Abstract
UV irradiation, a known carcinogen, induces the formation of dipyrimidine dimers with the predominant lesions being cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone adducts (6-4PPs). The relative roles of the yeast translesion synthesis DNA polymerases Polζ and Polη in UV survival and mutagenesis were examined using strains deficient in one or both polymerases. In addition, photoreactivation was used to specifically remove CPDs, thus allowing an estimate to be made of the relative contributions of CPDs vs. 6-4PPs to overall survival and mutagenesis. In terms of UV-induced mutagenesis, we focused on the +1 frameshift mutations detected by reversion of the lys2ΔA746 allele, as Polζ produces a distinct mutational signature in this assay. Results suggest that CPDs are responsible for most of the UV-associated toxicity as well as for the majority of UV-induced frameshift mutations in yeast. Although the presence of Polη generally suppresses UV-induced mutagenesis, our data suggest a role for this polymerase in generating some classes of +1 frameshifts. Finally, the examination of frameshift reversion spectra indicates a hierarchy between Polη and Polζ with respect to the bypass of UV-induced lesions.
ULTRAVIOLET (UV) radiation derived from sunlight is a potent mutagen associated with the development of skin cancer in humans. Long-wave UVA (320–400 nm) damages DNA indirectly via the production of reactive oxygen species (Kozmin et al. 2005), while UVB and UVC (290–320 nm and 100–290 nm, respectively) directly induce the formation of covalent linkages between adjacent pyrimidines (Ravanat et al. 2001). The two major forms of such linkages are the abundant cyclobutane pyrimidine dimers (CPDs) and the less abundant pyrimidine (6-4) pyrimidone adducts (6-4PPs) (reviewed in Taylor 2006). The highly conserved nucleotide excision repair (NER) pathway (for a review, see Prakash and Prakash 2000) is the predominant pathway for repairing UV-induced pyrimidine dimers, and organisms that lack this pathway exhibit enhanced sensitivity to the lethal and mutagenic effects of UV (Friedberg et al. 1995). In humans, a lack of NER is associated with the disease xeroderma pigmentosum (XP), which is characterized by an exquisite sensitivity to UV light and an extraordinarily high incidence of skin cancer (Hoeijmakers 2001). In addition to the NER pathway, some organisms, including Saccharomyces cerevisiae, have the ability to directly reverse CPDs enzymatically in a process called photoreactivation (PR). In PR, a DNA photolyase binds to a CPD and utilizes the energy associated with 365–385 nm light to cleave the cyclobutane ring and regenerate the two adjacent pyrimidine bases (reviewed in Sancar 2006). Because 6-4PPs cannot be similarly reversed in yeast, the relative effects of CPDs and 6-4PPs on survival and mutagenesis can be deduced by measuring these biological endpoints in the presence vs. absence of PR.
UV-induced lesions cannot be bypassed by the replicative DNA polymerases and, if not removed by NER or reversed by photolyase, have the potential to block the progress of the replication fork. Such replication-blocking lesions can be dealt with by one of two general tolerance/bypass pathways: template switching or translesion synthesis (TLS) (Barbour and Xiao 2003). In template switching the lesion-blocked primer strand uses the complementary strand of the sister chromatid as a template to bypass the damage. This can either involve annealing between the nascent leading and lagging strands at the replication fork or occur by homologous recombination with the duplex sister being invaded by the blocked nascent strand. As an alternative to template switching, TLS uses specialized DNA polymerases with generally low fidelity and low processivity to insert and/or extend nucleotides across from a lesion (Rattray and Strathern 2003). This pathway allows the cell to bypass lesions and continue replication, but often at the expense of increased mutagenesis.
S. cerevisiae has three TLS polymerases: Polζ, Rev1, and Polη (Rattray and Strathern 2003). Polζ, a complex of the Rev3 and Rev7 proteins, is required for >90% of induced mutagenesis and hence has been designated as an error-prone polymerase (Lawrence 2002). In vitro, purified Polζ bypasses lesions such as CPDs in a highly mutagenic manner (Nelson et al. 1996), but bypasses other lesions such as thymine glycols in a relatively error-free manner (Johnson et al. 2003). Polζ appears to be most efficient, however, at extending from a nucleotide inserted across from a lesion or extending mismatched termini (Prakash and Prakash 2002). This unique extension activity has led to the view that Polζ may work primarily in conjunction with other DNA polymerases in lesion bypass (Prakash and Prakash 2002). With regard to the bypass of CPDs vs. 6-4PPs in vivo, a transformation-based gap-filling assay has indicated a central role for Polζ in bypassing 6-4PPs, but a relatively minimal role in bypassing CPDs (Gibbs et al. 2005). Rev1, a member of the Y family of DNA polymerases, is generally thought to be required for in vivo Polζ activity, although there have been sporadic reports suggesting that there may be exceptions to this generality (Baynton et al. 1999). In addition, Rev1 has an independent deoxycytidyl transferase activity, specifically incorporating dCMP opposite abasic sites in vitro (Nelson et al. 1996; Haracska et al. 2002).
Polη is encoded by the RAD30 gene and, like Rev1, is a member of the Y family of DNA polymerases (McDonald et al. 1997; Roush et al. 1998). Polη was originally characterized on the basis of its unique ability to replicate past thymine–thymine dimers with high fidelity in vitro (Johnson et al. 1999), and yeast studies support a role for Polη in the error-free bypass of CPDs. Polη's role in the bypass of 6-4PPs is more controversial with some studies suggesting a predominant role for Polη in the bypass of 6-4PPs (Bresson and Fuchs 2002) and others suggesting only a minor role for this TLS polymerase (Gibbs et al. 2005). A lack of Polη in humans is associated with a variant form of XP in which the UV damage normally bypassed in an error-free manner by hPolη is presumably bypassed by a much more mutagenic translesion polymerase (Kannouche and Stary 2003). In spite of its original designation as an error-free TLS polymerase, Polη has very low fidelity when copying undamaged DNA templates (Washington et al. 1999) and is important for UV-induced mutagenesis in at least some yeast assays (Zhang and Siede 2002).
