Abstract
To date, the field of ancient DNA has relied almost exclusively on mitochondrial DNA (mtDNA) sequences. However, a number of recent studies have reported the successful recovery of ancient nuclear DNA (nuDNA) sequences, thereby allowing the characterization of genetic loci directly involved in phenotypic traits of extinct taxa. It is well documented that postmortem damage in ancient mtDNA can lead to the generation of artifactual sequences. However, as yet no one has thoroughly investigated the damage spectrum in ancient nuDNA. By comparing clone sequences from 23 fossil specimens, recovered from environments ranging from permafrost to desert, we demonstrate the presence of miscoding lesion damage in both the mtDNA and nuDNA, resulting in insertion of erroneous bases during amplification. Interestingly, no significant differences in the frequency of miscoding lesion damage are recorded between mtDNA and nuDNA despite great differences in cellular copy numbers. For both mtDNA and nuDNA, we find significant positive correlations between total sequence heterogeneity and the rates of type 1 transitions (adenine → guanine and thymine → cytosine) and type 2 transitions (cytosine → thymine and guanine → adenine), respectively. Type 2 transitions are by far the most dominant and increase relative to those of type 1 with damage load. The results suggest that the deamination of cytosine (and 5-methyl cytosine) to uracil (and thymine) is the main cause of miscoding lesions in both ancient mtDNA and nuDNA sequences. We argue that the problems presented by postmortem damage, as well as problems with contamination from exogenous sources of conserved nuclear genes, allelic variation, and the reliance on single nucleotide polymorphisms, call for great caution in studies relying on ancient nuDNA sequences.
WHEN an organism dies, its DNA starts degrading at a rate that is believed to be highly dependent on the environment and subsequent conditions of storage (Pääbo 1989; Lindahl 1993; Höss et al. 1996; Poinar et al. 1996; Hofreiter et al. 2001a; Smith et al. 2001; Pääbo et al. 2004; Willerslev et al. 2004a; Willerslev and Cooper 2005). Postmortem DNA is subject to degradation by microorganisms, soil invertebrates, and cellular nucleases, in addition to modifications by spontaneous chemical reactions such as hydrolysis and oxidation (Lindahl 1993; Hofreiter et al. 2001a; Gilbert and Hansen 2006). While some types of postmortem DNA modification block the extension of polymerase enzymes, thus rendering the molecules unsuitable as template for PCR, others, termed miscoding lesions, allow for amplification, but result in the incorporation of erroneous bases during PCR (Pääbo 1989; Lindahl 1993; Höss et al. 1996; Poinar et al. 1998; Hansen et al. 2001; Hofreiter et al. 2001b; Gilbert et al. 2003a,b; Banerjee and Brown 2004; Willerslev et al. 2004b; Mitchell et al. 2005; Gilbert and Hansen 2006). The most commonly observed miscoding lesions in ancient DNA (aDNA) studies are the transitions adenine → guanine (A → G), cytosine → thymine (C → T), G → A, and T → C (Hansen et al. 2001; Hofreiter et al. 2001b; Gilbert et al. 2003b). Although four different transitions have been observed, it has been argued that it is possible to differentiate the transitions into only two complementary groups, termed type 1 (TS1: A → G/T → C) and type 2 (TS2: C → T/G → A) transitions, caused putatively by the deamination of adenine to hypoxanthine (A → H) and the deamination of cytosine (and its homolog 5-methyl cytosine) to uracil (and thymine), respectively (Hansen et al. 2001; Hofreiter et al. 2001b; Gilbert et al. 2003b). For example, consider a single cytosine to uracil (C → U) deamination event on the light strand of mitochondrial DNA (mtDNA). The C → U event will be observed as a C → T transition on any light strand sequences amplified directly from the original damaged template, but any of the derived complementary heavy strand sequences will exhibit a G → A transition. Similarly, any sequences derived directly from the amplification of an A → H event will be exhibited as either an A → G or T → C transition. Intriguingly, even though miscoding lesions generating both type 1 and type 2 transitions are recorded for DNA in solution (deamination of cytosine with a rate ∼30–50 times higher than that for adenine; Lindahl 1993), it is still debated whether the deamination of adenine actually plays a role in the generation of type 1 events in aDNA or whether they are simply due to regular DNA polymerase errors (Hofreiter et al. 2001b; Gilbert et al. 2003b; Pääbo et al. 2004).
Although mtDNA is most frequently used in aDNA research, a number of recent studies have reported the successful retrieval of low- and single-copy nuclear DNA (nuDNA) sequences (Greenwood et al. 1999, 2001; Bunce et al. 2003; Huynen et al. 2003; Jaenicke-Despres et al. 2003; Orlando et al. 2003; Poinar et al. 2003; Noonan et al. 2005). Such studies represent important breakthroughs, as nuDNA can be used to answer previously intractable questions, such as the establishment of phenotypic characteristics (Bunce et al. 2003; Jaenicke-Despres et al. 2003) and the resolution of deep phylogenetic splits (Poinar et al. 2003). However, nuclear genes are typically 5000–10,000 times less abundant per cell than those of mitochondrial origin, which is probably the reason for ancient nuDNA being much more difficult to amplify than mtDNA from the same aDNA extracts (Poinar et al. 2003).
To date, studies that characterize DNA damage in fossil remains have been restricted to mtDNA (Höss et al. 1996; Hansen et al. 2001; Hofreiter et al. 2001a; Gilbert et al. 2003a,b; Threadgold and Brown 2003; Banerjee and Brown 2004; Gilbert et al. 2005). Although one study has reported substitutions in PCR-amplified ancient nuDNA sequences, the authors attribute the observed changes to mutagenic effects introduced under PCR reactions (Pusch et al. 2004), although this remains questionable (Serre et al. 2004a). Despite the lack of publications, the characterization of damage in ancient nuDNA remains important to the field. In particular, the fact that miscoding lesions can result in modification of the consensus sequence amplified from ancient mtDNA (Handt et al. 1996; Gilbert et al. 2003a; Banerjee and Brown 2004; Hebsgaard et al. 2005), along with the overall low abundance of nuDNA templates relative to mtDNA templates, carries the implication that sequences amplified from ancient nuDNA are at increased risk of containing sequence errors. Furthermore, nuDNA sequences generated from fossil remains often are single nucleotide polymorphisms (SNPs), and the determination of SNPs as real or as the result of postmortem damage is crucial.
