Abstract
The Smt3 (SUMO) protein is conjugated to substrate proteins through a cascade of E1, E2, and E3 enzymes. In budding yeast, the E3 step in sumoylation is largely controlled by Siz1p and Siz2p. Analysis of Siz− cells shows that SUMO E3 is required for minichromosome segregation and thus has a positive role in maintaining the fidelity of mitotic transmission of genetic information. Sumoylation of the carboxy-terminus of Top2p, a known SUMO target, is mediated by Siz1p and Siz2p both in vivo and in vitro. Sumoylation in vitro reveals that Top2p is an extremely potent substrate for Smt3p conjugation and that chromatin-bound Top2p can still be sumoylated, unlike many other SUMO substrates. By combining mutations in the TOP2 sumoylation sites and the SIZ1 and SIZ2 genes we demonstrate that the minichromosome segregation defect and dicentric minichromosome stabilization, both characteristic for Smt3p–E3-deficient cells, are mediated by the lack of Top2p sumoylation in these cells. A role for Smt3p-modification as a signal for Top2p targeting to pericentromeric regions was suggested by an analysis of Top2p–Smt3p fusion. We propose a model for the positive control of the centromeric pool of Top2p, required for high segregation fidelity, by Smt3p modification.
SUMO (small ubiquitin-like modifier) is a member of a growing family of ubiquitin-related proteins and is known to conjugate with RanGAP1, PML, IκBα, p53, yeast septin components, and other proteins (Hay et al. 1999; Hochstrasser 2000; Jentsch and Pyrowolakis 2000; Muller et al. 2001; Weissman 2001). The cells of higher eukaryotes have three SUMO paralogs: SUMO-1, SUMO-2, and SUMO-3 (Johnson 2004). In budding yeast, the sole SUMO-encoding gene SMT3 is essential for cell viability (Meluh and Koshland 1995). Common E1 and E2 enzymes are required to conjugate all the SUMO variants. The E1 enzymes Uba2p/Aos1p in Saccharomyces cerevisiae and SAE1/SAE2 in mammals form a transient thioester bond between the C-terminal glycine of SUMO and SAE2/Uba2p (Dohmen et al. 1995; Desterro et al. 1997; Johnson et al. 1997). SUMO is then transferred to the E2 conjugating enzyme Ubc9p (Johnson and Blobel 1997; Schwarz et al. 1998).
The SUMO E3 proteins have been characterized as cofactors required for substrate recognition by Ubc9p (Hochstrasser 2001; Jackson 2001). The S. cerevisiae Siz1p/Ull1p (Strunnikov et al. 2001; Takahashi et al. 2001a), has been shown to be the E3 factor specific for septin sumoylation (Johnson and Gupta 2001; Takahashi et al. 2001a,b). Several additional types of E3 factors have been found in mammalian cells (Pichler et al. 2002; Kagey et al. 2003), but their counterparts are not present in S. cerevisiae. Yeast cells lacking E3 (siz1/siz2 double deletion mutants) lose the bulk of detectable Smt3p conjugation, yet retain a wild-type growth rate (Johnson and Gupta 2001; Takahashi et al. 2003) after the 2μ plasmid is lost due to E3-dependent over-modification of its partition factors (Chen et al. 2005). The fact that the massive loss of SUMO conjugates in siz1/siz2 double mutants results in only negligible phenotypic changes underscores the technical difficulty of identifying physiologically important Smt3p substrates and unraveling the essential role of Smt3p for cell viability. As SMT3 is an essential gene, it is generally believed that some protein targets modified in the absence of Siz1p and Siz2p allow cells to survive. This modification either can potentially proceed by the conjugating activity of the E2 enzyme itself (Okuma et al. 1999) or may be catalyzed by some narrowly specialized SUMO E3 factors, such as the recently characterized Mms21p (Zhao and Blobel 2005).
The numerous biological roles of SUMO modification are dependent on the functions of the target proteins (Muller et al. 2001; Johnson 2004; Ulrich 2004). The pattern of Smt3p localization in yeast cells indicates that conjugated proteins are present in the nucleus (many targets) and in the bud neck (septins) (Johnson and Blobel 1999; Takahashi et al. 1999). Nuclear localization of Smt3p and the role of the SUMO-conjugation pathway in chromosome transmission fidelity (Biggins et al. 2001; Azuma et al. 2003) suggest that critically important targets of SUMO modification could be nuclear proteins. Indeed, recent identification of Smt3p substrates in vivo using proteomic approaches (Panse et al. 2004; Wohlschlegel et al. 2004; Zhou et al. 2004; Hannich et al. 2005) have demonstrated that numerous essential nuclear proteins are modified by SUMO.
The fact that mutations in enzymes removing SUMO from the conjugated targets result in severe loss of viability (Li and Hochstrasser 2000; Strunnikov et al. 2001) demonstrates that removal of Smt3p from its target is as important as conjugation. Budding yeast have two specialized SUMO isopeptidases: intranuclear, Smt4p(Ulp2p) (Li and Hochstrasser 2000; Strunnikov et al. 2001), and extranuclear, Ulp1p (Li and Hochstrasser 2003). These two enzymes are apparently strictly compartmentalized in the cell, as mistargeting of Ulp1p to the nucleus results in a severe phenotype (Panse et al. 2003). While the Ulp1p is an essential enzyme (Li and Hochstrasser 1999), carrying the bulk of Smt3p processing (Li and Hochstrasser 2003), the smt4 mutants are able to survive (Li and Hochstrasser 2000; Strunnikov et al. 2001), but probably due only to a trace of Ulp1p activity reaching the nucleus (Li and Hochstrasser 2003). Characterization of the SMT4 gene revealed a number of pathways controlled by Smt3p conjugation: SMT4 is a dosage suppressor of mutations in the genes encoding the chromosomal proteins MIF2, SMC2, PDS5 (Meluh and Koshland 1995; Strunnikov et al. 2001; Stead et al. 2003), and smt4 mutations are synthetically lethal with DNA-replication arrest (Bachant et al. 2002). The severe cellular defects of smt4 mutants can be attributed to over-sumoylation of many Smp3p targets. Thus, at present the negative impact of SUMO modification (Strunnikov et al. 2001; Bachant et al. 2002; Chen et al. 2005) has been documented to a much greater degree than its positive regulatory role.
Previously, we characterized SIZ1 and SIZ2 genes (Strunnikov et al. 2001) shown to encode the major SUMO E3 activity in yeast (Johnson and Gupta 2001). As double siz1/siz2 deletion results in elimination of approximately 99% of the SUMO conjugates (Johnson and Gupta 2001) and many chromosomal proteins are sumoylated in Siz+ cells (Zhou et al. 2004; Hannich et al. 2005), we became interested in assessing the potential role these two genes may play in chromosome segregation. Upon analysis of a number of potential substrates we found that Top2p modification is controlled by both Siz1p and Siz2p. While a previous study on Top2p in S. cerevisiae (Bachant et al. 2002) has uncovered that Top2p over-sumoylation results in precocious sister chromatid separation in kinetochore vicinity by an as yet unidentified mechanism (Bachant et al. 2002), the role Top2p sumoylation plays in the wild-type cells remains unknown. In higher eukaryotes and wild-type yeast cells only a very small fraction of topoisomerase II is sumoylated (Bachant et al. 2002; Azuma et al. 2003), making any direct analysis of this pool rather challenging. However, using a combination of siz1, siz2, and top2 mutants we show that SUMO E3 machinery specifically facilitates Top2p–Smt3p conjugation and demonstrate that both Top2p sumoylation and Siz1p/Siz2p activity have a previously uncharacterized positive regulatory role in transmission of genetic information. We demonstrate that the critical role of SUMO E3 in minichromosome segregation is likely limited to the Smt3p modification of the COOH-terminal tail of Top2p.
