Abstract
The self-compatible Coprinopsis cinerea homokaryon AmutBmut produces fruiting bodies without prior mating to another strain. Early stages of fruiting body development include the dark-dependent formation of primary hyphal knots and their light-induced transition to the more compact secondary hyphal knots. The AmutBmut UV mutant 6-031 forms primary hyphal knots, but development arrests at the transition state by a recessive defect in the cfs1 gene, isolated from a cosmid library by mutant complementation. A normal primordia phenotype was achieved when cfs1+ was embedded at both sides in at least 4.0 kb of native flanking DNA. Truncations of the flanking DNA lead to reduction in transformation frequencies and faults in primordia tissue formation, suggesting that the gene is also acting at later stages of development. The cfs1 gene encodes a protein highly similar to cyclopropane fatty acid synthases, a class of enzymes shown in prokaryotes and recently in a plant to convert membrane-bound unsaturated fatty acids into cyclopropane fatty acids. In C. cinerea 6-031, the mutant cfs1 allele carries a T-to-G transversion, leading to an amino acid substitution (Y441D) in a domain suggested to be involved in the catalytic function of the protein and/or membrane interaction.
THE heterothallic fungus Coprinopsis cinerea serves as a model organism to study fruiting body development in higher basidiomycetes. Fruiting bodies normally develop on the dikaryon (Kües 2000). However, the presence of two genetically distinct nuclei in the dikaryotic mycelium is a major drawback to performing genetic analysis on fruiting body development. The self-compatible homokaryon AmutBmut, having specific mutations in both mating-type loci (A43mut and B43mut), shows characteristics typical of the dikaryon, for example, formation of fused clamp cells at hyphal septa. It also gives rise to fruiting bodies without prior mating to another strain (Swamy et al. 1984; Walser et al. 2003). This special feature together with the ability to form unicellular haploid spores (oidia) provides us with an easily accessible genetic system (Kües et al. 2004). A series of developmental mutants have been generated from strain AmutBmut by UV- and REMI-mutagenesis (Granado et al. 1997; Lu et al. 2003; U. Kües, J. D. Granado and M. Aebi, unpublished results), which serves to isolate genes.
The development of fruiting bodies is a highly organized process, which requires the coordination between genetic, environmental, and physiological factors (Kües 2000). In the dark, upon nutritional depletion, single hyphae locally undergo intense branching to form microscopic primary hyphal knots. When kept in the dark, these develop into multicellular pigmented resting bodies called sclerotia (Kües et al. 2002a). Following a light signal, radial growth of primary hyphal knots and hyphal interaction lead to the formation of compact hyphal aggregates, secondary hyphal knots, which are specific fruiting body initials with undifferentiated hyphae. Once tissue differentiation initiates within the secondary hyphal knot, the structures are termed primordia. Cellular differentiation results in the formation of distinct cap and stipe tissues. Formation of the various tissues within the developing primordium needs altering dark and night phases and continues over 3 days, over which the size of the primordium enlarges (Boulianne et al. 2000; Kües 2000; Walser et al. 2003). In the mature primordium cap, induced by a further light signal, karyogamy occurs in specialized cells (basidia) present in the hymenium of the gills. Karyogamy is directly followed by meiosis. In parallel, the stipe elongates and the cap expands, giving rise to a fully developed fruiting body (Moore et al. 1979; Kües 2000).
So far, little is known about the genetic determinants that act in fruiting body initiation and formation. Induction of primary hyphal knots and the morphological transition from primary into secondary hyphal knots were shown to be regulated by the A and B mating-type genes (Kües et al. 1998, 2002b). The gene pcc1, encoding an HMG-box transcription factor, likely acts downstream of the A mating-type gene products and appears to negatively regulate fruiting body initiation (Murata et al. 1998). Onset of expression of two genes encoding fruiting body specific galectins (β-galactoside sugar-binding lectins) correlates with the formation of primary and secondary hyphal knots, respectively, and continues during primordia development (Boulianne et al. 2000; Bertossa et al. 2004). In addition, four genes have been identified that act in cap and stipe tissue formation and stipe elongation, respectively, including one for a potential photoreceptor (Muraguchi and Kamada 1998, 2000; Arima et al. 2004; Terashima et al. 2005). Next to the gene described in this study, a yet uncharacterized gene acting in fruiting body initiation has also been cloned (Clergeot et al. 2003).
Within our mutant collection derived from homokaryon AmutBmut, we identified two groups of mutants whose defects link to fruiting body initiation. Members of one group are termed pkn (primary knotless) mutants, because they do not form any primary hyphal knot in the dark. The other group of mutants is arrested at the transition from primary to secondary hyphal knots. Therefore, they are called skn (secondary knotless) mutants (U. Kües, Y. Liu and M. Aebi, unpublished). In this study, we isolated a gene that complemented the defect of fruiting body initiation in the skn1 UV-mutant 6-031. The predicted gene product is highly homologous to cyclopropane fatty acid synthases, a class of enzymes characterized before in bacteria and recently also in a plant.
MATERIALS AND METHODS
Fungal strains, culture conditions, and transformation:
C. cinerea strains were standardly grown at 37° on YMG/T complete medium and minimal medium (Granado et al. 1997) supplemented with p-aminobenzoate (PABA, 5 mg/liter) when required. Strain 6-031 (A43mut, B43mut, pab1-1, skn1) is a fruiting body initiation mutant generated from homokaryon AmutBmut (A43mut, B43mut, pab1-1) by UV-mutagenesis (U. Kües, J. D. Granado and M. Aebi, unpublished results). Monokaryons PG78 (A6, B42, pab1) and JV6 (A42, B42), both unrelated to homokaryon AmutBmut, and the AmutBmut co-isogenic monokaryons PS001-1 (A42, B42) and PS002-1 (A3, B1) were used in crosses (Kertesz-Chaloupková et al. 1998; P. Srivilai and U. Kües, unpublished results). Matings were performed on YMG/T plates by placing two mycelial blocks of inoculum 5 mm apart. For growth and induction of fruiting bodies, mating plates were incubated at 12-hr light/12-hr dark, 25°, 90% humidity under standard fruiting conditions (Granado et al. 1997). Randomly isolated basidiospores were germinated on YMG/T medium at 37°. Progeny of crosses were analyzed on minimal media for PABA auxotrophy. Presence of unfused and fused clamp cells, indicators of activated A and B mating-type pathways, respectively (Kües 2000), was determined by microscopy.
Monokaryon JV6 served to confirm mating types in A43mut, B43mut progenies from crosses with PS001-1 and PS002-1 that subsequently were submitted to fruiting tests. Frequencies of phenotypic distributions in A43mut, B43mut progenies were tested by a chi-square method. A skn1+, mat+, bad clone (PS-Mu1-3) and a skn1+, mat, bad clone (PS-Mu1-2) within the A42, B42 progeny of cross 6-031 × PS001-1 (defined by crosses with monokaryon PG78) were identified through mating with mutant 6-031. Mating of these two strains with A43mut, B43mut clones of the progeny 6-031 × PS001-1 that did not initiate fruiting identified homokaryon OU3-1 (A43mut, B43mut, pab1-1, skn1).
The F1 progeny of cross 6-031 × JV6 was randomly analyzed for fruiting ability by individually inoculating clones on YMG/T agar, growing them for 4 days at 37° in the dark, and subsequently transferring them to standard fruiting conditions. Dikaryons among the clones were identified by light inducing oidia production, germinating the spores on YMG/T agar, and analyzing pab-auxotrophy on minimal medium. For oidia induction of dikaryons, mutant 6-031 and other A43mut, B43mut strains, dark-grown cultures were exposed to light for 2 days (Kertesz-Chaloupková et al. 1998) and the number of oidia per plate was determined by a spectrophotometer.
DNA transformation was performed as described (Granado et al. 1997). For selecting PABA prototrophs in cotransformations, 1 μg of plasmid pPAB1-2 (Granado et al. 1997) was added. Upon germination on regeneration agar, transformants were individually transferred onto minimal medium for further growth. Subsequently, three or four individual transformants were inoculated on YMG/T agar per single petri dish and grown in the dark at 37° for 2 days to a colony size of 3–3.5 cm in diameter. To induce fruiting, plates were moved for 2 weeks to standard fruiting conditions. The number and size of primordia per transformant were scored. A small piece of gill tissue from primordia developed upon transformation with cosmid 40-5A was spread and nuclei in basidia stained with hematoxylin (Lu and Raju 1970).
