Abstract
Hsp70's are highly conserved essential protein chaperones that assist protein folding and prevent protein aggregation. They have modular structures consisting of ATPase, substrate-binding, and C-terminal domains. Substrate binding and release is regulated by ATP hydrolysis and nucleotide exchange, which in turn are regulated by cochaperones. Eukaryotes have constitutive (Hsc70) and stress-inducible (iHsp70) isoforms, but their functions have not been systematically compared. Using a yeast system to evaluate heterologous Hsp70's we find that primate Hsc70 supported growth but iHsp70 did not. Plant Hsc70 and iHsp70 counterparts behaved similarly, implying evolutionary conservation of this distinction. Swapping yeast and primate Hsp70 domains showed that (i) the Hsc70–iHsp70 distinction resided in the ATPase domain, (ii) substrate-binding domains of Hsp70's within and across species functioned similarly regarding growth, (iii) C-terminal domain function was important for growth, and (iv) Hsp70 functions important for cell growth and prion propagation were separable. Enzymatic analysis uncovered a correlation between substrate affinity and prion phenotype and showed that ATPase and protein-folding activities were generally similar. Our data support a view that intrinsic activities of Hsp70 isoforms are comparable, and functional differences in vivo lie mainly in complex interactions of Hsp70 with cochaperones.
HSP70 is a ubiquitous essential protein chaperone that assists protein folding and is involved in protein synthesis, protein transport across membranes, and dynamics of macromolecular assemblies such as microtubules and clathrin (Gao et al. 1991; Nelson et al. 1992; McClellan et al. 1998; Oka et al. 1998). Hsp70 also protects cells from stress by preventing protein aggregation and helping refold denatured proteins (see Mayer et al. 2001; Hartl and Hayer-Hartl 2002 for general reviews). Hsp70 has a high affinity for extended hydrophobic peptides and its diverse functions depend upon ATP-regulated binding and release of exposed hydrophobic surfaces on proteins.
Hsp70's have a modular structure consisting of a highly conserved amino-terminal ATPase domain, an adjacent well-conserved substrate-binding domain (SBD) that contains a hydrophobic pocket and a lid-like structure over the pocket, and a conserved but more variable C-terminal domain (CTD). In the ATP-bound state, the substrate-binding pocket is open and rapidly exchanges substrate. ATP hydrolysis induces closing of the the lid over the substrate-binding pocket, which stabilizes substrate binding. Return to the ATP-bound state restores the open conformation, facilitating substrate release. Intrinsic ATPase activity among Hsp70's is weak, and ATP hydrolysis is the rate-limiting step in the reaction cycle (Schmid et al. 1994; McCarty et al. 1995).
Fine tuning of this cycle is regulated by cochaperones. Hsp40's, universal partners of Hsp70's, stimulate ATP hydrolysis and coordinate it with substrate binding (Greene et al. 1998; Laufen et al. 1999; Wittung-Stafshede et al. 2003). Nucleotide exchange factors facilitate ADP release and subsequent rebinding of ATP (Kabani et al. 2002; Fan et al. 2003; Harrison 2003). In addition to regulating Hsp70 activity, cochaperones also recruit Hsp70 to various subcellular locations or protein complexes that require its chaperoning activity (Young et al. 2003). The CTD of many eukaryotic cytosolic Hsp70's contains a conserved octapeptide that mediates interactions with a variety of cochaperones, including Hsp40's and others that contain tetratricopeptide repeat (TPR) protein interaction motifs (Freeman et al. 1995; Duina et al. 1996; Marsh et al. 1998; Scheufler et al. 2000). TPR cochaperones have been shown to stimulate ATPase of yeast cytosolic Hsp70 (Fan et al. 2003; Wegele et al. 2003b; Hainzl et al. 2004).
Although the key features of Hsp70's reaction cycle are highly conserved, different isoforms have evolved to perform specialized functions, such as the yeast Ssb subfamily that associates with ribosomes and assists translation (Nelson et al. 1992). There is a universal dependency on Hsp70 chaperone function in cellular processes during which proteins are incompletely folded, and while several Hsp70 isoforms are found in the cytosol, others reside in the endoplasmic reticulum and mitchondria. Among the cytosolic isoforms, some are constitutively expressed (Hsc70's) whereas others (iHsp70's) are expressed when cells are exposed to stress. Under conditions of stress the increased abundance of Hsp70 protects cells by binding exposed hydrophobic surfaces on partially unfolded proteins and preventing nonproductive interactions that lead to protein aggregation. Feedback regulation of iHsp70 expression helps maintain overall Hsp70 abundance at a level appropriate for the need of its chaperoning function (Craig and Gross 1991; Rieger et al. 2005).
Since the structure and enzymatic activities among Hsp70's are similar and the various isoforms differ mainly in how their expression is regulated, it is generally thought that induction of iHsp70 expression is a response to protect cells from protein misfolding problems by increasing the overall amount of Hsp70. It is also possible, however, that a specialized Hsp70 activity of inducible isoforms is important under adverse conditions. In line with this notion, a recent study showing that iHsp70 was specifically required for growth of cancer cells suggests that iHsp70 can function differently from Hsc70 (Rohde et al. 2005). Nonetheless, although there is considerable interest in Hsp70, there is very little understanding of differences in function of cytosolic Hsp70 isoforms.
The lack of a useful in vivo system has limited our ability to compare functions of Hsp70 isoforms within or across eukaryotic species. To compare and dissect functions of Hsp70's we developed a yeast system that can be used to assess functional complementation by any Hsp70 isoform regarding both growth and prion propagation. Using this system we identified and characterized functional differences among Hsp70's that provide new insight into Hsp70 function.
MATERIALS AND METHODS
Yeast strain, plasmids, and growth conditions:
Strain G400-1C (MATa kar1-1 SUQ5 ade2-1 his3Δ202 leu2Δ1 trp1Δ63 ura3-52 ssa1∷KanMx ssa2∷HIS3 ssa3∷TRP1 ssa4∷ura3-f1/pRDW10) (Jones and Masison 2003) was used. Plasmids pRDW10 (Jung et al. 2000) and pN1 (Schwimmer and Masison 2002) are single-copy URA3- and LEU2-based vectors, respectively, with the SSA1 coding region and 500 bp of 5′- and 3′-flanking DNA. Primate HSC70 (Saguinus oedipus, the New World monkey cotton-top tamarin, GenBank accession no. AF142571) and iHSP70 (AF142572) (Wu et al. 2000) were PCR amplified using primers with 5′ NdeI and SphI sites and products were used to replace SSA1 from the initiator ATG to a site 280 bp 3′ of the translation terminator in pN1, creating pCCC and pPPP, respectively. Arabidopsis thaliana iHSP70 (AJ002551) was cloned by RT-PCR from mRNA prepared from heat-shocked plants, provided by Charles Guy (Sung et al. 2001), and cloned similarly, creating pAtP70. A. thaliana HSC70-1 (X74604) was amplified from cDNA and cloned similarly, creating pAtC70. Amino acid identities of Hsp70 isoforms used in this study are shown in Table 1.
