Abstract
While screening for genes that reverse the sporulation-deficient phenotype of the ras1Δ diploid Schizosaccharomyces pombe strain, we identified zds1. This gene shares sequence homology with the ZDS1 and ZDS2 genes from Saccharomyces cerevisiae, which appear to be involved in multiple cellular events. Expression of Zds1 in ras1Δ diploid cells elevated their sporulation rate from 0.3 to 11.2%. Expression of the Zds1 C-terminal region increased the sporulation rate further (to 21.9%) while introduction of the Zds1 N-terminal region had no effect. zds1 expression did not induce sporulation in strains with mutations in genes participating in the downstream MAP kinase cascade. The zds1-disrupted strain is sensitive to CaCl2, and this effect is suppressed by the C-terminal region of Zds1. The growth of the zds1Δ strain is markedly inhibited by cold temperatures, while its viability decreased in the stationary phase. Moreover, the zds1Δ strain is round in shape and very sensitive to zymolyase, and its cell wall becomes thicker than that of wild type. Thus, zds1 must be required to maintain cell wall integrity. The Zds1–GFP fusion protein localized to the cytosol, the septum, and the cell cortex. Its localization in the septum was dependent on its C-terminal region. Overexpression of the C-terminal region of Zds1 induced multi-septa and abnormal zygotes. We propose that the C-terminal region is the functional domain of Zds1 while the N-terminal region is a negative regulatory region. Thus, Zds1 is involved in multiple cellular events in fission yeast, including sexual differentiation, Ca2+ tolerance, cell wall integrity, viability in the stationary phase, and cell morphology.
THE fission yeast Schizosaccharomyces pombe proliferates continuously when it has abundant nutrients but arrests its cell cycle progression in the G1 phase upon depletion of glucose. Heterothallic cells of an opposite mating type, namely h− and h+, start to develop sexually through processes that include conjugation, meiosis, and sporulation. Homothallic cells (h90) switch frequently between the h− and h+ mating types. The sexual differentiation process is induced by the key transcription factor Ste11, which regulates the transcription of many genes involved in meiosis, including mat1-Pm, mat1-Mm, ste6, and mei2 (Sugimoto et al. 1991; Yamamoto et al. 1997). Mei2 is an RNA-binding protein that is negatively regulated by Pat1 protein kinase (Watanabe et al. 1997). Pat1 is inhibited by Mei3, whose expression is induced by the pheromone signaling pathway (Li and McLeod 1996). In an alternative regulatory pathway, Pat1 can be inhibited by a truncated version of Sla1, an ortholog of the mammalian La protein (Tanabe et al. 2003, 2004).
The sexual differentiation that precedes meiosis is regulated by the cAMP pathway, the stress responsive pathway, and the pheromone signaling pathway (Yamamoto 2003). The cAMP pathway signals the nutrient conditions, mainly the glucose levels, to the cell. When glucose (or nitrogen) is abundant, the heterotrimeric-type guanine nucleotide-binding protein (Gpa2) becomes activated; this subsequently activates adenylyl cyclase (Isshiki et al. 1992) to generate cAMP from ATP (Kawamukai et al. 1991). When cAMP is abundant, it associates with the regulatory subunit Cgs1, and the catalytic protein kinase Pka1 is released (Maeda et al. 1994). Sterility caused by higher cAMP levels is reversed by the moc1–moc4 genes (Kawamukai 1999; Goldar et al. 2005a,b). Pka1 phosphorylates the inhibitory zinc-finger protein Rst2, which otherwise induces the expression of ste11 (Higuchi et al. 2002).
The stress-responsive pathway involves the histidine-to-aspartate phosphorylation relay and a mitogen-activated protein kinase (MAPK) cascade. This MAPK pathway consists of Wis4/Wik1/Wak1 (MAPKKK), Win1 (MAPKKK), Wis1 (MAPKK), and Phh1/Sty1/Spc1 (MAPK) (Toone and Jones 2003). Sty1 phosphorylates the bZIP-type transcription factor Atf1 (Takeda et al. 1995), which induces the transcription of the ste11 gene.
The pheromone signaling pathway is initiated by the binding of mating pheromone to the pheromone receptor, which activates the receptor-coupled G protein Gpa1 (Obara et al. 1991). The signal is then transmitted to a MAPK cascade—a process that depends on the oncoprotein homolog Ras1 (Yamamoto et al. 1997). The Ras1 protein recruits the MAPKK kinase Byr2 (Wang et al. 1991) to the membrane, where it is activated. In the process of Byr2 activation, dimerization of Byr2 through Ste4 is proposed (Barr et al. 1996). Byr2 is maintained in an inactive form by an intermolecular interaction (Tu et al. 1997) or binding with the 14-3-3 homologs Rad24 and Rad25 (Ozoe et al. 2002). Activated Byr2 phosphorylates the MAPK kinase Byr1, thereby activating it, which in turn activates the MAP kinase Spk1 via a typical MAP kinase activation mechanism (Toda et al. 1991; Yamamoto et al. 2004). Mutations that block the function of any component of the pheromone signaling pathway cause fission yeast to become sterile, which indicates that this pathway is essential for the execution of sexual differentiation. Ras1 is also involved in another pathway, namely the morphological pathway. GTP-bound Ras1 interacts with Scd1, a putative guanine nucleotide exchange factor for the small G protein Cdc42, which is involved in regulating cell morphology (Chang et al. 1994). GTP-bound Cdc42 interacts with the p21-type kinase Shk1 (Marcus et al. 1995; Ottilie et al. 1995). Scd1, Cdc42, and Shk1, together with Scd2, form a quaternary complex that facilitates the smooth activation of Shk1 (Endo et al. 2003).
Previously, we reported the existence of S. pombe sam mutants, which undergo mating and sporulation without requiring nitrogen starvation. We characterized these mutants and found that two (sam3 and -9) are dominant while seven (sam1, -2, -4 ∼ -8) are recessive (Katayama et al. 1996). On the basis of this characterization, we isolated two suppressor genes, msa1 and msa2 (Jeong et al. 2004a,b). To further investigate the role that ras1 plays in sporulation and to identify the gene that confers the hypersporulating phenotype to the sam mutants, we generated a genome library from the sam9 mutant. We used this library to transform a ras1-deficient strain of S. pombe that cannot sporulate and screened out zds1 (zillion different screens) as the gene that reversed this phenotype. Although this was determined not to be identical to the sam9 gene, the fact that it bore homology to two genes from Saccharomyces cerevisiae, namely, ZDS1 and ZDS2, encouraged us to further investigate its functions. As their names suggest, ZDS1 and ZDS2 of S. cerevisiae have been independently identified by many different screenings of various gene types. This suggests that these genes may play roles in multiple signaling pathways. Thus, ZDS1 was isolated as a multicopy suppressor of sensitivity to the calmodulin inhibitor TFP in the ssd1Δ strain (Tsuchiya et al. 1996) as well as a multicopy suppressor of the hsl1 and hsl7 mutants (Ma et al. 1996), of cdc28-1N (Yu et al. 1996), and of a tif1 mutant (tif1 encodes the translation initiation factor eIF-4A) (Schwer et al. 1998). ZDS2 was isolated as a multicopy suppressor of sin4, a gene that confers resistance to the anticancer agent cisplatin (Burger et al. 2000). ZDS1 and ZDS2 were also isolated as (i) multicopy suppressors of temperature-sensitive mutations of a yeast mRNA-capping enzyme (CEG1) (Schwer and Shuman 1996), (ii) negative regulators of Cdc42p (Bi and Pringle 1996), (iii) multicopy suppressors of a cka2 mutant (cka2 encodes the α′-subunit of casein kinase II) (Bandhakavi et al. 2003), (iv) suppressors of the camptothecin-hypersensitive trf4 mutant (Walowsky et al. 1999), (v) genes that stabilize short linear centromeric plasmids (Roy and Runge 1999), (vi) multicopy suppressors of an rhc21 mutant (rhc21 encodes a component of the cohesin complex) (Heo et al. 1999), (vii) genes that interact with the Bcy1p N-terminal domain (Griffioen et al. 2001), and (viii) multicopy suppressors of a 1, 3-β-glucan synthase mutant (Sekiya-Kawasaki et al. 2002). In addition, Zds1p and Zds2p were reported to be involved in transcriptional silencing and longevity (Roy and Runge 2000). Despite these frequent appearances in various different roles, however, the exact functions of the proteins encoded by ZDS1 and ZDS2 are as yet unclear.
We thus investigated the function of zds1 in the ras1-deficient diploid strain of S. pombe and found that it is involved in a signaling pathway and in sexual differentiation. Zds1 is also required to maintain cell wall integrity, calcium tolerance, cell viability in the stationary phase, and low temperature growth, which suggests that it has multiple roles. This is the first characterization of a ZDS1 and ZDS2 orthologous gene, other than those of S. cerevisiae.
MATERIALS AND METHODS
Strains and media:
The S. pombe strains used in this study are listed in Table 1. S. pombe was grown in YEA-rich medium (0.5% yeast extract, 3% glucose, 75 mg/liter adenine), YES-rich medium (0.5% yeast extract, 3% glucose, 225 mg/liter adenine, histidine, leucine, uracil, and lysine hydrochloride) or pombe minimum (PM) synthetic medium (Alfa et al. 1993), supplemented with 75 mg/liter adenine, leucine, or uracil when necessary. SPA medium [1% glucose, 0.1% KH2PO4, 0.1% vitamin mixture (Alfa et al. 1993), 3% agar] was used to culture S. pombe when the sporulation rate had to be measured. To assay CaCl2 sensitivity, PMA plates without phosphate were made by substituting 14.6 mm sodium acetate for sodium phosphate and adjusting the pH to 5.5. Escherichia coli strains DH10B and DH5α were used for plasmid manipulation. E. coli was grown in LB (1% polypepton, 0.5% yeast extract, 1% NaCl, pH 7.2).
