Abstract
Aspergillus nidulans is an important experimental organism, and it is a model organism for the genus Aspergillus that includes serious pathogens as well as commercially important organisms. Gene targeting by homologous recombination during transformation is possible in A. nidulans, but the frequency of correct gene targeting is variable and often low. We have identified the A. nidulans homolog (nkuA) of the human KU70 gene that is essential for nonhomologous end joining of DNA in double-strand break repair. Deletion of nkuA (nkuAΔ) greatly reduces the frequency of nonhomologous integration of transforming DNA fragments, leading to dramatically improved gene targeting. We have also developed heterologous markers that are selectable in A. nidulans but do not direct integration at any site in the A. nidulans genome. In combination, nkuAΔ and the heterologous selectable markers make up a very efficient gene-targeting system. In experiments involving scores of genes, 90% or more of the transformants carried a single insertion of the transforming DNA at the correct site. The system works with linear and circular transforming molecules and it works for tagging genes with fluorescent moieties, replacing genes, and replacing promoters. This system is efficient enough to make genomewide gene-targeting projects feasible.
GENE targeting, which involves integration of transforming sequences into a genome by homologous recombination, is an enormously useful technique. It can be used to delete genes entirely, to replace one allele of a gene with another, to replace a gene's normal promoter with a regulatable promoter, and to tag genes with epitope tags or fluorescent proteins. These approaches have been facilitated by the development of fusion PCR (Kuwayama et al. 2002; Yang et al. 2004; Yu et al. 2004). Fusion PCR allows transforming fragments to be created with no ligation and if the genome of the organism of interest has been sequenced, the gene to be targeted does not need to be cloned. DNA fragments required for targeting can be amplified from genomic DNA using PCR primers based on the genomic sequence.
The filamentous fungus Aspergillus nidulans is an important experimental organism. Significant findings in a number of areas have resulted from work with A. nidulans, and it serves as an important model organism for the genus Aspergillus, which includes commercially important fermentation organisms as well as serious pathogens. Gene targeting is possible in A. nidulans, but homologous recombination during transformation is not particularly efficient. Substantial stretches of homologous DNA are required to obtain useful frequencies of homologous integration and, even then, the majority of transforming sequences integrate heterologously. For example, in the study of Yu et al. (2004), gene targeting with 29 different fusion PCR products with flanking sequences ranging from 480 bp to 4.3 kb resulted in correct gene-targeting frequencies ranging from 0 to 40% (20% or less in 24/29 cases). In addition, integration into multiple sites often occurs and transforming linear DNA fragments may circularize before integration. As a result, in most cases many transformants must be analyzed to identify one carrying a correct, single homologous targeting event. The A. nidulans genome has now been sequenced (Galagan et al. 2005) and the development of a more efficient gene-targeting system not only would facilitate current research with A. nidulans, but also would make possible genomewide gene-targeting projects (e.g., genomewide gene tagging or deletion).
We now report the development of a very efficient gene-targeting system for A. nidulans. Our approach is based on the results of Ninomiya et al. (2004) who found that the deletion of genes required for nonhomologous end joining DNA repair (homologs of the human KU70 and KU80 genes) increases the frequency of gene replacement in Neurospora crassa. We have identified and deleted the A. nidulans KU70 homolog. This deletion has little or no effect on growth or sensitivity to mutagens, but it dramatically improves gene targeting. We have developed heterologous selectable markers that can be used for gene targeting in A. nidulans, and we have used our system for gene tagging; gene replacement, including replacement of essential genes (gene replacement/heterokaryon rescue); promoter replacement; and targeted integration of circular molecules. In all cases, the great majority of transformants carried a single correct integration. The system is efficient enough to allow genomewide gene-targeting projects. Our data in combination with the data of Ninomiya et al. (2004) suggest that deletion of Ku homologs may be a generally useful strategy for improving gene targeting.
MATERIALS AND METHODS
Strains:
A. nidulans strains used in this study are listed in Table 1. Strains have been deposited at the Fungal Genetics Stock Center (http://www.fgsc.net/).
TABLE 1.
A. nidulans strains used in this study
| Strain | Genotype |
|---|---|
| KJ12 | wA3; argB2; pyroA4 |
| KJ15 | fwA1; pyrG89; argB2 |
| MH1046 | yA1, pabaA1; argB2; pyroA4, nkuA∷bar |
| LO1161 | pyrG89, pabaA1; md2A∷pyrG; riboB2 |
| LO1180 | wA3; pyrG∷pyroA; pyroA4 |
| SO451 | wA3; pyrG89; argB2; pyroA4, nkuA∷argB; sE15 |
| TN02 | wA3; argB2; pyroA4, nkuA∷argB |
| TN02A1 | yA1, pabaA1; argB2; nkuA∷argB |
| TN02A7 | pyrG89; pyroA4, nkuA∷argB; riboB2 |
| TN02A25 | pyrG89; argB2; pabaB22, nkuA∷argB; riboB2 |
| TN12 | pyrG89; nkuB∷A. fumigatus riboB; pyroA4, nkuA∷argB; riboB2 |
Media:
The inducing medium for the alcA promoter was solid minimal medium [6 g/liter NaNO3, 0.52 g/liter KCl, 0.52 g/liter MgSO4·7H2O, 1.52 g/liter KH2PO4, 9 g/liter fructose, 1 ml/liter trace element solution (Cove 1966), 15 g/liter agar, pH adjusted to 6.5 with NaOH before autoclaving] supplemented with 1 mg/ml (8.9 mm) uracil, 2.442 mg/ml (10 mm) uridine, 2.5 μg/ml riboflavin, 1μg/ml para-aminobenzoic acid, and 0.5 μg/ml pyridoxine with 6.25 mm threonine added as an inducer. YAG (5 g/liter yeast extract, 20 g/liter d-glucose, 15 g/liter agar) supplemented with 1 mg/ml uracil, 2.442 mg/ml (10 mm) uridine, and 2.5 μg/ml riboflavin was used as a repressing medium.
Methyl methanesulfonate (MMS) sensitivity tests were carried out on solid minimal medium with 10 mg/ml d-glucose as a carbon source and appropriate nutritional supplements [1.0 mg/ml uracil, 2.442 mg/ml (10 mm) uridine, 2.5 μg/ml riboflavin, 1 μg/ml para-aminobenzoic acid, 0.5 μg/ml pyridoxine, 0.5 mg/ml l-arginine]. MMS was obtained from Sigma-Aldrich.