MATERIALS AND METHODS
Microbiological and genetic methods:
Escherichia coli strains Top10 and BL21(DE3) were used for plasmid propagation and recombinant protein production, respectively. Yeast media and genetic techniques were performed as described (Guthrie and Fink 1991). The S. cerevisiae strains were of S288C and W303 backgrounds (Table 1). For all genetic tests an isogenic set of strains was used and experiments were repeated for both S288C and W303. Minichromosome stability, i.e., the fraction of cells in the population containing minichromosomes under selective conditions, was assayed essentially as described (Strunnikov et al. 1993). Briefly, exponential cultures of the strains harboring the YCplac111 (CEN4, LEU2), YCplac33 (CEN4, URA3) (Gietz and Sugino 1988), pPRS425 (2μ replication origin, LEU2), pRS415 (CEN6, LEU2) (Sikorski and Hieter 1989), pAS255 (cen3-BCT1, ARS1 TRP1 URA3), or pIA1 (URA3, 2μ) (P. Hieter, personal communication) plasmids and bearing different combinations of siz1/siz2 and/or top2 mutations were grown in minimal medium lacking uracil or leucine, respectively. Culture aliquots were plated on four YPD plates. The resulting colonies were analyzed for minichromosome presence by replica plating onto synthetic medium lacking uracil or leucine. The transmission efficiency (stability) of the conditional dicentric minichromosome pAS72 (LEU2 URA3 ARS CEN6 pGAL:CEN3, A. Strunnikov, unpublished data) was determined in a similar way, except that the log-phase cultures were first grown at 30° in selective medium containing 2% raffinose, 1% galactose (v/v) as a carbon source and then incubated in YPD for 4 hr before being plated.
TABLE 1.
S. cerevisiae strains
| Strains | Relevant genotype | Source |
|---|---|---|
| BY4733 | MATahis3 leu2 met15 trp1 ura3 | ATCC |
| 4bAS399 | MATahis3 leu2 lys2 met15 ura3 siz1-Δ∷kanMX siz2-Δ∷kanMX | This work |
| 12cAS399 | MATα his3 leu2 lys2 ura3 siz1-Δ∷kanMX siz2-Δ∷kanMX | This work |
| 924–YPH499 | MATaade2 his3 leu2 lys2 trp1 ura3 HF:SMT3∷LEU2 | This work |
| 924–4bAS399 | MATahis3 leu2 lys2 met15 ura3 siz1–Δ∷kanMX siz2-HF:SMT3∷LEU2 | This work |
| W303–1A | MATaade2 ura3 trp1 leu2 his3 can1 | R. Rothstein |
| YPH499 | MATaade2 his3 leu2 lys2 trp1 ura3 | P. Hieter |
| YPH499bp1 | MATaade2 his3 leu2 lys2 trp1 ura3 bar1-Δ pep4∷HIS3 SMC4:6His:3HA∷URA3 | This work |
| BY4733bp5 | MATahis3 leu2 met15 trp1 ura3 pep4∷HIS3 bar1-Δ∷LEU2 PDS5:6His:3HA∷URA3 | This work |
| EY0987/SPC42:mRFP | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 SPC42:mRFP∷kanMX | E. O'Shea |
ATCC, American Type Culture Collection.
Chromosome III loss rate assay was based on the ability of diploid strains to mate with both MATα and MATa tester strains if chromosome segregation was impaired. Loss of heterozygosity of both the MATa/MATα and leu2/LEU2 loci was considered a chromosome loss event. The details of the assay are as described (Gerring et al. 1990).
DNA constructs:
The SIZ1 overexpression construct was created by amplifying the galactose-controlled promoter and the marker from pFA6a–His3MX6–pGAL1–3HA (Longtine et al. 1998) with the specific primers fusing SIZ1 ORF to pGAL. The PCR product was directly used in yeast transformation to replace the genomic copy of the SIZ1 promoter.
The integrative construct to replace the SMT3 gene with the polyhistidine and FLAG-tagged SMT3 (HF-SMT3) expressed from the native promoter was constructed on the basis of the pHF-SMT3 plasmid (Johnson et al. 1997). The LEU2 marker was inserted into the unique MluI site in the SMT3 promoter and the resulting vector pAS924 was used to transform yeast after digestion with NcoI and BglII. The smt4, siz1, and siz2 deletions were as reported (Takahashi et al. 2000, 2001b).
To generate the tagged and mutagenized TOP2 replacement vectors a genomic copy of TOP2 gene was amplified by PCR (primers: GCAACTGCAGTACCTAACGGTGCTTTCGG and GCGCGTCGACATCCTCTTCATTGAACGAAAC) and the PCR products were cloned into the PstI-SalI restriction sites of pTS901IU (5xHA URA3) (Sasaki et al. 2000) to produce pYT1033 (URA3 TOP2:HA). The top2:HA (ΔC) vector pYT1035, was constructed similarly, but using a truncated primer (GCGCGTCGACAATTTTTTTGCCCTTTCTAGCA). The ΔC allele was designated top2–Δ200. The top2 3xKR triple mutant allele (top2–201) encoding the substitution mutations of K1220R, K1246R, K1277R was obtained by PCR-based site-directed mutagenesis (primer pairs: CAAAAAAATTAGGTTAGAGGATAA/TTATCCTCTAACCTAATTTTTTTG, CTACAAAGATTAGAAAAGAGAAAAC/GTTTTCTCTTTTCTAATCTTTGTAG, TTTCGACATAAGGAAAGAAGATA/TATCTTCTTTCCTTATGTCGAAA) and the PCR product was cloned into pTS901IU to produce pYT1034 (URA3 top2:HA 3xKR). Plasmid pML251 (top2-SNM:HA∷KanMX) was used to replace a single genomic copy of TOP2 gene (Bachant et al. 2002) after the marker was changed to URA3 to give pYT1032 (URA3 top2-SNM:HA). The plasmids for the COOH-terminal GFP-tagging of the different versions of TOP2 pYT1026, pYT1027, and pYT1028 were analogous to (respectively) pYT1033, pYT1034, and pYT1032, except they had the COOH-terminal fusions to the GFP-encoding sequence and LEU2 markers. The TOP2:SMT3:HA fusion plasmid pYT1051 (Figures 2B and 5D) was constructed by inserting the SMT3 gene in-frame into the SpeI site in pYT1033. The TOP2:SMT3:GFP fusion plasmid pYT1101 (Figure 5A) was constructed by inserting the SMT3 gene in-frame into the SpeI site in pYT1026.
Figure 2.