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SP66 | h90 leu1.32 ade6.216 | Laboratory stock |
| SP870 | h90 leu1.32 ade6.210 ura4-D18 | Laboratory stock |
| SPRN1 | h90 leu1.32 ade6.210 ura4-D18 ras1-ΔBglII-NheI | Laboratory stock |
| SPRN1DA | h90 leu1.32 ade6.210 ura4-D18 ras1-ΔBglII-NheI/h90 leu1.32 ade6.216 ura4-D18 ras1-ΔBglII-NheI | Laboratory stock |
| SPSA | h90 leu1.32 ade6.210 ura4-D18 byr2∷ura4∷ADE2 | Laboratory stock |
| SPSUD | h90 leu1.32 ade6.210 ura4-D18 byr2∷ura4/h90 leu1.32 ade6.210 ura4-D18 byr2∷ura4 | Laboratory stock |
| SPBUD | h90 leu1.32 ade6.210 ura4-D18 byr1∷ura4/h90 leu1.32 ade6.210 ura4-D18 byr1∷ura4 | Laboratory stock |
| SPKUD | h90 leu1.32 ade6.210 ura4-D18 spk1∷ura4/h90 leu1.32 ade6.210 ura4-D18 spk1∷ura4 | Laboratory stock |
| HS430RU | h90 ade6.216 ura4-D18 sam9 ras1∷ura4 | Laboratory stock |
| MY6010 | h90 leu1.32 ade6.210 ura4-D18 zds1∷ura4 | This study |
| MY6013 | h90 leu1.32 ade6.210 ura4-D18 zds1:GFPS65A≪LEU2 | This study |
DNA manipulation:
General procedures for DNA manipulation, Southern hybridization, and PCR were performed as previously described (Sambrook et al. 1989).
Isolation of a fission yeast homolog of ZDS1 and ZDS2:
We constructed a genomic library as follows: The HS430RU (sam9) genome was completely digested with either HindIII or SacI and fractionated in a 4–40% sucrose gradient. Fragments >4 kb were cloned into the low-copy vector pYC11-ars 3002 + 3 (Dubey et al. 1994; Tanabe et al. 2003) that had been digested with HindIII or SacI. The S. pombe strain SPRN1DA, which lacks ras1 and cannot sporulate, was transformed by this genomic library, grown for 5 days at 30°, and incubated for 2 days at 25°. The transformants were then exposed to iodine vapor for 1 min and the colonies that were stained brown (indicating their ability to sporulate) were picked and expanded. One of the plasmids obtained from the positive strains, named pSAMH601, was sequenced using the ABI Prism 377 DNA sequencer. Database homologs of the sequence were sought. pSAMH601 was found to contain the zds1 gene that bears homology to ZDS1 and ZDS2.
Gene disruption:
The zds1-disrupted strain was constructed as follows: the 5.1-kb HindIII–HincII fragment that contains the zds1 region was obtained from pSAMH601. This fragment was inserted into the HindIII–SmaI site of pBluescript II SK+ and the resulting plasmid, named pSAMH601HIII-II, was digested with PstI. The PstI-digested 1.8-kb ura4 gene was then inserted into the PstI site of pSAMH601HIII-II. The wild-type S. pombe strain SP870 (h90) (Kawamukai et al. 1992) was transformed with the ApaI–SacII fragment from the resulting plasmid (pSAMH601P-ura4) and stable Ura+ transformants were selected to obtain the zds1Δ strain. The proper integration of the zds1 disruption was verified by Southern blot analysis (data not shown) and the resulting strain was named MY6010.
Plasmid construction:
The plasmids pREP1–zds1 and pREP41–zds1, which bear the zds1 gene under the control of a thiamine-repressible promoter, were constructed as follows: the zds1 region of pSAMH601 was amplified by PCR using the SAMH601F and SAMH601R primers (Table 2). The PCR product was digested with SalI and BamHI and respectively inserted into pREP1 and pREP41, which differ only in the strength of the promoter (the promoter is weaker in pREP41) (Maundrell 1990; Basi et al. 1993). The sequence of zds1 was verified.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| SAMH601F | 5′-ATGTCGAC (SalI) AGAATTC (EcoRI) ATGTCTAGTTCTTCAGTT-3′ |
| SAMH601R | 5′-ATGTCGAC (SalI) GGATCC (BamHI) TTTAGCATTCTTTGAAAC-3′ |
| zds1-GFP5 | 5′-ATGAATTC (EcoRI) GCGGCCGC (NotI) ATGTCTAGTTCTTCAGTT-3′ |
| zds1-GFP3 | 5′-ATGAATTC (EcoRI) GCGGCCGC (NotI) TTTTAGCATTCTTTGAAAC-3′ |
| PH1–179 | 5′-AATTTAAAGCGGCCGC (NotI) CGGTAGATGCATCTTGTTC-3′ |
| PH1–226 | 5′-TTTTAAAAGCGGCCGC (NotI)CTAGGGAACGAATGTGCTC-3′ |
| PH1–567 | 5′-ATATGCATGCGGCCGC (NotI) CATAACCCTCAGTTTTATT-3′ |
| PH1–669 | 5′-ATAAATTTGCGGCCGC (NotI)CACCTTTGTTAGCAGTGGA-3′ |
| PH1–817 | 5′-GGCATATTGCGGCCGC (NotI)CACTCCAGTAATAATCCTT-3′ |
| PH194–938 | 5′-AATCGAAAGCGGCCGC (NotI) ATGACTGGTTGGAAATCGTTT-3′ |
| PH239–938 | 5′-TTTTAAAAGCGGCCGC (NotI) ATGGCTGACGATGCTGTAATT-3′ |
| PH573–938 | 5′-ATGCAGCTGCGGCCGC (NotI) ATGCGTGAGGAAAAGAATTAT-3′ |
| PH682–938 | 5′-TAAATTTTGCGGCCGC (NotI)ATGAAGAAAACCAAGAGGTCC-3′ |
Restriction enzyme sites are underlined.
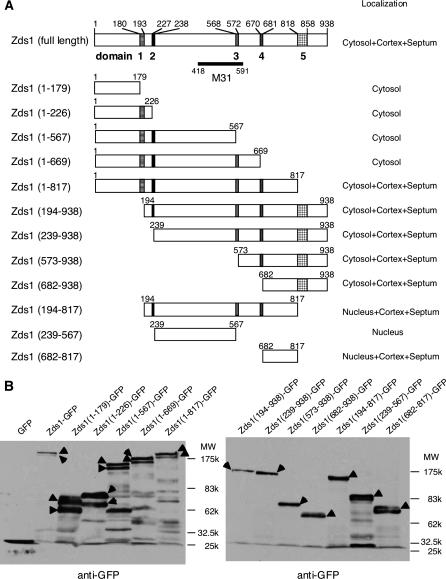
The plasmids pSLF172L Zds1–GFP and pSLF272L Zds1–GFP, which bear the zds1 gene fused with the green fluorescent protein (GFP) gene under the control of a thiamine-repressible promoter, were constructed as follows: the zds1 region of pSAMH601 was amplified by PCR using the zds1–GFP5 and zds1–GFP3 primers (Table 2). The PCR product was digested with NotI and inserted into pSLF172L GFPS65A and pSLF272L GFPS65A, which differ only in the strength of the promoter (it is weaker in pSLF272L GFPS65A) (Forsburg and Sherman 1997). The sequence of zds1 was verified. Various plasmids used in domain analysis were constructed in a similar way, employing the primers listed in Table 2. For example, pSLF172L Zds1(1–179)–GFP and pSLF272L Zds1(1–179)–GFP were constructed by inserting the PCR fragment that had been amplified using the zds1–GFP5 and PH1–179 primers into the NotI site of pSLF172LGFPS65A and pSLF272L GFPS65A, respectively. The other 11 constructs shown in Figure 5A were made in a similar manner.
Figure 5.
(A) Domain analysis of Zds1. Domain 1 (180–193) contains a sequence that is homologous to a region in S. cerevisiae Zds1p and Zds2p, domain 2 (227–238) contains a putative NLS, domain 3 (568–572) contains a region that is homologous to one in Zds2p, domain 4 (670–681) contains a Ser-rich sequence, and domain 5 (818–858) contains a putative functional domain. The M31 region is reported to localize to the nucleus (Sawin and Nurse 1996). (B) Western blot analysis of various Zds1–GFP fusion proteins. SP66 (wild-type) cells were transformed with the various plasmids shown in A and cultured to midlog phase. The proteins were then extracted. Arrowheads show various Zds1–GFP fusion proteins.
GFP tagging:
GFP carrying the S65A mutation was fused to the C terminus of the zds1 gene in the integration vector pYC11, a derivative of pBluescript KS(+) that retains the LEU2 marker (Takahashi et al. 1992). The wild-type S. pombe strain SP870 was transformed with pYC11–zds1–GFP, and stable leu+ integrants were selected and verified by Southern blot analysis (data not shown). The resulting strain was named MY6013.