TABLE 1.
Percentage of amino acid identity of Hsp70 isoforms
|
S. oedipus
|
A. thaliana
|
|||
|---|---|---|---|---|
| Hsp70 isoform | Hsc70 | iHsp70 | Hsc70 | iHsp70 |
| Ssa1p | 75 | 70 | 71 | 71 |
| S.o. Hsc70 | 77 | 76 | 76 | |
| S.o. iHsp70 | 70 | 69 | ||
| A.t. Hsc70 | 90 | |||
S. oedipus is the New World monkey cotton-top tamarin. S.o. Hsc70 is identical to human Hsc70 (Hsp70-8); S.o. iHsp70 is 96% identical to human iHsp70 (Hsp70-6).
A. thaliana is the plant thale cress. S.o., S. oedipus. A.t., A. thaliana.
Hybrid genes were constructed by first subcloning BamHI-SphI fragments from plasmids pN1, pCCC, and pPPP into plasmid pGEM-T (Promega, Madison, WI). The Quick-Change kit (Stratagene, La Jolla, CA) was used to make site-specific nucleotide mutations without altering peptide coding to introduce XbaI and XhoI sites at conserved codons corresponding to L391 and E540, respectively, of SSA1. After swapping NdeI–XbaI (ATPase domain), XbaI–XhoI (SBD), and XhoI–SphI (CTD) fragments, hybrid genes were returned to pN1 as BamHI–SphI fragments. L391 is at the end of a conserved stretch of leucine residues between the ATPase and peptide-binding domains. E540 is at a putative hinge point on the helix that extends across the substrate-binding pocket (James et al. 1997). Structures of all alleles amplified by PCR were confirmed by sequencing.
Rich medium (YPAD) contains 1% yeast extract, 2% peptone, 400 mg/liter adenine, and 2% glucose. 1/2YPD, which is limiting for adenine and enhances red color of [psi−] cells, is similar but lacks supplemented adenine and contains 0.5% yeast extract. Synthetic media were as described (Sherman 1994). Counterselection medium containing 5-fluoro-orotic acid (5-FOA) was used as described (Boeke et al. 1984). Briefly, G400-1C cells transformed by plasmids carrying Hsp70 isoforms and the LEU2 selectable marker were grown on medium lacking leucine and containing uracil, which relieves selective pressure for maintenance of the URA3 marker on pRDW10. Transformants were then transferred to medium containing 5-FOA, which kills cells that express URA3. Only cells having lost pRDW10 during growth on the uracil-containing medium will gow on 5-FOA. Yeast were grown at 30° unless indicated othwerwise.
Thermotolerance:
Aliquots of 650 μl were removed from cultures grown at 30° in YPAD to an OD600 = 0.15–0.3 and transferred to 13 × 100-mm borosilicate tubes. A zero-time sample of 100 μl was removed and transferred to a test tube on ice. The remaining cultures were placed in a 52° circulating water bath and 100-μl aliquots were removed periodically and transferred to tubes on ice. Samples were then titered on YPAD plates, which were incubated for 5 days at 30°.
Luciferase reactivation in vivo:
Reactivation of thermally denatured luciferase was done as described (Parsell et al. 1994; Jung and Masison 2001). Briefly, cells expressing a thermolabile form of bacterial luciferase (Escher et al. 1989) were shifted to 37° for 30 min to induce expression of heat-shock proteins and subsequently to 44° for 1 hr to cause thermal denaturation of luciferase. Cultures were then incubated at 23° and recovery of luciferase activity was monitored over time using a Zylux FB15 luminometer. Cyclohexamide was added to the cultures 10 min prior to the end of the 44° incubation to prevent synthesis of luciferase during the recovery period.
Protein expression and purification:
The Pichia pastoris expression system (Invitrogen, San Diego) was used to subclone and express Hsp70's. Selective purification of cloned Hsp70 by ultrasonication of lysate, DEAE-Sephacel, butyl-Sepharose, and gel filtration chromatography was done as described (Wegele et al. 2003a). The Hsp40 protein Ydj1p was expressed in Escherichia coli BL21(DE3) from plasmid pET11aYdj and purified with DEAE and hydroxyapatite chromatography as described (Lu and Cyr 1998; Fan et al. 2005). Sephadex G-50 (Fung et al. 1996) or charcoal (Ha et al. 1997) treatment was used to remove nucleotides from protein preparations.
Luciferase refolding in vitro:
Refolding of guanidinum-HCl denatured firefly luciferase (Promega) in buffer that contained Hsp70, with or without a twofold molar excess of Hsp40 (Ydj1p), was performed as described (Lu and Cyr 1998). Aliquots of the refolding reaction were mixed with luciferase assay reagent (Promega) and luminescence was measured in a Zylux FB15 luminometer. Identical reactions without Hsp70 were done to determine background refolding.
ATP hydrolysis assays:
Hsp70 protein (0.5 μm) in buffer Y (25 mm Hepes/KOH, pH 7.4, 5 mm MgCl2, 50 mm KCl, and 50 μg/ml bovine serum albumin) was incubated with 100 μm [α-32P]ATP and 2-μl aliquots were collected every 5 min and dried on polyethyleneimine-thin layer chromatography plates. Samples were resolved with a mixture of 0.5 m lithium chloride and 0.5 m formic acid. Data were quantified using a Fuji film FLA-3000 phosphoimager and background hydrolysis was subtracted. When Ydj1 was included, samples were analyzed according to Lu and Cyr (1998).
Peptide synthesis and labeling:
Peptide GOKTUG (GLQLSLTARG), a modified form of Pep70 (Fung et al. 1996), was synthesized by Biosource and purified by reversed phase chromatography. Purity was determined by mass spectroscopy. Peptide labeling by acrylodan (6-acrylol-2-dimethylaminonaphtalene) and removal of excess dye were performed according to Han and Christen (2004).
Determination of dissociation constants:
Experiments were performed in buffer Y (above). Measurements were done in a Perkin Elmer (Norwalk, CT) LS 50 spectrophotometer. Acrylodan-labeled peptide (50–200 nm) was titrated with increasing concentrations of Hsp70 protein (25 nm–30 μm) in the presence of 1 mm ATP/ADP (500 μl total volume). Excitation and emission wavelengths were set to 370 and 500 nm, respectively. Emission spectra of peptide-Hsp70 complexes were recorded between 420 and 620 nm with 0.4 and 0.2 slitwidths. Each set of experiments was performed twice. Fluorescence signals were corrected for background, inner-filter, and dilution effects. Dissociation constants (Kd) were determined by using the equation
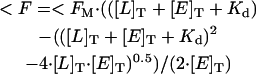 |
(Gisler et al. 1998), where <F is the increase in fluorescence at a given concentration and <FM is the maximum fluorescence change, and L and E are total concentrations of peptide and Hsp70, respectively.