In this study we investigate the frequency and types of miscoding lesions in various mtDNA and nuDNA markers across a variety of fossil specimens of different ages preserved under different environmental conditions. In addition, we discuss some of the factors that need to be considered when amplifying, sequencing, and interpreting nuclear data from archival and fossil remains.
MATERIALS AND METHODS
We analyzed previously unreported DNA sequences extracted from permafrost-preserved bones of the woolly rhinoceros (Coelodonta antiquitatis, n = 2) and the lion (Panthera leo spelaea, n = 6), as well as temperate-preserved bones of the pig (Sus scrofa, n = 5) and the female moa (Dinornis robustus, n = 4). Not included in the comparative study were amplification results from one woolly rhinoceros, two pigs, and 28 lion specimens yielding mtDNA sequences but no nuDNA sequences and from three woolly rhinoceroses and 88 lion specimens yielding neither mtDNA nor nuDNA sequences. Additionally, published mtDNA and nuDNA clone sequences from desert-preserved ground sloth coprolites and permafrost-preserved woolly mammoth and woolly rhinoceros bones and teeth (n = 6) were included in the analyses (Poinar et al. 1996, 2003; Greenwood et al. 1999, 2001; Orlando et al. 2003). Published sequences from ancient human remains were not included due to the high risk of contamination in such studies (Cooper and Poinar 2001; Hofreiter et al. 2001b; Pääbo et al. 2004; Willerslev and Cooper 2005). In total the data set comprised mtDNA and nuDNA sequences from 23 specimens (for sample details, see Table 1 and supplemental material at http://www.genetics.org/supplemental/).
TABLE 1.
Sample details
| Ageb (YBP)
|
Specimen typec
|
Sequence dataf
|
Sequence regiong
|
|||||
|---|---|---|---|---|---|---|---|---|
| Sample namea | Environmentd | Speciese | mtDNA | NuDNA | mtDNA | NuDNA | ||
| Mp-Wrangel | 4590 ± 50 | Bone | Permafrost | Mammuthus primigenius | 4/556 | 14/1596 | cyt b | Micro-satellites |
| Mp-Alaska8460 | 13,775 ± 145 | Tooth | Permafrost | Mammuthus primigenius | 14/2098i | 131/7252 | cyt b | vWf,a2ab,irib, numt, micro- satellite |
| Mp-Siberia | 26,000 ± 1600 | Bone | Permafrost | Mammuthus primigenius | 4/556 | 10/1140 | cyt b | Micro-satellites |
| SC7400 | 60–70,000h | Tooth | Temperate cave | C. antiquitatis | 43/7463i | 4/680 | cyt b | Numt |
| SC81205 | 40–45,000h | Tooth | Temperate cave | C. antiquitatis | 10/1820i | 9/1260 | 12S | Numt |
| PIN 3342-103 | >49000 | Bone | Permafrost | C. antiquitatis | 47/11697i | 12/3552 | 12S, cyt bj | Numt |
| PIN 3100-169 | 43700 ± 1000 | Bone | Permafrost | C. antiquitatis | 6/3842 | 3/837 | 12Sj | Numt |
| Ua11835 | 19875 ± 215 | Cropolite | Varm cave | Nothrotheriops shastensis | 76/6671 | 115/6813 | 12S | 28S,vW, Crem, PCLB4 |
| RB91 | 12,450 ± 60 | Bone | Permafrost | P. leospelaea | 10/1440 | 5/720 | atp8 | Numt |
| RB75 | 12,090 ± 80 | Bone | Permafrost | P. leospelaea | 16/2304 | 2/288 | atp8 | Numt |
| RB42 | >50,600 | Bone | Permafrost | P. leospelaea | 9/1296 | 9/1296 | atp8 | Numt |
| RB44 | 54,100 ± 1800 | Bone | Permafrost | P. leospelaea | 8/1152 | 24/3456 | atp8 | Numt |
| RB41 | 46,200 ± 1500 | Bone | Permafrost | P. leospelaea | 9/1296 | 3/432 | atp8 | Numt |
| RB46 | >61,500 | Bone | Permafrost | P. leospelaea | 7/1008 | 14/2016 | atp8 | Numt |
| GL92 | 91 | Bone | Museum | S. scrofa | 8/1712 | 6/1038 | D-loop | CD45 |
| GL71 | 86 | Bone | Museum | S. scrofa | 7/1493 | 8/1384 | D-loop | CD45 |
| GL55 | 46 | Bone | Museum | S. scrofa | 6/1284i | 6/1038 | D-loop | CD45 |
| GL45 | 98 | Bone | Museum | S. scrofa | 6/1284 | 7/1211 | D-loop | CD45 |
| GL76 | 50 | Bone | Museum | S. scrofa | 8/1712 | 8/1384 | D-loop | CD45 |
| Moa716 | 700–5000h | Bone | Temperate cave | D. robustus | 15/3031 | 15/615 | D-loop | Kw1,j adh |
| Moa237 | 700–5000 | Bone | Temperate cave | D. robustus | 15/3031 | 16/656 | D-loop | Kw1, adh |
| Moa660 | 613 ± 90 | Bone | Temperate cave | D. robustus | 14/2828 | 15/615i | D-loop | Kw1, adh |
| Moa799 | 1–3000h | Bone | Dry swamp | D. robustus | 16/3232 | 32/2138 | D-loop | Kw1,j adh |
YBP, years before present.
Data sources: Moa716, Moa237, Moa660, and Moa799 are from Bunce et al. (2003) and data generated in this study; Mp-Wrangel, Mp-Sibiria, and MP-Alaska8460 are from Greenwood et al. (1999, 2001) and data generated in this study; Ua11835 is from Poinar et al. (1998, 2003); SC81205 and SC7400 are from Orlando et al. (2003); GL92, GL71, GL55, GL45, and GL76 are from Larson et al. (2005) and this study; RB41, RB42, RB44, RB46, RB75, and RB91 are from this study; PIN3342-103 and PIN3100-169 are also from this study.