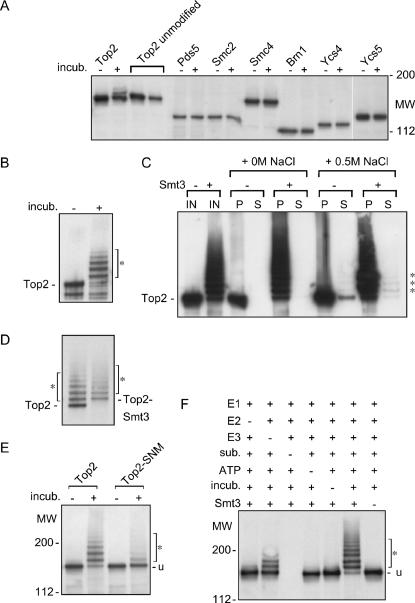
Top2p is modified in a SUMO E3 dependent manner. (A) Chromatin-bound Top2p is a sumoylation substrate in vitro. All putative targets are HA-tagged. Reaction mixtures with 1 μl of chromatin purified from yeast cells BY4733/pYT1033 (Top2), BY4733bp5 (Pds5), YPH499bp1 (Smc4), YPH499bp2 (Smc2), YPH499bp6 (Brn1), BY4733bp4 (Ycs4), and YPH499bp5 (Ycs5) were incubated at 37° for 120 min (incub. +) and subjected to immunoblotting with anti-HA antibodies. Identical reaction mixtures held on ice (incub. −) were used as negative controls. (B) Purified Top2p is modified by Smt3p in vitro. A total of 4.4 μg purified Top2p (Vaughn et al. 2005) was subjected to sumoylation in vitro (incub. +) or left on ice (incub. −) (see materials and methods) at 37° for 60 min. Western blotting was done with anti-Top2p antibodies. (C) Smt3p-modified Top2p remains strongly associated with chromatin. Top2p was modified in chromatin context in vitro as described in materials and methods. The mock reaction (Smt3 −) was carried out in the absence of recombinant Smt3p. IN, reaction before extraction. Extraction of Top2p after sumoylation reaction was performed for 30 min at 4° with EBX or EBX + 0.5 m NaCl buffers. Chromatin (P) and soluble fractions (S) were separated by centrifugation and analyzed by Western blotting. The Top2p–Smt3p conjugates are marked with an asterisk. (D) Top2p is modified by Smt3p in vitro. A total of 5 μl of chromatin from BY4733/pYT1033 (Top2) or BY4733/pYT1051 (Top2–Smt3) were incubated with the in vitro sumoylation reaction mix (see materials and methods) at 37° for 60 min and subjected to immunoblotting with anti-HA antibodies. The characteristic mobility shift caused by sumoylation corresponds to the shift generated by Smt3p fusion. The Top2p–Smt3p conjugates are marked with an asterisk. (E) The COOH-terminal consensus sumoylation sites of Top2p are the primary targets of Smt3p conjugation in vitro. Reaction mixtures with 5 μl of chromatin purified from yeast cells BY4733/pYT1033 (Top2) and BY4733/pYT1032 (Top2–SNM), containing HA-tagged Top2p, were incubated at 37° for 60 min and subjected to immunoblotting with anti-HA antibodies. (F) Top2p–Smt3p conjugation in vitro is stimulated by SUMO E3. A total of 5 μl of chromatin (sub.) from 4bAS399/pYT1033 (Siz−) was incubated (incub.) with the in vitro sumoylation reaction mix (see materials and methods) at 37° for 60 min and subjected to immunoblotting with anti-HA antibodies. Combinations of the following proteins and cofactors were used: E1, 4.5 μg Uba2p, 5.2 μg Aos1p; E2, 0.75 μg Ubc9p; E3, 4.5 μg Siz1pΔ440; ATP (10 mM) and 2.9 μg of 6xHis–Smt3p. The unmodified Top2p band is indicated by “u.” The Top2p–Smt3p conjugates are indicated with an asterisk. Multiple modified forms of Top2p were formed only in the presence of Smt3p.
Figure 5.
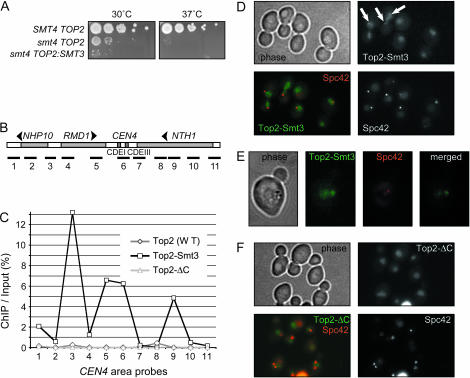
Constitutive sumoylation results in pericentromeric targeting of Top2p. (A) Synthetic interaction between smt4-Δ and TOP3:SMT3 fusion. The same-concentration (106 cells/ml) cultures of three strains were plated on YPD plates in serial 10-fold dilutions and incubated at 30° (permissive for smt4-Δ) and 37° (nonpermissive for smt4-Δ) temperatures for 48 hr. Integrated TOP2 variants produce Top2p–HA fusions. SMT4 TOP2: W303 with the wild-type SMT4 gene transformed with pYT1033. smt4 TOP2 and smt4 TOP2:SMT3 are W303 with smt4 deletions, transformed with pYT1033 or pYT1051, respectively. As smt4-Δ results in massive lethality, even at permissive temperature, the starting dilutions have different number of growing colonies, as compared to Smt4+. (B) Layout of the PCR probes used for ChIP analysis of Top2p binding to the CEN4 region. (C) Smt3p fusion to Top2p tail results in Top2p enrichment at the CEN4 pericentromeric region. The W303 strains transformed with pYT1033 (TOP2:HA), pYT1051 (TOP2:SMT3:HA) or pYT1035 (TOP2-ΔC:HA), all replacing the wild-type TOP2 gene, were subjected to ChIP analysis using the PCR probes shown in B. ChIP analysis was as described (Strunnikov et al. 2001; Wang et al. 2004). (D–F) Fusion to Smt3p changes localization of Top2p–GFP in the nucleus. Spc42p-mRFP was used to mark SPB in diploid strains expressing both the wild-type Top2p and a corresponding Top2p–GFP fusion: TOP2:SMT3:GFP (D and E show maximal resolution) and TOP2-ΔC:GFP (E). Twenty optical Z-sections with 0.2-μm spacing were combined to compose the images (2 × 2 binning, 1-sec exposure per frame, except in E: no binning, 3-sec exposure per frame). Arrows point to clustered Top2p–Smt3p–GFP staining in mitotic cells.
The dicentric minichromosome pAS72 was constructed from pRS415 (CEN6, LEU2) by inserting pGAL:CEN3 and URA3 into the polylinker region. The resulting minichromosome behaves as dicentric in dextrose-containing media and is functionally monocentric in galactose media.
Biochemical methods:
All chromatin fractions were prepared and verified by micrococcal nuclease digestion as described by Liang and Stillman (1997). For preparation of the yeast lysates and immunoblot analysis, the cells were collected, washed, resuspended in 2% SDS, and disrupted with glass beads using a TOMY shaker. The resulting lysates were boiled, supplemented with the LDS loading buffer (Invitrogen), and separated onto 4–12% Bis–Tris or 3–8% Tris–acetate NUPAGE gradient gels (Invitrogen). After Western blotting the specific protein reactive bands were visualized with ECL (Amersham Pharmacia).
The SUMO conjugation assay was performed as described by Takahashi et al. (2003). Briefly, the components of the conjugation reaction: 6xHis–Smt3p, GST–Uba2p, GST–Aos1p, Ubc9p, and Siz1p–Δ440 proteins were expressed and purified from E. coli and then used in the reaction mixture containing substrate. In most cases ∼5 μl of chromatin (Liang and Stillman 1997) was prepared from the strains with the HA-tagged target protein. The reaction mixture (20 μl) was incubated in the presence of 10 mm ATP at 37° (or on ice, as a control) for 1 or 2 hr. Top2p–HA chromatin was used in each experiment with other protein as a positive control. The Top2p modification reaction was saturated after 2 hr. Reaction aliquots of 5 μl were boiled and subjected to immunoblotting. The untagged Top2p for Figure 2B was purified in J. L. Nitiss's laboratory, as described (Vaughn et al. 2005). It was detected on Western blots using specific anti-Top2p antibody (TopoGEN).
To purify the in vivo Smt3p-conjugated proteins by IMAC, a 50-ml culture of yeast cells with a genomic copy HF-SMT3 was harvested, cells were disrupted by glass beads (10 min) in 500 μl of lysis buffer (0.1 m Tris pH 8.0, 6 m guanidine chloride, 0.5 n NaCl), and the extract was clarified by centrifugation at 20,000 × g for 30 min. The clarified protein extract was incubated in the batch mode with nickel-charged NTA resin (QIAGEN) for 6 hr at room temperature. The resin was then packed into a 2-ml Bio-Rad disposable column and the extract was passed one more time through the open column. The column was washed once with 10-column volumes of 0.1 m Tris pH 7, 6 m guanidine chloride, 0.5 m NaCl, and once with 0.1 m Tris pH 6, 6 m urea, 0.5 m NaCl. Then the bound proteins were eluted with stripping buffer (20 mm Tris pH 7, 40 mm EDTA, 2% SDS). The flow-through fraction was diluted 10-fold with water and proteins were precipitated with 10% TCA.