Western blotting:
Approximately 1 × 108 cells were harvested after growth in the appropriate medium, washed twice with H2O, dissolved in 100 μl of dH2O, and boiled at 95° for 5 min. Subsequently, 120 μl of 2× Laemmli buffer (4% SDS, 20% glycerol, 0.6 m β-mercaptoethanol, 8 m urea, 0.12 m Tris-HCl, pH 6.8) was added and the samples were vigorously vortexed with acid-washed glass beads for 3 min. After removing the glass beads, the samples were sonicated for 1 min and heated at 95° for 5 min. Large debris was removed by centrifugation at 16,000 × g for 1 min. Each sample was analyzed by SDS-polyacrylamide gel electrophoresis with a 7.5% polyacrylamide gel and then transferred to Immobilon transfer membranes (Millipore, Bedford, MA) by using a wet-type transfer system. To detect GFP fusion proteins, membranes were incubated with an anti-GFP monoclonal antibody (Roche) diluted 1:2000 in 5% dry milk in TBS-T (20 mm Tris, 137 mm NaCl, 0.2% Tween 20). Membranes were washed three times by TBS-T for 5 min per wash and then incubated with horseradish–peroxidase-conjugated anti-mouse secondary antibody (Promega, Madison, WI) diluted 1:3000 in 5% dry milk in TBS-T. After the membrane was washed, the secondary antibodies were detected with the ECL system as described by the manufacturer (Amersham, Buckinghamshire, UK).
Fluorescence microscopy:
SP66 (h90, wild type) or MY6010 (h90, zds1Δ) cells harboring various plasmids containing GFP-fused zds1 were grown to log phase at 30° in PMA liquid medium. Zds1–GFPs were observed in living cells by staining the cells with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) and observing them under the BX51 (Olympus) microscope (all observations except those in Figure 7C) or Eclipse 80i (Nikon) microscope (Figure 7C). Fluorescence images were taken with a digital camera VB-6000/6010 (Keyence) connected to the microscope.
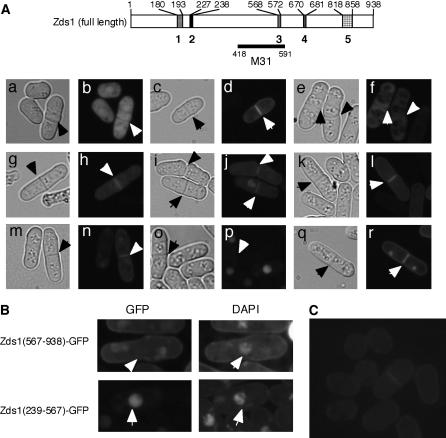
Figure 7.
The localization of various Zds1–GFP fusion proteins. Cells were cultured in PMA containing thiamine at 30° until they reached the stationary phase. The cells (at a density of 1 × 105 cells/ml) were then inoculated into PMA without thiamine and further cultures at 30°. After 18 hr, the cells were observed under a fluorescent microscope. (A) MY6010/pSLF172L GFPS65A (a and b), MY6010/pSLF172L Zds1–GFP (c and d), MY6010/pSLF172L Zds1(1–669)–GFP (e and f), MY6010/pSLF172L Zds1(1–817)–GFP (g and h), MY6010/pSLF272L Zds1(194–938)–GFP (i and j), MY6010/pSLF172L Zds1(239–938)–GFP (k and l), MY6010/pSLF172L Zds1(573–938)–GFP (m and n), MY6010/pSLF172L Zds1(239–567)–GFP (o and p), and MY6010/pSLF172L Zds1(682–817)–GFP (q and r). Phase-contrast microscopic photographs (a, c, e, g, i, k, m, o, and q) and fluorescent microscopic photographs (b, d, f, h, j, l, n, p, and r) were taken. (B) Localization of Zds1ΔNΔC–GFP in nuclei. Cells were stained with DAPI and observed under a fluorescent microscope. (C) Localization of the Zds1–GFP fusion protein in the zds1–GFP integrated strain, MY6013. Cells were cultured in YES at 30° until they reached the stationary phase. They were then cultured in PM plus adenine and uracil until the midlog phase, washed with H2O and then with nitrogen-free PM medium, and resuspended in nitrogen-free PM medium. After 6 hr, the cells were observed by fluorescent microscopy.
Electron microscopy:
SP66 (h90,wild type) or MY6010 (h90, zds1Δ) cells were grown in PM plus adenine and leucine (PMAL) medium at 30° for 72 hr and fixed with 2% glutaraldehyde in 0.1 m potassium phosphate (pH 7.2) for 2 hr. They were then postfixed with 1.7% potassium permanganate at 4° overnight, embedded in agarose, and stained with 1% uranyl acetate for 2 hr. The cells were dehydrated through a graded series of ethanol (60–100%) and then substituted with QY-1. The samples were embedded in Quetol653 resin and ultra-thin sections were stained with uranyl acetate and Sato's lead solution and viewed with a JEM-1200EX (Jeol, Tokyo) microscope at 80 kV.
Zymolyase assay to assess cell wall integrity:
Cells were grown at 30° in PMA with or without thiamine liquid medium until the cell density reached 1 × 107 cells/ml, after which the cells were collected by centrifugation at 1700 × g for 5 min. The cells were dissolved in TE and incubated at 30° for 180 min with 0.1 or 0.3 mg/ml zymolyase 20T (Seikagaku Kogyo). The degree of lysis was evaluated by measuring the absorbance at OD595.
RESULTS
Isolation of zds1, a fission yeast ortholog of ZDS1 and ZDS2:
To study the regulation of sexual differentiation in S. pombe, we previously obtained and characterized nine mutants, denoted as sam, that have a hypersporulating phenotype (Katayama et al. 1996). One of these, the sam9 mutant, is a dominant type. To isolate this gene, we constructed a genomic library from the sam9 mutant and screened for genes that cause the Spo− phenotype of the ras1− S. pombe diploid to revert to sporulation competence. In this screening, we obtained genes that are already known to stimulate sexual differentiation, namely, byr2, mei2, and ras1. We also identified a plasmid containing an ORF composed of 938 amino acids that has sequence homology to S. cerevisiae Zds1p (29% identity) and Zds2p (31% identity). S. cerevisiae ZDS1 and ZDS2 have been previously obtained in many different screenings. These include searches for a suppressor of cdc28-1N and a negative regulator of Cdc42p (Bi and Pringle 1996; Yu et al. 1996). We thus named the gene zds1 (S. pombe Zds1 ortholog). However, since the zds1 sequence including upstream and downstream regions from the sam9 mutant is identical to SPAC31F12.01, which was reported in the NCBI database, we concluded that zds1 is not the sam9 allele. Nevertheless, because the functions of S. cerevisiae ZDS1 and ZDS2 are still unclear and no one so far has studied their homolog(s) in S. pombe, we further characterized zds1. Comparison of the amino acid sequence of Zds1 with those of S. cerevisiae Zds1p and Zds2p revealed that the C-terminal region of Zds1 is well conserved while the N-terminal region is less conserved (Figure 1). The significance of this sequence similarity will be discussed later.
Figure 1.
Comparison of the amino acid sequence of S. pombe (Sp) Zds1 with those of the Zds1p and Zds2p from S. cerevisiae. The putative nuclear localization signal is indicated by the underlined letters. The consensus amino acids are indicated by boldface type.
Effect of zds1 expression in Ras1–MAPK gene-deficient cells:
We constructed plasmids that express zds1 under thiamine-repressible nmt1 and mutated nmt1 promoter, which differ in the strength of their activity, and denoted them as pREP1–zds1 and pREP41–zds1, respectively. The pREP1–zds1 plasmid expresses zds1 at full levels when thiamine is removed from the medium while the pREP41 plasmid expresses zds1 at weaker levels under the same conditions. These plasmids were used to transform ras1Δ diploid cells. During thiamine deficiency, the ras1Δ cells containing pREP1–zds1 and pREP41–zds1 sporulated at rates of 11.2 and 2.9%, respectively, whereas cells harboring the vector alone sporulated at a rate of 0.3% (Table 3) under conditions in which wild-type cells sporulated at a rate of 50%. As ras1 is required for both mating and sporulation (Wang et al. 1991), we next used h90 ras1Δ haploid cells to determine whether zds1 can revert its mating deficiency. SPRN1 (h90 ras1Δ) haploid cells expressing zds1 did not mate to form spores (data not shown). Thus, zds1 can cause ras1-deficient cells to recover their ability to sporulate, but not to mate, and this effect is dependent on its expression level.
TABLE 3.
zds1 expression reverses the sporulation deficiency of the ras1− strain but has no effect on byr2−, byr1−, and spk1− sporulation
| Sporulation rate (%)
|
|||
|---|---|---|---|
| Host | Plasmid | +Thiamine | −Thiamine |
| SPRN1DA (ras1−/ras1−) | pREP1 | 0.1 | 0.3 |
| pREP1-zds1 | 0.7 | 11.2 | |
| pREP41 | 0.1 | 0.7 | |
| pREP41-zds1 | 0.2 | 2.9 | |
| SPSUD (byr2−/byr2−) | pREP1 | 0 | 0 |
| pREP1-zds1 | 0 | 0 | |
| SPBUD (byr1−/byr1−) | pREP1 | 0 | 0 |
| pREP1-zds1 | 0 | 0 | |
| SPKUD (spk1−/spk1−) | pREP1 | 0 | 0 |
| pREP1-zds1 | 0 | 0 | |
The strains were streaked onto SPA plate and grown at 30° for 2 days. About 1000 cells were counted under the microscope.