Most studies of UV-induced mutagenesis in yeast have focused on base substitutions in either forward or reverse mutation assays (Kunz et al. 1987; McDonald et al. 1997; Yu et al. 2001; Zhang and Siede 2002; Kozmin et al. 2003). In forward mutation spectra of the CAN1 and SUP4-o genes, for example, >95% of the UV-induced events were base substitutions (Armstrong and Kunz 1990; Kozmin et al. 2003). One yeast study that compared induced reversion frequencies of cyc1 missense vs. frameshift alleles concluded, however, that UV-induced frameshifts may constitute 10–20% of the total mutations (Lawrence et al. 1984). In the current study, a reversion assay is used to focus specifically on the production of UV-induced frameshift mutations. These experiments employ the chromosomal lys2ΔA746 frameshift allele, which reverts by acquisition of a compensatory +1 frameshift mutation within a defined 150-bp reversion window (Harfe and Jinks-Robertson 1999). Because Polζ produces a unique mutational signature in this assay system, appropriate mutant strains can be used to deduce the relative roles of Polζ and Polη in UV-induced frameshift mutagenesis. In addition, survival and mutagenesis were assessed both in the presence and in the absence of photoreactivation, thereby allowing the relative contributions of CPDs and 6-4PPs to be determined. These studies reveal dose-dependent differences in survival and mutagenesis and provide further evidence that Polζ and Polη play partially redundant roles in the bypass of CPDs and 6-4PPs.
MATERIALS AND METHODS
Media and growth conditions:
Yeast strains were grown nonselectively in YEP medium (1% yeast extract, 2% Bacto-peptone, 250 mg/liter adenine, 2% agar for plates), which was supplemented with 2% dextrose (YEPD). Selective growth was on synthetic complete medium containing 2% dextrose (SCD) and lacking the appropriate nutrient (Sherman 1991). Canavanine-resistant mutants in the forward mutation assay were identified on SCD–Arg plates supplemented with 60 μg/ml canavanine. All growth was at 30°.
Strain construction:
Mutant strains were derived from SJR922 (MATα ade2-101oc his3ΔNco lys2ΔA746; Harfe and Jinks-Robertson 1999) by lithium acetate transformation (Gietz and Woods 2002). The rev3Δ strain, SJR1272, was constructed using a PCR-generated rev3Δ∷kan fragment (Swanson et al. 1999). SJR1463 contains the rad30Δ∷HIS3 allele and was constructed by transformation with SpeI-digested pJM82 (McDonald et al. 1997). The rev3Δrad30Δ double mutant was created by introducing rev3Δ∷kan into SJR1463.
UV survival and mutagenesis:
Cultures inoculated from single colonies were grown nonselectively in 5 ml YEPD to saturation (∼2 × 108 cells/ml). Cells were washed with H2O, resuspended in 1–1.5 ml of H2O, and 100 μl of the appropriate cell dilution were plated on YEPD and the appropriate SCD-based selective media to assess cell survival and mutagenesis, respectively. YEPD was used to assess UV-induced cell killing, with the survival data being indistinguishable from those obtained when using SCD medium. Within 1 hr of plating, cells were exposed to the specified dose of 254 nm UV light using a UV crosslinker (Ultra Lum). Immediately after UV exposure the plates were wrapped in aluminum foil to prevent photoreactivation. For photoreactivation experiments, plates were exposed to 365 nm of UVA light for 20 min (11 J/m2/sec), using a long-wave UV transilluminator (Spectroline). Because longer exposures to UVA (up to 60 min) did not improve survival, we assume that all of the CPDs that were capable of being photoreversed had been removed within the 20 min used in our experiments. Colonies arising on YEPD and SCD plates were counted after 2 and 3 days of incubation, respectively. Each data point corresponds to the mean of four to six independent survival/mutagenesis experiments, and error bars represent the standard deviation.
lys2ΔA746 reversion spectra:
Genomic DNA was isolated from purified Lys+ colonies using a glass bead lysis procedure. A portion of the LYS2 locus containing the reversion window was amplified by PCR and DNA sequence analysis of PCR-amplified genomic fragments was performed by Macrogen (Seoul, South Korea), using the primer 5′-CGCAACAATGGTTACTCT. Sequences were analyzed using the Sequence Manager software (DNASTAR, Madison, WI) licensed from BIMCORE at Emory University.
RESULTS AND DISCUSSION
The relative roles of the yeast TLS polymerases Polζ and Polη in survival and mutagenesis following UVC irradiation were determined using a wild-type (WT) strain, a strain lacking Polζ (rev3 mutant), a strain lacking Polη (rad30 mutant), and a strain simultaneously lacking both Polζ and Polη (rev3 rad30 double mutant). Because rev1 and rev3 mutants typically exhibit indistinguishable UV-induced toxicity/mutagenesis and Rev1 and Polζ are generally assumed to act in concert to bypass UV-induced lesions (Lawrence 2002), we did not examine rev1 mutants in our studies. Cells were grown to saturation in nonselective liquid medium, plated selectively or nonselectively as appropriate, and then irradiated with varying doses of UV up to 80 J/m2. Two different assays were used to assess mutagenesis. First, the forward mutation frequency at the CAN1 locus was measured to assay a very broad range of mutation types. Second, the level of frameshift mutagenesis was assessed using the lys2ΔA746 allele, which specifically detects net +1 frameshift mutations (Harfe and Jinks-Robertson 1999). This particular allele was used because of the very distinctive signature attributable to Polζ in the corresponding reversion spectra, thus allowing a detailed assessment of Polζ-dependent mutagenesis. Specifically, Polζ is required for the production of “complex” reversion events, which are defined as frameshift mutations accompanied by one or more closely spaced base substitutions (Harfe and Jinks-Robertson 2000; Minesinger and Jinks-Robertson 2005). Finally, the relative contributions of the two major classes of UV damage, CPDs and 6-4 PPs, to UV-associated killing and mutagenesis were estimated following the specific reversal of CPDs by PR.
Survival following UV irradiation:
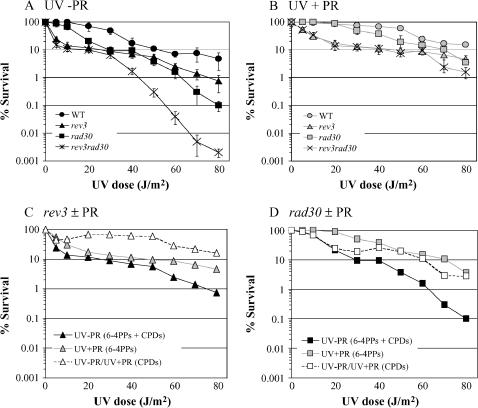
Some studies have reported that a Polζ-deficient, rev3 mutant is more sensitive to UV light than is a Polη-deficient, rad30 mutant (Kozmin et al. 2003), while others concluded the reverse (McDonald et al. 1997; Yu et al. 2001). These results can be reconciled by the survival curves shown in Figure 1A, where there is a clear reversal in the relative sensitivities of the rev3 and rad30 strains at a UV dose of ∼40 J/m2. The rev3 strain was thus more sensitive to UV irradiation than was a rad30 strain at low UV doses, while the relative sensitivities of the mutants were reversed at the higher UV doses (see also McDonald et al. 1997). We suggest that the greater survival-promoting role for Polη at higher UV doses may partially reflect the documented UV induction of RAD30 transcription (McDonald et al. 1997; Roush et al. 1998). Either a dose-dependent induction or the requirement for a threshold level of UV damage for RAD30 induction could, in principle, account for the greater dependence on Polζ for tolerating lesions produced by low UV doses.