Microscopy:
To generate strains expressing the GFP-tagged Top2p, the corresponding GFP-fusion plasmids were digested with SpeI or AvrII/BlnI and transformed into the W303-1A, YPH499bp, and BY4733 strains. The transformed cells were grown at low density in selective medium at 23°, washed in 0.5% PBS, and analyzed by fluorescent microscopy using a Zeiss AxioVert fluorescent microscope with a cooled CCD camera and Z-axis scanning capability. Coexpression of Top2p–GFP and Spc42p–mRFP fusions was achieved by crossing MATa strains expressing the Top2p fusions to EY0987/SPC42:mRFP (Huh et al. 2003).
RESULTS
The Smt3p E3 is required for minichromosome transmission fidelity:
We previously obtained evidence that implicated the SIZ1 and SIZ2 genes in chromosome metabolism (Strunnikov et al. 2001). In addition, as chromatin proteins were shown to compose a significant fraction of all the SUMO targets in yeast (Wohlschlegel et al. 2004; Zhou et al. 2004; Hannich et al. 2005) and Siz1p/Siz2p are responsible for the bulk of sumoylation in yeast (Johnson and Gupta 2001; Takahashi et al. 2001b), analysis of Siz1p/Siz2p function in chromatin might uncover the mechanism facilitating SUMO control of chromosome segregation in mitosis (Biggins et al. 2001). Although Siz1p, a SUMO E3, was previously found to be important for sumoylation of septins (Johnson and Gupta 2001; Takahashi et al. 2001a, 2001b), the physiological importance of septin modification was found to be negligibly small (Johnson and Blobel 1999). The role of Siz1p and Siz2p, as SUMO E3, in repressing amplification of 2μ plasmid has recently been documented (Chen et al. 2005), but their role in chromosome segregation is as yet uncharacterized.
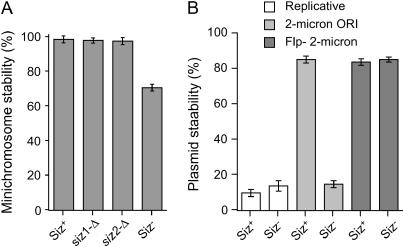
To investigate whether Siz1p and Siz2p may potentially play a role in the chromosome cycle in budding yeast, we first analyzed the intracellular localization of Siz1p and Siz2p using chromatin fractionation. Both Siz1p and Siz2p were found to be enriched in chromatin after fractionation (Liang and Stillman 1997) (data not shown). Thus, chromatin proteins may be the primary target of Siz1p and Siz2p E3 activity. Therefore, we assessed chromosome transmission fidelity in Siz− (siz1-Δ siz2-Δ) cells. Using diploid strains heterozygous in both the MATa/MATα and LEU2/leu2 loci of chromosome III, we established that chromosome III loss in a Siz− diploid strain is indistinguishable from a Siz+ diploid strain (data not shown). We also did not find any destabilization of chromosome III harboring translocation of rDNA (Freeman et al. 2000) in Siz− diploids. At the same time, we detected a notable destabilization of circular centromeric plasmids (minichromosomes) in the siz1-Δ siz2-Δ cells: the Siz− strains had a 30% decrease in minichromosome transmission fidelity, as compared to Siz+ cells (Figure 1A). To determine whether this minichromosome loss was a result of missegregation or impaired replication, we assessed the stability of noncentromeric plasmids in both the Siz+ and Siz− strains. The Siz− strains showed no difference in the stability of acentric ARS plasmids (Figure 1B). While plasmids containing the 2μ plasmid origin were extremely unstable in Siz− cells, this was due to the lack of endogenous 2μ plasmid (data not shown), which is lost in siz1-Δ siz2-Δ cells as a result of deregulation of the FLP gene (Chen et al. 2005). When a full-length Flp− 2μ plasmid was used, no difference was observed between its stability in Siz+ and Siz− strains (Figure 1D). Therefore, minichromosome destabilization in Siz− is likely due to missegregation. The fact that loss of E3 activity (and the ensuing massive loss of SUMO conjugation) has a negative impact on segregation of minichromosomes indicates that SUMO E3 has a previously unknown positive role in chromosome transmission fidelity. This positive regulatory pathway could bear greater physiological relevance than the previously reported negative role of over-sumoylation in chromosome segregation (Li and Hochstrasser 2000; Biggins et al. 2001; Strunnikov et al. 2001; Bachant et al. 2002).
Figure 1.
Minichromosome maintenance phenotype of SUMO E3 mutants. (A) Siz− mutants destabilize mitotic transmission of minichromosomes. YCplac111 stability was determined in the Siz+ (BY4733) and Siz− (4bAS399) strains at 30° as described in materials and methods. (B) Siz− mutants do not destabilize mitotic transmission of acentric plasmids. The stability of pAS255 (replicative), pRS426 (2μ ORI), and pIA1 (Flp− 2μ) plasmids in the Siz+ (BY4733) and Siz− (4bAS399) strains was determined at 30° as described in materials and methods.
Top2p modification by Smt3p is promoted by E3 both in vivo and in vitro:
The data in Figure 1 can be interpreted to indicate E3 activity is required for the Smt3p modification of a chromatin protein, with a role in accurate segregation of sister chromatids. Thus, we tested a sample of putative Smt3p targets, including Pol30p and all the tagged subunits of cohesin (Kagansky et al. 2004) and condensin (Freeman et al. 2000) in an in vitro sumoylation system composed of recombinant Smt3p, E1, E2, and E3 enzymes (Takahashi et al. 2003). In addition, Top2p over-sumoylation was previously shown to have a negative impact on pericentromeric cohesion (Bachant et al. 2002), prompting us to analyze Top2p as a potential target of the SUMO E3 activity responsible for the Siz− segregation defect (Figure 1A). Most of the proteins we tested showed no propensity for Smt3p modification, while being bound to chromatin, in the presence of either Siz1p or Siz2p in the reaction mix, with the exception of Top2p (Figure 2A and data not shown). The Smt3p modification of chromatin-bound Top2p was readily detected (Figure 2A), suggesting that Top2p may be a potential mediator of the E3 role in chromatin.
The use of an in vitro SUMO modification system allowed us to circumvent the low abundance of Top2p–Smt3p conjugates in vivo (Bachant et al. 2002). While the soluble Top2 protein was also shown to be a potent SUMO substrate in vitro (Figure 2B), the Smt3p modification of Top2p did not alter the Top2p affinity to chromatin (Figure 2C) as judged by unchanged resistance to salt extraction. Thus, we routinely used Top2p-containing chromatin as a substrate to make the in vitro sumoylation system a better approximation of the in vivo situation. The identity of the modified Top2p bands as Smt3p conjugates was confirmed by two experiments. First, we showed that direct fusion of Top2p to Smt3p produces the same electrophoretic shift (Figure 2D). Second, the “sumo-no-more mutant” top2-SNM (Bachant et al. 2002), lacking consensus sumoylation sites in the Top2p tail, showed only marginal modification by Smt3p upon prolonged incubation in vitro (Figure 2E).