The bar gene and selection:
The source of the bar cassette was Mogens Trier Hansen (Novozymes A/S, Bagsvaerd, Denmark). The bar gene encoding glufosinate resistance was taken from the plasmid pBP1T (Straubinger et al. 1992). The bar gene was then placed between the amdS promoter [containing the I9 and the I66 mutations that give increased expression (see Hynes and Davis 2004)] and the Aspergillus niger glucoamylase terminator.
Glufosinate was prepared by chloroform extraction of the commercial herbicide Basta (Hoechst Schering AgrEvo GmbH), which contains 200 g/liter glufosinate–ammonium. The yellowish aqueous phase separated from the chloroform layer containing the blue dye added to this preparation by the manufacturers was taken for use. The aqueous phase was stored in the cold until used and was added at 25 μl/ml of 1% glucose minimal medium with 10 mm ammonium tartrate as sole nitrogen source.
Polymerase chain reaction:
Several polymerase chain reaction (PCR) polymerases and PCR procedures were used in the participating laboratories. All polymerase chain reactions were performed according to manufacturer's instructions. In some cases PCR was carried out as described by Yang et al (2004). In other cases AccuPrime Pfx DNA Polymerase (Invitrogen, Carlsbad, CA) was used to amplify shorter DNA fragments. AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen, San Diego) was then used in the fusion PCR to obtain the final products. In other cases, Pfu DNA polymerase (Promega, Madison, WI) was used for amplification of short fragments and Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) was used for long fragments.
Transformation of A. nidulans:
Transformation was carried out as described previously (Andrianopoulos and Hynes 1998; Jung et al. 2000; Yang et al. 2004).
Southern hybridizations:
A. nidulans genomic DNA was isolated as described by Oakley et al. (1987) or Lee and Taylor (1990). Southern hybridization was performed in dried agarose gels (Oakley et al. 1987) or as described by Yang et al. (2004) or on Hybond N+ membranes (Amersham, Buckinghamshire, UK) after alkaline capillary transfer with 0.4 m NaOH.
Microscopy:
Images were taken using a 1.3 numerical aperture planfluor objective on an Olympus IX71 inverted microscope equipped with a mercury light source as well as a Uniblitz electronic shutter, a Prior z-axis drive, and a Hamamatsu Orca ER cooled CCD camera controlled by Slidebook software (Intelligent Imaging Innovations, Denver) on an Apple PowerMac G4 computer. Cells were grown and observed using four-chamber Lab-Tek chambered coverglasses (Nalge Nunc International, Naperville, IL). Imaging was through the coverslips at the bottom of the chambers as described previously (Horio and Oakley 2005).
RESULTS
Identification and deletion of the A. nidulans homolog of KU70:
We identified the A. nidulans KU70 homolog by carrying out a blast search of the A. nidulans genome database (http://www.broad.mit.edu/annotation/fungi/aspergillus/) with the human KU70 cDNA sequence. The search revealed a single KU70 homolog (AN7753.2 in the A. nidulans genome database, blast value 1e-52). We have designated this gene nkuA.
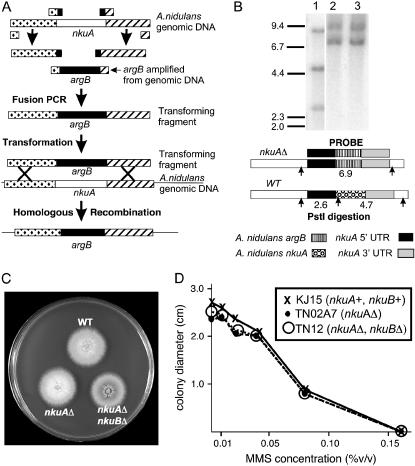
We deleted nkuA by replacing it with the A. nidulans argB gene (Upshall et al. 1986). We created, by fusion PCR, a fragment in which argB was flanked on each side by 2000 bp of the sequence that flanks nkuA in the genome (Figure 1). This fragment was transformed into strain KJ12. Ten argB+ transformants were screened by Southern hybridizations and two showed a pattern diagnostic for the replacement of nkuA by argB (Figure 1). We will use the abbreviation nkuAΔ for this replacement, which is more completely designated nkuA∷argB.
Figure 1.
Deletion and initial characterization of nkuA, the KU70 homolog of A. nidulans. (A) Deletion strategy. PCR was used to amplify regions flanking nkuA as well as the argB gene from genomic DNA. The PCR primers were synthesized with “tails” such that the nkuA flanking fragments anneal to the argB fragment during fusion PCR. Fusion PCR creates a fragment containing nkuA flanking sequences surrounding argB and transformation with this fragment can lead to replacement of nkuA with argB. (B) Verification of the replacement of nkuA with argB by Southern hybridization. The positions of DNA size standards (in kilobases) are at the left. The probe consists of nkuA flanking sequences surrounding argB. In wild-type DNA cut with PstI (lane 1), the probe hybridizes to bands of 2.6 and 4.7 kb at the nkuA locus (as shown in the diagram below) as well as an 8.8-kb fragment containing the wild-type argB gene. Lanes 2 and 3 show miniprep DNA from two putative nkuA replacement transformants. Homologous recombination replacing nkuA with argB converts the two fragments at the nkuA locus to a single 6.9-kb fragment as shown in the diagram. Since the mutant argB allele (argB2) is not a deletion, the probe also recognizes the argB gene in the transformants. DNA produced by this miniprep procedure usually has slightly reduced mobility compared to the CsCl purified wild-type DNA, presumably due to contaminating materials not removed by the miniprep procedure (Oakley et al. 1987). (C) Growth of strains carrying replacements of nkuA or nkuA and nkuB relative to a wild-type control. Growth rates are similar in all strains. The wild-type strain is KJ12, the nkuAΔ strain is TN02, and the nkuAΔ, nkuBΔ strain is TN12. The nkuAΔ, nkuBΔ double mutant appears smaller than the other strains because of the darker spore color but the colony diameter is actually about the same. (D) Growth rates on MMS of strains carrying replacements of nkuA or nkuA and nkuB as well as a nkuA+, nkuB+control strain. Replacement of nkuA and nkuB does not significantly affect MMS sensitivity.
Since KU70 is involved in nonhomologous end-joining repair of double-strand breaks in phylogenetically diverse organisms (reviewed by Hopfner et al. 2002; Lisby and Rothstein 2004) and in telomere maintenance in some organisms (reviewed by Hande 2004), we examined nkuAΔ strains for growth defects and increased sensitivity to mutagens. Growth and conidiation appeared normal in the initial nkuAΔ transformants and in segregants of crosses that carried nkuAΔ (Figure 1). NkuAΔ strains crossed readily to other strains and, thus, apparently are not defective in meiosis.