DNA and RNA techniques:
An indexed genomic cosmid library derived from homokaryon AmutBmut was transformed into mutant 6-031 and screened for cosmids that were able to restore fruiting ability in this strain, following a SIB-selection procedure. The pab1+ wild-type gene of C. cinerea present in the cosmid backbone was used as a selection marker. Cosmid DNAs from 60 pools of each 96-well microtiter dish-arranged E. coli clones, and from subpools and individual clones of microtiter dish 40 were isolated (Bottoli et al. 1999).
Cloning was performed by standard methods (Sambrook et al. 1989). Plasmids were propagated in E. coli strain XL1-Blue (Stratagene, La Jolla, CA). Derivative pSphA of cosmid 40-5A is a ligation product between a 16-kb SphI fragment (13-kb genomic DNA + 3-kb cosmid backbone) and a 7.5-kb SphI fragment (2-kb genomic DNA + 5.5-kb cosmid backbone) in their natural order. NotI fragments of cosmid 40-5A were cloned into the NotI site of pBC SK (+) (Stratagene). Plasmids pNotB5 and pNotB7 contained the same DNA insert but in opposite orientation. The insert in pNotB5 was sequenced on both strands by primer walking (Microsynth, Balgach, Switzerland). Sequences were assembled with the program DNASTAR and analyzed with OMIGA 2.0, BLAST (NCBI). The whole sequence but 32 bp originating from the linker of the cosmid was submitted to GenBank (AF338438).
pNotB5 and pNotB7 gave rise to the following pBC SK (+) subclones: p5SmaCS and p5BamCS (used in Northern analysis) carry gene arf1 on a 1.4-kb NotI–SmaI and a 3.5-kb NotI–BamHI fragment, respectively. p5EcoCS and p5XbaCS contain arf1 and a truncated cfs1+ gene on a 3.8-kb NotI–EcoRI and a 5.5-kb NotI–XbaI fragment, respectively. p5SpeCS includes both arf1 and cfs1+ on a 7-kb SpeI fragment, whereas cfs1+ is truncated in p5KpnCS on the shorter 4.4-kb NotI–KpnI fragment. p7XbaCS carries truncated cfs1+ and kin1 copies on a 5-kb XbaI–NotI fragment. p7SpeCS contains a truncated kin1 on a 3.5-kb SpeI–NotI fragment. Subclones constructed in pBluescript KS (−) (Stratagene) were as follows: pPvu8.5 contains an 8.5-kb PvuII fragment covering the complete cfs1+ gene and the 3′ end of kin1. pBam3.5 and pSmaSpe5.5 carry cfs1+ on a 3.5-kb BamHI and a 5.5-kb SmaI–SpeI insert, respectively. pEco4.4 contains truncated cfs1+ and kin1 copies on a 4.4-kb EcoRI fragment. Furthermore, the 8.5-kb PvuII fragment, 3.5-kb BamHI fragment, and 4.4-kb EcoRI fragment were also cloned into pPAB1-2 containing the C. cinerea pab1+ gene, resulting in pPvu8.5-pab, pBam3.5-pab, and pEco4.4-pab, respectively.
A cut-and-shut strategy using NcoI and plasmid pNotB7 resulted in p7NcoCSΔcfs with a deletion in cfs1+ (Δbp 5296–5476). Similarly, p5BstCSΔkin with a deletion in kin1 (Δbp 8502–8619) were created from plasmid p5NotB5 by using BstEII. An AatII deletion (Δbp 6567–6803) in p5SpeCS yielded p5SpeCSΔgtl. An NruI deletion in arf1 (Δbp 676–972) in p5SmaCS gave rise to p5SmaCSΔarf, from which the SmaI insert was cloned into pSmaSpe5.5 to generate p5SpeCSΔarf. The insertion of a 3.5-kb SpeI fragment from pNotB7 at the SpeI site in p5SpeCSΔarf resulted in pNotB5Δarf. The T-to-G transversion found in the cfs1 allele of mutant 6-031 was introduced into plasmid p5SpeCS by exchanging a 1-kb PCR-amplified StuI–NdeI fragment with the wild-type sequence, yielding p5SpeCS/6-031. pNotB5/6-031 distinguishes from pNotB5 by the same T-to-G transversion.
Genomic DNA of C. cinerea strains was isolated from powdered lyophilized mycelium (Zolan and Pukkila 1986). Two overlapping fragments containing the cfs1 allele of mutant 6-031 were independently amplified six times from genomic DNA with specific primers (a 3.1-kb fragment using primers 5′ TCAAGTCGGGTCGGTAGAAG 3′ and 5′ TTTGTTTCGGAGCTTGACTG 3′ and a 1.1-kb fragment using primers 5′ GGACGCTTCAAGATTAGATC 3′ and 5′ CTCTGAAGGAATCGCTCTTG 3′) and sequenced using a ABI PRISM DNA Sequencing Kit and a Model 373A DNA sequencer (Perkin-Elmer). Sequences of PCR products separately amplified with the same primer set were identical. Presence of the same sequence in p5SpeCS/6-031 has also been verified by sequencing.
Southern blot analysis was performed with 10 μg of genomic DNA per sample following basic protocols (Sambrook et al. 1989). Total RNA of strain AmutBmut was extracted with a guanidinium isothiocyanate procedure (Chomczynski and Sacchi 1987) from powdered lyophilized C. cinerea mycelia or tissues of different fruiting stages. Poly(A)+ RNA was isolated with the Oligotex mRNA Midi kit (QIAGEN). Per sample, 10 μg of total RNA or 2.5 μg of poly(A)+ RNA were used for Northern blot analysis (Sambrook et al. 1989). Hybridization signals in Southern and Northern blot analyses were produced with DNA fragments labeled with [α-32P]dCTP by random primed DNA labeling (Boehringer Mannheim).
The 5′ and 3′ cDNA ends of the cfs1+ gene were determined with the 5′/3′ RACE kit (Roche Molecular Biochemicals) following the instructions of the manufacturer. Poly(A)+ RNA from 5-mm-sized primordia of homokaryon AmutBmut was used for cDNA synthesis. In the 5′ RACE, a cfs1 specific primer sp1 (5′ ACAATGCACAGGAGTACATC 3′) was employed to synthesize the first strand cDNA. Two cfs1-specific primers, sp2 (5′ GCAATGGCATTGAGTCGAG 3′) and sp3 (5′ TAGACGATAGGGTCATCTCC 3′), were applied in subsequent PCR reactions. In the 3′ RACE, two cfs1-specific primers, sp4 (5′ GATTTTGCCCTCAAGCCAC 3′) and sp5 (5′ CAATTCGAGCCTGCCCAG 3′), were used. RACE products were cloned into pBluescript KS (−) by T/A cloning (Marchuk et al. 1991) and sequenced with a Model 373A DNA sequencer. The full coding length of the cfs1+ cDNA was obtained by PCR, using the two primers cfsATG (5′ ATGCCGGCCCACCACCACCCTTC 3′) and cfsREV (5′ CGCCGAGGCCGCCGTGTAAACAC 3′). For sequencing, the PCR product was cloned into the EcoRV site in the β-galactosidase gene of pBluescript KS (−) via T/A cloning, resulting in construct pYL28 having the cfs1+ cDNA inserted in frame to the β-galactosidase gene.
Computer analysis of protein sequences:
Proteomics tools provided by ExPaSy Molecular Biology Server (Swiss Institute of Bioinformatics, Geneva) were used to perform protein pattern and profile searches (InterPro), transmembrane region detection (TMpred and TMHMM), and secondary structure predication (PSA and PSIpred). Hydrophilicity profile was calculated with Goldman/Engelman/Steitz parameters in OMIGA 2.0.
RESULTS
Morphological and genetic analysis of UV-mutant 6-031:
UV-mutant 6-031 has a growth rate (8 mm/day on YMG/T agar at 37°) and a mycelial morphology indistinguishable from its progenitor strain AmutBmut. Like homokaryon AmutBmut (Kertesz-Chaloupková et al. 1998; Badalyan et al. 2004), the mutant forms fused clamp cells at the hyphal septa and produces ∼109 oidia/plate in a light-dependent manner, indicating that mating-type functions in mutant 6-031 are not affected. Mutant 6-031 forms primary hyphal knots in the dark that mature into sclerotia when cultures are further kept in the dark (not shown). However, primary hyphal knots do not develop into secondary hyphal knots under standard fruiting conditions, suggesting that mutant 6-031 has a specific defect in fruiting body initiation (skn1). As in the case of the wild type, mutant 6-031 is also not able to initiate fruiting body development in constant dark, in constant light, at other temperatures, or on minimal medium.