Age of specimen based on museum records, accelerator mass spectrometer dating, or on stratigraphy.
Type of specimen.
Type of original preservation environment.
Species name of specimen.
Sampling effort. The numbers of mitochondrial or nuclear clones/the total length of the sequences in base pairs. The average number of clones is very similar for mtDNA (16; SD 17) and nuDNA (20; SD 33).
Name of the mitochondrial and nuclear sequences studied.
Age of specimen based on stratigraphy.
Results were independently replicated.
Results were quantified by real time PCR.
Bone samples were collected, DNA extracted, and PCR amplified following established aDNA protocols: Using a Dremel tool, ∼1-cm3 fragments ∼0.5 cm in depth were removed from the bones. A Braun Mikrodismembrator was used to grind samples. Grinding equipment (stainless-steel balls and cups, rubber washers) was thoroughly bleached between each use. Decalcification was done in 5–30 vol of 0.5 m EDTA (pH 8) overnight at room temperature. The sediment was collected by centrifugation and digested with 0.25 mg/ml proteinase K/8 mg/ml DTT overnight at 50°–55°. The samples were extracted twice with phenol and once with chloroform, and the DNA was recovered and up-concentrated with Centricon-30 (Amicon, Beverly, MA) devices. The proofreading polymerase Platinum Taq Hifidelity (Invitrogen, San Diego) was used in PCR amplification to minimize the generation of DNA polymerase errors that can mimic errors caused by miscoding lesions (Hansen et al. 2001). PCR amplifications were performed in 25-μl volumes adding 0.02- to 1-μl DNA extracts, 0–2 mg/ml bovine serum albumin (BSA), 10 mm Tris·HCl, 1.5 mm MgCl2, 50 mm KCl (pH 8.3), 0.8 mm dNTPs, 1 mm of each primer, and 1 unit of DNA polymerase. Thermal cycling conditions were typically 40 cycles of 95°/52°–66°/68° (30–90 sec each) for mtDNA amplifications and 40–50 cycles of 95°/52°–55°/68° (45–60 sec each) for nuDNA amplifications. BSA was added to the PCR to overcome inhibition. Additionally, dilutions (1:10–1:50) of the DNA extracts were attempted. For primer details and amplification conditions, see Table 2. The amplification products were cloned and sequenced following established procedures (Willerslev et al. 1999).
TABLE 2.
Primers and PCR conditions
| Species/genea | Primer nameb | Primer sequence (5′–3′)c | Annealing temperature/ cyclesd | Sourcee |
|---|---|---|---|---|
| Woolly rhino/12S rDNA | 21pheF | AAAGCAAGGCATTGAAAATGCCTAGATGA | 55°/×40 | Tougard et al. (2001) |
| 684-12Sr | GGCGGTATATAGGCTGAATT | |||
| Woolly rhino/12S rDNA | 650-12sF | CCGATAAACCCCGATAAACC | 56°/×40 | This study |
| 1085valR | TGAAATCTCCTGGGTGTAAGC | |||
| Woolly rhino/Cytochrome b | 14228glu-2F | ACCAATGACATGAAAAATCATCGTT | 52°/×40 | This study |
| 14596cytbR | TTTCAGGTTTCTAGGAAGGTGT | |||
| Woolly rhino/Cytochrome b | 14279cytbF | ATGACTAACATCCGCAAATCCC | 66°/×40 | This study |
| 15107cytbR | GGGATGGATCGTAGGATTGCGTA | |||
| Woolly rhino/Cytochrome b | 14912cytbF | CCAACATAGACAAAATCCC | 52°/×40 | This study |
| 15446thrR | CCTTTTCTGGTTTACAAGACC | |||
| Woolly rhino/Real time PCR 1 | WR_QPCR_415F | ACGTCCTACCATGAGGCCAA | 55°/×40 | This study |
| WR_QPCR_484R | GGGATAGCTGAGAGAAGGTTTGTG | |||
| Woolly rhino/ Real time PCR 2 | WR_QPCR_411F | GGCTACGTCCTACCATGAGGC | 55°/×40 | This study |
| WR_QPCR_476R | TGAGAGAAGGTTTGTGATGACTGTG | |||
| Woolly rhino/Numt-1 | 66WR-PseudoF | GTAAGCATATGGTAAGCAC | 55°/×50 | This study |
| 405WR-PseudoR | GCTACACTTTGGTTTATCCAACTCC | |||
| Pig / Mt. control region | L15387 | CTCCGCCATCAGCACCCAAAG | 56°/×40 | Larson et al. (2005) |
| H648n | GCTYATATGCATGGGGACT | |||
| Pig / CD45 | PigEx9f | GAGAAATACATGGATATCCCTG | 56°/×40 | This study |
| PigEx9r | CTGGAGGTGTCTCTAAGAGG | |||
| Lion / ATP8 and num. | ATP8_1F | GCCACAGTTAGATACATC | 56°/×40 | I. Barnes, personal communication |
| ATP8_3R | GAGGTGAATAGATTTTCGTTC | |||
| Moa / Mt. control region | 185fm-CR | GTACATTCCCTGCATTGGCTC | 55–60°/×40 | Bunce et al. (2003) |
| 294rm-CR | GCGAGATTTGAACAGTACG | |||
| Moa / Mt. control region | 262fm-CR | GCGAAGACTGACTAGAAGC | 55–60°/×40 | Bunce et al. (2003) |
| 441rm-CR | CGCATACCGGGTCTGTTTATGC | |||
| Moa / KW1 gene (sex linked) | Kw1-185f | GGCYRYTGCCTCAGAAATTACAG | 52–55°/×40–50 | Bunce et al. (2003) |
| Kw1-260r | ATGCTATACTGCTTTAACAGA | |||
| Moa / ADH gene | Adh-230f | GAGGAATTAGCTYRTTAGCTGTC | 52–55°/×40–50 |
Bunce et al. (2003)
|
| Adh-290r | GGTTAACACTTTGCCAGTGG |
Specimen name and sequence region amplified.
Primer name.
Primer sequence.
Annealing temperature/ no. of PCR cycles.
Primer source.