In N. crassa, deletion of the KU70 homolog mus-51 results in increased sensitivity to the mutagens MMS, ethyl methanesulfonate (EMS), and bleomycin (Ninomiya et al. 2004). The increased sensitivity to MMS and EMS was somewhat surprising in that these are point mutagens that do not cause double-strand breaks. We tested an nkuAΔ strain for growth sensitivity to MMS over a range from 0.01 to 0.16% and found no difference in growth of the nkuAΔ strain relative to the nkuA+ control (Figure 1). Similarly, we found no inhibition of an nkuAΔ strain relative to an nkuA+ control on 0.5, 1.0, and 2.0 μg/ml bleomycin. This was somewhat surprising because although bleomycin has a complex mechanism of action (Hecht 2000), it does cause double-strand breaks (Povirk et al. 1977). We also tested growth of an nkuAΔ strain on 15 mm hydroxyurea, a DNA replication inhibitor, and found no inhibition. Finally, we tested the growth of an nkuAΔ strain on 25 μg/ml camptothecin, a topoisomerase I inhibitor that can cause DNA breakage during replication (Hsiang et al. 1985). The nkuAΔ strain showed no inhibition of growth relative to a wild-type control, but a positive control strain carrying a deletion of uvsB [a kinase with a central role in DNA repair (De Souza et al. 1999; Hofmann and Harris 2000)] showed, as expected, a significant reduction of growth relative to the nkuAΔ and wild-type control strains. The fact that nkuAΔ does not enhance the sensitivity to compounds that cause double-strand breaks indicates that A. nidulans has an efficient break repair system independent of nkuA.
Cloning of selectable markers from Aspergillus fumigatus:
Selectable markers from A. nidulans have a disadvantage in gene-targeting experiments in that homologous recombination (or gene conversion in the case of point mutations in selectable marker genes) can occur at the chromosomal copy of the selectable marker as well as at the targeted locus. Initial experiments indicated that this was a concern when targeting genes in nkuAΔ strains (A. Osmani and S. A. Osmani, unpublished data). To eliminate this concern, we have cloned the A. fumigatus homologs of the A. nidulans riboB and pyroA genes to use as selectable markers. We have also used the previously cloned A. fumigatus pyrG gene (Weidner et al. 1998) and we have constructed a vector that confers resistance to glufosinate (phosphinothricin).
We partially sequenced the A. nidulans riboB gene that has previously been cloned (Oakley et al. 1987), searched the A. nidulans database, and found that the riboB gene corresponds to AN0670.2. We then carried out a blast search of the A. fumigatus genome (http://tigrblast.tigr.org/er-blast/index.cgi?project=afu1) and identified the riboB homolog (Afu1g13300, blast value 5.1 e-106). We used the sequence information to design PCR primers to amplify the gene from A. fumigatus genomic DNA and cloned the fragment into pBlueScript SK+, creating plasmid pTN2. Similarly, we identified Afu5g08090 as the A. fumigatus pyroA homolog through a blast search with the published pyroA sequence (Osmani et al. 1999) (blast value 1.2 e-87), amplified the gene by PCR, and cloned the amplified fragment into pBlueScript SK+, creating plasmid pTN1. The A. fumigatus genes complement A. nidulans riboB2 and pyroA4 mutations but in numerous experiments (discussed subsequently and in our unpublished data) have not directed integration at these loci.
When plasmids pTN1 and pTN2 carrying the A. fumigatus pyroA and riboB genes (and no A. nidulans sequences) were used to transform nkuAΔ strains, only small, abortive colonies were obtained. These did not continue to grow when subcultured onto selective medium. When an nkuAΔ strain was transformed with a plasmid carrying A. fumigatus pyrG, larger, nearly normal colonies were obtained. Conidia from these colonies did not grow, however, to form colonies on selective medium. Stable transformants thus were not obtained with any of the three plasmids. Our interpretation is that nkuAΔ prevents heterologous integration into the genome such that the transforming plasmids are eventually lost. The fact that the colonies obtained with the plasmid carrying A. fumigatus pyrG are much larger than the colonies obtained with the other plasmids is interesting and merits further study.
We have also used a vector (pMT1612) conferring glufosinate resistance, in which the bar (glufosinate resistance) gene of Streptomyces hygroscopicus from plasmid pBP1T (Straubinger et al. 1992) is downstream of the A. nidulans amdS promoter [containing the I9 and I66 mutations that give increased expression (Hynes and Davis 2004)] and upstream of the A. niger glucoamylase terminator (M. T. Hansen, personal communication).
Using this plasmid (or other circular plasmids containing this bar cassette as the selectable marker) results in strong resistant transformants observable on a background of “abortive” weaker resistant transformants that do not form stable resistant colonies when picked to new selective media. Thus transient expression of the bar gene from nonintegrated plasmids would seem to be sufficient to give glufosinate resistance initially. Unstable transformants are not seen with linear fragments containing bar.
NkuAΔ dramatically improves the frequency of correct gene targeting:
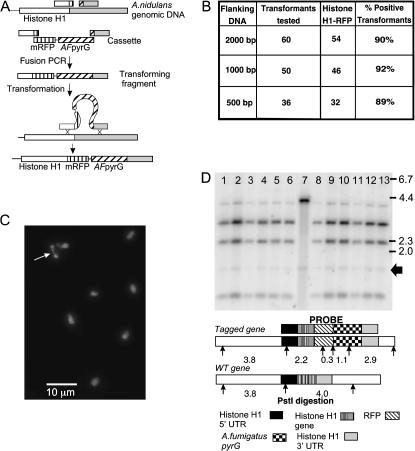
We initially tested the effects of nkuAΔ on gene targeting using a linear DNA fragment generated by fusion PCR to create a C-terminal histone H1-monomeric red fluorescent protein (mRFP) (Campbell et al. 2002; Toews et al. 2004) fusion. We chose this test system because histone H1-mRFP fusions are easily scored by fluorescence microscopy. The strategy and results are shown in Figure 2. We initially transformed a control nkuA+ strain (LO 1180) and an nkuAΔ strain (TN02A7) with a fusion PCR product consisting of the mRFP and A. fumigatus pyrG flanked on each side by ∼2000 bp of DNA from the histone H1 gene and from the histone H1 3′ untranslated region. The fusion PCR product was purified from an agarose gel. In the nkuA+ strain, we obtained 15 transformants, 10 of which had red fluorescent nuclei, indicating that they have a correct histone H1 mRFP fusion. This is an unusually high frequency of homologous integration, among the highest frequencies that we have obtained in an nkuA+ strain. One of the transformants grew poorly and was not used in subsequent experiments. We examined the remaining 9 histone H1 mRFP-positive transformants by Southern hybridizations and found that 7 had extra bands of hybridization in addition to the bands expected for a single correct homologous integration. Thus, only 2 of 15 transformants had a correct single integration.