Crosses between strain 6-031 and the AmutBmut co-isogenic monokaryons PS001-1 (A42, B42) and PS001-2 (A3, B1) gave rise to mature fruiting bodies, indicating that mutant 6-031 carries a recessive defect in fruiting body initiation. Self-compatible A43mut, B43mut descendants of the crosses were subjected to a fruiting test (49 clones from cross PS001-1 × 6-031; 64 clones from cross PS002-1 × 6-031). A total of 65% of the A43mut, B43mut progeny of cross PS001-1 × 6-031 (i.e., 32/49 clones) and 59% of cross PS002-1 × 6-031 A43mut, B43mut progeny (i.e., 38/64 clones) did not form secondary hyphal knots, suggesting that there is one defect in fruiting body initiation in mutant 6-031 (skn1) that is not linked to the mating type genes (P < 0.05; for comparison of background failure in fruiting initiation: 6 and 5% of A43mut, B43mut progenies of parallel crosses PS001-1 × AmutBmut and PS002-1 × AmutBmut, respectively, did not initiate fruiting). Ten clones (20%) and 12 clones (19%) of the A43mut, B43mut progenies of crosses PS001-1 × 6-031 and PS002-1 × 6-031, respectively, arrested primordia development at the basidial stage of prekaryogamy, indicating that mutant 6-031 has a second unlinked recessive defect in primordia maturation (mat; P < 0.05). The remaining clones split into two further groups, one with white mushrooms by a failure in basidiospore formation (two clones, i.e., 4% of A43mut, B43mut progeny of cross PS001-1 × 6-031; seven clones, i.e., 11% of A43mut, B43mut progeny of cross PS002-1 × 6-031) and one with mushrooms carrying mature black basidiospores (five clones and seven clones, i.e., 11% of A43mut, B43mut progeny of cross PS001-1 × 6-031 and PS002-1 × 6-031, respectively). Therefore, a third unlinked recessive gene is present in mutant 6-031 that acts in basidiospore formation (bad; P < 0.05).
Identification of a cosmid able to restore fruiting body initiation in mutant 6-031:
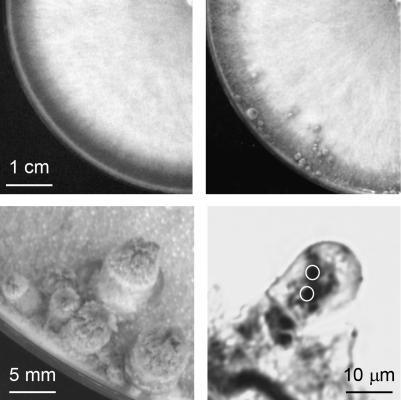
A genomic library of C. cinerea monokaryon AmutBmut (Bottoli et al. 1999) was employed to isolate DNA sequences that restored the fruiting initiation in mutant 6-031. In a first round of transformations using pools of 96 different cosmids, one transformant in a total number of 7948 (equivalent to the analysis of ∼45% of the entire library) formed primordia up to a size of 5–8 mm (Figure 1). In subsequent transformations dividing the positive pool 40 into subpools, 12 of 208 transformants of subpool 40-5 developed primordia, and in the final round using individual cosmids, 27 of 45 tested transformants of cosmid 40-5A. Basidia within these fully established primordia had either two distinct nuclei at the prekaryogamy stage (Figure 1) or no nucleus (data not shown).
Figure 1.
Cosmid 40-5A restores the defect in fruiting initiation in the Coprinopsis cinerea mutant 6-031 upon transformation. Mycelial morphology of mutant 6-031 before transformation (top left) and primordium formation after transformation (top right). Primordia of transformants are shown enlarged at the bottom left. Their basidia (bottom right) are in a stage of prekaryogamy, as indicated by the presence of the two nuclei (encircled) and their positions within the basidia (Kües 2000).
Transformation activities of subclones derived from cosmid 40-5A in strain 6-031:
Cosmid 40-5A with a 40-kb-sized insert of C. cinerea genomic DNA was digested with various restriction enzymes, and digestion mixtures were transformed into mutant 6-031. Digestions with BamHI, NotI, PvuII, or SphI still allowed initiation of fruiting body development in part of the transformants, unlike a number of other enzymes (EcoRI, EcoRV, KpnI, PstI, or XhoI). NotI divides cosmid 40-5A into three C. cinerea genomic DNA fragments (20, 10.5, and 8.9 kb, Figure 2A) plus an extra fragment representing the cosmid backbone (8.9 kb). NotI was chosen to construct pBC SK(+) subclones, which were cotransformed with plasmid pPAB1-2 into mutant 6-031. Plasmids pNotB5 and pNotB7 containing the same 10.5-kb insert fragment (NotI-B), restored fruiting body initiation (Figure 2). Some subclones of this C. cinerea genomic fragment (p5SpeCS, pSmaSpe5.5, pPvu8.5, and pBam3.5) were also active in fruiting body initiation. However, we noticed quantitative and qualitative variations in transformation activities, related to the length of transformed DNA fragments (Figure 2). The most effective constructs were cosmid 40-5A and its deletion derivative pSphA carrying the NotI-B fragment together with flanking DNA regions. Usually, 20–30% of the transformants of these cosmids initiated fruiting body development and developed primordia up to 5–8 mm in size. Four plasmids (pNotB5, pNotB7, p5SpeCS, and pSmaSpe5.5, containing a common 5.5-kb sequence) induced fruiting body initiation in 5–10% of the transformants, but the primordia formed were less developed and of a maximal size of only 2–5 mm. Normal cap and stipe differentiation were observed in these primordia. The reduction in percentage of transformants initiating fruiting might relate to the fact that cosmids 40-5A and pSphA carry the pab1+ selection marker, whereas the pBC SK(+) and pBluescript KS(−) constructs needed to be cotransformed with the pab1+ containing plasmid pPAB1-2. In contrast, this difference in the transformation procedure cannot account for the less-developed primordia obtained with the four plasmids. Moreover, upon cotransformation of plasmids pPAB1-2 and pPvu8.5 (having a 5.2-kb C. cinerea sequence in common with the former four plasmids), only 1–2% of transformants developed primordia, which were malformed and maximal 2–3 mm in size. In these primordia, the internal pileus trama tissue was missing. Plasmid pBam3.5 with a 3.5 kb BamHI fragment was the smallest construct regularly active in cotransformation, with 0.2–1% of transformants initiating fruiting but development arrested shortly after secondary hyphal knot formation at a size of ∼ 1 mm. When solely transforming constructs pPvu8.5-pab and pBam3.5-pab containing the pab1+ selection marker in addition to the 8.5-kb PvuII fragment or the 3.5-kb BamHI fragment, neither the transformation efficiency increased nor the primordia development improved in positive transformants obtained (not shown). The data suggest that the observed differences in transformation efficiency and degree of primordia maturation obtained with different C. cinerea fragments are not simply a result of variations in the transformation procedure.
Figure 2.
Identification of DNA fragments that restore fruiting body initiation in C. cinerea mutant 6-031. (A) The three C. cinerea genomic NotI fragments (A, B, C) present in cosmid 40-5A are shown, as well as the length and position of subfragments present in pSphA. The NotI-B fragment and subfragments were inserted into either pBC SK(+) or pBluescript KS(−) (see materials and methods). For transformation we used either 1 μg of cosmid DNA or 1 μg DNA of pBC SK(+)- or pBluescript KS(−)-based plasmids plus 1 μg of pPAB1-2 for cotransformation (experimental setup I). To equalize the absolute number of DNA molecules possibly acting in fruiting body initiation, 7 μg of cosmid 40-5A, 3.3 μg of pSphA, or 1 μg/7 kb DNA of pBC SK(+)- or pBluescript KS(−)-based plasmids plus 1 μg of pPAB1-2 were applied (experimental setup II). Per single experiment, between 32 and 248 transformants were obtained. Since percentages of transformants initiating fruiting were comparable between different transformations of the same DNA construct (not shown), transformants of different experiments were added. Transformation with 1 μg of pPAB1-2 and transformation with 1 μg of pPAB1-2 plus 1 μg of pBluecsript KS(−) or pBC SK(+) served as negative controls. From these control transformations, a total number of 1909, 215, and 132 transformants were obtained, respectively, and none of them initiated fruiting body formation. (B) The morphological progress in primordia development declines when reducing the length of DNA fragments in transformation. The phenomenon is indicated by the isolated primordia formed by the cosmid 40-5A, pSphA, pNotB5, pPvu8.5, and pBam3.5 transformants (top). Differentiation of cap and stipe tissues is normal in primordia induced by cosmid 40-5A, pSphA, and pNotB5. In contrast, the section through a primordium induced by plasmid pPvu8.5 shows that the internal pileus trama is missing (bottom).