Strict protocols were followed to minimize the risk of sample and extract contamination with exogenous sources of DNA, including the use of aDNA facilities (physically isolated from the laboratories where postamplification manipulation is performed), the incorporation of extraction and PCR blank controls at ratios of 1:5 and 1:1–2, respectively, and quantification of a few of the extracts by real time PCR using SYBR green detection chemistry (Applied Biosystems, Foster City, CA) (for details see supplemental material at http://www.genetics.org/supplemental/). Independent replication was carried out for a subset of results in dedicated aDNA facilities in Copenhagen and Oxford (Table 1).
As described earlier, either of the complementary DNA strands can act as templates for PCR, as any single miscoding lesion event can produce two observable phenotypes post-PCR. It has been argued that since the chemical events required to generate direct G → A and T → C transitions are biochemically unlikely, any G → A and T → C damage that is observed on a particular DNA strand must have originated on the complementary strand as C → U and A → H events, respectively (Gilbert et al. 2003b). Others have questioned this hypothesis due to the limited knowledge of DNA damage in fossil remains (Pääbo et al. 2004). We have therefore decided to follow the approach by Hansen et al. (2001) and Hofreiter et al. (2001a) and not distinguish between the different phenotypes of TS1 (A → G/T → C) and TS2 (C → T/G → A).
Total sequence heterogeneity (TSH) was calculated as the probability of observing transitions in a single position following the formula TSH = l/n, where l is the total number of observed substitutions and n is the total number of nucleotides examined (after Gilbert et al. 2003a; Table 3; Figure 1). Two sequences showing PCR artifacts in the form of “jumping PCR” are marked in the clone data sets (supplemental material S4–S40 at http://www.genetics.org/supplemental/) as CHI for chimeric. These sequences were not included in the analysis.
TABLE 3.
Total sequence heterogeneity
| Substitutionsa | A→G/T→C | A→T/T→A | A→C/T→G | C→T/G→A | C→G/G→C | C→A/G→T | Total |
|---|---|---|---|---|---|---|---|
| No. of substitutions in mtDNA | 125.00 | 33.00 | 21.00 | 294.73 | 20.05 | 31.58 | 525.36 |
| % | 23.79 | 6.28 | 4.00 | 56.10 | 3.82 | 6.01 | 100 |
| No. of substitutions in nuDNA | 42.00 | 9.00 | 3.00 | 218.10 | 0.00 | 6.35 | 278.44 |
| % | 15.08 | 3.23 | 1.08 | 78.33 | 0.00 | 2.28 | 100 |
The sum of the different substitutions for all 23 mitochondria and nuclear data sets. Substitutions originating from a C or a G have been corrected for the AT:GC ratio for each gene region.
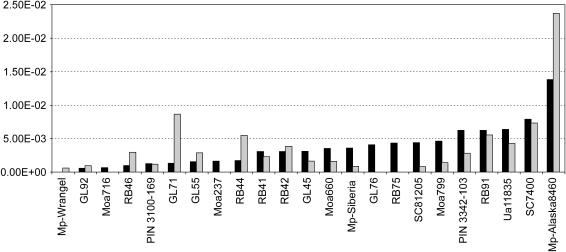
Figure 1.
Total sequence heterogeneity observed in the mtDNA and nuDNA clone sequences from the 23 specimens (see Table 1). Solid bars, mtDNA total sequence heterogeneity. Shaded bars, nuDNA total sequence heterogeneity.
Type 1 and type 2 transitions were calculated in a similar fashion, resulting in three values, pTSH, pTS1, and pTS2, for each of the 2 × 23 = 46 clone data sets. Transitions were scaled by multiplying with the AT:GC ratio of the specific gene region to compensate for nucleotide composition bias. Subsequently, pi, i = TSH, TS1, and TS2, were transformed using ri = −log(1 − pi), where ri = aiT, T is the age of the specimen, and ai is the average damage rate per unit time. The ri, i = TSH, TS1, and TS2, are referred to as the TSH, TS1, and TS2 rates, respectively.
Pearson's correlation coefficient ρ was calculated between (i) TSH and TS1 rates; (ii) TSH and TS2 rates; (iii) TS1 and TS2 rates; (iv) age of specimens and TSH rate; (v) age of specimens and TS1 rate; (vi) age of specimens and TS2 rate; (vii) age and the ratio of TS2 rate to that of TS1 (TS2/TS1; excluding cases where rTS1 = 0); and (viii) TSH rate and TS2/TS1 (excluding cases where rTS1 = 0). The correlation analysis was performed for all 46 clone sets and separately for the 23 nuclear clones and the 23 mitochondrial clones.
A t-test was performed to investigate whether TSH rates in nuDNA clones were significantly different from those in mtDNA clones. Data were further divided into three groups according to the environment of preservation: (i) permafrost specimens (n = 11); (ii) cave and dry swamp specimens (n = 7), which were originally excavated, although some had subsequently been stored in museums for years; and (iii) museum specimens (n = 5), which had always been stored in museums. Using t-tests it was investigated whether TSH rates, TSH rates divided by age, and the TS2/TS1 rates ratio differ among the three groups. Finally, it was investigated (using a chi-square test) whether the observed patterns of base transitions in mtDNA and nuDNA sequences were similarly distributed. All tests were performed at the 1% significance level.
RESULTS AND DISCUSSION
In this study nuclear and mitochondrial DNA were PCR amplified, cloned, and sequenced from a variety of faunal remains and combined with previously published clone data to compare the frequencies and types of miscoding lesion damage. As miscoding lesions are not the only factor that might influence intraclone heterogeneity, it is important to take into account other sources, such as innate DNA polymerase misincorporation errors, natural sequence heterogeneity, and variation in the number of starting template molecules for PCR (Hansen et al. 2001). We believe these factors are unlikely to have influenced the data in a significant fashion for the following reasons:
The majority of the clone sequences analyzed were generated using the proofreading DNA polymerase enzyme Platinum Taq High Fidelity, an enzyme with an innate error rate on good quality DNA of 2.0 × 10−6–6.5 × 10−7/nucleotide/cycle (Flaman et al. 1994; André et al. 1997). Although some have speculated that aDNA extracts might increase the misincorporation rates of some polymerases (e.g., Pusch and Bachmann 2004), our enzyme has been shown to retain its low misincorporation on DNA amplified from aDNA extracts (Gilbert et al. 2003b). Therefore, regular sequence errors are likely to account for only minor amounts of the calculated sequence heterogeneity, although we cannot completely exclude them from the data sets (e.g., errors generated during bacterial colony growth or sequencing).