RESULTS
Mammalian and plant Hsc70, but not iHsp70, supports yeast growth:
The yeast Ssa family of cytosolic Hsp70's consists of four functionally redundant isoforms (Ssa1p–Ssa4p). Ssa1p and Ssa2p are abundant, constitutively expressed, and 97% identical. Ssa1p expression is partially repressed under optimal conditions, but it becomes fully induced by stress or depletion of Ssa2p (Stone and Craig 1990). Ssa3p and Ssa4p are expressed only under nonoptimal growth conditions. Ssa Hsp70 function is essential for viability and any one of the four Ssap's can support yeast growth if sufficiently expressed (Werner-Washburne et al. 1987).
To test the ability of Hsp70's from other species to support yeast growth we used strain G400-1C, which lacks all chromosomal SSA genes and expresses SSA1 from plasmid pRDW10 (Jones and Masison 2003). This strain grows as well as wild type under optimal or stressful conditions, showing that Ssa1p provides all Hsp70 functions necessary for normal growth and stress protection. Since Ssa Hsp70 function is essential for viability, G400-1C is unable to grow if it loses pRDW10.
Strain G400-1C was transformed by plasmids carrying genes encoding the primate cytosolic Hsp70 isoforms Hsc70 (constitutive) or iHsp70 (inducible), and transformants were tested for their ability to grow on medium that kills cells expressing the URA3 gene, which is carried on pRDW10 (materials and methods). If the primate Hsp70's support yeast growth, then cells will be able to lose pRDW10 and survive on the counterselecting medium. The extent to which the primate proteins provide essential Hsp70 function is then reflected in the growth rate of cells having lost pRDW10 and expressing only the mammalian Hsp70. Cells transformed by the control plasmid expressing Ssa1p lost pRDW10 readily, those expressing Hsc70 lost pRDW10 less frequently, and those with the vector alone or expressing iHsp70 were unable to lose pRDW10. Thus, Hsc70 supported yeast growth but iHsp70 did not, which uncovers an important functional distinction between them. Cells supported by Hsc70 grew more slowly than those expressing Ssa1p (Figure 1B colonies, Table 2), indicating that mammalian and yeast Hsc70's also have significant functional differences.
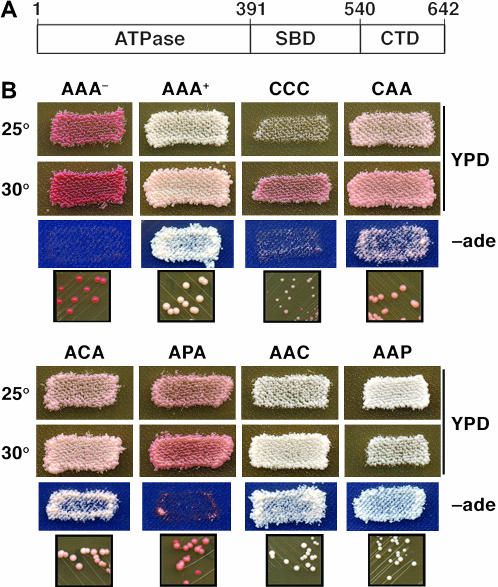
Figure 1.
Growth and [PSI+] prion phenotypes of cells expressing various Hsp70 isoforms. (A) Diagram of Ssa1p (AAA), indicating amino acid residue numbers at junctions of ATPase, substrate-binding (SBD), and C-terminal (CTD) domains. (B) Growth of strains is supported by the indicated Hsp70 isoform. AAA is yeast Ssa1p, and CCC and PPP are primate Hsc70 and Hsp70, respectively. The remaining isoforms are hybrids with ATPase, SBD, and CTD (from left to right) from the indicated Hsp70's. The relative ability of Hsp70's to support growth is reflected in the size of colonies in the small panels. The relative “strength” of [PSI+] propagation is reflected in the degree of red pigmentation and the relative rate of growth without adenine (−ade). As [PSI+] becomes weaker, cells become redder and less able to grow without adenine. The appearance of red colonies reflects mitotic loss of [PSI+]. AAA− and AAA+ exemplify typical [psi−] and [PSI+] phenotypes, respectively.
TABLE 2.
Growth and [PSI+] phenotype of cells supported by different Hsp70 isoforms
| Hsp70 isoforma
|
Growth rate
|
||
|---|---|---|---|
| (min/cell div)b | No. cells/15 hrc | [PSI+]d | |
| Unmodified | |||
| AAA | 113 | 250 | + + + + |
| CCC | 230 | 15 | + |
| PPP | Inviable | NA | |
| AtC70 | 169 | 40 | + + + + |
| AtP70 | Inviable | NA | |
| AAA substituted | |||
| CAA | 128 | 130 | + + + |
| PAA | Inviable | NA | |
| ACA | 113 | 250 | + + + |
| APA | 114 | 240 | + |
| AAC | 146 | 72 | + + + + + |
| AAP | 174 | 36 | + + + + + |
| Others | |||
| ACC | 144 | 76 | + + |
| APP | 226 | 16 | + + |
| CAC | 177 | 34 | + + + |
| PAP | Inviable | NA | |
| CCA | 129 | 126 | + |
| PPA | Inviable | NA | |
| CCP | 161 | 48 | + + |
| PPC | Inviable | NA | |
AAA is yeast Ssa1p, CCC and PPP are S. oedipus Hsc70 and Hsp70, respectively, and hybrid proteins have ATPase, substrate-binding, and C-terminal domains (from left to right) from the indicated proteins. AtC70 and AtP70 are A. thaliana Hsc70 and Hsp70, respectively.
Growth rate was measured for [psi−] cells grown in liquid YPAD at 30°. Inviable, allele does not support growth.
Number of cells expected after 15 hr if one cell was allowed to grow at the rate indicated. This number helps illustrate the degree to which Hsp70 isoforms support growth.
Relative [PSI+] phenotype was scored subjectively and includes color on YPD, growth without adenine, and mitotic stability (see Figure 1B). NA, not applicable as Hsp70 does not support growth.
The difference in the ability of Hsc70 and iHsp70 to support yeast growth could reflect a divergence of function related to the different conditions under which the isoforms are expressed. To test if the distinction in Hsc70 and iHsp70 function was a general one, we repeated the growth complementation test with Hsc70 and iHsp70 isoforms from the widely used plant model A. thaliana. As with their primate counterparts, and despite the fact that the plant iHsp70 is more similar to plant Hsc70 than the plant and primate Hsc70's are to each other (90% vs. 77%, respectively, Table 1), the plant Hsc70 supported yeast growth but the plant iHsp70 did not (Table 2). These results suggest that the basis for the distinction in function between these constitutively expressed and stress-inducible isoforms of Hsp70 is evolutionarily conserved.
To identify the structural domains responsible for the differences in function, we constructed hybrid proteins by separately substituting the ATPase, substrate-binding, and C-terminal domains of Ssa1p with those from primate Hsc70 and iHsp70 (see materials and methods, Figure 1A). On the basis of their three-domain structure, we refer to the Ssa1p, Hsc70, and iHsp70 proteins as AAA, CCC, and PPP, respectively. Hybrid proteins are named for the isoform domains they are derived from; e.g., the PAA hybrid has the ATPase domain of iHsp70 fused to the SBD and CTD of Ssa1p. We constructed additional hybrids in which single domains of CCC and PPP were replaced with those from AAA and hybrids in which the CTDs of CCC and PPP were switched. These additional hybrids (“others” in Table 2) were tested only for growth complementation and prion propagation (see below).