The 3.5-kb BamHI fragment present in pBam3.5 originated from the central region of the 10.5-kb NotI-B fragment (Figure 2). In a total of 498 clones obtained from cotransformation of pPAB1-2 and pEco4.4, a partial overlapping 4.4-kb EcoRI fragment gave rise to only one transformant (0.2%) able to initiate fruiting. Two more transformants with primordia of 200 tested clones were obtained from transforming mutant 6-031 with plasmid pEco4.4-pab, containing both the 4.4-kb EcoRI fragment and the C. cinerea pab1+ gene. Interestingly, primordia of these three transformants developed to a size and shape comparable to that of pNotB5 (not shown). Other plasmids carrying either C. cinerea inserts from the flanking regions of the 3.5-kb BamHI fragment (p5KpnCS, p5EcoCS, p5SmaCS, and p7SpeCS) or inserts splitting this fragment in half (p5XbaCS and p7XbaCS) were all negative in transformation.
The 3.5-kb BamHI fragment is linked to the skn1 mutation in strain 6-031:
Strains 6-031 and JV6 have a distinct BglII restriction fragment length polymorphism (RFLP) in the DNA region covered by the 3.5-kb BamHI fragment. Forty-six of 588 randomly isolated descendants of a cross between the strains initiated fruiting body development on YMG/T medium. Forty-one of these clones had the A43mut, B43mut mating type and the RFLP pattern of monokaryon JV6. Both parental patterns were detected in the remaining five clones, but analysis of the A mating-type-linked pab1-1 allele (Kües et al. 2001) in their oidia identified them as dikaryons (data not shown). Either the JV6 or the 6-031 RFLP pattern was found in 30 randomly isolated non-fruiting clones. The data suggest that the 3.5-kb BamHI fragment is linked to the fruiting initiation defect (skn1) in mutant 6-031.
Transformation activities of cosmid 40-5A and derived subclones in skn1, mat+, bad+ strain OU3-1:
To determine whether the quantitative and qualitative differences in complementation activity with different plasmids in mutant 6-031 were not triggered by the two other mutations present in the strain, mutant strain OU3-1 carrying only the skn1 mutation was selected from the A43mut, B43mut progeny of cross PS001-1 × 6-031.
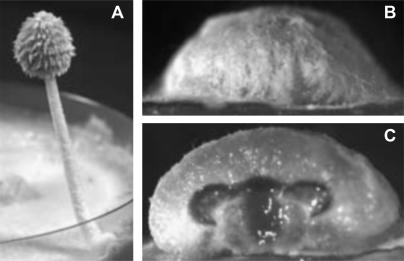
When transforming either cosmid 40-5A or its derivative pSphA into this strain (1 μg DNA per transformation), the defect in initiation was complemented in >40% of the transformants (61 positive/145 cosmid 40-5A transformants; 56 positive/131 pSphA transformants). Mature fruiting bodies with basidiospores were obtained in 18 and 13% of the cases, respectively (Figure 3A), showing that the arrest in primordia development in cosmid 40-5A and pSphA transformants of mutant 6-031 was due to its mat mutation.
Figure 3.
Fruiting behavior of transformants of homokaryon OU3-1. (A) Many transformants of cosmid 40-5A form mature fruiting bodies (∼4 cm). (B and C) pBam3.5 transformants form small, flat primordia (2 mm primordium shown) that lack inner pileus tissue.
Transformation efficiencies with strain OU3-1 generally were higher than those with mutant 6-031 and quantitative differences in transformation efficiencies were less pronounced. Nevertheless, in cotransformation of a pBC SK(+) or a pBluescript KS(−) derivative (usually 1 μg) and pPAB1-2 (1 μg), we again observed qualitative differences in complementation efficiency. Unlike for the cosmids, no mature fruiting bodies were obtained using any of the plasmid constructs. pNotB7 initiated fruiting in strain OU3-1 in 20% of cases (24 positive/118 total transformants) but development arrested latest at a primordia size of ∼5–6 mm. pSmaSpe5.5, pPvu8.5, and pBam3.5 were all less efficient in transformation with ∼15% transformants initiating fruiting (16 positive/110 pSmaSpe5.5 transformants, 24 positive/177 pPvu8.5 transformants, 29 positive/213 pBam3.5 transformants). Primordia development in the group of pSmaSpe5.5 transformants arrested at maximum sizes of 3–4 mm, and in the group of pPvu8.5 transformants arrested at a size of 2 mm. Most positive transformants of pBam3.5 had primordia of ≤1 mm in size, but eight transformants had primordia of 2–3 mm. Primordia from pPvu8.5 and pBam3.5 transformants were always malformed, being more flat than wild-type primordia. Moreover, they lacked inner tissues of the pileus (Figure 3, B and C). The results indicate again that pBam3.5 carries the gene for fruiting body initiation, although subsequent development does not follow the normal route. pBluescript KS(−) control transformants (51 clones) and control transformants of only pPAB1-2 (63 clones) gave no positive transformants. Transformants of p5SpecCS (350 ng DNA were used) formed normal-shaped primordia with final sizes of 2–3 mm with a transformation frequency of 6% (7 positives/105 transformants).
Characterization of the 10.5-kb NotI-B region:
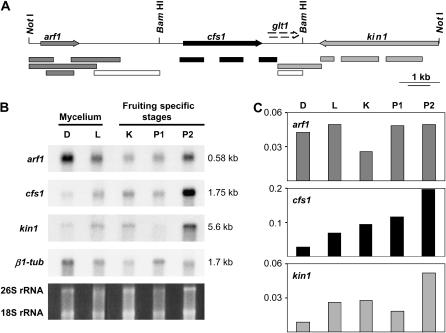
The quantitative and qualitative differences in complementation activities with different plasmids led us to sequence the whole genomic NotI-B region, which is 10,526 bp in size. A GenBank BLAST search with this sequence revealed four potential coding regions, whose deduced protein sequences showed highest similarities to the human ADP-ribosylation factor-like protein 2 ARL2 (67% identity and 82% similarity over a length of 185 aa; accession no. P36404), the CFA synthase of Escherichia coli (32% identity and 48% similarity over a length of 367 aa; accession no. P30010), the galacturonosyl transferase Cap1E in Streptococcus pneumoniae (32% identity and 49% similarity over a length of 91 aa; accession no. L36873), and the C-terminal part of the kinesin-like protein UNC-104 in Caenorhabditis elegans (32% identity and 51% similarity over a length of 271 aa; accession no. P23678) (Figure 4A). Whereas potential start codons are present on the NotI-B fragment for the potential ARL2- and cfa-like genes, the UNC-104-related sequence is incomplete and 5′-truncated by the NotI site. For the stretch of DNA translating in a Cap1E-like sequence, there is no obvious start codon. In accordance, a transcription analysis of the entire 10.5-kb NotI-B fragment detected three transcripts (Figure 4B), corresponding in location to the deduced coding regions for the ARL2-like protein (gene arf1), for the potential CFA (gene cfs1), and for the UNC-104-like kinesin (gene kin1). Weak transcripts for kin1 were detected in Northern blots of total RNA from homokaryon AmutBmut. When using poly(A)+ RNA to increase the sensitivity, transcripts for arf1, cfs1, and kin1 were well detected but we never observed a transcript for a further gene.
Figure 4.
Transcription profile of the NotI-B region. (A) Sequence analysis of pNotB5 revealed four potential C. cinerea open reading frames (indicated by the arrows arf1, cfs1, glt1, and kin1), of which one (kin1) is only partially present on the NotI-B fragment and one (glt1) has no obvious start codon. Three transcripts, arf1, cfs1, and kin1 were detected in Northern blot analysis of total RNA and poly(A)+RNA by various DNA probes (shown as differentially shaded boxes in relation to the respective transcript detected). No signal was detected with two probes (open boxes) in the intergenic arf1–cfs1 and cfs1–kin1 regions. (B) Gene expression was analyzed with poly(A)+RNA isolated from vegetative mycelium grown on YMG/T agar at 37° for 4 days in the dark (sample D) or 7 days in the light (sample L). Letter K denotes RNA isolated from cultures grown for 6 days at 37° in the dark that developed numerous primary hyphal knots within the aerial mycelium. P1 and P2 indicate RNA extracted from isolated 1-mm-sized primordia at a prekaryogamy stage and from isolated 1-cm-sized primordia undergoing meiotic divisions, respectively. Primordia were harvested at day 3 (P1) and day 5 (P2) after fully grown YMG/T cultures were incubated at 25° under standard fruiting conditions. The 26S and 18S rRNA are shown to indicate the quality of poly(A)+RNA samples. poly(A)+RNA was isolated from total RNA only once per developmental stage with comparable yields. The relative strength of signals in blots of poly(A)+RNA followed that of earlier total RNA blots. Total RNA has been analyzed before from two to three independent RNA isolations from dark and light incubated mycelium and developing primordia. Parallel blots from the same total and poly(A)+RNA isolations gave similar results. (C) Densitometric quantification of the transcripts shown in B. For calibration, the β1-tubulin (β1-tub) transcript per poly(A)+RNA was used as standard. Transcript levels are given in arbitrary units (transcripts/β1-tub transcript).