It is possible that the number of starting template molecules in the PCR reactions might influence the observed sequence heterogeneity. In the most extreme case, a PCR that starts off a single template molecule will not demonstrate any variation and hence appear damage free. However, for the sequence data generated in this study we are confident that the PCR reactions started from considerably more than a single template molecule, since dilutions up to 1/50 of the extracts prior to amplification still yielded amplification products for both the mtDNA and the nuDNA markers, the clones showed variation, and the PCR products quantified showed starting template numbers in the 102–104 range (supplemental material at http://www.genetics.org/supplemental/).
It seems unlikely that single-substitution heteroplasmy and recombination in the mtDNA data sets influence the observed heterogeneity due to both their reported rarity in coding regions of mammalian mtDNA and their apparent absence in complete mtDNA studies (including D-loop sequences) of pigs, moa, cats, mammoths, and rhinoceroses (Ghivizzani et al. 1993; Lopez et al 1996; Xu and Arnason 1997; Ursing and Arnason 1998; Lin et al. 1999; Cooper et al. 2001; J. Krause, personal communication).
However, we cannot exclude the possibility that adverse amplifications of nuclear alleles, pseudogenes, and other gene duplicates might contribute to the observed variation in the nuclear data sets (for a discussion see below and Greenwood et al. 1999, 2001). Therefore, with regard to the validity of the data, we believe that the observed heterogeneity in the mtDNA clones can be explained predominantly by damage, while the variation observed in the nuDNA data sets may have arisen from a combination of miscoding lesions and other factors. Despite this, only a minority of the specimens (8 of 23) exhibited higher TSH in the nuDNA sequences than in the mtDNA sequences (P = 17%, Figure 1). However, no significant differences in TSH levels between nuDNA and mtDNA sequences (P = 17%) were observed, suggesting that nuDNA miscoding lesion damage is less than or equal to that of mtDNA, despite a lower number of cellular copies. Intriguingly, nuDNA appears to be more limited by amplification length than mtDNA (Bunce et al. 2003; Poinar et al. 2003). This may represent a simple case of template quantity, whereby the abundance of mtDNA makes it more likely that long undamaged molecules exist. Alternatively, the nucleosome core (146 bp of nuDNA wrapped around a histone octamer) may represent a key “preservation unit” whereby strand breaks may be common in the linker regions between adjacent cores. Finally, the proximity of chromosomal proteins (e.g., histones) to the DNA increases the chance of forming protein–DNA crosslinks, structures that could easily act as polymerase blocks during amplification.
A significant difference was observed in the distributions of miscoding lesion “types” in the mtDNA and nuDNA sequences (P < 0.1%; Table 3). Overall the frequency of type 1 transitions (A → G/T → C) is lower in the nuDNA (15%) than in the mtDNA (24%) and the frequency of type 2 transitions (C → T/G → A) is higher (78% vs. 56%). This implies that different types of damage could occur at different rates in the mitochondria and the nucleus. The fact that mtDNA is not complexed with histone proteins could make it more susceptible to different types of oxidative and/or hydrolytic damage. However, at this stage we cannot rule out that the observed differences are due to other factors such as natural variation in the nuDNA.
Although a number of aDNA studies attribute the presence of type 1 transitions to postmortem damage (e.g., Hansen et al. 2001; Gilbert et al. 2003b), other studies (e.g., Hofreiter et al. 2001a; Pääbo et al. 2004) argue that their existence is an artifact of regular DNA polymerase errors. The positive correlations observed in this study between TSH and the number of type 1 (ρ = 0.48; Figure 2A) and type 2 transitions (ρ = 0.66; Figure 2B) and between TS1 and TS2 rates (ρ = 0.45; Figure 2C) make it difficult to explain the type 1 transitions solely on the basis of DNA polymerase errors. Furthermore, we find no obvious correlation between the rate of type 1 transitions and the type of DNA polymerase enzyme used (supplemental material at http://www.genetics.org/supplemental/). However, we do observe some discrepancy in the levels of type 1 relative to type 2 transitions compared to previous observations (Gilbert et al. 2003b). In this study, we observe a total of 167 and 512 (∼1:3) type 1 and type 2 events (Table 3), respectively (counts are adjusted for base composition), which is lower than the 177 and 366 (∼1:2) type 1 and type 2 events observed by Gilbert et al. (2003b). Additionally, only 7 of the 46 data sets investigated (both mtDNA and nuDNA) show more type 1 than type 2 transitions (supplemental material at http://www.genetics.org/supplemental/) compared to 26 of 65 human mtDNA data sets studied by Gilbert et al. (2003b). However, our observed type 1:type 2 ratio is higher than that reported by Hofreiter et al. (2001a). When distinguishing between consistent and singleton substitutions, Hofreiter et al. (2001a) report consistent changes to be only type 2 events, and the ratio for the remaining singletons to be 44 type 1 to 282 type 2 (∼1:6) (M. Hofreiter, unpublished data).
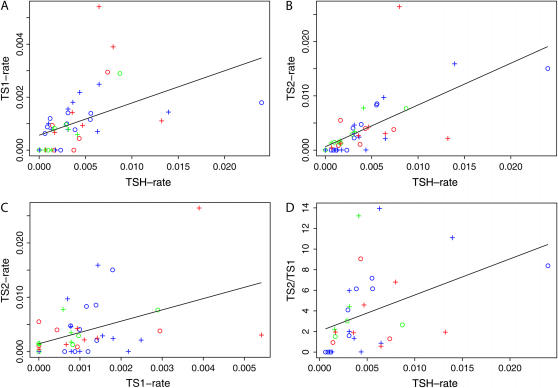
Figure 2.