ATPase domains of mammalian constitutive and inducible Hsp70's have distinct functions:
The PAA hybrid protein did not support yeast growth but CAA supported growth relatively well (Table 2 and compare colony size in Figure 1B). Therefore, a major difference between the constitutive and inducible primate Hsp70's is the function of the ATPase domain. All hybrid proteins containing the ATPase domain of iHsp70 (P − −) failed to support growth at both optimal (30°) and elevated but nonlethal (37°–39°) temperatures (Table 2 and data not shown). In contrast, all other Hsp70 hybrids, including APP, supported growth. Thus, activity of the mammalian iHsp70 ATPase domain was not compatible with Hsp70 functions required for yeast growth.
Substrate-binding domains of different Hsp70 isoforms function similarly in vivo:
Cells expressing Ssa1p hybrids with SBD substitutions (ACA and APA) grew at the wild-type rate (Table 2 and compare colony size in Figure 1B). Although substrate binding is crucial for Hsp70 functions, these results show that there are minimal differences in the ability of the different SBDs to interact directly with cellular substrates. Therefore, differences in Hsp70 function, both within and across species, are better explained by how this peptide-binding activity is regulated by the flanking domains.
Hsp70 C-terminal domain function is important for growth:
Cells expressing Ssa1p hybrids with substituted CTDs (AAC, AAP) grew but at significantly reduced rates (Table 2 and compare colony size in Figure 1B). Thus, although the Hsp70 substrate binding cycle is regulated primarily by ATP hydrolysis and nucleotide exchange, the C-terminal domain can significantly influence Ssa1p activities important for cell growth. Together with the observations that SBD functions are similar and that cells expressing CCA grew better than those expressing AAC (Table 2), these results show that the CTD is an important determinant of functional differences between yeast and mammalian Hsc70. The CTD is the most diverged region of Hsp70's and among eukaryotic cytosolic isoforms it is known to interact with a variety of cochaperones that regulate Hsp70 function (Freeman et al. 1995; Demand et al. 1998; Scheufler et al. 2000). These results might therefore reflect differences in specificity regarding such cochaperone interactions.
Differences in Hsp70 domain functions for growth and [PSI+] prion propagation:
The genotype of strain G400-1C allows straightforward monitoring of the propagation characteristics of the yeast prion [PSI+]. [PSI+] propagates in the cytoplasm as self-replicating aggregates, thought to be amyloid, of the translation release factor Sup35p (eRF3) (Wickner 1994; Stansfield et al. 1995; Zhouravleva et al. 1995; Patino et al. 1996). Aggregation of Sup35p in [PSI+] cells reduces the ability of Sup35p to function in translation termination, which suppresses the ade2-1 nonsense allele in certain yeast strains (Cox 1965). Nonsuppressed ade2-1 cells require adenine and are red on limiting adenine due to accumulation of an Ade2p substrate. By suppressing ade2-1, [PSI+] confers growth without adenine and a normal white colony color (Figure 1B, compare AAA− and AAA+).
A weakened [PSI+] phenotype is seen as partial red pigmentation on limiting adenine and reduced ability to grow without adenine. Spontantous loss of [PSI+] during mitosis, which gives rise to red colonies, also reflects weakened [PSI+] propagation. Conversely, a stronger [PSI+] phenotype is seen as enhanced growth without adenine and whiter colony color. Strain G400-1C has the typical [PSI+] phenotype of uniform white colonies and good growth without adenine (Figure 1B, AAA+), which shows that Ssa1p supports normal [PSI+] propagation as the only Ssap in the cell (Jones and Masison 2003). We showed earlier that a mutant form of Ssa1p weakens [PSI+], indicating that proper Hsp70 function is necessary for normal [PSI+] propagation (Jung et al. 2000; Jones and Masison 2003).
In addition to its reduced ability to support yeast growth, mammalian Hsc70 (CCC) had reduced ability to support [PSI+] propagation, which was seen as reduced growth without adenine, partial pigmentation of cells, and the appearance of red [psi−] colonies (Figure 1B and data not shown). The [PSI+] phenotypes of cells expressing the hybrid Hsp70's, however, showed that Hsp70 functions important for prion propagation were separable from those important for growth. Although cells expressing Ssa1p with substituted C-terminal domains (AAC, AAP) grew more slowy than those expressing Ssa1p, they displayed a stronger [PSI+] phenotype, as indicated by whiter colony color and increased relative growth rate without adenine (Figure 1B, Table 2, data not shown). Thus, the CTD substitutions had opposite effects on Hsp70 functions with regard to growth and prion propagation.
In contrast, cells expressing Ssa1p hybrids with mammalian SBDs (i.e., ACA and APA) grew normally but displayed a weakened [PSI+] phenotype (Figure 1B, Table 2), which was seen as increased pigmentation and, in cells expressing APA, spontaneous mitotic loss of [PSI+] (red colonies). [PSI+] propagation was more significantly impaired in cells expressing the APA hybrid than in cells expressing ACA. Since both of these hybrids supported growth as well as Ssa1p, this difference in magnitude of effect on [PSI+] is apparently unrelated to Hsp70 SBD activities required for essential cellular processes and reveals another qualitative distinction in functions of constitutive and inducible Hsp70 isoforms. Together, these data uncover differences in functions of constitutive and inducible Hsp70's with regard to both growth and prion propagation and are consistent with our earlier work showing that Hsp70 mutations that impair [PSI+] propagation are separable from those affecting cell growth (Jung et al. 2000; Jones and Masison 2003).
Hsp70 function in thermotolerance and refolding of thermally denatured protein:
In addition to acting in processes essential for cell growth under optimal conditions, Hsp70 protects cells from stress by preventing aggregation of partially unfolded proteins. When levels of stress are high enough to overcome the capacity of Hsp70 to prevent aggregation, survival of fungi, plants, and other organisms also requires an Hsp100-family chaperone (Hsp104 in Saccharomyces cerevisiae) that acts by resolubilizing aggregated proteins (Parsell et al. 1994; Glover and Tkach 2001). Hsp70 and its cochaperone Hsp40 cooperate with Hsp104 in protein disaggregation (Glover and Lindquist 1998). Yeast lacking Hsp104 grow normally under optimal conditions but die much more rapidly when exposed to a lethal stress. Hsp104 disaggregation activity is also essential for [PSI+] propagation (Chernoff et al. 1995). To further assess the function of the primate and hybrid proteins in vivo, we compared their ability to provide thermotolerance and to aid in the reactivation of a thermally denatured reporter protein. Although these processes in yeast are dependent upon Hsp104, such comparisons can reveal differences in the ability of Hsp70 to prevent protein aggregation and to cooperate with other chaperones in protein disaggregation.