We next assessed whether the expression of the zds1 gene can also overcome the sporulation-blocking effects of deficiency in byr2, which encodes a MAPKKK in the Ras1–MAPK pathway. SPSA (byr2Δ) and SPSUD (byr2Δ/byr2Δ) were thus transformed with pREP1–zds1 and pREP41–zds1, but neither plasmid caused the yeasts to recover their ability to sporulate (Table 3). We also tested the effect of pREP1–zds1 in diploid yeasts that are deficient in byr1, which encodes a MAPKK, or spk1, which encodes a MAPK, but again, no effect was observed (Table 3). Thus, the zds1 gene can reverse the sporulation-blocking effects of ras1 deficiency in diploid cells but cannot overturn the effects of mutations in genes operating downstream of Ras1 in the Ras1–MAPK pathway.
Growth and morphological phenotypes of zds1-disrupted cells:
We examined the function of zds1 by constructing zds1-disrupted cells. The zds1 disruptant was obtained by homologous recombination with the ura4 marker gene from the haploid S. pombe strain SP870. The success of the zds1 disruption was confirmed by Southern blot analysis (data not shown). We first assessed if zds1Δ cells are also sensitive to temperature. At 36.5°, the growth of zds1Δ cells on YEA and PMA plates did not differ from that of the wild-type cells (data not shown). At 16°, zds1Δ cells did not grow at all on YEA plates, but on the PMA plates they grew at rates similar to the wild-type cells (Figure 2A). However, when the zds1Δ cells were cultured at 16° on PMA plates for 14 days, the colonies became shiny and microscopic observation revealed cell lysis. Thus, zds1Δ cells are clearly sensitive to low temperature when grown in a rich medium. However, there was no obvious difference in the mating rates of the zds1Δ cells and the sporulation rates of the zds1Δ/zds1Δ cells compared with the wild-type strain (data not shown).
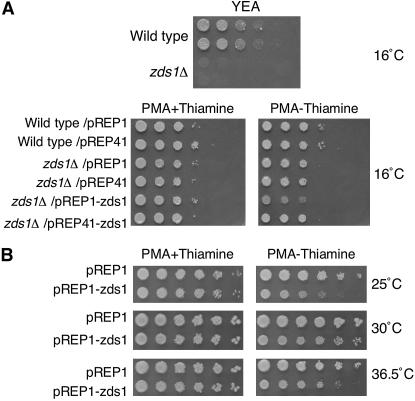
Figure 2.
Temperature sensitivity of zds1-disrupted and zds1-overexpressing cells. (A) Wild-type and zds1-disruptant strains or those harboring the indicated plasmids were grown for 14 days at 16° on the indicated plates and growth was observed. (B) Wild type S. pombe cells (SP66) that overexpress zds1 due to transformation with pREP1–zds1 were spotted onto PMA plates in the presence or absence of thiamine, absence of which induces the expression of zds1 from the plasmids. The cells were incubated at 25°, 30°, or 36.5° for 5 days and growth was observed.
We noted that transformation of zds1Δ cells with pREP1–zds1 appears to inhibit their growth somewhat (Figure 2A). We thus tested the effect of zds1 overexpression on wild-type cell growth. At 30°, the growth of zds1-overexpressing cells (wild-type cells transformed with pREP1–zds1 or pREP41–zds1) was slightly retarded compared to that of wild-type cells. This inhibition of growth was much more prominent at 25° and at 36.5° (Figure 2B). This effect of zds1 expression is also dependent on the promoter strength as the pREP41–zds1 plasmid, which has a weaker nmt41 promoter, had little effect on wild-type growth (data not shown). Thus, overexpression of zds1 in the wild-type strain also results in cold and high temperature sensitivity.
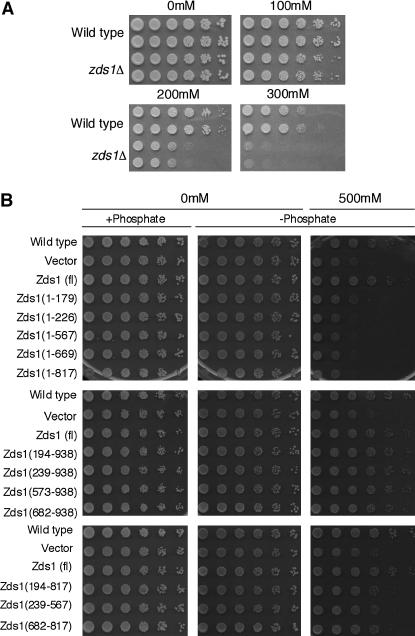
In S. cerevisiae, it has been reported that the zds1Δ mutation causes growth defects and results in an abnormal morphology when Ca2+ is present in the medium (Mizunuma et al. 1998). To test whether the S. pombe zds1Δ strain develops a similar phenotype, we cultured zds1Δ cells on YEA plates containing CaCl2. While zds1Δ cells grew as well as the wild-type cells in the absence of CaCl2, they grew more slowly in the presence of 100 mm CaCl2 (Figure 3A). Thus, like the S. cerevisiae zds1Δ mutant, S. pombe zds1Δ cells are sensitive to CaCl2.
Figure 3.
Calcium sensitivity of zds1-disrupted cells. (A) SP66 (h90, wild type) and MY6010 (h90, zds1Δ) cells were cultured at 30° in liquid medium until they reached log phase. They were concentrated to 2 × 107 cells/ml and then diluted sequentially fivefold (to the right direction). The cells were spotted on YEA plates containing 0–300 mm CaCl2 and incubated at 30° for 3 days (0 and 100 mm CaCl2), 4 days (200 mm CaCl2), or 7 days (300 mm CaCl2). (B) The C-terminal region of Zds1 suppresses the calcium sensitivity of zds1-disrupted cells. SP66/pSLF272L GFPS65A cells were spotted on the plate as a wild-type strain. All other strains were transformants of MY6010. All plasmids were derived from the vector pSLF272L GFPS65A. The transformants were cultured at 30° in liquid PMA medium containing thiamine until they reached log phase, after which they were washed to remove the thiamine and resuspended in H2O. The cell concentration was adjusted to 2 × 107 cells/ml and the cells were diluted sequentially fivefold (to the right direction). Cells were spotted on PMA (+phosphate), PMA containing sodium acetate instead of sodium phosphate (−phosphate; middle), or PMA (−phosphate; right) containing 500 mm CaCl2. These plates were incubated at 30° for 5 days (without CaCl2) or for 8 days (with CaCl2).
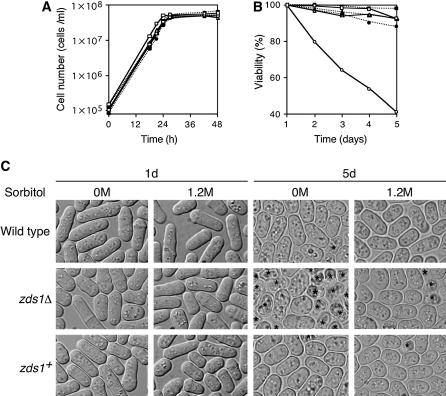
We next assessed the viability of zds1Δ cells. Cells of strains SP66 (h90, wild type) harboring pREP41, MY6010 (h90, zds1Δ) harboring pREP41, or pREP41–zds1 were incubated in liquid PMA medium at 30° for 5 days. We constructed growth curves for these three transformants, but there was no difference in growth ratio among them and they all reached the stationary phase at 24 hr (Figure 4A). Cells were further incubated for 4 days and their viabilities were measured. The viability of the zds1Δ cells harboring the vector alone decreased after incubation for 2–5 days (Figure 4B), while that of wild-type or zds1Δ cells that expressed zds1 on the plasmids did not. We then tested the suppressive effect of an osmotic stabilizer, sorbitol, on the viability loss of the zds1Δ mutant. The addition of 1.2 m sorbitol clearly suppressed the viability loss of the zds1Δ cells in the stationary phase (Figure 4B), indicating that zds1Δ has a defect in maintaining cell wall strength or overall cell rigidity. The zds1Δ cells were round after being cultured at 30° for 5 days in PMA (Figure 4C) but thereafter many dead cells were observed, which is presumably the cause of their low viability in the stationary phase. Notably, this morphological change was only partly suppressed by addition of 1.2 m sorbitol (Figure 4C).
Figure 4.
Low viability of zds1Δ in the stationary phase is suppressed under high osmotic condition. SP66 (h90, wild type)/pREP41, MY6010 (h90, zds1Δ)/pREP41, and MY6010/pREP41–zds1 (zds1+) were inoculated at a concentration of 1 × 105 cells/ml in PMA medium with or without 1.2 m sorbitol and were cultured at 30°. (A) Growth curve. Cell number was counted using the Cell Counter (Sysmex). (B) Viability in the stationary phase. Cells were cultured until the indicated days. Cells were stained with methylene blue and ∼1000 cells, including unstained cells (living cells) and stained cells (dead cells), were counted under a microscope. SP66/pREP41 (open square), MY6010/pREP41 (open circle), and MY6010/pREP41–zds1 (open triangle) were grown without sorbitol. SP66/pREP41 (solid square), MY6010/pREP41 (solid circle), and MY6010/pREP41–zds1 (solid triangle) were grown with 1.2 m sorbitol. (C) Cell morphology. Indicated cells were grown as in B for 1 day and 5 days and observed. Asterisks indicate dead cells.