Figure 1.
Survival as a function of UVC dose in the (A) absence or (B) presence of photoreactivation (PR). WT (•,  ), rev3 (▴,
), rev3 (▴,  ), rad30 (▪,
), rad30 (▪,  ), and rev3 rad30 (×) strains are shown. Error bars correspond to the standard deviation. (C and D) The relative toxicities of CPDs (open symbols) vs. 6-4PPs (shaded symbols) in the rev3 and rad30 mutants, respectively. The toxicity associated with CPDs at a given UV dose was calculated by dividing the UV − PR survival at that dose by UV + PR survival.
), and rev3 rad30 (×) strains are shown. Error bars correspond to the standard deviation. (C and D) The relative toxicities of CPDs (open symbols) vs. 6-4PPs (shaded symbols) in the rev3 and rad30 mutants, respectively. The toxicity associated with CPDs at a given UV dose was calculated by dividing the UV − PR survival at that dose by UV + PR survival.
As reported previously (McDonald et al. 1997; Yu et al. 2001), the rev3 rad30 double mutant was much more sensitive than either single mutant to UV doses exceeding 30 J/m2, suggesting functionally redundant roles for Polζ and Polη in the bypass of a common UV lesion. In contrast to the synergism between rev3 and rad30 observed at high UV doses, however, at the lowest doses where the rad30 mutant had begun to exhibit clear UV sensitivity (10–20 J/m2), the rev3 rad30 double mutant was no more sensitive than was the rev3 single mutant. The epistasis of rev3 to rad30 at low UV doses suggests that Polη and Polζ may act together to bypass some types of UV-induced lesions, and that Polζ additionally can promote UV survival in a manner that is independent of Polη.
The survival curves when UV exposure was followed by 20 min of photoreactivation (UV + PR) are shown in Figure 1B. Photoreactivation in the absence of prior UV irradiation had no effect on survival, and even at the highest UV dose used, the maximum recovery of viability was obtained within the 20-min period of PR (data not shown). Although we assume that PR removes the majority of, if not all, CPDs, some protection from removal may be provided by positioned, static nucleosomes (Thoma 1999). For the sake of discussion, it is assumed that lesions remaining after PR are composed predominantly of 6-4PPs, although other minor photoproducts (Taylor 2006) as well as residual CPDs may be present. The most notable feature of the UV + PR data is the coincidence of the rev3 and rev3 rad30 survival curves, with these strains being more sensitive to 6-4PPs than the WT and rad30 strains. These data suggest a requirement for Polζ in bypassing 6-4PPs, with little, if any, ability of Polη alone to bypass this lesion. These data are consistent with the demonstrated role of Polζ, but not of Polη, in 6-4PP bypass in in vivo gap repair assays (Gibbs et al. 2005) and contradict the idea that Polη might play a major, Polζ-independent role in the global bypass of 6-4PPs (Bresson and Fuchs 2002). If Polζ requires another polymerase for 6-4PP bypass, this role would presumably be filled by Rev1 or by one of the replicative DNA polymerases.
The effect of 6-4PPs on survival is reflected by the percentage of survival following the removal of CPDs by PR (Figure 1B, UV + PR). The effect of CPDs on survival at each UV dose can be similarly estimated as the ratio of the survival before PR to that after PR [(UV − PR)/(UV + PR)]. Although this type of calculation makes the simplifying assumption that the effects of UV-induced lesions are completely independent of each other, a partitioning of the UV-killing effects into those attributable primarily to CPDs vs. 6-4PPs is nevertheless informative. The dose-dependent survival profiles of the rev3 and rad30 single-mutant strains without photoreactivity (CPDs + 6-4PPs) and with photoreactivity (6-4PPs), as well as those obtained by taking the ratio of these two measured survival values (CPDs), are presented in Figure 1, C and D. In the rev3 mutant, where Polη is the only functional TLS polymerase, PR had little effect on survival, with 6-4PPs thus being more toxic than CPDs. In contrast, the rad30 mutant, which has Polζ as its only TLS polymerase, generally exhibited greater toxicity to CPDs than to 6-4PPs. These data are consistent with the ability of purified Polη to bypass CPDs in vitro (Johnson et al. 2001) and with the requirement for Polη in CPD bypass in in vivo gap-filling assays (Gibbs et al. 2005). These data also lend support to in vivo evidence suggesting that the bypass of most 6-4PPs requires Polζ activity (Gibbs et al. 2005).
UV-induced mutagenesis without photoreactivation:
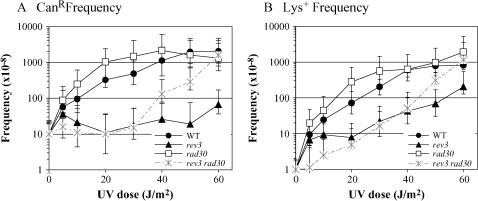
In Escherichia coli the majority of UV-induced mutations are base substitutions at dipyrimidine sites, with frameshifts typically composing <25% of the spectra (e.g., see Miller 1985; Schaaper et al. 1987). One of the very striking aspects of UV-induced frameshift mutagenesis in bacteria is that −1 events outnumber +1 events at least 10:1. In yeast, forward mutation studies have been done using either the SUP4-o ochre-suppressing tRNA gene (Kunz et al. 1987) or the CAN1 locus (Kozmin et al. 2003) as a target. As in bacterial cells, base substitutions predominate and occur mainly at dipyrimidine sites. Although UV-induced frameshift mutations appear to be proportionally less frequent in yeast than in bacterial cells, a similar strong bias for −1 as opposed to +1 events has been reported. In the current study, we have chosen to focus on UV-induced +1 frameshifts in yeast, primarily because of the unique mutational signature produced by Polζ in the lys2ΔA746 reversion assay (Harfe and Jinks-Robertson 2000). To allow comparison to other studies and to provide a context for the UV-induced frameshift mutagenesis, we also examined forward mutation at the CAN1 locus.