Figure 2.
mRFP tagging of histone H1. (A) Tagging strategy. PCR fragments containing the C terminus of histone H1 and a sequence 3′ to the histone H1 coding sequence were amplified from genomic DNA, and a cassette containing mRFP along with the A. fumigatus pyrG gene was amplified from a plasmid. Primers were used that had “tails” such that fusion PCR produced a molecule that carried the mRFP sequence fused in frame to the 3′-end of the histone H1 gene followed by the A. fumigatus pyrG gene and a sequence 3′ to the histone H1 gene. Targeted integration of the resulting linear molecule into the genome produces a full-length histone H1 gene fused in frame with mRFP. (B) Frequences of gene targeting in an nkuAΔ strain with linear fragments carrying different lengths of flanking DNA. Transformants were assayed for mRFP labeling of chromosomes by fluorescence microscopy. (C) Histone H1-mRFP-labeled chromosomes. A mitotic nucleus is designated with an arrow. (D) A Southern hybridization of DNA from a wild-type strain (lane 7) and 12 histone H1 mRFP-positive transformants (lanes 1–6, 8–13). Positions of DNA size standards (in kilobases) are at the right. As shown in the diagram below, digestion of wild-type DNA with PstI produces fragments of 3.8 and 4.0 kb recognized by the probe (arrows represent PstI sites). The region of overlap between the probe and the 3.8-kb fragment is small (360 bp). Hybridization of the probe to this fragment is weaker than hybridization to the 4.0-kb fragment, and it is barely visible below the 4.0-kb fragment in the wild type. Correct gene targeting leaves the 3.8-kb fragment intact but replaces the 4.0-kb fragment with fragments of 2.9, 2.2, 1.1, and 0.3 kb. The 0.3-kb band was very faint and is not present in the region of the gel shown. The bands of hybridization expected for correct gene targeting are present in all transformants. An additional faint band of ∼1.5 kb (arrow) is visible in the wild type and all transformants. We presume that this is from a gene with weak homology to the probe, perhaps another histone, and does not affect the interpretation of the Southern hybridization.
In the nkuAΔ strain, 54 of 60 transformants tested (90%) were histone H1 mRFP-positive (Figure 2). We carried out Southern hybridizations on 12 positive transformants and each carried a single correct integration (Figure 2). NkuAΔ thus increased the frequency of the desired targeting event from ∼13 to ∼90%.
Having established that gene targeting with a fragment with 2000 bp of flanking DNA is efficient in an nkuA strain, we wanted to determine if targeting is efficient with smaller flanking sequences. There are two advantages in being able to use small flanking regions. First, the fusion PCR is more efficient because the fragment is shorter. Second, because the flanks are shorter, there is less chance of PCR creating a mutation in the gene to be targeted or a nearby gene that overlaps with the 3′ flanking region. We consequently tested the efficiency of gene targeting in an nkuAΔ strain (TN02A7) using fusion PCR products with 1000- and 500-bp flanking regions. These products were not band purified from gels but were simply purified with Amicon YM30 filters to remove primers, nucleotides, etc. In both cases, ∼90% of the transformants were histone H1 mRFP positive (Figure 2). To determine if the transformants carried a single correct integration, we carried out Southern hybridizations on DNA from 10 transformants obtained with the fusion PCR product with 500-bp flanking regions. All transformants had a single correct homologous integration. In a transformation of an nkuA+ control (LO1180) with the fusion PCR product with 500-bp flanking regions, 19 of 50 transformants were histone H1 mRFP positive. Southern hybridizations were carried out on 12 of the histone H1 mRFP-positive transformants and 6 of them showed a single correct gene replacement. These data indicate that nkuAΔ dramatically decreases the frequency of nonhomologous integrations, resulting in much more efficient gene targeting. They also indicate that 500-bp flanking regions are large enough to allow efficient gene targeting in an nkuAΔ strain. It is worth noting that the histone H1-mRFP, nkuAΔ strains grew robustly, at a rate indistinguishable from controls, and that the histone H1 allowed visualization of chromatin and chromosomes through the cell cycle.
Although gene targeting was efficient with 500-bp flanking regions, fewer transformants were obtained than with 2000- and 1000-bp flanking regions. We also attempted to GFP tag An-nsp1, the A. nidulans homolog of the Saccharomyces cerevisiae NSP1 nucleoporin (De Souza et al. 2004), with a fragment containing 30-bp flanking regions. The advantage of such small flanking regions is that they can be synthesized as part of the PCR primer and this eliminates the need for fusion PCR. Few transformants were obtained and none carried a GFP-tagged An-nsp1. The picture that emerges is that nkuAΔ greatly facilitates gene targeting by decreasing the frequency of nonhomologous integration during transformation. Transformation is less efficient with smaller homologous flanking sequences, but 500 bp of flanking DNA is adequate to give acceptable numbers and frequencies of correct targeting events
An obvious question is whether nkuAΔ is useful for targeting other loci. We have now attempted to GFP tag the C termini of 28 genes of diverse functions in strains carrying nkuAΔ. In 24 cases, tagging was successful and in 4 cases it was unsuccessful. The unsuccessful cases are probably due to lethality of the fusion protein. We have used fusion PCR fragments with ∼500-bp flanks in 12 of the 28 cases. In 10 cases, the tagging was successful and in the 2 other cases the C-terminal GFP fusion was apparently lethal. Fusion PCR products with 500-bp flanks thus are effective for targeting a variety of genes, although it is often advantageous to use larger flanking sequences because they give higher transformation frequencies. Interestingly, even in instances in which the fusion proteins are lethal, nkuAΔ is advantageous in that many of the transformants are balanced heterokaryons (see below) and this can be a useful indication that the fusion is lethal. While we have not carried out a detailed analysis of each targeted gene, 114 transformants from 20 tagging experiments were examined by diagnostic PCR and 107 carried correct integration events (94%). We further examined 20 An-nsp1-GFP transformants by Southern hybridizations. All transformants examined carried a single correct integration event. (However, one transformant was not selected for further analysis because it grew poorly and thus might have carried an incorrect integration event.) We have also attempted to S tag (reviewed by Terpe 2003) the C termini of 14 proteins and were successful in each case. Forty-one transformants were examined by diagnostic PCR and 38 carried correct integrations (93%). NkuAΔ thus seems to be of great value in tagging a wide variety of genes.