Correlation analyses. Colors indicate the preservation environment: blue, permafrost; red, cave and swamp; and green, museum samples. + denotes mtDNA sequences and ○ denotes nuDNA sequences. (A) Type 1 transitions (TS1) as a function of TSH. (B) Type 2 transitions (TS2) as a function of TSH. (C) TS1 as a function of TS2. (D) TS2/TS1 as a function of TSH.
The correlation analyses show a clear overall bias toward type 2 transitions (in both nuDNA and mtDNA templates) with increasing levels of total sequence heterogeneity (ρ = 0.42; P = 0.7%; Figure 2D). This pattern has previously been reported for human D-loop sequences (Gilbert et al. 2003b), although it has been argued that in this case the pattern could be partially or completely explained by contamination, which is especially problematic in studies of human aDNA (Pääbo et al. 2004). Our results therefore confirm the general presence of a type 2 damage bias in mtDNA and demonstrate that this observation also holds true for nuDNA. As such, the results corroborate the previously noted observation (Gilbert et al. 2003b) that, under equal environmental conditions, type 2 transitions occur at a faster rate than type 1 transitions. This in turn indicates that deamination of cytosine and its homolog 5-methyl cytosine to uracil and adenine is the dominant type of miscoding lesion in both mtDNA and nuDNA sequences from fossil remains.
The observed lack of correlation between DNA damage and age of specimens was not unexpected (TSH: ρ = 0.29, P = 2.3%; TS1: ρ = 0.15, P = 16%; TS2: ρ = 0.14, P = 17%; TS2/TS1: ρ = −0.15, P = 2.1%; supplemental material at http://www.genetics.org/supplemental/) as numerous studies have demonstrated that preservation conditions rather than age determine rates of DNA degradation (Pääbo 1989; Lindahl 1993; Höss et al. 1996; Poinar et al. 1996; Kumar et al. 2000; Hofreiter et al. 2001a; Smith et al. 2001; Gilbert et al. 2003b; Pääbo et al. 2004; Willerslev et al. 2004a; Willerslev and Cooper 2005). More surprising is the apparent lack of significant differences among the three groups (permafrost, cave, and museum) for TSH rate and the ratio of TS2/TS1 (P-values >20%; Table 4), because these environments represent different temperature regimes of storage that are known to influence damage rates (Höss et al. 1996; Smith et al. 2001; Willerslev et al. 2004a). However, the museum group differed from the other two groups when comparing TSH rate/age (permafrost, P < 1%; cave, P < 1%), while there was no difference between the permafrost and the cave groups (P = 12%). This is likely due to long-term storage at room temperature after excavation of most of the mammoth specimens (Greenwood et al. 1999, 2001). Importantly, the museum samples (all museum samples are from pigs <130 years of age) show significantly higher overall sequence heterogeneity than the environmental samples, suggesting that the given museum storage conditions are not optimal for DNA preservation. Given that museum/herbarium material is an important source of aDNA, it is concerning that little research has been conducted to investigate how to maximize biomolecular preservation. If indeed DNA damage and/or degradation accumulates as the result of suboptimal storage conditions in museums (as has been observed empirically by many researchers), then alterations to current sample storage practices should be investigated.
TABLE 4.
Results of t-test
| Testa | t-value | P (%) |
|---|---|---|
| Permafrost vs. caves | 0.18 | 85 |
| Permafrost vs. museum | 1.31 | 20 |
| Caves vs. museum | 1.22 | 24 |
The total sequence heterogeneity for specimens preserved under different conditions (permafrost, caves, or museum storage) is compared.
In summary, this study demonstrates that there is no significant evidence for nuDNA sequences being more prone to miscoding lesions than mtDNA sequences despite the large discrepancy in cellular copy numbers. The data also suggest that deamination of cytosine (and 5-methyl cytosine) is the most frequent type of miscoding lesion in mtDNA and nuDNA sequences from fossil remains, although the rate at which they are affected may differ. Additionally, a bias toward deamination of cytosine relative to type 1 transitions with an overall increase in damage appears to apply to both ancient mtDNA and nuDNA sequences. Finally, we note that the findings reported in this study are limited by the sample size, and as more relevant data accumulate, future studies might be able to focus on other important questions, such as whether correlations exist between specimen type and state of DNA damage.
In conclusion, in the decade ahead, the field of aDNA will increasingly focus on nuclear genes from fossil and archival material. However, as the aDNA community embarks in this new direction, considerable care needs to be exercised to ensure that authentic sequences are generated. In the past two decades, many aDNA articles have been published in which the mtDNA data have turned out to be contaminated, pseudogenes, and/or modified by damage (for recent reviews see Pääbo et al. 2004; Willerslev and Cooper 2005). Therefore, we find it important to highlight briefly the variety of factors that need to be considered when amplifying and analyzing ancient nuDNA.
To gain a complete, unbiased appraisal of the damage spectrum in both ancient nuclear and mitochondrial DNA, further studies should attempt to generate PCR products of the same length with an equal number of starting template molecules. Additionally, when designing PCR assays for ancient nuclear loci, restricting amplicon size will both increase the chance of successful amplification and maximize the number of starting template molecules. Importantly, sequence heterogeneity in the clone data might in fact represent real allelic variation. In this study we cannot rule out that some sites interpreted as damaged might indeed represent real allelic differences (e.g., the fast-evolving CD45 gene seen in the pig data). For the moa data, however, one of the nuclear genes (kw1) is sex linked and therefore will have no allelic variation. Allelic variation in combination with low starting template numbers can also lead to cases of “allelic dropout” whereby one allelic form is amplified preferentially over another, which calls for reproducibility of results (Taberlet et al. 1996; Morin et al. 2001).
In the same way that nuclear mitochondrial insertions (numts) have caused the misinterpretation of mitochondrial phylogenies (see Willerslev and Cooper 2005), nuclear pseudogenes and gene duplications can cause problems in the interpretation of nuDNA sequences. Complete genomes are now available for a variety of organisms, making it possible to screen for the presence of duplicated genes. However, in cases where a genetic background is not well characterized, there is a considerable risk of coamplifying a functional duplicated gene or a nonfunctional pseudogene.