A dose-dependent increase in the frequency of UV-induced can1 mutants was observed in the WT background, with an ∼200-fold increase in can1 mutant frequency at the highest UV dose relative to the spontaneous (no UV) frequency (Figure 2A). Consistent with a role for Polη in the error-free bypass of UV damage, and hence the avoidance of UV-induced mutagenesis, the frequency of UV-induced can1 mutations was consistently elevated in a rad30 mutant relative to the WT strain. As expected, UV-induced mutagenesis was almost completely dependent on Polζ activity, as there was little induction of can1 mutants in the rev3 background. There was, however, an unexpected induction of can1 mutants at the highest UV doses in the rev3 rad30 double mutant. In the absence of Polη, there thus appears to be a Polζ-independent mechanism for generating UV-induced mutations that eliminate CAN1 activity. We considered the possibility that the corresponding mutants might harbor a complete loss of the CAN1 locus (Chen and Kolodner 1999), but because we were still able to PCR amplify the locus in the mutants examined (data not shown), this is unlikely. Whether the Polζ-independent can1 mutants reflect a novel mechanism of UV-induced mutagenesis, or might simply be a manifestation of the very low survival of the rev3 rad30 mutant at high UV doses, is not known.
Figure 2.
UV-induced mutagenesis. Frequencies of UV-induced forward mutation at CAN1 (A) and lys2ΔA746 reversion (B) in WT (•), rev3 (▴), rad30 (□), and rev3 rad30 (×) strains. UV-induced mutation frequencies were calculated by subtracting the frequency in the starting culture (0 J/m2) from that obtained after each specified UV dose. Error bars correspond to the standard deviation.
A stronger induction of Lys+ revertants than of can1 mutants was evident following UV irradiation; at a dose of 60 J/m2, the reversion frequency of the lys2ΔA746 allele was stimulated ∼800-fold (Figure 2B). The strong UV induction of frameshifts was not unique to the +1 events detected by the lys2ΔA746 reversion assay; a similar level of induction was observed when reversion of the lys2ΔBgl allele, which reverts via compensatory −1 frameshifts (Greene and Jinks-Robertson 1997), was examined (data not shown). Thus, in striking contrast to the situation in bacterial cells where −1 frameshifts greatly outnumber +1 frameshifts (Miller 1985; Schaaper et al. 1987), +1 and −1 frameshifts are induced at similar levels at the yeast LYS2 locus. Given the reported rarity of UV-induced frameshifts in yeast forward mutation assays, the slightly greater induction of Lys+ revertants than of can1 mutants was surprising. In the case of UV-induced mutations in SUP4-o, the rarity of frameshifts could be related to the nature of the target. The SUP4-o locus is quite small (89 bp) and contains no A/T runs >3 nt (Kunz et al. 1987), which is where most frameshifts accumulate in the lys2ΔA746 assay (see spectra below). In the case of the UV-induced can1 mutants, only 32 mutants were sequenced and these were isolated following a single UV dose that resulted in only 10–15% killing (Kozmin et al. 2003). We estimate that at a comparable level of killing in our WT strain (∼10 J/m2 of UV), the frequency of UV-induced Lys+ revertants likely would have been 5- to 10-fold lower than that of can1 mutants. On the basis of our analyses, however, we conclude that, as in bacterial cells, frameshift mutations can compose a sizeable fraction of UV-induced mutations in yeast.
As was seen in the CAN1 forward mutation assay, reversion of the lys2ΔA746 allele was elevated slightly in the rad30 background and was reduced in the rev3 background relative to the WT background. In contrast to the near complete dependency of can1 mutations on Polζ (Paulovich et al. 1998; Friedl et al. 2001; Stelter and Ulrich 2003), however, a significant induction of Lys+ revertants was detected in the rev3 mutant. These data are consistent with the earlier report that a small proportion of UV-induced his4 frameshifts occur independently of REV3 and REV1 (Lawrence et al. 1984). As observed in the CAN1 assay, there was an inexplicable induction of Lys+ revertants at high UV doses in the rev3 rad30 double mutant. This strong induction of Lys+ revertants also occurred in a rev3 rad30 rev1 triple mutant at high UV doses (data not shown), indicating that the residual frameshift mutagenesis is independent of all three yeast TLS polymerases.
UV-induced mutagenesis with photoreactivation:
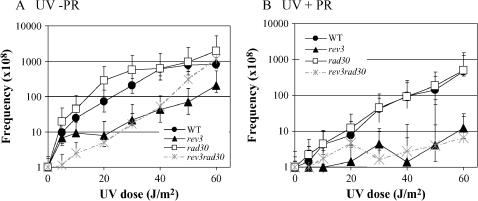
As with survival, UV-induced reversion of the lys2ΔA746 allele was examined in the presence vs. absence of PR. At a given UV dose, +1 frameshifts due to the combined action of CPDs and 6-4PPs are reflected by the Lys+ frequency in the absence of PR (Figure 3A), while mutagenesis due to 6-4PPs corresponds to the Lys+ frequency following the removal of CPDs by PR (Figure 3B). For the WT, rev3, rad30, and rev3 rad30 strains, a comparison of the frequency of Lys+ revertants before and after PR indicates that the majority of the UV-induced +1 frameshifts in each strain were due to CPDs. A similar conclusion concerning the relative contributions of CPDs and 6-4PPs to mutagenesis was reached in studies using the SUP4-o forward mutation assay (Armstrong and Kunz 1992) and in mammalian cells (Jans et al. 2005). Loss of Polη was associated with an increase in the UV-induced lys2ΔA746 reversion frequency in the absence of PR, but its loss had no effect on the frequency of Lys+ revertants when UV treatment was followed by PR. Polη thus appears to prevent the accumulation of +1 frameshifts due to CPDs, but not to other types of UV-induced lesions. Although this is consistent with a role for Polη specifically in the error-free bypass of CPDs (Johnson et al. 1999), analysis of individual classes of frameshifts suggests that bypass of some CPDs by Polη can be mutagenic as well (see below). Finally, in either the presence or absence of PR, the induction of Lys+ revertants was greatly reduced in a rev3 background, indicating a role for Polζ in the production of the majority of +1 frameshifts initiated by either CPDs or 6-4PPs. Although examination of mutagenesis in the absence vs. presence of PR clearly implicates CPDs as the lesion responsible for most in vivo frameshift mutagenesis, it should be noted that these results cannot be used to infer the relative mutagenic potential of a single CPD vs. 6-4PP.