NkuAΔ allows efficient gene targeting with suboptimal fusion PCR products:
Although fusion PCR is a remarkably useful technique, production of the correct, full-length product is often inefficient. In addition to the large, complete fusion PCR product, smaller incomplete products are present. In this case, the correct band must be purified from a gel or time and effort must be spent in optimizing PCR conditions for the particular fragment being amplified. We reasoned that if nonhomologous integration were reduced by nkuAΔ, the smaller fragments might not be a significant problem. They would integrate homologously, simply replacing chromosomal sequences with identical sequences generated by PCR.
To mRFP tag the C terminus of γ-tubulin, we generated a fusion PCR product that contained an mRFP/A. fumigatus pyrG cassette flanked by 500-bp sequences from the γ-tubulin gene and its 3′ untranslated region. The PCR fusion product also contained a prominent lower band and several additional bands in addition to the desired band (Figure 3). We transformed with this PCR product and examined transformants by fluorescence microscopy to determine if they carried a γ-tubulin mRFP fusion (easily scored because γ-tubulin localizes to the spindle pole body). Of 20 transformants tested, 13 contained a γ-tubulin mRFP fusion. These were analyzed by Southern hybridizations; 12 of the 13 contained a single correct integration and 1 contained a correct integration plus an additional band of hybridization. All γ-tubulin mRFP fusion strains grew indistinguishably from the wild type. These data demonstrate that nkuAΔ allows reasonably efficient targeting with suboptimal fusion PCR products. This can be a considerable practical advantage in that it can save the time required to optimize fusion PCR or purify the desired fragment.
Figure 3.
mRFP tagging of the γ-tubulin gene with a suboptimal PCR product. (A) The C-terminal tagging strategy is similar to the mRFP histone H1 tagging strategy discussed in Figure 2. (B) The fusion PCR product. Lane 1 is a HindIII digest of bacteriophage λ used as a molecular weight standard. Lane 2 is the fusion PCR product used for transformation. The 4.0-kb fragment designated with an arrow is the correct fusion PCR product. (C) Fluorescence microscopy of a transformant germling shows the spindle pole body localization (arrows) characteristic of γ-tubulin.
NkuAΔ facilitates promoter replacements, gene replacements, and heterokaryon gene replacements:
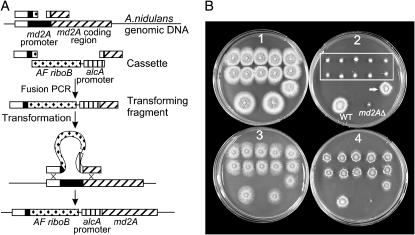
Encouraged by the results with C-terminal mRFP, GFP, and S tagging, we examined the utility of nkuAΔ strains for other types of gene targeting. It is often useful to regulate the expression of genes, and the alcA promoter of A. nidulans is highly regulatable, allowing gene expression to be turned down to very low levels or up to very high levels (Adams et al. 1988; Waring et al. 1989; Kennedy and Turner 1996). We tested the efficacy of fusion PCR and nkuAΔ for replacement of the promoter of the A. nidulans MAD2 homolog, md2A, with the alcA promoter. We used the A. fumigatus riboB gene as a selectable marker and 500-bp flanking sequences. The fusion PCR product was not band purified before transformation.
To test transformants for promoter replacements, we used the fact that deletion or underexpression of MAD2 homologs in many organisms causes hypersensitivity to antimicrotubule agents such as benomyl, and this is the case with a deletion of md2A in A. nidulans (Prigozhina et al. 2004). We tested 10 transformants for benomyl sensitivity on YAG medium, which represses the alcA promoter (Figure 4), and on a minimal medium containing threonine as an alcA inducer. In instances in which the md2A promoter was replaced by the alcA promotor, the transformant should be benomyl hypersensitive on repressing medium but not on inducing medium. We found that this was the case with 10 of 11 transformants tested (Figure 4). We carried out Southern blots on the 10 positive transformants and found that all 10 had a single correct replacement of the md2A promoter by the alcA promoter.
Figure 4.
Replacement of the md2A promoter with the alcA promoter. (A) Fusion PCR was used to create a linear molecule containing sequences upstream from the md2A promoter, the A. fumigatus riboB gene, the alcA promoter, and a portion of the md2A coding region. Correct integration of this fragment places the md2A coding region under control of the alcA promoter. (B) Benomyl sensitivity caused by repression of md2A expression. The top two rows of colonies (outlined by a rectangle on plate 2) are transformants in which the md2A promoter has been correctly replaced with the alcA promoter. A 13th transformant (arrow on plate 2) does not carry the correct promoter replacement. The two colonies at the bottom of plate 2 are a wild-type control and a strain in which the md2A gene has been deleted. Plates 1 and 2 contain YAG medium that represses the alcA promoter. Plate 1 contains no benomyl and growth of all strains is normal. Plate 2 contains 0.3 μg/ml benomyl. As shown previously (Prigozhina et al. 2004), the md2A deletion causes benomyl supersensitivity. Likewise, repression of the alcA promoter in the correctly targeted transformants causes benomyl supersensitivity due to reduced expression of md2A. Growth is reduced only slightly in the wild-type and the incorrectly targeted transformant. Plates 3 and 4 contain alcA-inducing medium. Plate 4 contains 0.3 μg/ml benomyl and the correctly targeted transformants grow nearly as well as the wild type because expression of md2A is induced. As expected, growth of the md2A deletant is greatly inhibited.
To further test the value of nkuAΔ for gene replacements, we targeted two A. nidulans genes, AN5843.2 and AN2667.2. For these experiments, we used the bar (glufosinate resistance) cassette as the selectable marker and transformed strain TN02. The transforming fragments in these cases were constructed by normal recombinant DNA techniques rather than by fusion PCR. For AN5843.2, we used 1967 bp of 5′ flanking DNA and 1127 bp of 3′ flanking DNA. Five transformants were screened by Southern hybridizations and all carried a correct gene replacement. For AN2667.2, we used 1152 bp of 5′ flanking DNA and 1721 bp of 3′ flanking DNA. Eight transformants were screened by Southern hybridizations and all carried the correct gene replacement. None of these targeted deletions resulted in an observable phenotype. Targeted deletion therefore is highly efficient in an nkuAΔ strain even when screening for an observable phenotype is not possible.