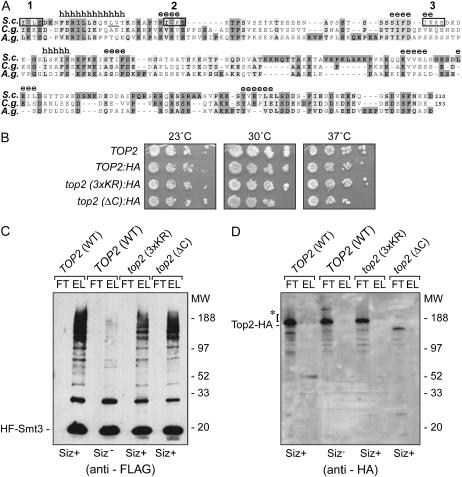
Chromatin-associated Top2p, even when isolated from siz1-Δ siz2-Δ cells, was found to be modified by Smt3p to some extent even in the absence of E3 (Figure 2F). However, the addition of recombinant Siz1p (or Siz2p, data not shown) to the system allowed conversion of virtually all Top2p into Smt3p-modified forms in an ATP-dependent fashion (Figure 2F). As the SUMO E2 Ubc9p is known to support limited E3-independent Smt3p conjugation in vitro (Okuma et al. 1999), we investigated the role of E3 in Top2p modification in vivo. We generated replacements of the wild-type TOP2 gene with the HA-tagged wild-type gene, the top2-3xKR, and top2-ΔC alleles (Figure 3A). The mutant alleles of TOP2 had no notable impact on overall cell growth or viability (Figure 3B). The 3xKR (top2–201) allele was a triple substitution of the SUMO-acceptor lysine residue (sites 1, 2, and 3, Figure 3A). This allele is different from the top2–SNM allele (Bachant et al. 2002), in which all the lysine residues (not just the acceptor residues) in the consensus sites were mutated, raising the possibility that the top2–SNM allele is defective in more than just Smt3p conjugation interference. The ΔC allele (top2–200) lacks the whole region encoding the COOH-terminal Top2p tail, as shown in Figure 3A, and the corresponding Top2p–ΔC protein was completely refractory to sumoylation in vitro (data not shown).
Figure 3.
Top2p tail is modified in vivo in a SUMO E3-dependent manner. (A) The COOH-terminal region of Top2p and the consensus sumoylation sites have limited conservation among yeast species. The S. cerevisiae Top2p tail region (residues 1219–1428) was aligned with the Candida albicans (C.g.) and Ashbya gossypii (A.g.) topoisomerases II and the secondary structure was predicted using the JPred package (Cuff and Barton 2000). h, alpha helix; e, beta-sheet. The SUMO consensus sites are marked with open boxes and numbered. The acceptor lysine residues are outlined. (B) Cell growth is not perturbed by the tagged top2 alleles ΔC and 3KR. Ten-fold serial dilutions from overnight cultures of wild-type (TOP2, TOP2:HA) and different top2 mutants were spotted onto YPD plates and incubated at the temperatures indicated. The tagged TOP2 allele replacement strains (W303-1A/pYT1033, W303-1A/pYT1034, and W303-1A/pYT1035) were generated after transformation with plasmids as described in materials and methods. The corresponding GFP-tagged strains (YPH499/pYT1026, YPH499/pYT1027, and YPH499/pYT1028) were also generated and showed no interference with cell proliferation or Top2p nuclear localization (data not shown). (C and D) Western blot analysis of Top2p sumoylation in vivo. Extracts from the strains 924-YPH499 (Siz+) or 924-4bAS399 (Siz−) expressing physiological levels of HF-Smt3p and carrying different top2 alleles were fractionated by IMAC. FT, flow through; EL, eluate. The wild type TOP2 gene and all top2 alleles were HA-tagged. HF–Smt3p conjugates eluted from the column were analyzed by Western blotting using anti-FLAG (C) and anti-HA (D) antibodies. The conjugated forms of Top2p (present only in Siz+ TOP2 cells) are indicated by an asterisk.
Only a small fraction of Top2p is sumoylated in wild-type cells (Bachant et al. 2002), making analysis of this pool quite challenging. To facilitate enrichment for SUMO conjugates we replaced the native SMT3 gene with a construct expressing the polyhistidine and FLAG-tagged Smt3 protein (HF–Smp3p), and introduced either a double siz1 siz2 deletion with the wild-type TOP2, with an HA tag (Siz− in Figure 3, C and D), or the HA-tagged top2 ΔC and 3xKR alleles (Siz+). IMAC fractionation, allowing enrichment for the HF–Smt3p conjugates, indicated that the combined deletions of SIZ1 and SIZ2 genes resulted in the loss of Top2p sumoylation (Figure 3, C and D). Elimination of a single SIZ gene did not result in a notable reduction of Top2p sumoylation (data not shown). E3-dependent SUMO modification of Top2p in vivo was limited to the COOH-terminal tail (Figure 3A) and, in particular, to the predicted consensus site lysines (K1220, K1246, and K1277) as shown by IMAC in the top2 cis mutants ΔC and 3xKR, which phenocopied the loss of Top2p sumoylation in the Siz− cells (Figure 3, C and D). Even though the Top2p–ΔC protein had a lower abundance than wild-type Top2p or Top2p–3xKR (Figure 3D), scaled up purifications did not reveal any sumoylation of this truncated protein (data not shown). Thus, the Top2p is modified by Smt3p in vivo predominately at the COOH-terminal sites, and the Siz1p and Siz2p play a critical, but mutually redundant role in catalyzing this modification.
The positive regulatory role of E3 in minichromosome transmission is mediated by Top2p:
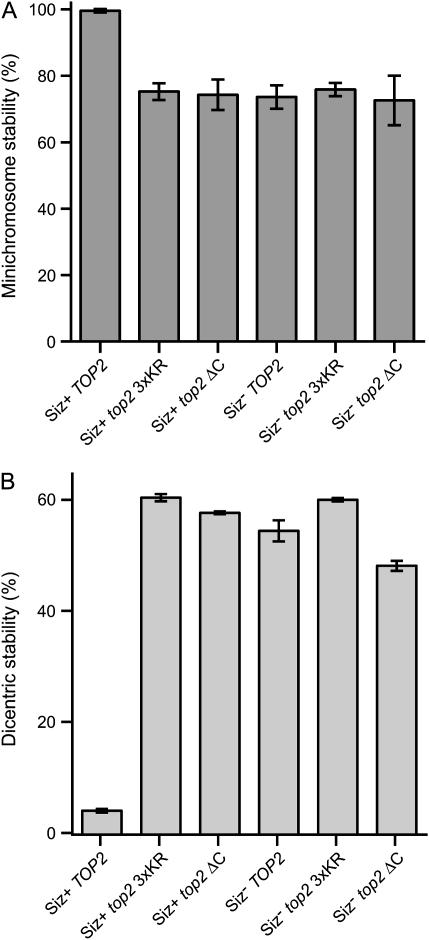
To test whether the role of E3 in minichromosome segregation (Figure 1) is linked to the Top2p function we analyzed minichromosome transmission in the top2 3xKR and ΔC mutants and compared it with SIZ1 and SIZ2 double deletion strains. Analysis of minichromosome stability revealed that both top2 mutants had a decrease in segregation fidelity, which was similar to the minichromosome destabilization in the siz1-Δ/siz2-Δ (Siz−) strain (Figure 4). Moreover, we found that the Siz− and top2 mutations were epistatic for minichromosome stability in triple-mutant strains (combining both top2 and siz mutations), as no additive decrease in minichromosome stability was observed in these mutants (Figure 4). Thus, we demonstrated that the positive regulatory role of the Smt3p E3 in chromosome stability is to modify the consensus sumoylation sites in the COOH tail of topoisomerase II.
Figure 4.