Figure 3.
Contributions of CPDs and 6-4PPs to +1 frameshift mutagenesis. UV-induced reversion of the lys2ΔA746 allele in WT (•), rev3 (▴), rad30 (□), or rev3 rad30 (×) strains in the absence (A) or presence (B) of photoreactivation (PR) is shown. The UV-induced reversion frequencies were calculated by subtracting the frequency in the starting culture (0 J/m2) from that obtained after each specified UV dose. Error bars correspond to the standard deviation.
UV-induced reversion spectra in a WT background:
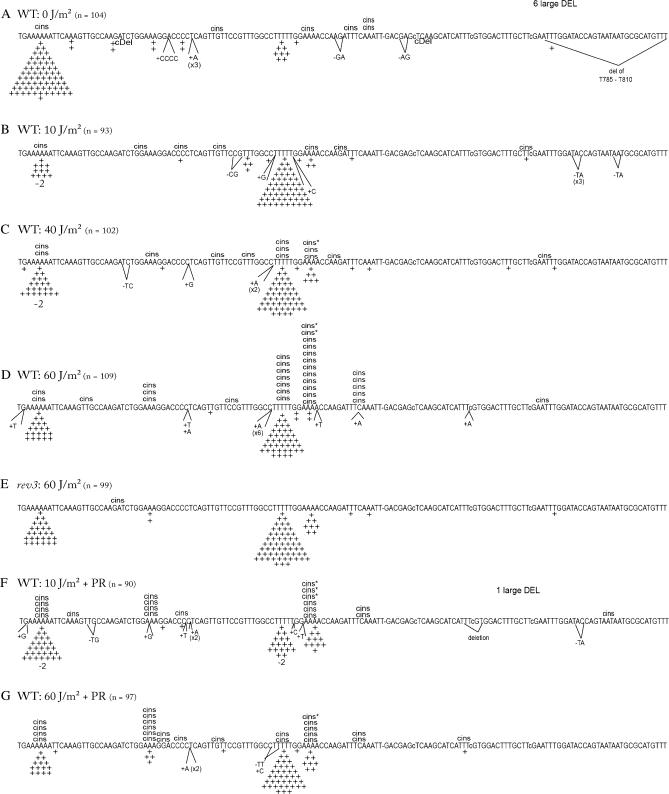
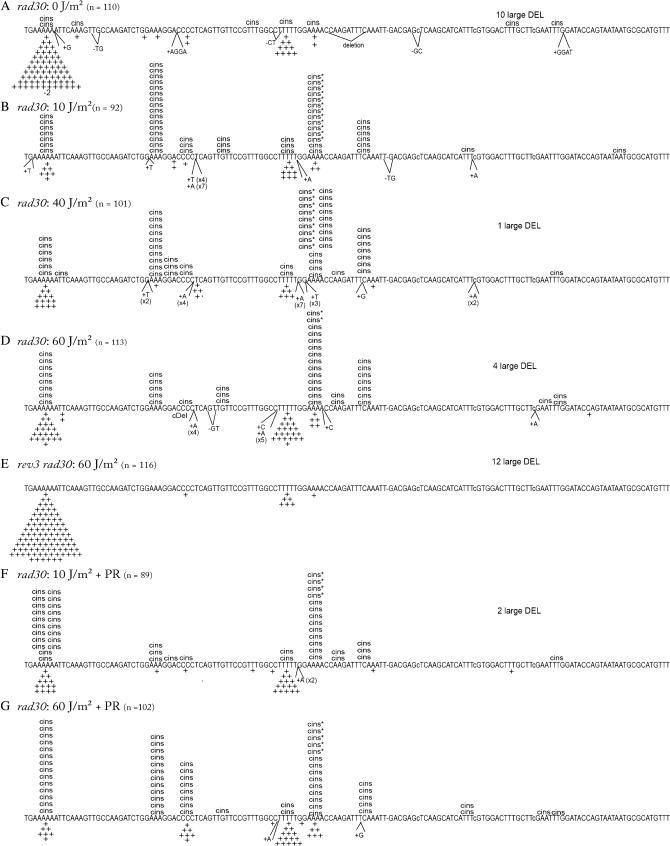
The lys2ΔA746 allele reverts by acquiring a compensatory +1 frameshift within an ∼150-bp reversion window defined by stop codons in the alternative reading frames. Our previous analyses of spontaneous lys2ΔA746 reversion have demonstrated that mutation spectra provide a wealth of information that cannot be discerned by simply measuring mutation rates (Minesinger and Jinks-Robertson 2005; Sabbioneda et al. 2005). The spectra of lys2ΔA746 reversion events as a function of UV dose were determined by sequencing ∼100 revertants isolated following irradiation with 0, 10, 40, or 60 J/m2 of UVC (Figure 4). In the absence of UV irradiation, 84% (87/104) of the reversion events were simple +1 frameshifts, and >90% of these (81/87) were in homopolymer runs >3 nt (Figure 4A). The two longest runs in the reversion window were hotspots for the simple +1 events; 76% of the +1 events were in the 6A run and 9% were in the 5T run. The strong 6A:5T bias most likely reflects the positive correlation between run length and the frequency of DNA polymerase slippage (Tran et al. 1997; Greene and Jinks-Robertson 2001). In addition to the predominant class of simple insertions, complex insertions (cins) in which the selected frameshift was accompanied by one or more base substitutions accounted for 6% of the spontaneous events. Finally, ∼6% of the revertants contained a 95- or 131-bp deletion with endpoints in 10- or 7-bp direct repeats, respectively. The large deletions were most evident among the spontaneous revertants and presumably reflect polymerase slippage during normal, replicative DNA synthesis.
Figure 4.
UV-induced lys2ΔA746 reversion spectra in a WT (RAD30) background. The sequence of the reversion window for the lys2ΔA746 allele is shown; the position of the nucleotide deleted to create the lys2ΔA746 allele is indicated by a dash and the nucleotides changed to extend the reversion window are indicated by lowercase letters (Harfe and Jinks-Robertson 1999). Pluses and minuses indicate 1-nt insertions and 2-bp deletions, respectively; complex insertions (cins) are indicated above the sequence. Complex events above the 4A run that are followed by an asterisk (cins*) indicate events where the frameshift event could have occurred at either the 5T run or the 4A run. The number of events created by the deletion of 95 or 131 bp with endpoints in 10- or 7-bp direct repeats, respectively, is indicated as “large DEL” above each spectrum. The total number of revertants sequenced (n) for each strain is indicated. The spontaneous WT spectrum (0 J/m2) was published previously (Harfe and Jinks-Robertson 2000).