Another extremely useful technique in A. nidulans is gene replacement/heterokaryon rescue (Osmani et al. 1988; Oakley et al. 1990; Martin et al. 1997; Jung et al. 2000). Replacement of essential genes with selectable markers during transformation is normally lethal, but balanced heterokaryons forming during transformation carry nuclei with the gene replacement as well as untransformed nuclei. These heterokaryons grow on the selection medium because the untransformed nuclei carry a functional copy of the targeted gene, while the transformed nuclei carry the selectable marker (replacing the targeted gene). Since conidia (asexual spores) are uninucleate, the conidia produced by a gene replacement heterokaryon will not grow to form colonies on selective medium. The phenotypes of the lethal gene disruptions or replacements can be determined, moreover, in the conidia produced by the heterokaryon. Gene replacement/heterokaryon rescue is not normally an efficient technique because two events must occur together—correct gene replacement and heterokaryon formation. On the other hand, if a transforming fragment integrates heterologously and does not disrupt an essential gene, the transformant can grow without heterokaryon formation. Thus there is a partial selection for heterologous integration.
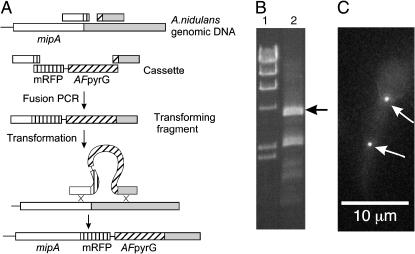
To determine if heterokaryon gene replacement is facilitated by nkuAΔ, we transformed an nkuAΔ strain with a fusion PCR fragment consisting of the A. fumigatus pyrG gene surrounded on each side by 1000 bp of mipA flanking DNA. In two experiments, 54 of 93 transformants tested (58%) were balanced heterokaryons.
The efficiency of gene targeting in nkuAΔ strains allows one to determine easily and rapidly if a gene is essential or not. If it is not essential, very few transformants will be heterokaryons. If it is essential, a substantial fraction of the transformants will be balanced heterokaryons in which the hyphae are able to grow on selective medium, but the conidia produced from the hyphae will not be able to grow on selective medium. We have used this approach in an ongoing project to examine the function of genes involved in nuclear transport. To date we have relatively complete data on 30 genes. In 18 cases, transformation with fusion PCR products designed to delete target genes produced high frequencies of balanced heterokaryons and were determined by diagnostic PCR to have deletions of the desired genes. These 18 target genes thus are essential. In 12 additional cases, heterokaryons were not produced and diagnostic PCR revealed that the target genes had been deleted. These genes thus are not essential. An added benefit of this approach is that the phenotype of lethal genes can be observed by germinating spores from the balanced heterokaryons.
Finally, we have found that cotransformation of two different transforming molecules (plasmid/linear, linear/linear) occurs in nkuAΔ strains. However, nkuAΔ does not improve the frequency of cotransformation (results not shown).
Gene targeting with circular plasmids:
Although fusion PCR is a powerful technique, it is sometimes advantageous to target plasmids for integration at particular sites in the genome. To determine if nkuAΔ facilitates homologous integration of plasmids, we transformed strain TN02A25 with a plasmid carrying the bar gene flanked by flanking sequences for the acuE (malate synthase) gene (AN6653.2) (1313 bp of the 5′ flank and 1123 bp of the 3′ flank). Five transformants were screened by Southern hybridization. All five showed homologous recombination at the acuE locus. In four cases, the plasmid integrated by homologous recombination in the 5′ flanking region, and in one case, crossing over in the 5′ and 3′ regions resulted in replacement of the acuE gene by bar. NkuAΔ, thus, as expected, facilitates targeting of circular molecules.
We have used this technique to create a strain in which nkuA is replaced by bar. Strain TN02A1, which carries a replacement of nkuA by the A. nidulans argB, was transformed with a plasmid carrying the bar cassette flanked by 2288 bp of nkuA 5′ flanking DNA and 2030 bp of nkuA 3′ flanking DNA. One transformant carried a replacement of nkuA∷argB by nkuA∷bar. In addition, transformation of this strain with a linear fragment also generated nkuA∷bar replacements and two of these were confirmed by Southern blot analysis (Figure 5). This strain (MH1046) is useful because the nkuA deletion can be followed in crosses by scoring for glufosinate resistance, eliminating the need for having argB2 in the strains to which the nkuAΔ strain is crossed.
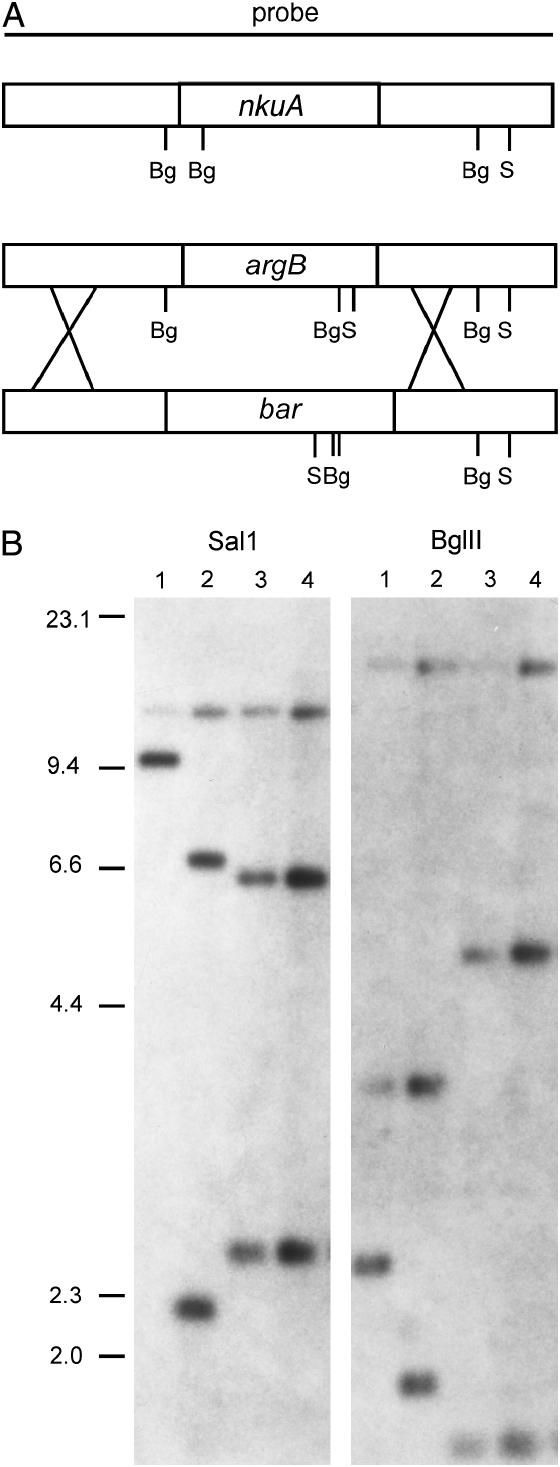
Figure 5.