The arf1 transcript is ∼0.58 kb in size (Figure 4B). arf1 is expressed in all developmental stages tested (mycelia grown in constant dark and constant light, dark-grown mycelium containing primary hyphal knots, 1-mm-sized primordia at a prekaryogamy stage of tissue differentiation, and 1-cm-sized primordia being after karyogamy in meiosis) at a similar level, but decreases slightly at the stage of primary hyphal knot formation (Figure 4C). The expression of the 1.75-kb-sized cfs1 transcript is low in mycelium growing in the dark compared to fully grown cultures with primary hyphal knots and compared to older cultures grown in constant light (Figure 4B). cfs1 transcription continues in developing primordia and is highest in mature primordia (Figure 4C). Gene kin1 has a 5.6-kb transcript (Figure 4B), which is poorly expressed in all stages except in primordia at the meiotic stage just about to undergo rapid stipe elongation and cap expansion (Figure 4C).
Gene cfs1 is essential for fruiting body initiation:
The transformation data of mutants 6-031 (shown in Figure 2) and OU3-1 did not yet allow us to definitely assign the fruiting restoring ability of the 10.5-kb NotI-B fragment to a single gene. The higher frequency of transformants initiating fruiting and the qualitative progress in primordia development with larger DNA fragments indicated that more than one of the cloned genes could contribute together or sequentially to fruiting body initiation. Southern blot analysis, using genomic DNA from mutant 6-031 and homokaryon AmutBmut digested with a number of restriction enzymes and three DNA probes that together covered the whole NotI-B fragment (a 4.4-kb NotI–KpnI fragment, a 3.5-kb BamHI fragment, and a 3.5-kb SpeI–NotI fragment), excluded the possibility of a large deletion in this region in mutant 6-031 (not shown). Moreover, each probe detected only single bands (not shown), indicating the single copy nature of all cloned genes.
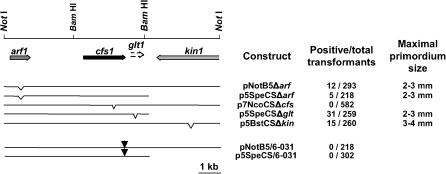
Next, we constructed plasmids with deletions within the coding region of arf1, cfs1, or kin1 and within the uncertain glt1 region (see material and methods) and tested them by transforming mutant 6-031. Only plasmid p7NcoCSΔcfs, carrying a deletion in cfs1+, lost the ability to restore fruiting body initiation in the mutant (Figure 5), defining cfs1 as the gene active in fruiting body initiation. Since interruptions within other genes did not fundamentally influence the transformation efficiency and since primordia formed were of normal shape and 2–4 mm in size as were those obtained with the unmutated NotI-B fragment in the same series of experiments (Figure 5), it is possible that the general chromosomal environment of gene cfs1 rather than the presence of other functional genes influences its expression and accounts for the developmental differences observed in our transformation experiments.
Figure 5.
Gene cfs1 is essential for fruiting body initiation. Plasmids pNotB5Δarf, p5SpeCSΔarf, p5SpeCSΔglt, and p5BstCSΔkin containing a deletion in arf1, kin1, or the glt1 region (positions of deletions within subcloned fragments are indicated by open triangles) still permitted initiation of fruiting body formation when transformed into mutant 6-031. Positive transformants developed 2–4 mm primordia with normal morphology. In contrast, a deletion in cfs1+ in plasmid p7NcoCSΔcfs abolished the ability to induce fruiting body formation. Likewise, plasmids pNotB5/6-031 and p5SpeCS/6-031 carrying the T-to-G transversion found in the cfs1 allele of mutant 6-031 (positions are marked by arrowheads) did not activate fruiting body formation in mutant 6-031. In all transformations, 1 μg DNA/7 kb length of the test plasmids plus 1 μg of pPAB1-2 have been used.
In the cfs1 allele from mutant 6-031, a single T-to-G transversion was found, which leads to a Y441D amino acid substitution in the C-terminus of the deduced protein sequence (Figure 6). When introducing this T-to-G transversion into the cloned NotI-B fragment and a smaller 7-kb NotI–SpeI fragment, neither of the two resulting plasmids, pNotB5/6-031 and p5SpeCS/6-031, was able to restore fruiting body initiation (Figure 5), indicating that the point mutation is indeed the cause of the inability to initiate fruiting body formation in mutant 6-031.
Figure 6.
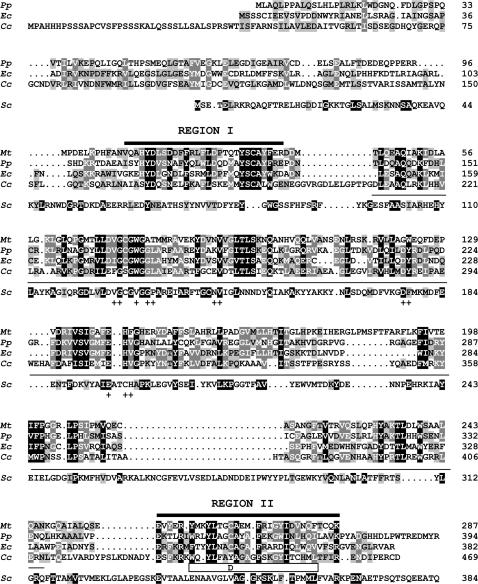
Sequence alignment of C. cinerea Cfs1 (Cc) to bacterial cyclopropane fatty acid synthases (Mt, Pp, Ec) and to yeast Δ24-sterol methyltransferase ERG6. Pp, protein ylp3 of the Gram− bacterium Pseudomonas putida (P31049); Ec, CFA of the Gram− bacterium E. coli (P30010); Mt, CFA1 of the Gram+ bacterium Mycobacterium tuberculosis (Q11195); Sc, ERG6 of yeast S. cerevisiae (S42003). In the ERG6 protein sequence, amino acids identical to the C. cinerea Cfs1 sequence are shaded in black, similar ones are shaded in gray. A line marks the structurally conserved SAM-dependent MTase fold. A “+” indicates those amino acids shown in other proteins to contact SAM (Fauman et al., 1999). The open box underneath the C-terminal end of the protein indicates potential membrane-interacting regions in Cfs1. The letter D in this box indicates the amino acid exchange found in Cfs1 of mutant 6-031 at position Y441. Region I and region II denote protein regions specific to cyclopropane fatty acid synthases.
The cfs1 gene encodes a protein highly similar to bacterial cyclopropane fatty acid synthases:
The cDNA of the cfs1+ gene is 1776 bp in size with a 66-bp-long nucleotide sequence upstream of the first start codon ATG and a 300-bp-long nucleotide sequence downstream of the stop codon TGA. Comparison of the genomic DNA and cDNA sequences revealed an open reading frame (ORF) of 1407 bp interrupted by 10 introns of 53–70 bp in size with typical C. cinerea 5′- and 3′-splice sites and branch-receptor sequences (Boulianne et al. 2000). The cfs1 promoter region contains a CAAT element 60–57 bp upstream of the transcription initiation site. No classical TATA box is found, but an AATAAAAA sequence is 37–30 bp upstream of the transcription initiation site. A total of 456 bp are upstream to the transcription initiation site in the 3.5-kb BamHI fragment having the smallest sequence regularly but inefficiently active in transformation (Figure 2). This sequence should mediate at least some promoter activity. Within the cfs1 coding region, 332 bp downstream of the start codon ATG in the second exon is the EcoRI restriction site used to construct plasmids pEco4.4 and pEco4.4-pab with 5′ truncated cfs1+ copies. Homologous recombination at the natural cfs1 locus within the C. cinerea 6-031 genome or in-frame integration into another gene might therefore explain the occasional transformants initiating primordia development obtained with these two plasmids (not shown).