Domain analysis of Zds1:
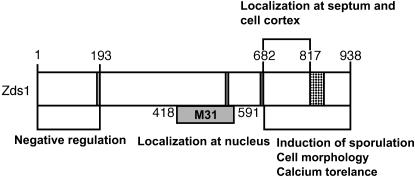
We next performed domain analysis of Zds1 to identify its functional domains. First, we searched for amino acid regions that are highly conserved between S. cerevisiae Zds1p, Zds2p, and S. pombe Zds1. We also assessed, by using the PSORT program (http://psort.nibb.ac.jp), whether some predicted signatures are present. On the basis of these analyses, Zds1 could be divided into five putative domains. Domain 1 (180–193) shares homology with Zds1p and Zds2p, domain 2 (227–238) contains a putative nuclear localization signal (NLS), domain 3 (568–572) shares homology with Zds2p, domain 4 (670–681) is a serine-rich region, and domain 5 (818–858) shares the highest homology with Zds1p and Zds2p. Domain 5 is presumed to be a functional domain because the same regions of S. cerevisiae Zds1p and Zds2p were determined to be functional domains (Schwer et al. 1998). After taking these five putative domains into account, 12 fragments containing different regions of Zds1 (Figure 5A) were amplified by PCR using the appropriate primer sets (Table 2). Those 12 fragments were inserted into the NotI site of pSLF172L GFPS65A or pSLF272L GFPS65A and the functions and subcellular localization of the resulting truncated Zds1 species were examined.
The C-terminal region of Zds1 is needed to suppress the Ca2+ sensitivity of zds1Δ cells:
We first investigated which region can suppress the CaCl2 sensitivity of zds1Δ. Thus, cells transformed with the various plasmids were spotted onto plates containing CaCl2. As Ca2+ reacts with phosphate to produce calcium phosphate, plates lacking phosphate and containing sodium acetate instead were prepared. All cells were grown on the plates lacking phosphate as well as on the plates containing phosphate (Figure 3B). When the cells were grown on plates containing 500 mm CaCl2, the zds1Δ cells did not grow well compared to wild-type cells (Figure 3B). However, zds1Δ cells expressing Zds1 (full length) grew as well as wild-type cells (Figure 3B). Although zds1Δ cells expressing Zds1ΔC or Zds1ΔNΔC did not grow even as well as zds1Δ cells, when they expressed Zds1ΔN, they grew as well as wild-type cells. Thus, we concluded that the CaCl2 sensitivity of zds1Δ cells is suppressed by the presence of the C-terminal region (682–938) that includes domain 5 of Zds1.
The N-terminal region of Zds1 is a negative regulatory region:
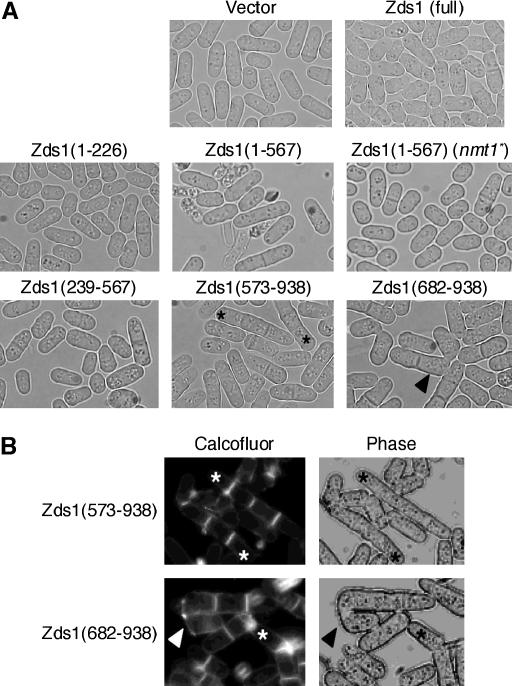
We next examined which region of Zds1 can suppress the abnormal morphology of zds1Δ cells. When full-length Zds1 was expressed in zds1Δ cells, the weakness of the zds1Δ cell wall was suppressed, although this suppression was incomplete (Figure 4). This incomplete suppression is probably due to the fact that high levels of Zds1 increased the number of ellipsoidal cells. Low-level expression of Zds1ΔC did not affect the morphology of zds1Δ cells [Figure 6A Zds1(1–567) (nmt1*)]. When Zds1(1–567), Zds1(1–669), or Zds1(1–817) were expressed at high levels, many dead cells appeared, even in the log phase (Figure 6A; data not shown). Dead cells were not observed if either Zds1(1–179) or Zds1(1–226) was expressed at high levels (Figure 6A; data not shown). Thus, the central region of Zds1 may have some function related to cell morphology. When Zds1ΔN was expressed in zds1Δ cells, multi-septated cells were observed (Figure 6A). Moreover, abnormal zygotes (that have the septum or are composed of more than three cells) were observed (Figure 6A). To observe multi-septated cells more clearly, they were stained with calcofluor. Multi-septated cells compose ∼11–12% of septated cells (Figure 6B). Calcofluor is believed to largely stain chitin, but apparently it also reacts with other epitopes since staining is observed in a chitin-defective mutant (Matsuo et al. 2004, 2005).
Figure 6.
The morphology of cells expressing part of Zds1. (A) Wild-type SP66 was transformed with various plasmids that contain various lengths of the zds1 gene. Each transformant was cultured in PMA medium at 30° for 24 hr. The strains tested were SP66 harboring pSLF172L GFPS65A, pSLF172L Zds1–GFP, pSLF172L Zds1(1–226)–GFP, pSLF172L Zds1(1–567)–GFP, pSLF272L Zds1(1–567)–GFP, pSLF172L Zds1(239–567)–GFP, pSLF172L Zds1(573–938)–GFP, and pSLF172L Zds1(682–938)–GFP. (B) MY6010 cells expressing Zds1(573–938) and Zds1(682–938) were stained with calcofluor white. Asterisks show multi-septated cells and arrowheads show the abnormal zygotes.
Expression of Zds1ΔN also increased mating efficiency. However, expression of Zds1ΔNΔC did not have any of these effects (Figure 6A). On the basis of these results, we believe that the C-terminal region (818–938) of Zds1 serves to maintain cell morphology and functions in the progression of sexual differentiation, while the N-terminal region (1–193) of Zds1 has a negative regulatory function. When the expression of these proteins was confirmed by Western blotting, Zds1 (full length) and Zds1ΔC appeared as two bands, while Zds1ΔN had only one band (Figure 5B). Since the N-terminal region (1–179) of Zds1 has many Ser residues, it is possible that this region is regulated by phosphorylation.
The C-terminal region of Zds1 is involved in sporulation:
We next investigated which region of Zds1 reverses the inability of ras1-deficient strains to sporulate. Thus, SPRN1DA (h90, ras1−/ras1−) strains that harbor the plasmids expressing various regions of Zds1 were grown on SPA plates to calculate sporulation efficiency. While Zds1ΔC-overexpressing SPRN1DA sporulated in a similar manner to the strain that harbored the vector (Table 4), Zds1ΔN-overexpressing SPRN1DA sporulated at a higher rate than the strain expressing full-length Zds1 (Table 4). This further supports the notion that the C-terminal region of Zds1 is involved in sporulation, while the N-terminal region of Zds1 has a negative regulatory function.
TABLE 4.
Sporulation efficiency of the truncated zds1 gene
| Plasmid | Sporulation rate (%) |
|---|---|
| pSLF172L GFP(S65A) | 0.3 |
| pSLF172L Zds1–GFP | 5.8 |
| pSLF172L Zds1(1–179)–GFP | 0.7 |
| pSLF172L Zds1(1–226)–GFP | 0.4 |
| pSLF172L Zds1(1–567)–GFP | 0.3 |
| pSLF172L Zds1(1–669)–GFP | 0.4 |
| pSLF172L Zds1(1–817)–GFP | 0.3 |
| pSLF172L Zds1(194–938)–GFP | 21.9 |
| pSLF172L Zds1(239–938)–GFP | 11.8 |
| pSLF172L Zds1(573–938)–GFP | 17.9 |
| pSLF172L Zds1(682–938)–GFP | 7.0 |
| pSLF172L Zds1(194–817)–GFP | 0.7 |
| pSLF172L Zds1(239–567)–GFP | 0.7 |
| pSLF172L Zds1(682–817)–GFP | 0.4 |
SPRN1DA (ras1−/ras1−) was transformed with the above plasmids, streaked onto SPA plate, and grown at 30° for 2 days. About 1000 cells were counted under the microscope.
Subcellular localization of Zds1:
We next observed the subcellular localization of Zds1(full length)–GFP and found that it localized to the cytoplasm, the cell cortex, and the septum (Figure 7A, c and d). The same Zds1 (full length)–GFP localization pattern (albeit with a weaker signal) was observed in the MY6013 strain, in which the genomic zds1 gene was fused with GFP in its C terminus (Figure 7C). Thus, the expression level does not affect the localization of Zds1–GFP (Figure 7C). Zds1(1–179)–GFP, Zds1(1–226)–GFP, Zds1(1–567)–GFP, and Zds1(1–669)–GFP were not able to localize to the septum or the cell cortex (Figure 7A, e and f; data not shown), but Zds1(1–817)–GFP was able to localize to these subregions (Figure 7A, g and h). In contrast, all Zds1ΔN–GFP proteins were able to localize to the septum and the cell cortex (Figure 7A, i–n). These results indicate that the C-terminal region ranging from residues 682–817 is important for the localization of Zds1 in both the septum and the cell cortex. In fact, Zds1(682–817)–GFP localized to the septum, the cell cortex, and the nucleus (mainly the nucleolus) (Figure 7A, q and r).