Construction of an nkuA deletion with the bar marker. (A) Strategy for replacement of the nku∷argB deletion with nkuA∷bar. The bar cassette was inserted to replace the region between −190 and +2296 bp of the nkuA gene. A linear fragment was transformed into the nkuA∷argB strain. TN02A1 and four glufosinate-resistant plasmids that had simultaneously become argB+ were recovered. (B) Southern blot analysis of two nkuA∷bar transformants. DNA was digested with SalI or BglII as indicated and the blot hybridized with a probe corresponding to the nkuA gene with 2470 and 2344 bp of flanking DNA as shown in A. Lane 1 contains wild-type (nkuA+) genomic DNA, lane 2 contains TN02A1 (nkuA∷argB) DNA, and lanes 3 and 4 contain DNA from two nkuA∷bar transformants. The restriction patterns are consistent with the predicted SalI and BglII sites as shown in A.
Identification and deletion of the A. nidulans homolog of KU80:
We identified the A. nidulans KU80 homolog by carrying out a blast search of the A. nidulans genome database with a human KU80 cDNA sequence (NCBI NP_066964). The search revealed a single KU80 homolog (AN4552.2, blast value 1e-32). We have designated this gene nkuB.
We deleted nkuB by replacing it with the A. fumigatus riboB gene. We created, by fusion PCR, a fragment in which A. fumigatus riboB was flanked on each side by ∼1000 bp of the sequence that flanks nkuB in the genome. This fragment was transformed into the nkuAΔ strain TN02A7. Transformants in which nkuB was replaced by riboB were identified by Southern hybridizations. For brevity, we will refer to this replacement as nkuBΔ. A more complete, but cumbersome, designation is nkuB∷A. fumigatus riboB.
The nkuAΔ, nkuBΔ double mutant grew at the same rate as the nkuA single-mutant strain and the parental strain (Figure 1). As with the nkuAΔ mutant, the nkuAΔ, nkuBΔ mutant did not show enhanced sensitivity to MMS (Figure 1).
To determine if gene targeting is enhanced in the nkuAΔ, nkuBΔ double mutant relative to the nkuAΔ single mutant, we transformed the double mutant with a fusion PCR product consisting of the mRFP and A. fumigatus pyrG flanked on each side by 2000 bp of DNA from the histone H1 gene and from the histone H1 3′ untranslated region (the same linear construct that we used to transform the nkuAΔ single mutant). Of 100 transformants, 87 were positive for histone H1 mRFP. This value is very similar to the value obtained with the nkuAΔ single mutant (Figure 2) and gene targeting thus does not appear to be enhanced in the double mutant relative to the single mutant.
DISCUSSION
We have developed a highly efficient gene targeting for A. nidulans that uses nkuAΔ, a deletion of a gene required for nonhomologous end joining, in combination with heterologous selectable markers. In numerous targeting experiments involving many different genes, ∼90% of transformants have been correctly targeted. This is true of GFP tagging, mRFP tagging, promoter replacement, and replacement of nonessential genes. It does not seem to be dependent on the transformation procedure since good results were obtained with three somewhat different transformation procedures. Replacement of essential genes rescued by the formation of balanced heterokaryons is somewhat less efficient, but is much more efficient than has been possible previously. Even transformation with crude PCR products containing multiple bands gives a majority of transformants with correct gene-targeting events. As importantly, integration events in additional to the correct targeting event are very rare. We have carried out Southern hybrizations on 83 transformants in nkuAΔ strains to date. Only one carried transforming DNA sequences in addition to the correct targeting event and this transformant was from a transformation with a crude fusion PCR preparation. While there will certainly be variations among organisms, our data, in combination with the data of Ninomiya et al. (2004), suggest that deletion of KU homologs may be a generally useful strategy for improving gene targeting.
The nkuAΔ strains transform well and grow robustly. In experiments to date, we have seen no evidence for interactions of nkuAΔ with tagged, deleted, or promoter replaced versions of a variety of genes involved in mitosis, nuclear transport, or cell cycle regulation. One would anticipate that nkuAΔ would interact with mutations in genes involved in DNA repair, of course, and if one is concerned that nkuAΔ will affect a process under study, it can be removed by a simple cross.
The gene-targeting system that we have developed is efficient enough that only a few transformants need to be obtained to be certain of having at least one with the correct targeting event. Assuming 90% to be the probability that each transformant carries a single correct targeting event, if one obtains even five transformants, then the probability that at least one will carry the desired targeting event is 0.9999. Since one need obtain only a few transformants to be certain of obtaining one with the correct integration, one can transform with a reduced number of protoplasts and a reduced amount of DNA. More usefully, it is quite practical to target 10 or more genes in a single transformation experiment with a protoplast preparation no larger than one usually uses to target a single gene in an nkuA+ strain. Fusion PCR, moreover, allows 10 or more transforming fragments to be generated quite quickly. The efficiency of gene targeting in the nkuAΔ strains also allows one to determine quickly if a targeting event is lethal. For example, if a particular GFP fusion is lethal, a large fraction of transformants will be balanced heterokaryons (discussed below).
The rapidity of fusion PCR, coupled with the efficiency of gene targeting in nkuAΔ strains, makes genomewide gene-targeting projects quite feasible. Indeed, if one can target 20 genes/day, one could target every gene in the A. nidulans genome in <500 working days. This should make genomewide gene tagging, promoter replacement, and gene knockout experiments feasible.
While the function of many genes in the A. nidulans genome can be guessed from sequence similarities, the functions of the majority of the genes are unknown. The combination of fusion PCR and nkuAΔ provide a powerful system for knocking out these genes. A. nidulans has particular advantages, moreover, with respect to gene knockouts. The presence of a high frequency of balanced heterokaryons (the conidia of which do not grow to form colonies on selective medium) among gene knockout transformants is a strong indication that a gene is essential. In addition, one can usually determine the phenotype of the knockout by observing the germination and growth of spores produced by the heterokaryon. Likewise, the existence of excellent regulatable promoters in A. nidulans (Adams et al. 1988; Waring et al. 1989; Kennedy and Turner 1996; Pachlinger et al. 2005) makes it possible, in principle, to repress or overexpress essentially every gene in the genome.
Acknowledgments
We thank Morgens Trier Hansen for pMT1612 and David Askew (University of Cincinnati College of Medicine) for the A. fumigatus genomic DNA. This work was supported by funding from the Australian Research Council to M.J.H. and from the National Institutes of Health to S.A.O. and B.R.O.