Reversion of the lys2ΔA746 allele was stimulated 24-fold following UV irradiation at a dose of 10 J/m2 (see Figure 2B), and this was accompanied by a very striking reversal in the relative numbers of simple insertions in the 6A vs. 5T run (Figure 4B). Whereas there was an 8:1 bias in favor of simple insertions within the 6A run among spontaneous revertants, there was a 5:1 bias in favor of 5T run insertions among the UV-induced revertants. Irradiation with 40 J/m2 of UV stimulated the Lys+ frequency 560-fold, and the UV-associated bias for simple insertions in the 5T rather than the 6A run persisted (Figure 4C). Relative to the simple insertions, an enhanced accumulation of complex insertions also began to be evident at the 40 J/m2 UV dose, with complex insertions increasing to 13% of the total reversion events. The shift toward complex insertions within the reversion spectrum was much more evident at the 60 J/m2 UV dose, with these events accounting for 29% (31/109) of the total UV-induced revertants sequenced (Figure 4D). Whereas the spontaneous complex events were scattered throughout the reversion window, the UV-induced events clustered at hotspots that presumably reflect the abundances of the underlying UV lesions, the efficiency of NER, and/or the relative efficiencies of the lesion bypass/tolerance mechanisms. To confirm that the complex events induced by 60 J/m2 UV were dependent on the presence of Polζ, a spectrum was also obtained from a rev3 mutant. As expected, complex events were virtually eliminated in the rev3 mutant (Figure 4E), with only one event being observed out of the 99 revertants sequenced. Finally, spectra were obtained after allowing time for photoreactivation to specifically reverse CPDs. PR was carried out after the 10- and 60-J/m2 UV exposures, and the corresponding spectra are presented in Figure 4, F and G.
UV-induced reversion spectra in a rad30 background:
The UV-induced reversion spectra generated in the absence of Polη (rad30 mutant) are presented in Figure 5. As with the WT strain, spectra were obtained following irradiation with 0, 10, 40, or 60 J/m2 of UVC and when the 10- and 60-J/m2 doses were followed by photoreactivation. In the absence of UV, the lys2ΔA746 reversion spectrum of the rad30 mutant (Figure 5A) was similar to that of the WT strain (Figure 4A), with a strong bias for simple events in the 6A relative to the 5T run, and only a small percentage of the total frameshifts being complex. The UV-induced spectra generated in the rad30 background (Figure 5, B–D) were strikingly different, however, from those generated in the presence of Polη (Figure 4, B–D). First, there were roughly equivalent numbers of UV-induced simple events in the 6A and 5T runs in the rad30 mutant, while events in the 5T run were much more frequent in the WT background. Second, there was a dramatic increase in the proportion of complex events in the rad30 mutant, with complex events outnumbering the simple events at each UV dose examined. As in the WT strain, the complex events in the rad30 mutant were dependent on the presence of Polζ, as these events were not evident when REV3 was deleted (Figure 5E).
Figure 5.
UV-induced lys2ΔA746 reversion spectra in a rad30 background. See Figure 4 legend for details.
Comparison of WT and rad30 spectra:
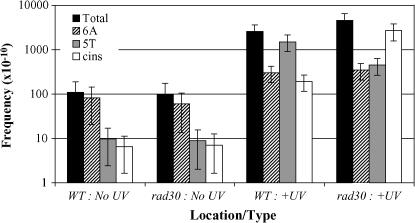
To facilitate the analysis of the frameshift reversion data, Figure 6 presents the frequencies of the three most prominent classes of +1 frameshifts: simple events in the 6A run, simple events in the 5T run, and complex events occurring throughout the reversion window. The frequency of a given class of event was derived by multiplying the measured Lys+ reversion frequency by the percentage of the relevant event in the corresponding spectrum. Only the frequencies following exposure of the WT and rad30 strains to the 10-J/m2 dose of UV are shown, as no additional insight was gained when the higher UV doses were examined. With respect to spontaneous Lys+ revertants, the frequencies of +1 events in 6A and 5T runs, as well as the frequencies of complex events, were indistinguishable in the WT and rad30 strains. Upon exposure of the WT strain to UV, the proportion of events that were complex did not increase significantly, but, as noted previously there was a striking reversal in the distributions of +1 frameshifts between the 6A and 5T runs. This translates into an ∼180-fold induction by UV of simple insertions in the 5T run, but only a 4-fold induction of insertions in the 6A run. This dramatic difference could reflect the relative abundances of the primary lesions at or near these runs, the relative efficiency of lesion removal by NER, and/or the propensity of a given lesion to generate +1 frameshifts as opposed to other types of mutations. If one assumes that the UV-induced lesions that lead to the frameshifts are within the runs, it is interesting to note that the underlying dipyrimidine lesions in the 6A run would be on the transcribed (noncoding) strand, while those in the 5T run would be on the nontranscribed (coding) strand. Transcription-coupled NER would be expected to result in more efficient repair of lesions on the transcribed than on the nontranscribed strand (Friedberg et al. 1995), and this could at least partially account for the strong bias for UV-induced insertions to occur in the 5T run.
Figure 6.
Frequencies of specific classes of +1 frameshift events in WT and rad30 strains. Solid bars correspond to the overall lys2ΔA746 reversion frequencies, hatched bars to the frequencies of simple insertions in the 6A run, shaded bars to the frequencies of simple insertions in the 5T run, and open bars to the frequencies of complex events. The reversion frequency (and standard deviation) for each type of event was calculated by multiplying the total frequency by the percentage of the specific event in the correponding spectrum. The UV data were generated at a dose of 10 J/m2.