The cfs1+ ORF encodes a polypeptide of 469 amino acid residues with a predicted molecular mass of 52 kD. The Cfs1 protein has an overall high identity to a number of bacterial CFA synthases (Figure 6), a particular subfamily of the S-adenosyl-l-methionine (SAM)-dependent C-methyltransferases (MTases) (Fauman et al. 1999). NCBI database searches for related proteins among others also identified a functional enzyme from the plant Sterculia foetida (AAM33848) and potential but not yet characterized eukaryotic protein products from the filamentous ascomycetes Neurospora crassa (EAA32979) and Magnaporthe grisea (EAA50372), the basidiomycete Ustilago maydis (XP_751900.1), the plant Arabidopsis thaliana (BAB02771.1), the worm C. elegans (T18571), and the slime mold Dictostelium discoideum (EAL65538). In contrast, no homologous human or any other mammal sequence seems to exist in the GenBank databases. Within the fungi, no closely related sequences were found in the ascomycete yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae. In the yeast S. cerevisiae, the most similar protein is ERG6, a Δ24-sterol-C-methyltransferase (31% identity and 48% similarity over a length of 236 aa; 19% identity and 33% similarity over the whole protein length; S42003; Figure 6).
Consistent with a function as SAM-dependent MTase, a SAM-binding motif is present in Cfs1 between aa 209 and 338 (Figure 6). Protein profile programs (PSA and PSIpred) predict this region to adopt the typical 6 helices-7 strands configuration of SAM-binding domains (referred to as a SAM-dependent MTase fold) (Fauman et al., 1999). All but one amino acid shown to contact SAM in structurally characterized SAM-dependent MTases (Fauman et al., 1999) are conserved in Cfs1 (Figure 6). Both programs TMpred and TMHMM predict potential transmembrane domains in the C-terminus of the C. cinerea Cfs1 protein (aa 435–453 and aa 437–456), closely behind the SAM-binding motif. The Y441D amino acid substitution in mutant 6-031 is localized in this region (Figure 6). This substitution overturns the prediction of a transmembrane domain and gives rise to higher hydrophilicity (Goldman/Engelman/Steitz prediction) at the C-terminal region (not shown). Overall, the wild-type Cfs1 protein appears to be hydrophilic without any long hydrophobic region.
DISCUSSION
In this study, we characterized the first mutant of the basidiomycete C. cinerea with a specific defect in fruiting body initiation at the transition stage from primary to secondary hyphal knots. Through mutant complementation, we cloned the cfs1+ gene that encodes a potential CFA synthase. Transcription of the gene is low during growth of the mycelium compared to fully grown mycelium susceptible to a light signal for induction of secondary hyphal knot formation. Transcription is also higher in older cultures grown in constant light, indicating a possible age effect in gene expression. When transforming the subcloned cfs1+ gene into the fruiting defective mutants 6-031 and OU3-1, we observed quantitative and qualitative differences in complementation activities with cfs1+-carrying DNA fragments of different length. For efficient complementation, a DNA fragment of >10 kb was needed. Smaller DNAs could induce formation of the unstructured secondary hyphal knots but subsequent development arrested early in primordia formation and, in some instances, lead to defects in tissue differentiation. Such effects on primordia development are not expected if the cfs1 gene is needed only in induction of secondary hyphal knots. High expression of the gene during primordia development further supports that the gene also functions in later stages of development.
Interruptions within the open reading frames of the neighboring genes (arf1, kin1, and the non-transcribed glt1 region downstream to the cfs1 gene, Figure 5) present in the 10.5-kb active NotI-B fragment had no major effect on the complementation activity of cfs1+. This indicates as a likely cause for the differences in transformation activity that the larger chromosomal environment plays a role in proper cfs1 gene expression. Transcription of arf1 is somewhat decreased at the onset of cfs1 expression during primary hyphal knot formation, and kin1 is specifically transcribed at subsequent stages of primordia formation when cfs1 transcription is the highest. Possibly, the transcription profiles of arf1 and kin1 influence the expression of cfs1 during early and later stages of fruiting body development.
Structure of the Cfs1 protein:
Within the C. cinerea Cfs1 protein, we identified a potential SAM-binding domain, a SAM-dependent MTase fold (Fauman et al. 1999). CFA synthases are C-MTases that in bacteria transfer a methylene group from SAM to a C-atom in unsaturated membrane localized phospholipids, thereby forming a cyclopropane ring (Grogan and Cronan 1997). Plant and fungal Δ24-sterol C-MTase are the closest related to CFA synthases, possibly because their enzyme activities are both linked to lipid bilayers (Taylor and Cronan 1979; Leber et al. 1994). Low homology is found over the whole protein length between ERG6 of S. cerevisiae and Cfs1 of C. cinerea, with the highest conservation in the SAM-binding motif (Figure 6). In ERG6, directly at the N-terminal end of the SAM-dependent MTase fold is a sequence (DFYEYGWGSSFHFS; residues 77–92) referred to as region I that is highly specific to all Δ24-sterol C-MTases, has sterol binding activity, and forms an α-helix with a loop structure that targets into the substrate pocket (Nes et al. 1998, 2004). This sequence is not present in the family of CFA synthases (Figure 6). Instead, a highly specific sequence occupies the corresponding position (region I consensus: V/I85XXH88Y100D100V/L/I92S77N/D100D/N65F65F/Y100XL/I73W/F85L92D77P54S/T69M/L77T/S58Y100S/T100C85A92Y/F54F/W100E38R/K38; see supplemental material at http://www.genetics.org/supplemental/). In the crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis, this CFA synthase signature sequence adopts a helix-loop-helix-loop structure that coats the surface of a hydrophobic tunnel from the entrance to active SAM-binding sites. The alkyl chains of model substrates enter these pockets in a U-shaped manner, thereby contacting amino acids from region I (Huang et al. 2002).
In E. coli, CFA synthase is a soluble protein found in the cell cytoplasm that uses SAM as a soluble and unsaturated fatty acid (UFA)-containing phospholipid as an insoluble substrate while transiently accessing to both inner and outer leaflets of intact UFA-containing membranes (Taylor and Cronan 1979). The substrate C-double bond, positioned at 9–11 carbon units from the glycerol backbone of the phospholipid molecule, is located deeply within the hydrophobic core of the membrane bilayer (Gally et al. 1979; Seelig and Seelig 1980). Inhibitor studies with sulfhydryl-modifying reagents and C-terminal truncation (50 aa) suggested that the C-terminus and possibly C354 within the C-terminus have a role in catalysis or interaction with the membrane (Wang et al. 1992). However, changing C354 to an alanine or serine did not result in loss of function (Grogan and Cronan 1997; Courtois et al. 2004). The C-terminal region (region II in Figure 6, consensus: XV/M/L/I50XXQ/E73XXXR/K65V/M/L/I54Y/W/F100XXY96L/M73XXC69A58XXF100K/R58XG58XL/I/V81D/N58V/L73XQ77V/M/L/I62T50XK/R96; see supplemental material at http://www.genetics.org/supplemental/) is nevertheless important, since the Y441D substitution in our mutant 6-031 (corresponding to A353 in E. coli Cfa) resulted in a loss of function. Computer programs predict the wild-type Cfs1 of C. cinerea being a cytoplasmic protein, like the CFA synthase of E. coli, with two transmembrane domains in the C-terminus. Possibly the C-terminus functions in transiently anchoring the Cfs1 protein to the membrane and/or represents part of the catalytic domain. In the crystallized M. tuberculosis enzymes, the C-terminal end with region II forms an α-helix and a β-sheet. C269 within this α-helix (corresponding to C354 in E. coli Cfa) is in the vicinity of the active site, while the β-sheet dangles away from the site (Huang et al. 2002).
Occurrence of cyclopropane fatty acid synthases and their products:
CFA synthases have been found in a broad range of bacteria, with cis-9,10-methylenehexadecanoic acid (17CFA), cis-9,10-methyleneoctadecanoic acid [MOA, dihydrosterculic acid (DHSA), C19], and cis-11,12-methyleneoctadecanoic acid (lactobacillic acid, C19) being characteristic bacterial CFAs (for review see Grogan and Cronan 1997). In E. coli, the CFA synthase is not essential for growth under an assortment of experimental conditions but improves survival in low-pH environments (Chang and Cronan 1999). In other bacteria, the production of CFAs also relates to stress conditions. As a consequence of CFA production, membrane properties, in particular membrane fluidity, alter with enhanced bacterial stress tolerance (Couto et al. 1996; Sajbidor 1997; Chang and Cronan 1999). Phospholipids containing CFAs have a broader transition temperature range and increased rigidity than those containing UFAs, which confers more resistance of the membrane lipid matrix to environmental perturbations (Dufourc et al. 1984; Perly et al. 1985).