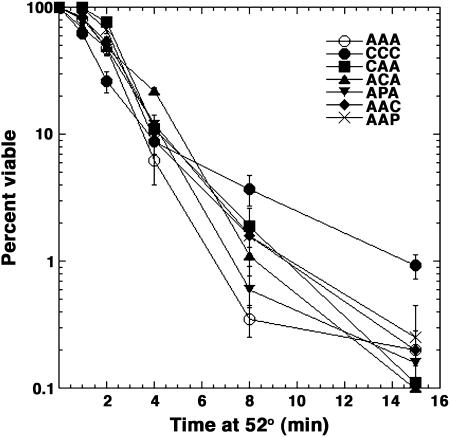
After exposure to a lethal temperature for 15 min, a time when survival depends mainly on protein disaggregation, all strains except those expressing CCC had a fraction of viable cells similar to that of the stain expressing AAA (Figure 2). Survival of cells expressing CCC was reproducibly three to fivefold higher than that of cells expressing AAA, indicating that it performed well in the disaggregation process. In the first 2 min after the heat shock, when survival relies more on prevention of protein aggregation, the percentage of survivors in cells expressing CCC was reproducibly lower than that of cells expressing AAA while the other strains survived normally. These data suggest that as components of the Hsp104 disagregation machinery the different isoforms functioned at least as well as wild type and that CCC might be less efficient than AAA at preventing protein aggregation in vivo.
Figure 2.
Thermotolerance of cells expressing various Hsp70 isoforms. Survival of cells exposed to the lethal temperature of 52°, expressed as a percentage of viable cells in the preexposure culture, is plotted as a function of time.
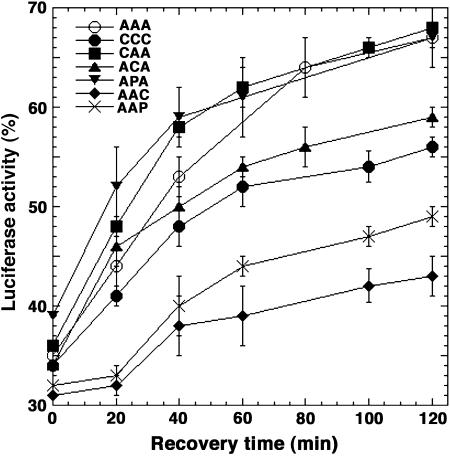
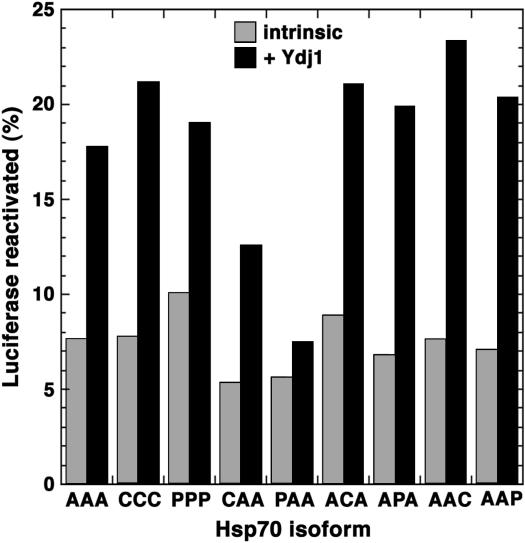
To compare functions of the different Hsp70's in protein disaggregation in vivo we monitored reactivation of a thermolabile bacterial luciferase after exposing cells to elevated but nonlethal temperature (Figure 3). ACA and CCC showed modestly reduced ability to function in this assay. The opposite result regarding CCC in the thermotolerance assay might reflect a difference in the ability of CCC to cooperate with Hsp104 in disaggregating proteins important for cell growth and a specific substrate not normally expressed in yeast. The strains expressing AAC and AAP were least efficient in reactivating luciferase, which reflects a reduced ability of these isoforms to function with Hsp104 in disaggregating this substrate. Although an altered interaction with Hsp104 is not likely to explain the growth defect of cells expressing AAC or AAP, it might affect the process of breaking prion aggregates in a way that could explain the improved [PSI+] phenotype in these cells.
Figure 3.
Reactivation of thermally denatured bacterial luciferase in cells expressing various Hsp70 isoforms. After causing thermolabile luciferase to aggregate by exposing cultures to elevated but nonlethal temperature, recovery of luciferase activity, expressed as a percentage of that in the preexposure cultures, was monitored as a function of time.
Enzymatic activities of purified Hsp70's:
In addition to its structure, the enzymatic functions of Hsp70 are highly conserved. The ability of the primate, plant, and hybrid proteins to support growth implies that such activities of these various Hsp70's are comparable. Nevertheless, the variation in phenotype of cells expressing the different isoforms reflects functional differences among them. To test if the different phenotypes of cells expressing the various Hsp70's were attributable to specific differences in intrinsic or stimulated Hsp70 activity we measured enzymatic activities of purified Hsp70's alone or combined with Ydj1p, which is one of the two major cytosolic Hsp40's of yeast known to regulate Ssa1p.
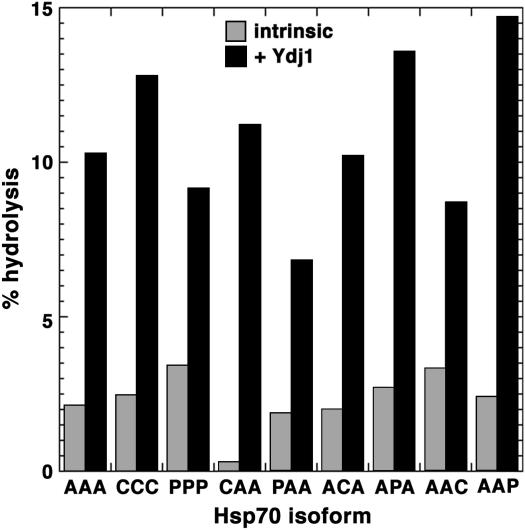
Figure 4 shows amounts of ATP hydrolyzed in 10 min by the unmodified Hsp70 isoforms and those containing single substitutions of Ssa1p domains. CAA stands out as having the lowest intrinsic activity but it has an Hsp40-stimulated activity similar to that of Ssa1p (AAA). AAP had the highest stimulated activity, which might account for its reduced ability to support growth. However, cells expressing AAC had a phenotype similar those expressing AAP but the AAC protein had near normal ATPase activity. Additionally, PPP, which did not support growth, had intrinsic and Hsp40-stimulated ATPase activity similar to that of wild type. Therefore, the variation in growth and prion phenotypes cannot be explained simply by differences in intrinsic or Hsp40-stimulated ATPase activity. Moreover, it appears unlikely that the inability of PPP to support growth was due solely to a major difference in such ATPase activity.
Figure 4.