Whereas the distributions of the spontaneous mutation types in the WT vs. rad30 strain were indistinguishable, those produced following UV irradiation were very different. First, the strong bias for the UV-induced events in the WT strain to occur in the 5T run rather than in the 6A run was not present in the rad30 mutant. If one compares the frequencies of these events in the WT and rad30 strains, the frequency of the 5T run events was 5-fold less in the rad30 than in the WT strain, while the frequency of the 6A run events was unaffected by loss of Polη. The +1 events that occurred in the 5T run thus were largely dependent on the presence of Polη, while those occurring in the 6A run were not. The second notable difference between the WT and rad30 spectra was the very striking increase in the proportion of Polζ-dependent complex frameshifts upon loss of Polη, from 8% in the WT strain to 59% in the rad30 mutant. This translates into a 380-fold induction of Polζ-dependent complex events in the absence of Polη, but only a 33-fold induction in its presence. Thus, in a rad30 mutant, UV lesions that are normally bypassed by Polη are instead bypassed by Polζ to give the signature complex frameshifts. We suggest that these data reflect an underlying hierarchy in the utilization of Polζ vs. Polη in lesion bypass, which could be related to the nature of the underlying lesion, the local sequence context surrounding a lesion, and/or the location of a lesion on the coding vs. noncoding strand of LYS2. An important goal of future studies will be to identify the factors that regulate the use of Polη vs. Polζ in lesion bypass.
Further insight into the Polη-dependent bypass of UV-induced lesions was obtained by calculating the frequencies of the individual classes of frameshifts derived from CPDs vs. 6-4PPs (Table 1). The frequency of a given event attributable to 6-4PPs was calculated by multiplying the UV + PR Lys+ frequency by the proportion of the event in the corresponding UV + PR reversion spectrum. The frequency of the event due to CPDs was then calculated by subtracting the UV + PR frequency from the UV − PR frequency. In the WT strain, the largest class of UV-induced frameshifts was the simple +1 events in the 5T run, and >90% of these were attributable to CPDs. CPDs were also the major contributor to the 5T run events in the rad30 mutant, but, as noted above, events in the 5T run no longer dominated the spectrum. Whereas CPDs and 6-4PPs were equal contributors to the complex frameshifts and the 6A run +1 events in the WT background, CPDs contributed more to these frameshifts than did 6-4PPs in the rad30 strain. Together, these data are consistent with the role of Polη in the nonmutagenic bypass of CPDs; in its absence, CPDs are more likely to produce +1 frameshifts.
TABLE 1.
Frequencies of specific mutation types following exposure to 10 J/m2 of UVC
| Frequencies (×10−8)
|
|||||
|---|---|---|---|---|---|
| Strain | UV lesion | All events | 6A run | 5T run | Complex |
| WT | CPDs + 6-4PPs | 25.6 ± 10.2 | 3.02 ± 1.2 | 15.1 ± 6.0 | 1.92 ± 0.7 |
| 6-4PPs only | 5.06 ± 1.23 | 1.57 ± 0.4 | 0.96 ± 0.2 | 1.07 ± 0.3 | |
| CPDs only | 20.3 ± 10.2 | 1.45 ± 1.2 | 14.2 ± 6.0 | 0.85 ± 0.7 | |
| rad30 | CPDs + 6-4PPs | 45.8 ± 18.7 | 3.48 ± 1.4 | 4.48 ± 1.8 | 26.9 ± 11.0 |
| 6-4PPs only | 5.31 ± 1.9 | 0.83 ± 0.3 | 0.90 ± 0.3 | 2.74 ± 1.0 | |
| CPDs only | 40.4 ± 18.7 | 2.79 ± 1.4 | 3.74 ± 1.8 | 24.6 ± 11.0 | |
A more complicated picture of lesion bypass by Polη emerges if one compares the contributions of a given lesion to each specific class of frameshift in the WT vs. rad30 background. As noted above, for example, most of the 6A run events were generated in response to CPDs in the rad30 mutant, while CPDs and 6-4PPs were equal contributors in the WT strain. A closer examination of the 6A run data reveals not only that Polη had the expected effect of reducing the CPD-generated +1 events (the frequency of these events increased twofold in the rad30 mutant), but also that Polη unexpectedly promoted the occurrence of +1 events derived from 6-4PPs (the frequency of these events decreased twofold in the rad30 mutant). In relation to a possible participation by Polη in the “error-prone” bypass of 6-4PPs to generate +1 frameshifts, it has been suggested that the RAD30-dependent reversion of the arg4-17 nonsense allele, which reverts primarily via C-to-T transitions, likely reflects the mutagenic bypass of a 6-4PP (Zhang and Siede 2002). In addition, Polη has been shown to be required for the mutagenic bypass of a site-specific (6-4)TT engineered into a duplex plasmid (Bresson and Fuchs 2002), although a contradictory result was obtained using an in vivo gap-filling assay (Gibbs et al. 2005). The data presented here extend the mutagenic potential of 6-4PPs to include the production of some types of UV-induced frameshift events by Polη. This particular role of Polη must be relatively minor, however, as this TLS polymerase did not contribute to overall survival in the UV + PR experiments.
In contrast to the equivalent contributions of 6-4PPs and CPDs to the 6A run and complex events in a WT background, >90% of the events in the 5T run were derived from CPDs. As noted above, loss of Polη was accompanied by the expected increase in the CPD-associated 6A run events, but the frequency of the 5T run events decreased upon loss of Polη. Thus, in the context of the 5T run, Polη promotes rather than antagonizes the production of simple +1 events derived from the bypass of CPDs. That Polη can have opposing roles in the CPD-associated frameshift mutagenesis that occurs in closely linked homopolymer runs (i.e., the 6A and 5T runs) illustrates the importance of examining mutation spectra in conjunction with measuring mutation frequencies.
Conclusions:
The contributions of the yeast TLS polymerases Polζ and Polη to survival and frameshift mutagenesis following UVC irradiation were examined using WT, rev3, rad30, and rev3 rad30 strains. The specific removal of the CPDs by PR further allowed an estimation of the relative contributions of CPDs vs. other types of UV-induced lesions, which correspond primarily to 6-4PPs, to overall survival and mutagenesis. Consistent with an induction of RAD30 transcription by UV, the relative contributions of Polζ and Polη to survival were dose dependent, with Polζ being more important at low UV doses and Polη at high doses. As previously reported, synergism was evident at high doses in the rev3 rad30 double mutant, suggesting functional redundancy of Polζ and Polη. At low UV doses, however, deletion of REV3 was epistatic to that of RAD30, suggesting that there may be joint action of Polη and Polζ in bypass of some types of UV lesion bypass. Finally, the relative toxicities of CPDs and 6-4PPs were influenced by which TLS polymerase was functional.