Epistatic interaction between the Smt3p-conjugation-deficient top2 mutations and SUMO E3 deficiency in the control of minichromosome stability. (A) Transmission efficiency of the pRS415 minichromosome. Minichromosome stability was measured (see materials and methods) at 30° in the wild-type (BY4733) and siz1/siz2 mutant (4bAS399) strains with different top2 variants (pYT1033, pYT1034, or pYT1035). (B) SUMO E3 mutants and top2 Smt3p-conjugation-deficient mutations stabilize dicentric minichromosomes. Transmission efficiency of pAS72, a conditional dicentric minichromosome, was measured at 30° in the wild-type (BY4733) and siz1/siz2 (4bAS399) strains with top2 variants (pYT1033, pYT1034, or pYT1035) as described in materials and methods.
As the siz1-Δ/siz2-Δ, top2 3xKR, and ΔC mutations destabilized only the centromere-containing minichromosomes (Figures 1 and 4 and data not shown), the chromatin immunoprecipitation (ChIP) analysis showed that the Top2p protein is present in yeast centromeres (see Figure 5), and Top2p over-sumoylation has been shown to disrupt centromere cohesion (Bachant et al. 2002), we investigated whether the E3-dependent Top2p sumoylation is involved in kinetochore function. Therefore, we tested the same (Figure 4A) set of mutants for stable maintenance of dicentric minichromosomes. Stabilization of dicentric chromosomes has been shown to be a reliable and sensitive genetic assay for testing kinetochore proficiency, as dicentric minichromosomes are not stably maintained in yeast cells unless kinetochore function is compromised by cis or trans mutations (Mythreye and Bloom 2003). To eliminate minichromosome rearrangement as a possible pathway allowing dicentric minichromosomes to be stabilized, we used a conditional dicentric minichromosome, where one of the kinetochores is inactivated by potently inducible transcription prior to the experiment. As shown in Figure 4B, the top2 3xKR and the siz1-Δ/siz2-Δ mutations were epistatic in their ability to stabilize dicentric minichromosomes, as well as in the monocentric minichromosome segregation phenotype (Figure 4A). Thus it is likely that the SUMO E3-dependent regulatory pathway, which facilitates positive regulation of Top2p function in minichromosome segregation, mainly controls the function of the Top2p pool located at the centromeres.
Chromosomal address of sumoylated Top2p:
We hypothesized that a specific physiological level of Top2p sumoylation is required to target it to centromeric regions. Testing this hypothesis is, however, technically challenging due to the above-mentioned difficulties in locating a small sumoylated fraction of a given protein in the cell. To overcome this technical problem we utilized a Top2p–Smt3p fusion as an in situ model of sumoylated Top2p. This approach is based on recent data on physiologically relevant replacement of the isopeptide bond-conjugated ubiquitin with peptide-bond ubiquitin fusions (Ciechanover and Ben-Saadon 2004; Saeki et al. 2004). Thus, we inserted the SMT3 ORF into integrative TOP2 constructs so that an in-frame Top2p(core)–Smt3p–Top2(tail) fusion is produced, with Smt3p inserted between Leu-1235 and Val-1236 of the native Top2p sequence (underlined in Figure 3A). This fusion places the Smt3p constitutively between the first and the second sumoylation sites in the Top2p tail. Haploid strains carrying the integrated HA-tagged and GFP-tagged versions of TOP2:SMT3 fusions were viable and the Top2p–Smt3p–GFP fusions localized throughout the nucleus (data not shown). Moreover, the Top2p–Smt3p appear to be mimicking the sumoylated Top2p in vivo, as such a fusion resulted in almost complete growth inhibition in the smt4-Δ background (Figure 5A). Both HA and GFP-tagged TOP2:SMT3 fusions also resulted in a notable mitotic delay (Figure 5D and data not shown) in the corresponding cell populations, indicating that Smt3p fusion to all of the Top2p molecules in the cell may be detrimental to proliferation.
To test whether Top2p–Smt3p fusion is enriched at the centromeres, as predicted by genetic analysis (Figures 1 and 4) for sumoylated Top2p, we conducted ChIP analysis of the HA-tagged Top2p–Smt3p. Chromatin extracted from the strains expressing Top2p–Smt3p–HA, Top2p–HA and Top2p–ΔC–HA was subjected to immunoprecipitation with anti-HA antibody and analyzed by PCR as described (Strunnikov et al. 2001; Wang et al. 2004). The PCR probes were designed to tile the 5.5-kb region centered at the CEN4 core sequence (Figure 5B). While Top2p–HA displayed only minimal enrichment at the pericentromeric loci compared to Top2p–ΔC–HA, the Top2p–Smt3p-HA fusion was significantly and reproducibly enriched in the CEN4 vicinity (Figure 5C). As it is not known whether Top2p has specific enrichment sites in the genome, it is difficult to determine whether the observed enrichment of binding represents a strictly pericentromeric phenomenon or whether the whole Top2p–Smt3p pool becomes more concentrated at the defined genomic loci. However, ChIP analysis of a randomly selected set of genomic sites (according to Wang et al. 2005) did not reveal any enrichment for Top2p–Smt3p (data not shown), suggesting that sumoylated Top2p likely has a propensity to be enriched at the centromeric regions, as compared to the unmodified form, consistent with our genetic results (Figure 4).
To address the above mentioned caveat and to mimic the wild-type situation, where only a fraction of Top2p is sumoylated, we crossed the haploid strains with integrated HA-tagged and GFP-tagged TOP2:SMT3 fusions to the wild-type TOP2 strains. The resulting strains had both the constitutively modified (fused to Smt3p) Top2p and the wild-type Top2p, with only the fusion form detectable by either GFP or HA tags. Analysis of Top2p–Smt3p–GFP localization in the strain expressing Spc42p-mRFP, an SPB marker, revealed that the fusion, while still diffusely localized to the nucleus, forms distinctive areas of concentration next to spindle pole bodies in mitotic cells (Figure 5D, arrows, and 5E). This localization of the modified Top2p pool is consistent with it being enriched around the centromeric regions. The peri-SPB GFP enrichment was not observed, however, when Top2p–ΔC–GFP (Figure 5F) or wild-type Top2p–GFP (not shown) fusions were investigated in the similarly constructed diploid strains. This result suggests that the peri-SPB enrichment of Top2p–Smt3p–GFP is mediated by Smt3p.
DISCUSSION
Chromosomal function of Siz1p/Siz2p:
The SUMO E3 proteins appear to serve as specificity factors directing the sumoylation event to specific targets in eukaryotic cells (Johnson 2004; Muller et al. 2004). In budding yeast, the Siz1 and Siz2 proteins are localized in the nucleus and are required for the bulk of sumoylation (Johnson and Gupta 2001; Takahashi et al. 2001b). However, the siz1 siz2 double mutants are viable, indicating that the majority of Smt3p-conjugation events are not required for the essential housekeeping functions of the cell. In this work we demonstrated that SUMO E3 is required for minichromosome transmission fidelity. This suggests that certain proteins required for normal chromosome dynamics may be functionally impaired by the lack of E3-dependent sumoylation. Even though we were unable to detect a significant destabilization of the relatively short chromosome III or a longer, rDNA-containing, chromosome, we did find a notable increase in mitotic recombination for distal chromosomal markers (not shown), which suggests an additional role of SUMO E3 in chromatin.
A concurrent study has established that the Siz1 and Siz2 proteins are involved in inhibiting amplification of 2μ plasmids by virtue of promoting the inhibitory sumoylation of two plasmid-encoded proteins (Chen et al. 2005). This exemplifies the negative regulatory role of Smt3p in chromatin. The Loc− chromosomal phenotype (with an inability to separate sister chromatids in mitosis) of the smt3–331 mutation (Biggins et al. 2001) also suggests that Smt3p hyperconjugation has a negative impact on chromosome segregation, as this mutant displays accumulation of sumoylated proteins (A. Strunnikov, unpublished data) in a manner similar to smt4 mutants (Li and Hochstrasser 2000; Strunnikov et al. 2001) lacking isopeptidase activity. In contrast, we can view destabilization of minichromosome transmission in siz1/siz2 as a demonstration of a positive regulatory role of Smt3p in segregation of chromosomal material in budding yeast. Establishing the fact that SUMO E3 factors are required for the fidelity of minichromosome transmission allowed us to identify Top2p as an Smt3p target protein likely mediating this E3 role in chromosome segregation.