Intrinsic and Hsp40-simulated ATPase activity of various Hsp70 isoforms. The percentages of ATP hydrolyzed by the different Hsp70 proteins in 10 min in the absence (intrinsic, shaded bars) or presence (+ Ydj1, solid bars) of the Ssa1p Hsp40 partner Ydj1p are shown.
We then tested the ability of purified Hsp70's, alone or combined with Ydj1p, to refold denatured luciferase in vitro. Figure 5 shows relative levels of reactivation of chemically denatured luciferase after 15 min of incubation with Hsp70. Here again the differences in enzymatic activity generally were modest. The hybrids with the lowest refolding activity were CAA and PAA. PAA was least able to refold luciferase, which might account for its inability to support growth. However, PPP, which also failed to support growth, functioned as well as wild-type Ssa1p in this assay. Therefore, even where differences in ability to refold denatured luciferase were detectable they did not correleate with strain phenotypes.
Figure 5.
Intrinsic and Hsp40-simulated refolding of chemically denatured firefly luciferase by various Hsp70 isoforms. Reactivation of denatured luciferase after incubating with Hsp70 for 15 min in the absence (intrinsic, shaded bars) or presence (+ Ydj1, solid bars) of the Ssa1p Hsp40 partner Ydj1p is shown as a percentage of that present before denaturing. Background activity measured in samples without Hsp70 has been subtracted.
To determine if the Hsp70 isoforms differed in affinity for substrate, we measured their binding affinities (Kd) using a fluorescently labeled peptide. Hsp70 exchanges substrate rapidly when in the open ATP-bound form, whereas it releases substrate much more slowly when in the closed ADP-bound state. Overall we found that the Hsp70's bound peptide ∼10- to 20-fold more tightly when associated with ADP than with ATP (Table 3), which is in line with previously published data (Greene et al. 1995). In comparing the Kd's of the ADP-bound Hsp70's that supported growth, those that improved [PSI+] propagation (AAC and AAP) had the lowest affinity for peptide, while the proteins that had the greatest inhibitory effect on [PSI+] (CCC and APA, Table 2 and Figure 1B) bound peptide most tightly. These data agree with our earlier work showing that alterations that enhance Hsp70 substrate binding inhibit [PSI+] propagation (Jones et al. 2004). Thus, it is possible that the effects of the different hybrids on [PSI+] propagation can be explained by differences in affinity of the isoforms for substrates.
TABLE 3.
Hsp70 substrate-binding affinities
Abundance of Hsp70 and Hsp104:
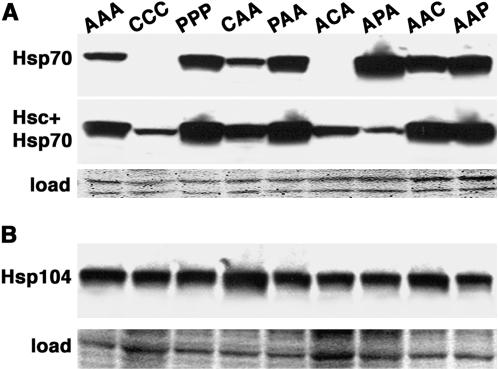
It was possible that differences in Hsp70 abundance in cells expressing the various isoforms contributed to the different phenotypes we observed. We therefore compared relative abundance of Hsp70 by Western analysis, using antibodies that recognize iHsp70 or both Hsc70 and iHsp70 (Figure 6A). Although abundance of Hsp70 varied among half the strains, the only appreciable differences compared with AAA were a modest reduction of ACA and more significant reduction of CCC. Aside from CCC there was no correlation between Hsp70 abundance and any of the phenotypes assessed. Therefore, a difference in abundance might explain the exceptional phenotypes of cells expressing CCC, but it is unlikely to explain differences among strains expressing the other isoforms.
Figure 6.
Western analysis to compare the abundance of Hsp70 and Hsp104 in cells expressing various Hsp70 isoforms. (A) Blots containing identical samples from lysates of cells expressing the various Hsp70 isoforms, indicated at top, were probed with antibody recognizing iHsp70 (top row) or both Hsc70 and iHsp70 (middle row). (B) Samples from similar lysates were probed with anti-Hsp104 antibody (top row). The bottom rows in both A and B show portions of membranes used for the blots stained by amido black as loading and transfer controls.
As Hsp104 plays a major role in both thermotolerance and luciferase reactivation, we also compared abundance of Hsp104 (Figure 6B). When taking into account the amount of total protein in each lane, the only strains with notable differences in Hsp104 abundance were those expressing CCC, ACA, and APA, which had slightly less Hsp104, and CAA that had an elevated amount. Therefore, except for cells expressing CAA, differences in Hsp104 expression are not likely to explain the observed differences in thermotolerance or luciferase reactivation in vivo.
DISCUSSION
We developed a yeast system that allows us to test the ability of any Hsp70 isoform to functionally replace yeast Hsp70 and used it to show that growth of budding yeast can be supported by Hsp70 from species as diverse as mammals and plants. We further found that constitutive and inducible isoforms of primate cytosolic Hsp70 function differently, a distinction that we show is conserved in plants. Using hybrid proteins we identified Hsp70 structural domains that confer this and other functional differences. Among these differences we found that Hsp70 functions important for prion propagation were separable from those important for growth. Biochemical analysis of the Hsp70 isoforms showed that, aside from substrate affinities, differences in enzymatic activities were not considerable, nor did they correlate with phenotype, suggesting that the observed phenotypic differences were due to complex regulation of Hsp70 functions in the cell.
Hsp70 interacts with numerous cofactors and is a major component of several different protein-folding machines in the cell. The fact that yeast growth can be supported by our collection of diverse Hsp70 proteins is remarkable as it demonstrates conservation of Hsp70's many functional interactions, including those involving communication between Hsp70 domains as well as between Hsp70 and its cochaperones. The ability of Ydj1p to stimulate ATPase and protein refolding of the various Hsp70's to similar levels in vitro is a clear demonstration of this conservation for an Hsp70–Hsp40 interaction. We attributed the inability of primate iHsp70 to support yeast growth to its ATPase domain, but its ATPase activity in vitro was similar to that of Ssa1p. Therefore, it is likely that differences in vivo are influenced by interactions with other cofactors, possibly those coinduced with iHsp70.
Craig and colleagues found that substrate-binding domains were not responsible for differences between the functionally distinct Ssa and Ssb Hsp70 subfamilies of yeast, leading them to propose that peptide-binding activity per se plays little or no role in specifying Hsp70 function in vivo (James et al. 1997). Our finding that hybrid Ssa1 proteins containing primate SBDs from either Hsc70 or iHsp70 supported growth as well as Ssa1p shows that such functional redundancy of cytosolic Hsp70 SBDs extends both widely across eukaryotic species and between mammalian Hsp70 isoforms. Thus, the functional differences among Hsp70's appear to lie more in how the SBD-substrate interactions are regulated than in the physical properties of the different SBDs. In fact, we find that affinity of the different Hsp70 isoforms for substrate was determined mostly by the domains flanking the SBD. ATPase and C-terminal domains are known to interact with a variety of cochaperones, which adds to the complexity of this regulation in vivo. Our data therefore are consistent with the view that specificity of Hsp70 function in vivo is dictated by various cochaperone interactions (James et al. 1997; Young et al. 2003; Mayer and Bukau 2005) and suggest either that the SBD has little influence on the specificity of cochaperone interaction or that such SBD–cochaperone interactions are highly conserved.