Finally, contamination has been a major problem in the field of aDNA (Pääbo et al. 2004; Willerslev and Cooper 2005). When amplifying mitochondrial templates, a contaminating sequence can often be identified; for example, a D-loop sequence from contemporary humans is readily distinguishable from that of Neanderthals (e.g., Serre et al. 2004b). When amplifying highly conserved nuclear targets, contamination will not be so easy to spot and can easily be mistaken for allelic variation, and consequently it might be difficult to distinguish between exogenous and endogenous DNA sequences.
We advocate that as the field of aDNA moves into amplifying nuclear targets, factors such as damage, amplicon size, allelic variation, low template copy numbers, and contamination need to be taken into account.
Acknowledgments
We are grateful to Hendrik Poinar, Johannes Krause, Tina Brand, and Martin B. Hebsgaard for help and discussion; Andrei V. Sher and Ross Macphee for providing the woolly rhinoceros and mammoth samples; and Michael Hofreiter for providing unpublished sequence data. J.B. and E.W. were supported by the Carlsberg Foundation of Denmark, the National Science Foundation of Denmark, and the Wellcome Trust. C.W. was supported by the Danish Cancer Society and the Carlsberg Foundation. R.B. was supported by the Biotechnology and Biological Sciences Research Council and Natural Environment Research Council.
References
- André, P., A. Kim, K. Khrapko and W. G. Thilly, 1997. Fidelity and mutational spectrum of Pfu DNA polymerase on a human mitochondrial DNA sequence. Genome Res. 7: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, M., and T. Brown, 2004. Non-random DNA damage resulting from heat treatment: implications for sequence analysis of ancient DNA. J. Archaeol. Sci. 31: 59–63. [Google Scholar]
- Bunce, M., T. H. Worthy, T. Ford, W. Hoppitt, E. Willerslev et al., 2003. Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 425: 172–175. [DOI] [PubMed] [Google Scholar]
- Cooper, A., and H. N. Poinar, 2001. Ancient DNA: do it right or not at all. Science 289: 1139. [DOI] [PubMed] [Google Scholar]
- Cooper, A., C. Lalueza-Fox, S. Anderson, A. Rambaut, J. Austin et al., 2001. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409: 704–707. [DOI] [PubMed] [Google Scholar]
- Flaman, J. M., T. Frebourg, V. Moreau, F. Charbonnier, C. Martin et al., 1994. A rapid PCR fidelity assay. Nucleic Acids Res. 22: 3259–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghivizzani, S. C., S. L. Mackay, C. S. Madsen, P. J. Laipis and W. W. Hauswirth, 1993. Transcribed heteroplasmic repeated sequences in the porcine mitochondrial DNA D-loop region. J. Mol. Evol. 37: 36–37. [DOI] [PubMed] [Google Scholar]
- Gilbert, M. T. P., and A. J. Hansen, 2006. Post-mortem damage in aDNA: implications and assessing aDNA quality, in Molecular Markers, PCR, Bioinformatics and Ancient DNA—Technology, Troubleshooting and Applications, edited by G. Dorado. Science Publishers, New York (in press).
- Gilbert, M. T. P., E. Willerslev, A. J. Hansen, I. Barnes, L. Rudbeck et al., 2003. a Distribution patterns of post-mortem damage in human mitochondrial DNA. Am. J. Hum. Genet. 72: 32–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, M. T. P., A. J. Hansen, E. Willerslev, I. Barnes, L. Rudbeck et al., 2003. b Characterisation of genetic miscoding lesions caused by post-mortem damage. Am. J. Hum. Genet. 72: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, M. T. P., B. A. Shapiro, A. Drummond and A. Cooper, 2005. Post mortem DNA damage hotspots in Bison (Bison bison and B. bonasus) provide supporting evidence for mutational hotspots in human mitochondria. J. Archaeol. Sci. 32: 1053–1060. [Google Scholar]
- Greenwood, A. D., C. Capelli, G. Possnert and S. Pääbo, 1999. Nuclear DNA sequences from late Pleistocene megafauna. Mol. Biol. Evol. 16: 1466–1473. [DOI] [PubMed] [Google Scholar]
- Greenwood, A. D., F. Lee, C. Capelli, R. DeSalle, A. Tikhonov et al., 2001. Evolution of endogenous retrovirus-like elements of the woolly mammoth (Mammuthus primigenius) and its relatives. Mol. Biol. Evol. 18: 840–847. [DOI] [PubMed] [Google Scholar]
- Handt, O., M. Krings, R. H. Ward and S. Pääbo, 1996. The retrieval of ancient human DNA sequences. Am. J. Hum. Genet. 59: 368–376. [PMC free article] [PubMed] [Google Scholar]
- Hansen, A., E. Willerslev, C. Wiuf, T. Mourier and P. Arctander, 2001. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 18: 262–265. [DOI] [PubMed] [Google Scholar]
- Hebsgaard, M. B., M. J. Phillips and E. Willerslev, 2005. Geologically ancient DNA: Fact or artefact? Trends Microbiol. 13: 212–220. [DOI] [PubMed] [Google Scholar]
- Hofreiter, M., V. Jaenicke, D. Serre, A. von Haeseler and S. Pääbo, 2001. a DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 29: 4793–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter, M., D. Serre, H. N. Poinar, M. Kuch and S. Pääbo, 2001. b Ancient DNA. Nat. Rev. Genet. 2: 353–360. [DOI] [PubMed] [Google Scholar]
- Höss, M., P. Jaruga, T. H. Zastawny, M. Dizdaroglu and S. Pääbo, 1996. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 24: 1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen, L. C., D. Millar, R. P. Scofield and D. M. Lambert, 2003. Nuclear DNA sequences detect species limits in ancient moa. Nature 425: 175–178. [DOI] [PubMed] [Google Scholar]
- Jaenicke-Despres, V., E. S. Buckler, B. D. Smith, M. T. P. Gilbert, A. Cooper et al., 2003. Early allelic selection in maize as revealed by ancient DNA. Science 302: 1206–1208. [DOI] [PubMed] [Google Scholar]