In eukaryotes, CFAs have only sporadically been reported, while the chemical structures are far more diverse. For example, cyclopropyl hydroxy-eicosanoids were described in a red alga (Nagle and Gerwick 1990), cyclopropanated C19 straight-chain fatty acid (cladocroic acid) in a sponge (D'Auria et al. 1993), cyclopropane containing eicosanoid (C20) in a soft coral (White and Jensen 1993), and CFAs with 17, 18 and 19 C-atoms in females and eggs of millipedes (Oudejans and van der Horst 1978). DHSA has been identified in trypanosomatid protozoa and CFA synthase activity has been demonstrated (Li et al. 1993). DHSA synthesis from oleoyl phospatidylcholines has recently also been observed in tobacco cells after transformation of a gene from the plant Sterculia foetida (Bao et al. 2003). DHSA and cis-9,10-methyleneheptadecanoic acid occur in roots of S. foetida (Kaimal and Lakshminarayana, 1970). Biological roles of CFAs in eukaryotes have still to be clarified. Functions in cold hardiness and drought resistance in plants and desiccation tolerance in millipedes have been discussed (Grogan and Cronan 1997). In slime molds, CFA levels have been linked to cellular aggregation and cAMP metabolism (Saito and Ochiai 1998; Matsuoka et al. 2004). These latter observations can be of special interest to our study since in C. cinerea there is an increase in levels of cAMP at the onset of fruiting and cAMP has been shown in particular mutants to induce fruiting (Kües et al. 2004).
To our knowledge, no cyclopropanated moiety has so far been reported in higher fungal lipids. However, in the higher basidiomycetes, the unsaturated linoleic acid is the major constituent of fatty acids (Solberg 1989; Bonzom et al. 1999; Sakai and Kajiwara 2004). The related oleic acid is shown in E. coli to be a substrate for the action of CFA synthase (Marinari et al. 1974) and preliminary expression studies suggest that the E. coli and C. cinerea enzymes are at least partially interchangeable in function (S. Loos, M. Aebi and U. Kües, unpublished results).
In this study, we show that the C. cinerea cfs1 gene is superfluous for vegetative mycelial growth, but essential for fruiting body development. As in bacteria, the physical properties of cellular membranes may alter through production of CFAs by Cfs1 and this could be the trigger to initiate fruiting body morphogenesis in the fungus. Accordingly, in feeding experiments membrane-interactive compounds such as sucrose esters of fatty acids, plant saponins, and cerebrosides induced fruiting body development in various basidiomycetes (Kawai 1989; Oita and Yanagi 1993; Mizushina et al. 1998; Magae et al. 2004). From such studies, membrane alteration has been postulated to be a stress signal that promotes the fungus to shift from vegetative to reproductive growth.
Acknowledgments
We are very grateful to Ben Lu for help with nuclear staining. P.S. holds a Ph.D. studentship from the Mahasarakham University, Thailand. Parts of this work were financed by the Swiss National Science Foundation (grants 31-46940.96, 31-46940.96/2 and 31-59157.99) and the Eidgenössische Technische Hochschule-Zürich. The lab in Göttingen was funded by the Deutsche Bundesstiftung Umwelt.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. AF338438.
References
- Arima, T., M. Yamamoto, A. Hirata, S. Kawano and T. Kamada, 2004. The eln3 gene involved in fruiting body morphogenesis of Coprinus cinereus encodes a putative membrane protein with a general glycosyltransferase domain. Fungal Genet. Biol. 41: 805–812. [DOI] [PubMed] [Google Scholar]
- Badalyan, S. M., E. Polak, R. Hermann, M. Aebi and U. Kües, 2004. Role of peg formation in clamp cell fusion in homobasidiomycete fungi. J. Basic Microbiol. 44: 167–177. [DOI] [PubMed] [Google Scholar]
- Bao, X., J. J. Thelen, G. Bonaventura and J. B. Ohlrogge, 2003. Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J. Biol. Chem. 278: 12846–12853. [DOI] [PubMed] [Google Scholar]
- Bertossa, R. C., U. Kües, M. Aebi and M. Künzler, 2004. Promoter analysis of cgl2, a galectin encoding gene transcribed during fruiting body formation in Coprinopsis cinerea (Coprinus cinereus). Fungal Genet. Biol. 41: 1120–1131. [DOI] [PubMed] [Google Scholar]
- Bonzom, P. M. A., A. Nicolaou, M. Zloh, W. Baldeo and W. A. Gibbons, 1999. NMR lipid profile of Agaricus bisporus. Phytochemistry 50: 1311–1321. [Google Scholar]
- Bottoli, A.P. F., K. Kertesz-Chaloupková, R. P. Boulianne, J. D. Granado, M. Aebi et al., 1999. Rapid isolation of genes from an indexed genomic library of C. cinereus in a novel pab1+ cosmid. J. Microbiol. Methods 35: 129–141. [DOI] [PubMed] [Google Scholar]
- Boulianne, R. P., Y. Liu, M. Aebi, B. C. Lu and U. Kües, 2000. Fruiting body development in Coprinus cinereus: regulated expression of two galectins secreted by a non-classical pathway. Microbiology 146: 1841–1853. [DOI] [PubMed] [Google Scholar]
- Chang, Y. Y., and J. E. Cronan, Jr., 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol. Microbiol. 33: 249–259. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and N. Sacchi, 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162: 156–159. [DOI] [PubMed] [Google Scholar]
- Clergeot, P.-H., G. Ruprich-Robert, Y. Liu, S. Loos, P. Srivilai et al., 2003. Mutants in initiation of fruiting body development of the basidiomycete Coprinus cinereus. Fungal Genet. Newsl. 50(Suppl.): 68. [Google Scholar]
- Courtois, F., C. Guérard, X. Thomas and O. Ploux, 2004. Escherichia coli cyclopropane fatty acid synthase. Mechanistic and site-directed mutagenic studies. Eur. J. Biochem. 271: 4769–4778. [DOI] [PubMed] [Google Scholar]
- Couto, J. A., N. Rozes and T. Hogg, 1996. Ethanol-induced changes in the fatty acid composition of Lactobacillus hilgardii, its effects on plasma membrane fluidity and relationship with ethanol tolerance. J. Appl. Bacteriol. 81: 126–132. [Google Scholar]
- D'Auria, M. V., L. G. Paloma, L. Minale, R. Riccio and A. Zampella, 1993. Metabolites of the New Caledonian sponge Cladocroce incurvata. J. Nat. Prod. 56: 418–423. [Google Scholar]
- Dufourc, E. J., I. C. Smith and H. C. Jarrell, 1984. Role of cyclopropane moieties in the lipid properties of biological membranes: a 2H NMR structural and dynamical approach. Biochemistry 23: 2300–2309. [Google Scholar]
- Fauman, E. B., R. M. Blumenthal and X. D. Cheng, 1999. Structure and evolution of AdoMet-dependent methyltransferases, pp. 1–38 in S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions, edited by X. D. Cheng and R. M. Blumenthal. World Scientific, Singapore.
- Gally, H. U., G. Pluschke, P. Overath and J. Seelig, 1979. Structure of Escherichia coli membranes. Phospholipid conformation in model membranes and cells as studied by deuterium magnetic resonance. Biochemistry 18: 5605–5610. [DOI] [PubMed] [Google Scholar]
- Granado, J. D., K. Kertesz-Chaloupková, M. Aebi and U. Kües, 1997. Restriction enzyme-mediated DNA intergration in Coprinus cinereus. Mol. Gen. Genet. 256: 28–36. [DOI] [PubMed] [Google Scholar]
- Grogan, D. W., and J. E. Cronan, Jr., 1997. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol. Mol. Biol. Rev. 61: 429–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., C. V. Smith, M. S. Glickman, W. R. Jacobs, Jr. and J. C. Sacchettini, 2002. Crystal structure of myolic acid cyclopropane synthase from Mycobacterium tubercolosis. J. Biol. Chem. 277: 11559–11569. [DOI] [PubMed] [Google Scholar]
- Kaimal, T. N. B., and G. Lakshminarayana, 1970. Fatty acid compositions of lipids isolated from different parts of Ceiba pentandra, Sterculia foetida and Hydnocarpus wightiana. Phytochem. 9: 2225–2229. [Google Scholar]
- Kawai, G., 1989. Molecular species of cerebrosides in fruiting bodies of Lentinus edodes and their biological activity. Biochim. Biophys. Acta 1001: 185–190. [DOI] [PubMed] [Google Scholar]
- Kertesz-Chaloupková, K., P. J. Walser, J. D. Granado, M. Aebi and U. Kües, 1998. Blue light overrides repression of asexual sporulation by mating type genes in the basidiomycete Coprinus cinereus. Fungal Genet. Biol. 23: 95–109. [DOI] [PubMed] [Google Scholar]
- Kües, U., 2000. Life history and developmental processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kües, U., J. D. Granado, R. Hermann, R. P. Boulianne, K. Kertesz-Chaloupková et al., 1998. The A mating type and blue light regulate all known differentiation processes in the basidiomycete Coprinus cinereus. Mol. Gen. Genet. 260: 81–91. [DOI] [PubMed] [Google Scholar]
- Kües, U., T. Y. James, R. Vilgalys and M. P. Challen, 2001. The chromosomal region containing pab-1, mip, and the A mating type locus of the secondary homothallic homobasidiomycete Coprinus bilanatus. Curr. Genet. 39: 16–24. [DOI] [PubMed] [Google Scholar]
- Kües, U., E. Polak, A. P. F. Bottoli, M. Hollenstein, P. J. Walser et al., 2002. a Vegetative development in Coprinus cinereus, pp. 133–164 in Molecular Biology of Fungal Development, edited by H. D. Osiewacz. Marcel Dekker, New York.