Although the regulation of SBD function was more important than SBD structure regarding growth complementation, we found that isoforms AAA, ACA, and APA, which differ only in their SBD, had different [PSI+] phenotypes. [PSI+] therefore represents a diagnostic that is useful for identifying differences in Hsp70 function that are subtle enough to be undectable by their effects on cell growth. Our current data do not distinguish if such effects are due to altered interactions of Hsp70 with amyloid forms of Sup35p as a substrate or with other components of the chaperone machinery such as Hsp40 or Hsp104. Nevertheless, our finding that SBD substitutions weakened prion propagation without detectably affecting cell growth points to Hsp70 as a possible target for therapeutic intervention in amyloid diseases.
Our data show that the Hsp70 C-terminal domain plays an important role in Hsp70 functions required for cell growth. Although Hsp40 interacts with both the C-terminal and ATPase domains of Hsp70 (Freeman et al. 1995; Gassler et al. 1998; Greene et al. 1998), it is not known if Hsp70 activity is regulated by a specific interaction between these two domains. While it has been shown that the combined SBD and CTD can influence activity of the ATPase domain (Lopez-Buesa et al. 1998), if a specific cooperation between the ATPase and C-terminal domains exists, our data showing that ACA and APA support growth as well as AAA implies that the intervening SBD does not strongly influence such cooperation.
The CTD is the least conserved region of Hsp70 but most eukaryotic cytosolic Hsp70's from yeast to mammals end with a conserved octapeptide [GPT(I/V)EEVD] that is known to physically interact with TPR motifs. TPRs are structural domains involved in protein–protein interactions that define, among other subclasses, a family of Hsp70 and Hsp90 cochaperones (Sikorski et al. 1990; Blatch and Lassle 1999; Scheufler et al. 2000). Earlier we showed that this octapeptide on Ssa1p is important for yeast growth, although it is not essential for viability, and that some TPR cochaperones can bind, albeit weakly, to Hsp70 that lacks the octapeptide (Jones et al. 2004). Smith and colleagues also showed that deleting the octapeptide from human Hsp70 reduced Hop1–Hsp70 interactions and that extended deletion of CTD residues abolishes the interaction, indicating that CTD residues other than the octapeptide affect TPR cochaperone interaction (Carrigan et al. 2004). The yeast and mammalian Hsp70's analyzed here differ in several CTD residues, including a conservative change in the fourth position of the octapeptide. Whereas Ssa1 has valine, the mammalian Hsp70's have isoleucine. Thus, sequences both within and near the octapeptide likely affect the specificity or affinity of TPR cochaperone interactions, which in turn affect functions important for growth or prion propagation. Our system will be useful for identifying and characterizing Hsp70–cochaperone interactions and the mechanisms underlying their physiological effects.
Although humans have at least six cytosolic Hsp70's, one of which is strongly and constitutively expressed in all cells and another that is strictly stress inducible, the degree of functional redundancy or divergence among them is unknown. Our finding that constitutive and inducible cytosolic isoforms of Hsp70 are functionally distinct regarding cell growth suggests that iHsp70 lacks an activity required for growth under normal conditions. The conservation of this difference in Hsc70/iHsp70 function in plants testifies to its significance and raises the possibility that a specific Hsp70 function is important when cells are exposed to stress. In support of these notions, Jäättelä and colleagues found that viability of both normal and cancer cells depends upon Hsc70 and that cancer cells have an additional requirement for iHsp70 (Nylandsted et al. 2000; Rohde et al. 2005). Therefore, depending on conditions, survival can hinge on the particular cytosolic Hsp70 isoform being expressed. Additionally, in numerous studies assessing effects of Hsp70 overproduction little or no reason is given for the choice of Hsp70 isoform used, presumably because Hsp70's are considered functionally equivalent. Our data suggest that the outcome of such experiments might be influenced by the particular Hsp70 isoform used.
Acknowledgments
We thank Charles Guy (University of Florida) for Arabidopsis mRNA, Douglas Cyr (University of North Carolina) for YDJ1 plasmids, Rene Chun and John Adams (Cedars-Sinai Medical Center, California) for construction of pCCC and pPPP, and Gary Jones (University of Ireland, Maynooth) for construction of pAtC70. This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Diabetes, Digestive, and Kidney diseases.
References
- Blatch, G. L., and M. Lassle, 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays 21: 932–993. [DOI] [PubMed] [Google Scholar]
- Boeke, J. D., F. LaCroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Carrigan, P. E., G. M. Nelson, P. J. Roberts, J. Stoffer, D. L. Riggs et al., 2004. Multiple domains of the co-chaperone Hop are important for Hsp70 binding. J. Biol. Chem. 279: 16185–16193. [DOI] [PubMed] [Google Scholar]
- Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov and S. W. Liebman, 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268: 880–884. [DOI] [PubMed] [Google Scholar]
- Cox, B. S., 1965. “Ψ” a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20: 505–521. [Google Scholar]
- Craig, E. A., and C. A. Gross, 1991. Is hsp70 the cellular thermometer? Trends Biochem. Sci. 16: 135–140. [DOI] [PubMed] [Google Scholar]
- Demand, J., J. Luders and J. Hohfeld, 1998. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 18: 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina, A. A., J. A. Marsh and R. F. Gaber, 1996. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast 12: 943–952. [DOI] [PubMed] [Google Scholar]
- Escher, A., D. J. O'Kane, J. Lee and A. A. Szalay, 1989. Bacterial luciferase alpha beta fusion protein is fully active as a monomer and highly sensitive in vivo to elevated temperature. Proc. Natl Acad. Sci. USA 86: 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. Y., S. Lee and D. M. Cyr, 2003. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaperones 8: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. Y., H. Y. Ren, P. Lee, A. J. Caplan and D. M. Cyr, 2005. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. J. Biol. Chem. 280: 695–702. [DOI] [PubMed] [Google Scholar]
- Freeman, B. C., M. P. Myers, R. Schumacher and R. I. Morimoto, 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14: 2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, K. L., L. Hilgenberg, N. M. Wang and W. J. Chirico, 1996. Conformations of the nucleotide and polypeptide binding domains of a cytosolic Hsp70 molecular chaperone are coupled. J. Biol. Chem. 271: 21559–21565. [DOI] [PubMed] [Google Scholar]
- Gao, B. C., J. Biosca, E. A. Craig, L. E. Greene and E. Eisenberg, 1991. Uncoating of coated vesicles by yeast hsp70 proteins. J. Biol. Chem. 266: 19565–19571. [PubMed] [Google Scholar]
- Gassler, C. S., A. Buchberger, T. Laufen, M. P. Mayer, H. Schroder et al., 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA 95: 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisler, S. M., E. V. Pierpaoli and P. Christen, 1998. Catapult mechanism renders the chaperone action of Hsp70 unidirectional. J. Mol. Biol. 279: 833–840. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and S. Lindquist, 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94: 73–82. [DOI] [PubMed] [Google Scholar]