- Kumar, S. S., I. Nasidze, S. R. Walimbe and M. Stoneking, 2000. Brief communication: discouraging prospects for ancient DNA from India. Am. J. Phys. Anthropol. 113: 129–133. [DOI] [PubMed] [Google Scholar]
- Larson, G., K. Dobney, U. Albarella, M. Fang, E. Matisoo-Smith et al., 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307: 1618–1621. [DOI] [PubMed] [Google Scholar]
- Lin, C. S., Y. L. Sun, C. Y. Liu, P. C. Yang, L. C. Chang, et al., 1999. Complete nucleotide sequence of pig (Sus scrofa) mitochondrial genome and dating evolutionary divergence within Artiodactyla. Gene 236: 107–114. [DOI] [PubMed] [Google Scholar]
- Lindahl, T., 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715. [DOI] [PubMed] [Google Scholar]
- Lopez, J. V., S. Cevario and S. J. O'Brien, 1996. Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics 33: 229–246. [DOI] [PubMed] [Google Scholar]
- Mitchell, D., E. Willerslev and A. Hansen, 2005. Damage and repair of ancient DNA. Mutat. Res. 571: 265–276. [DOI] [PubMed] [Google Scholar]
- Morin, P. A., K. E. Chambers, C. Boesch and L. Vigilant, 2001. Quantitative polymerase chain reaction analysis of DNA from noninvasive samples for accurate microsatellite genotyping of wild chimpanzees (Pan troglodytes verus). Mol. Ecol. 10: 1835–1844. [DOI] [PubMed] [Google Scholar]
- Noonan, J. P., M. Hofreiter, D. Smith, J. R. Priest, N. Rohland et al., 2005. Genomic sequencing of Pleistocene cave bears. Science 309: 597–599. [DOI] [PubMed] [Google Scholar]
- Orlando, L., J. A. Leonard, A. Thenot, V. Laudet, G. Guerin et al., 2003. Ancient DNA analysis reveals woolly rhino evolutionary relationships. Mol. Phylogenet. Evol. 28: 76–90. [DOI] [PubMed] [Google Scholar]
- Pääbo, S., 1989. Ancient DNA: extraction, characterization, molecular cloning and enzymatic amplification. Proc. Natl. Acad. Sci. USA 86: 1939–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pääbo, S., H. Poinar, D. Serre, V. Jaenicke-Despres, J. Hebler et al., 2004. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38: 645–679. [DOI] [PubMed] [Google Scholar]
- Poinar, H. N., M. Höss, J. L. Bada and S. Pääbo, 1996. Amino acid racemization and the preservation of ancient DNA. Science 272: 864–866. [DOI] [PubMed] [Google Scholar]
- Poinar, H. N., M. Hofreiter, G. S. Spaulding, P. S. Martin, A. B. Stankiewicz et al., 1998. Molecular coproscopy: dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science. 281: 402–406. [DOI] [PubMed] [Google Scholar]
- Poinar, H. N., M. Kuch, G. McDonald, P. Martin and S. Pääbo, 2003. Nuclear gene sequences from a late pleistocene sloth coprolite. Curr. Biol. 13: 1150–1152. [DOI] [PubMed] [Google Scholar]
- Pusch, C. M., and L. Bachmann, 2004. Spiking of contemporary human template DNA with ancient DNA extracts induces mutations under PCR and generates nonauthentic mitochondrial sequences. Mol. Biol. Evol. 21: 957–964. [DOI] [PubMed] [Google Scholar]
- Pusch, C. M., M. Broghammer, G. J. Nicholson, A. G. Nerlich, A. Zink et al., 2004. PCR-induced sequence alterations hamper the typing of prehistoric bone samples for diagnostic achondroplasia mutations. Mol. Biol. Evol. 21: 2005–2011. [DOI] [PubMed] [Google Scholar]
- Serre, D., A. Hofreiter, M. Pääbo, S. 2004. a Mutations induced by ancient DNA extracts? Mol. Biol. Evol. 21: 1463–1467. [DOI] [PubMed] [Google Scholar]
- Serre, D., A. Langaney, M. Chech, M. Teschler-Nicola, M. Paunovic et al., 2004. b No evidence of Neanderthal mtDNA contribution to early modern humans. PLoS Biol. 2: E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C. I., A. T. Chamberlain, M. S. Riley, A. Cooper, C. B. Stringer et al., 2001. Neanderthal DNA: Not just old but old and cold? Nature 10: 771–772. [DOI] [PubMed] [Google Scholar]
- Taberlet, P., S. Griffin, B. Goossens, S. Questiau, V. Manceau et al., 1996. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24: 3189–3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgold, J., and T. A. Brown, 2003. Degradation of DNA in artificially charred wheat seeds. J. Archaeol. Sci. 30: 1067–1076. [Google Scholar]
- Tougard, C., T. Delefosse, C. Hanni and C. Montgelard, 2001. Phylogenetic relationships of the five extant Rhinoceros species (Rhinocerotidae, Perissodactyla) based on mitochondrial cytochrome b and 12S rRNA genes. Mol. Phylogenet. Evol. 19: 34–44. [DOI] [PubMed] [Google Scholar]
- Ursing, B. M., and U. Arnason, 1998. The complete mitochondrial DNA sequence of the pig (Sus scrofa). J. Mol. Evol. 47: 302–306. [DOI] [PubMed] [Google Scholar]
- Willerslev, E., and A. Cooper, 2005. Ancient DNA. Proc. Biol. Sci. 272: 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev, E., A. J. Hansen, B. Christensen, J. P. Steffensen and P. Arctander, 1999. Diversity of Holocene life forms in fossil glacier ice. Proc. Natl. Acad. Sci. USA 96: 8017–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev, E., A. J. Hansen and H. N. Poinar, 2004. a Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol. Evol. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Willerslev, E., A. J. Hansen, R. Rønn, T. B. Brand, I. Barnes et al., 2004. b Long-term persistence of bacterial DNA. Curr. Biol. 14: R9–R10. [DOI] [PubMed] [Google Scholar]
- Xu, X., and U. Arnason, 1997. The complete mitochondrial DNA sequence of the white rhinoceros, Ceratotherium simum, and comparison with the mtDNA sequence of the Indian rhinoceros, Rhinoceros unicornis. Mol. Phylogenet. Evol. 7: 189–194. [DOI] [PubMed] [Google Scholar]