Top2p tail is a potent SUMO E3 substrate:
Several essential chromosomal proteins in budding yeast have been reported to have SUMO-modifications in vivo: Top2p, Pol30p, Pds5p, and Ycs4p (Bachant et al. 2002; Hoege et al. 2002; Stead et al. 2003; D'amours et al. 2004). However, the experimental evidence for the biological role of sumoylation in most of these cases has proven to be inconclusive. Recent proteome-wide analyses (Panse et al. 2004; Wohlschlegel et al. 2004; Zhou et al. 2004; Hannich et al. 2005) of Smt3p targets in S. cerevisiae suggest that modifications of some of the previously reported targets (e.g., Pds5p and Ycs4p) cannot be detected by these techniques (Wohlschlegel et al. 2004). This agrees with our data in vitro (Figure 2A), suggesting that these proteins are poor substrates for Smt3p conjugation. Many other chromosomal proteins, including the condensin subunits Brn1p, Smc4p, and Smc2p and some cohesin subunits, have been shown by proteomic approaches to be Smt3p substrates in vivo (Wohlschlegel et al. 2004). Retesting these proteins for modification in vitro (Figure 2A) and in vivo (not shown), using the Smt3p “fingerprint” technique (Panse et al. 2004), failed to detect significant modifications, suggesting that these proteins are also poor substrates. In the case of PCNA (Pol30p) the E3-dependence of its sumoylation (Hoege et al. 2002; Haracska et al. 2004) was demonstrated in vitro (Stelter and Ulrich 2003) and confirmed by us (Y. Takahashi, unpublished data), yet in the chromatin context Pol30p showed no modification in vitro (data not shown). These data suggest that while many chromosome proteins can be modified by SUMO in vivo, such a modification, in many cases, is incompatible with the chromatin association of these proteins. Thus, one can hypothesize that for many proteins sumoylation serves as an inhibitor of chromatin association.
In contrast, our analysis of chromatin-bound Top2p indicates that it is by far the most potent acceptor of Smt3p conjugation among the chromosomal SUMO targets tested. We demonstrated that Top2p can be sumoylated in vitro and elucidated the key role of SUMO E3 in this modification (Figures 2 and 3). We established that Top2p sumoylation is not inhibited in the chromatin-bound forms, making this substrate unique among other SUMO targets, particularly PCNA, and suggesting a high degree of functional specialization of Top2p sumoylation. Mutations of the three consensus-site lysine residues in the Top2p tail largely abolish the ability of Top2p to be modified by Smt3p in vivo and greatly inhibit the in vitro modification reaction (Figure 2E). Deletion of the whole Top2p tail (Top2p–ΔC) eliminates the residual nonspecific modification in vitro (data not shown). These results confirm the role of these sites in Top2p modification (Bachant et al. 2002) and establish that the modification of the Top2p tail is mediated by SUMO E3.
The function of Siz1p/Siz2p in minichromosome transmission is to modify Top2p:
While in higher eukaryotes the effect of UBC9 depletion on chromosome segregation is evident in vitro (Azuma et al. 2003) but not in vivo (Hayashi et al. 2002), disruption of SUMO E2 function in yeast cells impairs mitotic chromosome segregation (Dieckhoff et al. 2004). We established that depletion of the major SUMO E3 activity (Siz1p and Siz2p) also results in an in vivo segregation defect (Figures 1 and 4). Moreover, we found that the Top2p tail deletion and the triple lysine-to-arginine residue mutation at the SUMO acceptor sites have a destabilizing effect on minichromosome transmission similar and epistatic to the SUMO E3 double mutants. While previously overmodification of the Top2p SUMO-target sites in smt4 mutants was shown to impair pericentromeric cohesion, the elimination of these modification sites by point mutations resulted in a practically undetectable cohesion phenotype (Bachant et al. 2002). Thus, the role of Smt3p modification of Top2p in wild-type yeast cells remains obscure.
The results showing that the simultaneous loss of Siz1p and Siz2p activity is epistatic to the top2 SNM alleles and yields an unmodified Top2p suggest that the role of SUMO E3 in chromosome segregation could be limited to the Smt3p modification of a specific Top2p subpopulation. As sumoylation of topoisomerase II in S. cerevisiae (data not shown) and vertebrates (by SUMO-2) (Azuma et al. 2003) peaks in mitosis, it is conceivable that the sumoylated pool of Top2p plays an important role in mitotic chromosome segregation. As the sumoylated pool of Top2p is very small (Figure 3C), it is likely that this subset of Top2p molecules participates in centromere–kinetochore dynamics. Indeed, utilizing a novel approach of modeling sumoylated proteins by direct fusion of targets to SUMO (constitutive SUMO modification), we were able to show that the modified pool of Top2p is enriched at the centromeres (Figure 5C). The exact function of this pool is still unknown, but our genetic data (Figure 4) and disruption of pericentromeric sister chromatid cohesion by hypersumoylation of Top2p (Bachant et al. 2002) suggest that Top2p, when sumoylated at the physiological level, is involved in establishing or maintaining the bipolar kinetochore orientation.
What molecular mechanism can be responsible for the role played by sumoylated Top2p both at the centromere in general and in sister chromatid cohesion in particular? As chromatin-bound Top2p can be readily modified in vitro (Figure 2). it is conceivable that mitotic activation (Johnson and Gupta 2001; Takahashi et al. 2001a) of Siz1p and/or Siz2p activity results in a localized Top2p sumoylation at the centromeric regions. In turn, the SUMO moiety at the Top2p tail could contribute to cohesion by stabilizing the Top2p dimer at the loci that hold two sister chromatids together. As the sumoylation sites in the Top2p tail are situated close to the DNA-release gate in the dimer (Champoux 2001), one can speculate that SUMO-modified tails may delay release of the DNA strands after the enzymatic topoisomerase II reaction is complete; thus, allowing a cohesion mechanism alternative to cohesin clamp.
In this report we demonstrate that the major S. cerevisiae SUMO E3 function in minichromosome transmission is in the same pathway as Smt3p modification of the Top2p tail. While it is formally possible that in vivo there is another Siz1p/Siz2p substrate protein that bridges the Siz1p/Siz2p E3 activity and Top2p modification, all of these data can be explained by direct sumoylation of Top2p by Siz1p/Siz2p, as occurs in vitro (Figure 2). Thus, it is concluded that the important role played by Siz1p and Siz2p in mitotic segregation is embodied by the small pool of Smt3p-modifid Top2p, probably localized in the vicinity of the centromere.
Acknowledgments
We thank J. Nitiss, S. Elledge, D. Bachant, S. Gasser, V. Guacci, T. Sasaki, L. Freeman and A. Kagansky for research materials; Esther Lee and Tiffany Williams for technical help; and A. Hinnebusch, M. Dasso, A. Arnaoutov and N. Dhillon for helpful discussion and comments on the manuscript. This work was supported by the National Institutes of Health (NIH)/Department of Health and Human Services and, in part, by a Grant-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan to Y.K. Y.T. is a Japan Society for the Promotion of Science Research Fellow in Biomedical and Behavioral Research at NIH.