It has been reported previously, on the basis of random observations of fused GFP, that the 418- to 591-amino-acid region of Zds1 (called the M31 region) localizes to the nucleus (Sawin and Nurse 1996). Indeed, we found that although full-length Zds1 is not nuclearly localized (Figure 7A, d), Zds1(194–817)–GFP and Zds1(239–567)–GFP are (Figure 7A, p). Intriguingly, however, Zds1(239–938)–GFP, which contains the M31 region, does not localize to the nucleus. In addition, Zds1(1–567)–GFP, Zds1(1–669)–GFP, Zds1(1–817)–GFP, and Zds1(194–938)–GFP, which have both the NLS and the M31 region (full or in part) also do not localize to the nucleus. Collectively, all these results indicate that the putative NLS is not required and that deletion of the 1–193 and the 818–938 region is required for the nuclear localization of Zds1. The significance of the nuclear localization of part of Zds1 is not clear at this moment. The fluorescence of Zds1(194–938)–GFP, Zds1(573–938)–GFP and Zds1(682–938)–GFP formed large round bright spots (Figure 7A, j and n; data not shown). The Zds1 (573–938)–GFP spots differed significantly from the DAPI-stained nuclei and also differed from FM4-64-stained vacuoles (Figure 7B).
Thus, Zds1 localizes to many cellular compartments, including the cytoplasm, the cell cortex, the nucleus, and the septum, when part of it is expressed. Since the various functions of Zds1 seem to be separable, as shown in detail below, it may be that the localizations and functions of the Zds1 regions correlate with each other.
Susceptibility of the zds1Δ- and zds1-overexpressing strains to zymolyase:
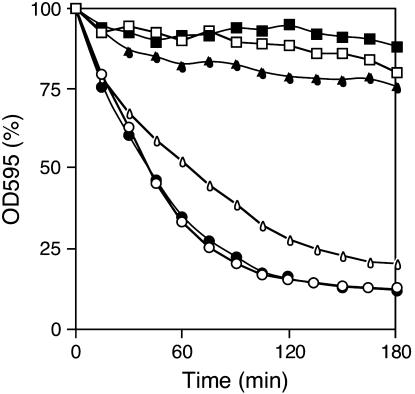
Since the zds1Δ cell colonies became shiny and dead cells were observed more frequently than with wild-type cells, and the viability of zds1Δ cells decreased in the stationary phase, we suspected that zds1Δ cell walls may be weak. We thus tested the zymolyase susceptibility of the zds1 disruptant. Wild-type and zds1Δ cells were incubated with 0.1 mg/ml zymolyase 20T for 180 min and their degree of lysis was evaluated by examining the absorbance at OD595. The OD595 of the wild-type cells was 80% that of the OD595 of treated cells after 180 min incubation with zymolyase. In contrast, the OD595 of zds1Δ cells decreased to ∼60% after 30 min and subsequently to ∼10% after 180 min (Figure 8).
Figure 8.
Susceptibility of the zds1Δ strain to zymolyase. Cells were cultured in PMA medium at 30° to a density of 1 × 107 cells/ml and then harvested and resuspended in TE (pH 8.0). Cells were incubated with 0.1 mg/ml zymolyase 20T at 30° for the indicated time. SP66/pREP41 (open square), MY6010/pREP41 (open circle), and MY6010/pREP41–zds1 (open triangle) were grown in the presence of thiamine. SP66/pREP41 (solid square), MY6010/pREP41 (solid circle), and MY6010/pREP41-zds1 (solid triangle) were grown without thiamine.
Similarly, when the zds1-disrupted MY6010 cells harboring pREP41–zds1 were cultured with thiamine (which represses the promoter driving the zds1 gene), the OD595 decreased at the same rate as was observed with the zds1Δ cells. However, when MY6010 cells harboring pREP41–zds1 were cultured without thiamine (which induces zds1 expression), the OD595 was similar to that of wild-type cells. Thus, zds1 deficiency enhances the ability of zymolyase to lyse the cell wall, indicating that zds1 is needed to maintain cell wall strength. We then tested cells bearing plasmids harboring various zds1Δ deletion mutants for their susceptibility to zymolyase. Whereas zds1Δ cells expressing full-length Zds1 did not lyse, zds1Δ cells expressing Zds1ΔC [Zds1(1–817)], Zds1ΔN [Zds1(194–938)–GFP], or Zds1ΔNΔC [Zds1(194–817)] lysed in a manner similar to zds1Δ cells (data not shown). That only full-length Zds1 suppresses the zymolyase sensitivity of zds1Δ indicates that both the N- and C-terminal regions of Zds1 are required to suppress the zymolyase sensitivity of zds1Δ.
Electron microscopic observation of zds1Δ cells:
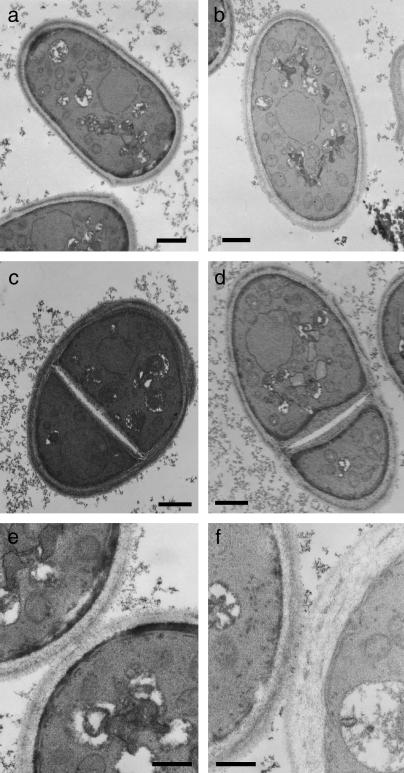
As mentioned above, zds1Δ cells show a round morphology when they are cultured for a long time in minimal medium, and the percentage of dead cells becomes high. We thus cultured wild-type cells and zds1Δ cells in PMAL medium at 30° for 72 hr and observed them by transmission electron microscopy (TEM) to determine whether they have an abnormal cellular structure. The cell wall of zds1Δ cells was a little thicker than that of wild-type cells (Figure 9, a and b). The septum of zds1Δ cells was also thicker than that of wild-type cells; in particular, the thickness of the secondary septum was irregular (Figure 9, c and d). The secondary septa of wild-type and zds1Δ cells were 151 and 275 nm on average, respectively. Cell wall thickness was also observed in zds1Δ cells immediately after their separation (Figure 9f). The dead cells of zds1Δ also tend to have thicker cell walls (data not shown). These observations indicate that Zds1 is involved in proper cell wall formation. This finding is supported by the observations regarding the zymolyase sensitivity of zds1Δ.
Figure 9.
Electron micrographs of zds1-disrupted cells. Wild-type (SP66) and zds1-disrupted cells were cultured in PMAL medium at 30° for 72 hr and observed by TEM. Wild-type cells (a, c, and e), zds1-disrupted cells (b, d, and f), and septating cells of wild-type (c and e) and zds1-disrupted cells (d and f) were observed. The cells were observed at a magnification of 5000 (a and b), 6500 (c and d), and 15,000 (e and f). Bars, 1 μm in a, b, c, and d; 500 nm in e and f.
DISCUSSION
Zds1 is involved in sexual differentiation and Ca2+ tolerance:
We identified a novel gene, which we named zds1, as a suppressor of the sterility caused by the deletion of ras1 in a diploid strain. The zds1 gene suppressed the sterile phenotype of ras1− but not that of the other disruptants involved in the Ras1–MAPK signaling pathway, including byr2, byr1, and spk1. This suggests that Zds1 is positioned upstream of Byr2. Zds1 shares ∼30% identity with Zds1p and Zds2p of S. cerevisiae, and its C-terminal region shows particularly strong homology with the C-terminal regions of Zds1p and Zds2p (Figure 1). This C-terminal region is not only a functional domain of S. cerevisiae Zds1p and Zds2p (Schwer et al. 1998), but also, as shown in this study, a functional domain of S. pombe Zds1. Notably, overexpression of the C-terminal region of Zds1 induced the ras1− diploid strain to sporulate at a much higher rate than when full-length Zds1 was overexpressed (Table 4). This suggests that the N-terminal region of Zds1 negatively regulates its C-terminal functional domain. Consistent with this suggestion, the sterile phenotype caused by mutation in the cgs1 gene, which encodes a regulatory subunit of cAMP-dependent kinase (Maeda et al. 1994), was also partly reverted by overexpression of the C-terminal region of Zds1, but not by the full-length Zds1 (data not shown). This result also suggested that Zds1 does not regulate the upstream component of the cAMP pathway but, rather, regulates the Ras1–MAPK signaling pathway. Localization of the C-terminal region of Zds1 in the cell cortex also indicated that Zds1 functions in the signaling of the Ras–MAPK pathway at the cell cortex. It should be noted that while Zds1 is involved in the sexual differentiation of S. pombe, it has not been shown that Zds1p and Zds2p are similarly involved in the sexual differentiation of S. cerevisiae. This apparent difference might reflect distinct Ras functions in these two yeasts. The Ras protein is involved in sexual differentiation of S. pombe whereas the two Ras proteins are involved in cell growth of S. cerevisiae (Marcus et al. 1993).