- Kües, U., P. J. Walser, M. J. Klaus and M. Aebi, 2002. b Influence of activated A and B mating type pathways on developmental processes in the basidiomycete Coprinus cinereus. Mol. Genet. Genomics 268: 262–271. [DOI] [PubMed] [Google Scholar]
- Kües, U., M. Künzler, A. P. F. Bottoli, P. J. Walser, J. D. Granado et al., 2004. Mushroom development in higher basidiomycetes; implications for human and animal health, pp. 431–470 in Fungi in Human and Animal Health, edited by R. K. S. Kushwaha. Scientific Publishers, Jodhpur, India.
- Leber, R., E. Zinser, G. Zellnig, F. Paltauf and G. Daum, 1994. Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast 10: 1421–1428. [DOI] [PubMed] [Google Scholar]
- Li, R. X., S. Ganguli and R. A. Pascal, Jr., 1993. Synthesis of sulfur-substituted phosphatidylethanolamines and inhibition of protozoan cyclopropane fatty acid synthase. Tetrahedron Lett. 34: 1279–1282. [Google Scholar]
- Lu, B. C., and N. B. Raju, 1970. Meiosis in Coprinus. II. Chromosome pairing and the lampbrush diplotene stage of meiotic prophase. Chromosoma 29: 305–316. [DOI] [PubMed] [Google Scholar]
- Lu, B. C., N. Gallo and U. Kües, 2003. White-cap mutants and meiotic apoptosis in the basidiomycete Coprinus cinereus. Fungal Genet. Biol. 39: 82–93. [DOI] [PubMed] [Google Scholar]
- Magae, Y., T. Nishimura and S. Ohara, 2004. 3-O-alkyl-D-glucose derivatives induce fruit bodies of Pleurotus. Mycol. Res. 109: 374–376. [DOI] [PubMed] [Google Scholar]
- Marchuk, D., M. Drumm, A. Saulino and F. S. Collins, 1991. Construction of T-vectors, a rapid and general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 19: 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinari, L. A., H. Goldfine and C. Panos, 1974. Specificity of cyclopropane fatty acid synthesis in Escherichia coli. Utilization of isomers of monounsaturated fatty acid. Biochemistry 13: 1978–1983. [DOI] [PubMed] [Google Scholar]
- Matsuoka, S., H. Kuwayama, D. Ikeno, M. Oyama and M. Maeda, 2004. Defect in peroximal multifunctional enzyme MFE1 affects cAMP relay in Dictyostelium. Dev. Growth Differ. 46: 195–199. [DOI] [PubMed] [Google Scholar]
- Mizushina, Y., L. Hanashima, T. Yamaguchi, M. Takemura, F. Sugawara et al., 1998. A mushroom fruiting body-inducing substance inhibits activities of replicative DNA polymerases. Biochem. Biophys. Res. Commun. 249: 17–22. [DOI] [PubMed] [Google Scholar]
- Moore, D., M. M. Y. Elhiti and R. D. Butler, 1979. Morphogenesis of the carpophore of Coprinus cinereus. New Phytol. 83: 695–722. [Google Scholar]
- Muraguchi, H., and T. Kamada, 1998. The ich1 gene of the mushroom Coprinus cinereus is essential for pileus formation in fruiting. Development 125: 3133–3141. [DOI] [PubMed] [Google Scholar]
- Muraguchi, H., and T. Kamada, 2000. A mutation in the eln2 gene encoding a cytochrome P450 of Coprinus cinereus affects mushroom morphogenesis. Fungal Genet. Biol. 29: 49–59. [DOI] [PubMed] [Google Scholar]
- Murata, Y., M. Fujii, M. E. Zolan and T. Kamada, 1998. Molecular analysis of pcc1, a gene that leads to A-regulated sexual morphogenesis in Coprinus cinereus. Genetics 149: 1753–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle, D. G., and W. H. Gerwick, 1990. Isolation and structure of constanolactones A and B, new cyclopropyl hydroxyeicosanoids from the temperate red alga Constantinea simplex. Tetrahedron Lett. 31: 2995–2998. [Google Scholar]
- Nes, W. D., B. S. McCourt, W. X. Zhou, J. Ma, J. A. Marshall et al., 1998. Overexpression, purification, and stereochemical studies of the recombinant (S)-adenosyl-L-methionine: delta 24(25)- to delta 24(28)-sterol methyl transferase enzyme from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 353: 297–311. [DOI] [PubMed] [Google Scholar]
- Nes, W. D., P. Jayasimha, W. Zhou, R. Kanagasabai, C. Jin et al., 2004. Sterol methyltransferase: functional analysis of highly conserved residues by site-directed mutagenesis. Biochemistry 43: 569–576. [DOI] [PubMed] [Google Scholar]
- Oita, S., and S. O. Yanagi, 1993. Stimulation of Schizophyllum commune fruit body formation by inhibitor of membrane function and cell wall synthesis. Biosci. Biotechnol. Biochem. 57: 1270–1274. [Google Scholar]
- Oudejans, R. C. H. M., and D. J. van der Horst, 1978. Cyclopropane fatty acids in millipedes: their occurrence and metabolism. Abh. Naturwiss. Ver. Hamburg 21/22: 345–348. [Google Scholar]
- Perly, B., I. C. Smith and H. C. Jarrell, 1985. Effects of replacement of a double bond by a cyclopropane ring in phosphatidylethanolamines: a 2H NMR study of phase transitions and molecular organization. Biochemistry 24: 1055–1063. [DOI] [PubMed] [Google Scholar]
- Saito, T., and H. Ochiai, 1998. Fatty acid composition of the cellular slime mold Polysphondylium pallidum. Lipids 33: 327–332. [DOI] [PubMed] [Google Scholar]
- Sajbidor, J., 1997. Effect of some environmental factors on the content and composition of microbial membrane lipids. Crit. Rev. Biotechnol. 17: 87–103. [DOI] [PubMed] [Google Scholar]
- Sakai, H, and S. Kajiwara, 2004. Membrane lipid profile of an edible basidiomycete Lentinula edodes during growth and cell differentiation. Lipids 39: 67–73. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Seelig, J., and A. Seelig, 1980. Lipid conformation in model membranes and biological membranes. Q. Rev. Biophys. 13: 19–61. [DOI] [PubMed] [Google Scholar]
- Solberg, Y., 1989. A literature review of the lipid constituents of higher fungi, new investigation of Agaricus species. Int. J. Mycol. Lichenol. 4: 137–154. [Google Scholar]
- Swamy, S., I. Uno and T. Ishikawa, 1984. Morphogenetic effects of mutations at the A and B incompatibility factors in Coprinus cinereus. J. Gen. Microbiol. 130: 3219–3224. [Google Scholar]
- Taylor, F. R., and J. E. Cronan, Jr., 1979. Cyclopropane fatty acid synthase of Escherichia coli: stabilization, purification, and interaction with phospholipid vesicles. Biochemistry 18: 3292–3300. [DOI] [PubMed] [Google Scholar]
- Terashima, K., K. Yuki, H. Muraguchi, M. Akiyama and T. Kamada, 2005. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics 171: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser, P. J., R. Velagapudi, M. Aebi and U. Kües, 2003. Extracellular matrix proteins in mushroom development. Recent Res. Devel. Microbiol. 7: 381–415. [Google Scholar]
- Wang, A. Y., D. W. Grogan and J. E. Cronan, Jr., 1992. Cyclopropane fatty acid synthase of Escherichia coli: deduced amino acid sequence, purification, and studies of the enzyme active site. Biochemistry 31: 11020–11028. [DOI] [PubMed] [Google Scholar]
- White, J. D., and M. S. Jensen, 1993. Biomimetic synthesis of a cyclopropane containing eicosanoid from the coral Plexaura homomalla. Assignment of relative configuration. J. Am. Chem. Soc. 115: 2970–2971. [Google Scholar]
- Zolan, M. E., and P. J. Pukkila, 1986. Inheritance of DNA methylation in Coprinus cinereus. Mol. Cell. Biol. 6: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]