Note added in proof: After the online-ahead-of-print publication of this manuscript, two articles were published that indicate that deletion of KU homologs also improves the frequency of gene targeting in Aspergillus fumigatus (M. E. da Silva Ferreira, M. R. Kress, M. Savoldi, M. H. Goldman, A. Hartl et al., 2006, The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot. Cell 5: 207–211; S. Krappmann, C. Sasse and G. H. Braus, 2006, Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5: 212–215).
References
- Adams, T. H., M. T. Boylan and W. E. Timberlake, 1988. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54: 353–362. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos, A., and M. J. Hynes, 1988. Cloning and analysis of the positively acting regulatory gene amdR from Aspergillus nidulans. Mol. Cell. Biol. 8: 3532–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird et al., 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99: 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove, D. J., 1966. The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113: 51–56. [DOI] [PubMed] [Google Scholar]
- De Souza, C. P. C., X. S. Ye and S. A. Osmani, 1999. Checkpoint defects leading to premature mitosis also cause endoreduplication of DNA in Aspergillus nidulans. Mol. Biol. Cell 10: 3661–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, C. P. C., A. H. Osmani, S. Hashmi and S. A. Osmani, 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14: 1973–1984. [DOI] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, C. Cuomo, L.-J. Ma, J. Wortman et al., 2005. Sequencing and comparative analysis of Aspergillus nidulans. Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Hande, M. P., 2004. DNA repair factors and telomere-chromosome integrity in mammalian cells. Cytogenet. Genome Res. 104: 116–122. [DOI] [PubMed] [Google Scholar]
- Hecht, S. M., 2000. Bleomycin: new perspectives on the mechanism of action. J. Nat. Prod. 63: 158–168. [DOI] [PubMed] [Google Scholar]
- Hofmann, A. F., and S. D. Harris, 2000. The Aspergillus nidulans uvsB gene encodes an A M-related kinase required for multiple facets of the DNA damage response. Genetics 154: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner, K.-P., C. D. Putnam and J. A. Tainer, 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12: 115–122. [DOI] [PubMed] [Google Scholar]
- Horio, T., and B. R. Oakley, 2005. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell 16: 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang, Y. H., R. Hertzberg, S. Hecht and L. F. Liu, 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260: 14873–14878. [PubMed] [Google Scholar]
- Hynes, M. J., and M. A. Davis, 2004. Regulation of the amdS gene in Aspergillus nidulans, pp. 421–435 in The Mycota, Vol. 3: Biochemistry and Molecular Biology, edited by R. Brambl and G. A. Marzluf. Springer-Verlag, Berlin and Heidelberg.
- Jung, M. K., Y. Ovechkina, N. Prigozhina, C. E. Oakley and B. R. Oakley, 2000. The use of beta-D-glucanase as a substitute for Novozym 234 in immunofluorescence and protoplasting. Fungal Genet. Newsl. 47: 65–66. [Google Scholar]
- Kennedy, J., and G. Turner, 1996. δ-(L-α-aminoadipyl)-L-cysteinyl-D-valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol. Gen. Genet. 253: 189–197. [DOI] [PubMed] [Google Scholar]
- Kuwayama, H., S. Obara, T. Morio, M. Katoh, H. Urushihara et al., 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30: e2. [DOI] [PMC free article] [PubMed]
- Lee, S. B., and J. W. Taylor, 1990. Isolation of DNA from fungal mycelia and single spores, pp. 282–287 in PCR Protocols: A Guide to Methods and Applications, edited by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White. Academic Press, San Diego/New York/Berkeley,CA/ Boston/London/Sydney/Tokyo/Toronto.
- Lisby, M., and R. Rothstein, 2004. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 16: 328–334. [DOI] [PubMed] [Google Scholar]
- Martin, M. A., S. A. Osmani and B. R. Oakley, 1997. The role of γ-tubulin in mitotic spindle formation and cell cycle progression in Aspergillus nidulans. J. Cell Sci. 110: 623–633. [DOI] [PubMed] [Google Scholar]
- Ninomiya, Y., K. Suzuki, C. Ishii and H. Inoue, 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 101: 12248–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. R., C. E. Oakley, Y. Yoon and M. K. Jung, 1990. Gamma tubulin is a component of the spindle-pole-body that is essential for microtubule function in Aspergillus nidulans. Cell 61: 1289–1301. [DOI] [PubMed] [Google Scholar]
- Oakley, C. E., C. F. Weil, P. L. Kretz and B. R. Oakley, 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53: 293–298. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., D. B. Engle, J. H. Doonan and N. R. Morris, 1988. Spindle formation and chromatin condensation in cells blocked at interphase by mutation of a negative cell cycle control gene. Cell 52: 241–251. [DOI] [PubMed] [Google Scholar]
- Osmani, A. H., G. S. May and S. A. Osmani, 1999. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J. Biol. Chem. 274: 23565–23569. [DOI] [PubMed] [Google Scholar]
- Pachlinger, R., R. Mitterbauer, G. Adam and J. Strauss, 2005. Metabolically independent and accurately adjustable Aspergillus sp. expression system. Appl. Environ. Microbiol. 71: 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk, L. F., W. Wubker, W. Kohnlein and F. Hutchinson, 1977. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 4: 3573–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigozhina, N. L., C. E. Oakley, A. M. Lewis, T. Nayak, S. A. Osmani et al., 2004. γ-Tubulin plays an essential role in the coordination of mitotic events. Mol. Biol. Cell 15: 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straubinger, B., E. Straubinger, S. Wirsel, G. Turgeon and O. Yoder, 1992. Versatile fungal transformation vectors carrying the selectable bar gene of Streptomyces hygroscopicus. Fungal Genet. Newsl. 39: 82–83. [Google Scholar]
- Terpe, K., 2003. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60: 523–533. [DOI] [PubMed] [Google Scholar]
- Toews, M. W., J. Warmbold, S. Konzack, P. Rischitor, D. Veith et al., 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (Gateway). Curr. Genet. 45: 383–389. [DOI] [PubMed] [Google Scholar]
- Upshall, A., 1986. Genetic and molecular characterization of argB+ transformants of Aspergillus nidulans. Curr. Genet. 10: 593–599. [DOI] [PubMed] [Google Scholar]
- Waring, R. B., G. S. May and N. R. Morris, 1989. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene 79: 119–130. [DOI] [PubMed] [Google Scholar]
- Weidner, G., C. d'Enfert, A. Koch, P. C. Mol and A. A. Brakhage, 1998. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5′-monophosphate decarboxylase. Curr. Genet. 33: 378–385. [DOI] [PubMed] [Google Scholar]
- Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies et al., 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3: 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.-H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Dominguez et al., 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981. [DOI] [PubMed] [Google Scholar]