According to data collected by the Sanger Institute (http://www.sanger.ac.uk/Projects/S_pombe/), the mRNA expression level of Zds1 (SPAC31F12.01) is constant during the cell cycle but is highly induced during meiosis I. This is consistent with our observation that Zds1 is involved in sexual differentiation. Consistent with this hypothesis, we observed that Zds1–GFP is localized to the surface of spores at the earlier stage of spore formation (data not shown).
Disruption of the zds1 gene resulted in calcium sensitivity, similar to that observed for the S. cerevisiae zds1Δ strain (Mizunuma et al. 1998). It was reported that in S. cerevisiae, Zds1p negatively regulates Swe1p (a negative regulator of Cdc28p), which is activated by Ca2+ (Mizunuma et al. 1998). In S. cerevisiae zds1Δ cells, Swe1p was activated to induce G2 delay in the presence of Ca2+. As a result, S. cerevisiae zds1Δ cells showed a growth defect and formed elongated buds (Mizunuma et al. 1998). In the zds1Δ cells of S. pombe, exogenous Ca2+ also induced growth inhibition (Figure 3A) and increased cell mass (data not shown) in a manner similar to that of S. cerevisiae Zds1. We also showed that this CaCl2 sensitivity of zds1Δ cells was suppressed by the presence of the C-terminal region of the protein. Thus, the C-terminal region is a functional domain involved not only in the induction of sexual differentiation, but also in Ca2+ tolerance. However, we do not know how these two seemingly unrelated phenotypes are interconnected through the function of Zds1.
Zds1 is involved in cell wall strength and morphology:
zds1Δ cells showed round-cell morphology (Figure 4) and electron microscopy indicated that they have thicker cell walls than wild-type cells (Figure 9). This became clearer when cells entered the stationary phase. This may be because polarized growth slows down in the stationary phase, which may enhance the abnormal cell morphology caused by zds1 deletion. The septum, in particular the secondary septum, of zds1Δ cells was irregular (Figure 9). This correlates with the zymolyase sensitivity that these cells exhibit (Figure 8) and with their loss in viability in the stationary phase (Figure 4). Thus, these results suggested that cell wall integrity was impaired by zds1 deletion. We speculated that as the cell wall became thicker, the density of cell wall contents might be reduced so that zymolyase can attack glucan more easily. A similar observation was made with the rho4 mutant, whose cell wall is thicker and more sensitive to glucanase (Nakano et al. 2003). Notably, full-length Zds1 is needed to suppress the round-cell morphology and zymolyase sensitivity of zds1Δ cells. This reflects the importance of the whole structure of Zds1, but it is in contrast to the observation that the functional domain required for sexual differentiation and Ca2+ sensitivity is limited to the C terminus of Zds1. The requirement for the whole structure of Zds1 in maintaining cell wall integrity suggested that the N-terminal domain does not solely exist as a negative regulator of the C-terminal functional domain. Thus, the N-terminal domain might be necessary to keep Zds1 in a certain conformation or to interact with other proteins.
Domain analysis of Zds1 and its functional correlation:
Our Zds1 domain analysis and the correlating localizations of these domains are summarized in Figure 10. Thus, full-length Zds1–GFP localizes to the cytosol, the septum, and the cell cortex, but is not found in the nucleus. Localization of Zds1–GFP in the cell cortex presumably relates to the involvement of Zds1 in cell morphology and/or cell wall integrity. Localization of Zds1–GFP in the septum presumably relates to its role in cell separation as we observed that overexpression of truncated Zds1 induced multi-septated cells (Figure 6). The 682–817 region of Zds1 is required for localization to the septum and the cell cortex (Figure 7A). It was previously shown that the M31 region of Zds1 (residues 418–591) localizes to the nucleus (Sawin and Nurse 1996) and we found that this nuclear localization required the deletion of both the N- and C-terminal regions (Figure 7A). However, the significance of this observation is not yet clear. Intriguingly, Zds1 localizes to the bud neck, bud tip, and cytoplasm while Zds2p localizes to the nucleus of S. cerevisiae (Bi and Pringle 1996). Since the Zds1p and Zds2p proteins display some common roles, such as the suppression of various mutants (Bi and Pringle 1996; Heo et al. 1999; Schwer and Shuman 1996; Griffioen et al. 2001; Bandhakavi et al. 2003) but also have distinct roles, such as in silencing (Roy and Runge 2000), it is plausible that the different localization patterns of S. cerevisiae Zds1p and Zds2p may reflect their different functions. Thus, the disparate localizations of the truncated S. pombe Zds1 proteins may reflect various functions of the protein that we do not yet understand. The central region of Zds1 does not show homology to Zds1p and Zds2p or to other proteins, and overexpression of this region had no effect on the morphological change and Ca2+ sensitivity of zds1Δ cells or the localization of the protein (Figure 3B, Figure 6, and Figure 7). Overexpression of Zds1(1–179) or Zds1(1–226) also had no effect on these functions. However, overexpression of Zds1(1–567), Zds1(1–669), or Zds1(1–817) did induce many dead cells in the log phase (Figure 6). Thus, while the central region of Zds1 may not have any obvious function on its own, it may be important in the sense that it connects the N terminus to the C terminus.
Figure 10.
Summary of the Zds1 domain analysis conducted in this study. The N-terminal region serves to inhibit the functional domain of Zds1, which is the C-terminal region. The latter domain induces sexual development and calcium tolerance. The role of the central region is obscure but, interestingly, its sole expression induces nuclear localization.
In Western blot analysis, full-length Zds1–GFP and Zds1ΔC–GFP appeared as two bands whereas Zds1ΔN appeared as only one band (Figure 5B). This suggests that Zds1 is modified at its N-terminal region. Since the N-terminal region has many Ser residues (Figure 1), it may be modified by phosphorylation.
Differences between S. pombe Zds1 and S. cerevisiae Zds1p and Zds2p:
S. pombe Zds1 and S. cerevisiae Zds1p and Zds2p have roles in common as well as some more specific functions. For example, both S. pombe zds1Δ and S. cerevisiae zds1Δ cells show calcium sensitivity (Figure 3) (Mizunuma et al. 1998) while S. pombe zds1Δ and S. cerevisiae zds1Δzds2Δ cells show cold sensitivity (Schwer et al. 1998). However, while Zds1 is involved in sexual differentiation (Tables 3 and 4), the involvement of S. cerevisiae Zds1p or Zds2p in sexual differentiation has not been observed. Moreover, Zds1 acts as a positive regulator in cell morphology while S. cerevisiae Zds1 and Zds2 act as negative regulators (Bi and Pringle 1996). These differences may reflect different systems of growth and cell division in S. pombe and S. cerevisiae. ZDS homologs can be found in related species of yeasts but not in animals and plants. Since ZDS homologs exist only in organisms that have glucan, on the basis of our observations (Figure 8) and reports in S. cerevisiae (Sekiya-Kawasaki et al. 2002), we suspect that participation of zds1 and related genes in glucan metabolism is probably of prime importance.