- Glover, J. R., and J. M. Tkach, 2001. Crowbars and ratchets: hsp100 chaperones as tools in reversing protein aggregation. Biochem. Cell Biol. 79: 557–568. [PubMed] [Google Scholar]
- Greene, L. E., R. Zinner, S. Naficy and E. Eisenberg, 1995. Effect of nucleotide on the binding of peptides to 70-kDa heat shock protein. J. Biol. Chem. 270: 2967–2973. [DOI] [PubMed] [Google Scholar]
- Greene, M. K., K. Maskos and S. J. Landry, 1998. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc. Natl. Acad. Sci. USA 95: 6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, J. H., U. Hellman, E. R. Johnson, L. Li, D. B. McKay et al., 1997. Destabilization of peptide binding and interdomain communication by an E543K mutation in the bovine 70-kDa heat shock cognate protein, a molecular chaperone. J. Biol. Chem. 272: 27796–27803. [DOI] [PubMed] [Google Scholar]
- Hainzl, O., H. Wegele, K. Richter and J. Buchner, 2004. Cns1 is an activator of the Ssa1 ATPase activity. J. Biol. Chem. 279: 23267–23273. [DOI] [PubMed] [Google Scholar]
- Han, W., and P. Christen, 2004. cis-effect of DnaJ on DnaK in ternary complexes with chimeric DnaK/DnaJ-binding peptides. FEBS Lett. 563: 146–150. [DOI] [PubMed] [Google Scholar]
- Harrison, C., 2003. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaperones 8: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl, F. U., and M. Hayer-Hartl, 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858. [DOI] [PubMed] [Google Scholar]
- James, P., C. Pfund and E. A. Craig, 1997. Functional specificity among Hsp70 molecular chaperones. Science 275: 387–389. [DOI] [PubMed] [Google Scholar]
- Jones, G. W., and D. C. Masison, 2003. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, G. W., Y. Song, S. Chung and D. C. Masison, 2004. Propagation of yeast [PSI+] prion is impaired by factors that regulate Hsp70 substrate binding. Mol. Cell. Biol. 24: 3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, G., and D. C. Masison, 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43: 7–10. [DOI] [PubMed] [Google Scholar]
- Jung, G., G. Jones, R. D. Wegrzyn and D. C. Masison, 2000. A role for cytosolic hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani, M., J. M. Beckerich and J. L. Brodsky, 2002. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22: 4677–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufen, T., M. P. Mayer, C. Beisel, D. Klostermeier, A. Mogk et al., 1999. Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl Acad. Sci. USA 96: 5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Buesa, P., C. Pfund and E. A. Craig, 1998. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc. Natl. Acad. Sci. USA 95: 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Z., and D. M. Cyr, 1998. The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J. Biol. Chem. 273: 5970–5978. [DOI] [PubMed] [Google Scholar]
- Marsh, J. A., H. M. Kalton and R. F. Gaber, 1998. Cns1 is an essential protein associated with the Hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in Cpr7Δ cells. Mol. Cell. Biol. 18: 7353–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. P., and B. Bukau, 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62: 670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M. P., D. Brehmer, C. S. Gassler and B. Bukau, 2001. Hsp70 chaperone machines. Adv. Protein Chem. 59: 1–44. [DOI] [PubMed] [Google Scholar]
- McCarty, J. S., A. Buchberger, J. Reinstein and B. Bukau, 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249: 126–137. [DOI] [PubMed] [Google Scholar]
- McClellan, A. J., J. B. Endres, J. P. Vogel, D. Palazzi, M. D. Rose et al., 1998. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol. Biol. Cell 9: 3533–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne and E. A. Craig, 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71: 97–105. [DOI] [PubMed] [Google Scholar]
- Nylandsted, J., M. Rohde, K. Brand, L. Bastholm, F. Elling et al., 2000. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. USA 97: 7871–7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, M., M. Nakai, T. Endo, C. R. Lim, Y. Kimata et al., 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273: 29727–29737. [DOI] [PubMed] [Google Scholar]
- Parsell, D. A., A. S. Kowal, M. A. Singer and S. Lindquist, 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372: 475–478. [DOI] [PubMed] [Google Scholar]
- Patino, M. M., J. J. Liu, J. R. Glover and S. Lindquist, 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273: 622–626. [DOI] [PubMed] [Google Scholar]
- Rieger, T. R., R. I. Morimoto and V. Hatzimanikatis, 2005. Mathematical modeling of the eukaryotic heat-shock response: dynamics of the hsp70 promoter. Biophys. J. 88: 1646–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, M., M. Daugaard, M. H. Jensen, K. Helin, J. Nylandsted et al., 2005. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 19: 570–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder et al., 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101: 199–210. [DOI] [PubMed] [Google Scholar]
- Schmid, D., A. Baici, H. Gehring and P. Christen, 1994. Kinetics of molecular chaperone action. Science 263: 971–973. [DOI] [PubMed] [Google Scholar]
- Schwimmer, C., and D. C. Masison, 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22: 3590–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., 1994. Getting started with yeast. Methods Enzymol. 194: 3–21. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., M. S. Boguski, M. Goebl and P. Hieter, 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60: 307–317. [DOI] [PubMed] [Google Scholar]
- Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski et al., 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14: 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, D. E., and E. A. Craig, 1990. Self-regulation of 70-kilodalton heat shock proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 1622–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, D. Y., E. Vierling and C. L. Guy, 2001. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 126: 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele, H., M. Haslbeck and J. Buchner, 2003. a Recombinant expression and purification of Ssa1p (Hsp70) from Saccharomyces cerevisiae using Pichia pastoris. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 786: 109–115. [DOI] [PubMed] [Google Scholar]
- Wegele, H., M. Haslbeck, J. Reinstein and J. Buchner, 2003. b Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278: 25970–25976. [DOI] [PubMed] [Google Scholar]
- Werner-Washburne, M., D. E. Stone and E. A. Craig, 1987. Complex interactions among members of an essential subfamily of Hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7: 2568–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner, R. B., 1994. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 264: 566–569. [DOI] [PubMed] [Google Scholar]
- Wittung-Stafshede, P., J. Guidry, B. E. Horne and S. J. Landry, 2003. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry 42: 4937–4944. [DOI] [PubMed] [Google Scholar]
- Wu, S., S. Ren, H. Chen, R. F. Chun, M. A. Gacad et al., 2000. Intracellular vitamin D binding proteins: novel facilitators of vitamin D-directed transactivation. Mol. Endocrinol. 14: 1387–1397. [DOI] [PubMed] [Google Scholar]
- Young, J. C., J. M. Barral and F. U. Hartl, 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28: 541–547. [DOI] [PubMed] [Google Scholar]
- Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov et al., 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14: 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]