spatially distributed populations. J. Theor. Biol. 185: 165–171. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka, H., and G. Glaser, 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53: 43–70. [DOI] [PubMed] [Google Scholar]
- Faguy, D. M., and W. F. Doolittle, 2000. Horizontal transfer of catalase-peroxidase genes between archaea and pathogenic bacteria. Trends Genet. 16: 196–197. [DOI] [PubMed] [Google Scholar]
- Frank, S. A., 1994. Spatial polymorphism of bacteriocins and other allelopathic traits. Evol. Ecol. 8: 369–386. [Google Scholar]
- Funnell, B. E., and G. J. Phillips, 2004. Plasmid Biology. ASM Press, Washington, DC.
- Gerdes, K., A. P. Gultyaev, T. Franch, K. Pedersen and N. D. Mikkelsen, 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31: 1–31. [DOI] [PubMed] [Google Scholar]
- Gerdes, K., S. K. Christensen and A. Lobner-Olesen, 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3: 371–382. [DOI] [PubMed] [Google Scholar]
- Haig, D., 1997. Social genes, pp. 284–304 in Behavioral Ecology: An Evolutionary Approach, Ed. 4, edited by J. R. Krebs and N. B. Davies. Blackwell Scientific, Oxford.
- Handa, N., Y. Nakayama, M. Sadykov and I. Kobayashi, 2001. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction–modification gene complex. Mol. Microbiol. 40: 932–940. [DOI] [PubMed] [Google Scholar]
- Harada, Y., and Y. Iwasa, 1994. Lattice population-dynamics for plants with dispersing seeds and vegetative propagation. Res. Popul. Ecol. 36: 237–249. [Google Scholar]
- Iwasa, Y., M. Nakamaru and S. A. Levin, 1998. Allelopathy of bacteria in a lattice population: competition between colicin-sensitive and colicin-producing strains. Evol. Ecol. 12: 785–802. [Google Scholar]
- Keller, L., 1999. Levels of Selection in Evolution. Princeton University Press, Princeton, NJ.
- Kobayashi, I., 2004. Genetic addiction: a principle of gene symbiosis in a genome, pp. 105–144 in Plasmid Biology, edited by B. E. Funnell and G. J. Phillips. ASM Press, Washington, DC.
- Lehnherr, H., E. Maguin, S. Jafri and M. B. Yarmolinsky, 1993. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 233: 414–428. [DOI] [PubMed] [Google Scholar]
- Matsuda, H., N. Ogita, A. Sasaki and K. Sato, 1992. Statistical-mechanics of population—the lattice Lotka-Volterra model. Prog. Theor. Phys. 88: 1035–1049. [Google Scholar]
- Molin, S., and T. Tolker-Nielsen, 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14: 255–261. [DOI] [PubMed] [Google Scholar]
- Mongold, J. A., 1992. Theoretical implications for the evolution of postsegregational killing by bacterial plasmids. Am. Nat. 139: 677–689. [Google Scholar]
- Naito, T., K. Kusano and I. Kobayashi, 1995. Selfish behavior of restriction–modification systems. Science 267: 897–899. [DOI] [PubMed] [Google Scholar]
- Nakamaru, M., H. Matsuda and Y. Iwasa, 1997. The evolution of cooperation in a lattice-structured population. J. Theor. Biol. 184: 65–81. [DOI] [PubMed] [Google Scholar]
- Nakayama, Y., and I. Kobayashi, 1998. Restriction–modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. USA 95: 6442–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, D. P., and K. Gerdes, 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadykov, M., Y. Asami, H. Niki, N. Handa, M. Itaya et al., 2003. Multiplication of a restriction–modification gene complex. Mol. Microbiol. 48: 417–427. [DOI] [PubMed] [Google Scholar]
- Sato, K., H. Matsuda and A. Sasaki, 1994. Pathogen invasion and host extinction in lattice structured populations. J. Math. Biol. 32: 251–268. [DOI] [PubMed] [Google Scholar]
- Simonsen, L., 1990. Dynamics of plasmid transfer on surfaces. J. Gen. Microbiol. 136: 1001–1007. [DOI] [PubMed] [Google Scholar]
- Van Elas, J. D., J. Fry, P. Hirsch and S. Molin, 2000. Ecology of plasmid transfer and spread, pp. 175–206 in The Horizontal Gene Pool, edited by C. M. Thomas. Harwood Academic, Amsterdam.
- Zhang, J., Y. Zhang and M. Inouye, 2003. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 278: 32300–32306. [DOI] [PubMed] [Google Scholar]