Acknowledgments
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Alfa, C., P. Fantes, J. Hyams, M. McLeod and E. Warbrick, 1993. Experiments With Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bandhakavi, S., R. O. McCann, D. E. Hanna and C. V. Glover, 2003. Genetic interactions among ZDS1,2, CDC37, and protein kinase CK2 in Saccharomyces cerevisiae. FEBS Lett. 554: 295–300. [DOI] [PubMed] [Google Scholar]
- Barr, M. M., H. Tu, L. Van Aelst and M. Wigler., 1996. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol. Cell. Biol. 16: 5597–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi, G., E. K. Schmid and Maundrell, 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136. [DOI] [PubMed] [Google Scholar]
- Bi, E., and J. R. Pringle, 1996. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger, H., A. Capello, P. W. Schenk, G. Stoter, J. Brouwer et al., 2000. A genome-wide screening in Saccharomyces cerevisiae for genes that confer resistance to the anticancer agent cisplatin. Biochem. Biophys. Res. Commun. 269: 767–774. [DOI] [PubMed] [Google Scholar]
- Chang, E. C., M. Barr, Y. Wang, V. Jung, H. P. Xu et al., 1994. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell 79: 131–141. [DOI] [PubMed] [Google Scholar]
- DeVoti, J., G. Seydoux, D. Beach and M. McLeod, 1991. Interaction between ran1+ protein kinase and cAMP dependent protein kinase as negative regulators of fission yeast meiosis. EMBO J. 10: 3759–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, D. D., J. Zhu, D. L. Carlson, K. Sharma and J. A. Huberman, 1994. Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J. 13: 3638–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo, M., M. Shirouzu and S. Yokoyama, 2003. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 278: 843–852. [DOI] [PubMed] [Google Scholar]
- Forsburg, S. L., and D. A. Sherman, 1997. General purpose tagging vectors for fission yeast. Gene 191: 191–195. [DOI] [PubMed] [Google Scholar]
- Goldar, M. M., T. Nishie, Y. Ishikura, T. Fukuda, K. Takegawa et al., 2005. a Functional conservation between fission yeast moc1/ds23 and its two orthologs, budding yeast SDS23 and SDS24, and phenotypic differences in their disruptants. Biosci. Biotechnol. Biochem. 69: 1422–1426. [DOI] [PubMed] [Google Scholar]
- Goldar, M. M., H. T. Jeong, K. Tanaka, H. Matsuda and M. Kawamukai, 2005. b Moc3, a novel Zn finger type protein involved in sexual development, ascus formation, and stress response of Schizosaccharomyces pombe. Curr. Genet. 48: 345–355. [DOI] [PubMed] [Google Scholar]
- Griffioen, G., P. Branduardi, A. Ballarini, P. Anghileri, J. Norbeck et al., 2001. Nucleocytoplasmic distribution of budding yeast protein kinase A regulatory subunit Bcy1 requires Zds1 and is regulated by Yak1-dependent phosphorylation of its targeting domain. Mol. Cell. Biol. 21: 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo, S. J., K. Tatebayashi and H. Ikeda, 1999. The budding yeast cohesin gene SCC1/MCD1/RHC21 genetically interacts with PKA, CDK and APC. Curr. Genet. 36: 329–338. [DOI] [PubMed] [Google Scholar]
- Higuchi, T., Y. Watanabe and M. Yamamoto, 2002. Protein kinase A regulates sexual differentiation and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol. 22: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki, T., N. Mochizuki, T. Maeda and M. Yamamoto, 1992. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 6: 2455–2462. [DOI] [PubMed] [Google Scholar]
- Jeong, H. T., F. Ozoe, K. Tanaka, T. Nakagawa, H. Matsuda et al., 2004. a A novel gene, msa1, inhibits sexual differentiation in Schizosaccharomyces pombe. Genetics 167: 77–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H. T., Y. Oowatari, M. Abe, K. Tanaka, H. Matsuda et al., 2004. b A negative regulator (Msa2/Nrd1) interacts with a positive regulator (Cpc2) of sexual differentiation in Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68: 1489–1499. [DOI] [PubMed] [Google Scholar]
- Katayama, S., F. Ozoe, R. Kurokawa, K. Tanaka, T. Nakagawa et al., 1996. Genetic analysis of the sam mutations, which induce sexual development with no requirement for nutritional starvation in fission yeast. Biosci. Biotechnol. Biochem. 60: 994–999. [DOI] [PubMed] [Google Scholar]
- Kawamukai, M., 1999. Isolation of a novel gene, moc2, encoding a putative RNA helicase as a suppressor of sterile strains in Schizosaccharomyces pombe. Biochim. Biophys. Acta 1446: 93–101. [DOI] [PubMed] [Google Scholar]
- Kawamukai, M., K. Ferguson, M. Wigler and D. Young, 1991. Genetic and biochemical analysis of the adenylyl cyclase of Schizosaccharomyces pombe. Cell. Regul. 2: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai, M., J. Gerst, J. Field, M. Riggs, L. Rodgers et al., 1992. Genetic and biochemical analysis of the adenylyl cyclase-associated protein, cap, in Schizosaccharomyces pombe. Mol. Biol. Cell 3: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., and M. McLeod, 1996. Molecular mimicry in development: identification of ste11+ as a substrate and mei3+ as a pseudosubstrate inhibitor of ran1+ kinase. Cell 87: 869–880. [DOI] [PubMed] [Google Scholar]
- Ma, X. J., Q. Lu and M. Grunstein, 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10: 1327–1340. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Y. Watanabe, H. Kunitomo and M. Yamamoto, 1994. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 269: 9632–9637. [PubMed] [Google Scholar]
- Marcus, S., M. Wigler, H. P. Xu, R. Ballester, M. Kawamukai et al., 1993. RAS function and protein kinase cascades. Ciba Found. Symp. 176: 53–61. [DOI] [PubMed] [Google Scholar]
- Marcus, S., A. Polverino, E. Chang, D. Robbins, M. H. Cobb et al., 1995. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 92: 6180–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, Y., K. Tanaka, T. Nakagawa, H. Matsuda and M. Kawamukai, 2004. Genetic analysis of chs1+ and chs2+ encoding chitin synthases from Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 68: 1489–1499. [DOI] [PubMed] [Google Scholar]
- Matsuo, Y., K. Tanaka, H. Matsuda and M. Kawamukai, 2005. cda1+, encoding chitin deacetylase is required for proper spore formation in Schizosaccharomyces pombe. FEBS Lett. 579: 2737–2743. [DOI] [PubMed] [Google Scholar]
- Maundrell, K., 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265: 10857–10864. [PubMed] [Google Scholar]
- Mizunuma, M., D. Hirata, K. Miyahara, E. Tsuchiya and T. Miyakawa, 1998. Role of calcineurin and Mpk1 in regulating the onset of mitosis in budding yeast. Nature 392: 303–306. [DOI] [PubMed] [Google Scholar]
- Nakano, K., T. Mutoh, R. Arai and I. Mabuchi, 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells 8: 357–370. [DOI] [PubMed] [Google Scholar]
- Obara, T., M. Nakafuku, M. Yamamoto and Y. Kaziro, 1991. Isolation and characterization of a gene encoding a G-protein α subunit from Schizosaccharomyces pombe: involvement in mating and sporulation pathways. Proc. Natl. Acad. Sci. USA 88: 5877–5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottilie, S., P. J. Miller, D. I. Johnson, C. L. Creasy, M. A. Sells et al., 1995. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 14: 5908–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoe, F., R. Kurokawa, Y. Kobayashi, H. T. Jeong, K. Tanaka et al., 2002. The 14–3-3 proteins Rad24 and Rad25 negatively regulate Byr2 by affecting its localization in Schizosaccharomyces pombe. Mol. Cell. Biol. 22: 7105–7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, N., and K. W. Runge, 1999. The ZDS1 and ZDS2 proteins require the Sir3p component of yeast silent chromatin to enhance the stability of short linear centromeric plasmids. Chromosoma 108: 146–161. [DOI] [PubMed] [Google Scholar]
- Roy, N., and K. W. Runge, 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10: 111–114. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sawin, K. E., and P. Nurse, 1996. Identification of fission yeast nuclear markers using random polypeptide fusions with green fluorescent protein. Proc. Natl. Acad. Sci. USA 94: 15146–15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., and S. Shuman, 1996. Multicopy suppressors of temperature-sensitive mutations of yeast mRNA capping enzyme. Gene Expr. 5: 331–344. [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., P. Linder and S. Shuman, 1998. Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res. 26: 803–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono et al., 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-β-glucan synthase, in Saccharomyces cerevisiae. Genetics 162: 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, A., Y. Iino, T. Maeda, Y. Watanabe and M. Yamamoto, 1991. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes Dev. 5: 1990–1999. [DOI] [PubMed] [Google Scholar]
- Takahashi, K., S. Murakami, Y. Chikashige, H. Funabiki, O. Niwa et al., 1992. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol. Biol. Cell 3: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, T., T. Toda, K. Kominami, A. Kohnosu, M. Yanagida et al., 1995. Schizosaccharomyces pombe atf1+ encodes a transcription factor required for sexual development and entry into stationary phase. EMBO J. 14: 6193–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, K., N. Ito, T. Wakuri, F. Ozoe, M. Umeda et al., 2003. Sla1, a Schizosaccharomyces pombe homolog of the human La protein, induces ectopic meiosis when its C-terminus is truncated. Eukaryot. Cell 2: 1274–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, K., K. Tanaka, H. Matsuda and M. Kawamukai, 2004. Truncated Sla1 induces haploid meiosis through the Pat1-Mei2 system in fission yeast. Biosci. Biotechnol. Biochem. 68: 226–270. [DOI] [PubMed] [Google Scholar]
- Toda, T., M. Shimanuki and M. Yanagida, 1991. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 5: 60–73. [DOI] [PubMed] [Google Scholar]
- Toone, W. M., and N. Jones, 2003. Stress responses in S. pombe, pp. 57–72 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Tsuchiya, E., G. Matsuzaki, K. Kurano, T. Fukuchi, A. Tsukao et al., 1996. The Saccharomyces cerevisiae SSD1 gene is involved in the tolerance to high concentration of Ca2+ with the participation of HST1/NRC1/BFR1. Gene 176: 35–38. [DOI] [PubMed] [Google Scholar]
- Tu, H., M. Barr, D. L. Dong and M. Wigler, 1997. Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol. 17: 5876–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walowsky, C., D. J. Fitzhugh, I. B. Castaño, J. Y. Ju, N. A. Levin et al., 1999. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J. Biol. Chem. 274: 7302–7308. [DOI] [PubMed] [Google Scholar]
- Wang, Y., H. P. Xu, M. Riggs, L. Rodgers and M Wigler, 1991. byr2, a Schizosaccharomyces pombe gene encoding a protein kinase capable of partial suppression of the ras1 mutant phenotype. Mol. Cell. Biol. 11: 3554–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., S. Shinozaki-Yabana, Y. Chikashige, Y. Hiraoka and M. Yamamoto, 1997. Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386: 187–190. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M., 2003. Initiation of meiosis, pp. 297–309 in The Molecular Biology of Schizosaccharomyces pombe, edited by R. Egel. Springer-Verlag, Berlin.
- Yamamoto, M., Y. Imai and Y. Watanabe, 1997. Mating and sporulation in Schizosaccharomyces pombe, pp. 1037–1106 in The Molecular and Cellular Biology of the Yeast Saccharomyces, Vol. 3, edited by J. R. Pringle, J. R. Broach and E. W. Jones. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Yamamoto, T., Y. Chikashige, F. Ozoe, M. Kawamukai and Y. Hiraoka, 2004. Activation of the pheromone-responsive MAP kinase drives haploid cells to ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J. Cell Sci. 11: 3875–3886. [DOI] [PubMed] [Google Scholar]
- Yu, Y., Y. W. Jiang, R. J. Wellinger, K. Carlson, J. M. Roberts et al., 1996. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 16: 5254–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]