References
- Azuma, Y., A. Arnaoutov and M. Dasso, 2003. SUMO-2/3 regulates topoisomerase-II in mitosis. J. Cell Biol. 163: 477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner and S. J. Elledge, 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell 9: 1169–1182. [DOI] [PubMed] [Google Scholar]
- Biggins, S., N. Bhalla, A. Chang, D. L. Smith and A. W. Murray, 2001. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics 159: 453–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux, J. J., 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70: 369–413. [DOI] [PubMed] [Google Scholar]
- Chen, X. L., A. Reindle and E. S. Johnson, 2005. Misregulation of 2 microm circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 25: 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover, A., and R. Ben-Saadon, 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14: 103–106. [DOI] [PubMed] [Google Scholar]
- Cuff, J. A., and G. J. Barton, 2000. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins 40: 502–511. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., F. Stegmeier and A. Amon, 2004. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell 117: 455–469. [DOI] [PubMed] [Google Scholar]
- Desterro, J. M., J. Thomson and R. T. Hay, 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417: 297–300. [DOI] [PubMed] [Google Scholar]
- Dieckhoff, P., M. Bolte, Y. Sancak, G. H. Braus and S. Irniger, 2004. Smt3/SUMO and Ubc9 are required for efficient APC/C-mediated proteolysis in budding yeast. Mol. Microbiol. 51: 1375–1387. [DOI] [PubMed] [Google Scholar]
- Dohmen, R. J., R. Stappen, J. P. Mcgrath, H. Forrova, J. Kolarov et al., 1995. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 270: 18099–18109. [DOI] [PubMed] [Google Scholar]
- Freeman, L., L. Aragon-Alcaide and A. Strunnikov, 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring, S. L., F. Spencer and P. Hieter, 1990. The CHL1(CTF1) gene product of Saccharomyces cerevisiae is important for chromosome transmission and normal cell cycle progression in G2/M. EMBO J. 9: 4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and A. Sugino, 1988. New yeast—Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink (Editors), 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, New York.
- Hannich, J. T., A. Lewis, M. B. Kroetz, S. J. Li, H. Heide et al., 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 280: 4102–4110. [DOI] [PubMed] [Google Scholar]
- Haracska, L., C. A. Torres-Ramos, R. E. Johnson, S. Prakash and L. Prakash, 2004. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 4267–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, R. T., L. Vuillard, J. M. Desterro and M. S. Rodriguez, 1999. Control of NF-kappa B transcriptional activation by signal induced proteolysis of I kappa B alpha. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354: 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, T., M. Seki, D. Maeda, W. Wang, Y. Kawabe et al., 2002. Ubc9 is essential for viability of higher eukaryotic cells. Exp. Cell Res. 280: 212–221. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., 2000. Biochemistry: all in the ubiquitin family. Science 289: 563–564. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107: 5–8. [DOI] [PubMed] [Google Scholar]
- Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis and S. Jentsch, 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Jackson, P. K., 2001. A new RING for SUMO: wrestling transcriptional responses into nuclear bodies with PIAS family E3 SUMO ligases. Genes Dev. 15: 3053–3058. [DOI] [PubMed] [Google Scholar]
- Jentsch, S., and G. Pyrowolakis, 2000. Ubiquitin and its kin: How close are the family ties? Trends Cell Biol. 10: 335–342. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., 2004. Protein modification by sumo. Annu. Rev. Biochem. 73: 355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272: 26799–26802. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, E. S., and A. A. Gupta, 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., I. Schwienhorst, R. J. Dohmen and G. Blobel, 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16: 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagansky, A., L. Freeman, D. Lukyanov and A. Strunnikov, 2004. Histone tail-independent chromatin binding activity of recombinant cohesin holocomplex. J. Biol. Chem. 279: 3382–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey, M. H., T. A. Melhuish and D. Wotton, 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113: 127–137. [DOI] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 1999. A new protease required for cell-cycle progression in yeast. Nature 398: 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20: 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. J., and M. Hochstrasser, 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C., and B. Stillman, 1997. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 11: 3375–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Meluh, P. B., and D. Koshland, 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6: 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, S., C. Hoege, G. Pyrowolakis and S. Jentsch, 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2: 202–210. [DOI] [PubMed] [Google Scholar]
- Muller, S., A. Ledl and D. Schmidt, 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23: 1998–2008. [DOI] [PubMed] [Google Scholar]
- Mythreye, K., and K. S. Bloom, 2003. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J. Cell Biol. 160: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma, T., R. Honda, G. Ichikawa, N. Tsumagari and H. Yasuda, 1999. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 254: 693–698. [DOI] [PubMed] [Google Scholar]
- Panse, V. G., U. Hardeland, T. Werner, B. Kuster and E. Hurt, 2004. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 279: 41346–41351. [DOI] [PubMed] [Google Scholar]
- Panse, V. G., B. Kuster, T. Gerstberger and E. Hurt, 2003. Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5: 21–27. [DOI] [PubMed] [Google Scholar]
- Pichler, A., A. Gast, J. S. Seeler, A. Dejean and F. Melchior, 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120. [DOI] [PubMed] [Google Scholar]
- Saeki, Y., E. Isono, T. Oguchi, M. Shimada, T. Sone et al., 2004. Intracellularly inducible, ubiquitin hydrolase-insensitive tandem ubiquitins inhibit the 26S proteasome activity and cell division. Genes Genet. Syst. 79: 77–86. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., E. A. Toh and Y. Kikuchi, 2000. Yeast Krr1p physically and functionally interacts with a novel essential Kri1p, and both proteins are required for 40S ribosome biogenesis in the nucleolus. Mol. Cell. Biol. 20: 7971–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S. E., K. Matuschewski, D. Liakopoulos, M. Scheffner and S. Jentsch, 1998. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95: 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead, K., C. Aguilar, T. Hartman, M. Drexel, P. Meluh et al., 2003. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J. Cell Biol. 163: 729–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter, P., and H. D. Ulrich, 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A. V., V. L. Larionov and D. Koshland, 1993. SMC1: an essential yeast gene encoding a putative head-rod-tail protein is required for nuclear division and defines a new ubiquitous protein family. J. Cell. Biochem. 123: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov, A. V., L. Aravind and E. V. Koonin, 2001. Saccharomyces cerevisiae SMT4 encodes an evolutionarily conserved protease with a role in chromosome condensation regulation. Genetics 158: 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., M. Iwase, M. Konishi, M. Tanaka, A. Toh-e et al., 1999. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 259: 582–587. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., J. Mizoi, E. A. Toh and Y. Kikuchi, 2000. Yeast Ulp1, an Smt3-specific protease, associates with nucleoporins. J. Biochem. 128: 723–725. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., T. Kahyo, E. A. Toh, H. Yasuda and Y. Kikuchi, 2001. a Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276: 48973–48977. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., A. Toh-e and Y. Kikuchi, 2001. b A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275: 223–231. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., E. A. Toh and Y. Kikuchi, 2003. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133: 415–422. [DOI] [PubMed] [Google Scholar]
- Ulrich, H. D., 2004. How to activate a damage-tolerant polymerase: consequences of PCNA modifications by ubiquitin and SUMO. Cell Cycle 3: 15–18. [PubMed] [Google Scholar]
- Vaughn, J., S. Huang, I. Wessel, T. K. Sorensen, T. Hsieh et al., 2005. Stability of the topoisomerase II closed clamp conformation may influence DNA-stimulated ATP hydrolysis. J. Biol. Chem. 280: 11920–11929. [DOI] [PubMed] [Google Scholar]
- Wang, B. D., V. Yong-Gonzalez and A. V. Strunnikov, 2004. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle 3: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. D., D. Eyre, M. Basrai, M. Lichten and A. Strunnikov, 2005. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 25: 7216–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman, A. M., 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2: 169–178. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel, J. A., E. S. Johnson, S. I. Reed and J. R. Yates, III, 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279: 45662–45668. [DOI] [PubMed] [Google Scholar]
- Zhao, X., and G. Blobel, 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, W., J. J. Ryan and H. Zhou, 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 279: 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]