In terms of UV-induced mutagenesis, +1 and −1 frameshifts appeared to be induced to similar levels by UVC and to compose a significant proportion of the total induced mutations. In agreement with presumed error-free and error-prone bypass of UV-induced lesions by Polη and Polζ, respectively, reversion frequencies generally were elevated in the rad30 mutant and reduced in the rev3 mutant. A more detailed assessment of the contribution of Polη to +1 frameshift mutagenesis, however, was obtained by examining UV-induced frameshift spectra in WT and rad30 strains. While Polη generally prevented frameshift mutagenesis, as expected of an error-free TLS polymerase, it also appeared to promote the occurrence of some specific classes of events. Finally, an analysis of Polζ-dependent complex frameshifts suggests a hierarchy in the utilization of Polζ vs. Polη in the bypass of UV-induced lesions, a hierarchy that likely will extend to the bypass of other types of lesions. The complexities uncovered by the mutagenesis data reported here underscore the need to exercise caution when using a limited data set to deduce the bypass specificities of TLS polymerases in vivo.
Acknowledgments
We thank John McDonald for providing the pJM82 plasmid used in the construction of the rad30 strain, Brian Harfe for experimental contributions in the early phases of this study, and the members of the S.J.-R. laboratory for comments on the manuscript. Finally, we thank Nayun Kim, Gray Crouse, and Wolfram Siede for helping to clarify our thinking concerning the relative contributions of CPDs and 6-4PPs to survival. This work was supported by grant R01 GM064769 to S.J.-R. from the National Institutes of Health. A.L.A. was partially supported by Graduate Division of Biological and Biomedical Sciences at Emory University and National Institutes of Health training grant T32 GM-08367.
References
- Armstrong, J. D., and B. A. Kunz, 1990. Site and strand specificity of UVB mutagenesis in the SUP4-o gene of yeast. Proc. Natl. Acad. Sci. USA 87: 9005–9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, J. D., and B. A. Kunz, 1992. Photoreactivation implicates cyclobutane dimers as the major promutagenic UVB lesions in yeast. Mutat. Res. 268: 83–94. [DOI] [PubMed] [Google Scholar]
- Barbour, L., and W. Xiao, 2003. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 532: 137–155. [DOI] [PubMed] [Google Scholar]
- Baynton, K., A. Bresswon-Roy and R. P. P. Fuchs, 1999. Distinct roles for Rev1p and Rev7p during translesion synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 34: 124–133. [DOI] [PubMed] [Google Scholar]
- Bresson, A., and R. P. Fuchs, 2002. Lesion bypass in yeast cells: Pol η participates in a multi-DNA polymerase process. EMBO J. 21: 3881–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., and R. D. Kolodner, 1999. Gross chromosome rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat. Genet. 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Friedberg, E. C., G. C. Walker and W. Siede, 1995. DNA Repair and Mutagenesis. ASM Press, Washington, DC.
- Friedl, A. A., B. Liefshitz, R. Steinlauf and M. Kupiec, 2001. Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 486: 137–146. [DOI] [PubMed] [Google Scholar]
- Gibbs, P. E., J. McDonald, R. Woodgate and C. W. Lawrence, 2005. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics 169: 575–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Greene, C. N., and S. Jinks-Robertson, 1997. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins. Mol. Cell. Biol. 17: 2844–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene, C. N., and S. Jinks-Robertson, 2001. Spontaneous frameshift mutations in Saccharomyces cerevisiae: accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics 159: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska, L., S. Prakash and L. Prakash, 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J. Biol. Chem. 277: 15546–15551. [DOI] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 1999. Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe, B. D., and S. Jinks-Robertson, 2000. DNA polymerase ζ introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell 6: 1491–1499. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers, J. H., 2001. Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374. [DOI] [PubMed] [Google Scholar]
- Jans, J., W. Schul, Y. G. Sert, Y. Rijksen, H. Rebel et al., 2005. Powerful skin cancer protection by a CPD-photolyase transgene. Curr. Biol. 15: 105–115. [DOI] [PubMed] [Google Scholar]
- Johnson, R. E., S. Prakash and L. Prakash, 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Nature 283: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Johnson, R. E., L. Haracska, S. Prakash and L. Prakash, 2001. Role of DNA polymerase η in the bypass of a (6–4) TT photoproduct. Mol. Cell. Biol. 21: 3558–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. E., S. L. Yu, S. Prakash and L. Prakash, 2003. Yeast DNA polymerase ζ is essential for error-free replication past thymine glycol. Genes Dev. 17: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche, P., and A. Stary, 2003. Xeroderma pigmentosum variant and error-prone DNA polymerases. Biochimie 85: 1123–1132. [DOI] [PubMed] [Google Scholar]
- Kozmin, S. G., Y. I. Pavlov, T. A. Kunkel and E. Sage, 2003. Roles of Saccharomyces cerevisiae DNA polymerases Polη and Polζ in response to irradiation by simulated sunlight. Nucleic Acids Res. 31: 4541–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin, S., G. Slezak, A. Reynaud-Angelin, C. Elie, Y. de Rycke et al., 2005. UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 13538–13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz, B. A., M. K. Pierce, J. R. A. Mis and C. N. Giroux, 1987. DNA sequence analysis of the mutational specificity of UV light in the SUP4-o gene of yeast. Mutagenesis 2: 445–453. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. W., 2002. Cellular roles of DNA polymerase ζ and Rev1 protein. DNA Repair 1: 425–435. [DOI] [PubMed] [Google Scholar]
- Lawrence, C. W., T. O'Brien and J. Bond, 1984. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol. Gen. Genet. 195: 487–490. [DOI] [PubMed] [Google Scholar]
- McDonald, J. P., A. S. Levine and R. Woodgate, 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147: 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H., 1985. Mutagenic specificity of ultraviolet light. J. Mol. Biol. 182: 45–65. [DOI] [PubMed] [Google Scholar]
- Minesinger, B. K., and S. Jinks-Robertson, 2005. Roles of RAD6 epistasis group members in spontaneous Polζ-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 169: 1939–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. R., C. W. Lawrence and D. C. Hinkle, 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731. [DOI] [PubMed] [Google Scholar]
- Paulovich, A. G., C. D. Armour and L. H. Hartwell, 1998. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics 150: 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2000. Nucleotide excision repair in yeast. Mutat. Res. 451: 13–24. [DOI] [PubMed] [Google Scholar]
- Prakash, S., and L. Prakash, 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16: 1872–1883. [DOI] [PubMed] [Google Scholar]
- Rattray, A. J., and J. N. Strathern, 2003. Error-prone DNA polymerases: when making a mistake is the only way to get ahead. Annu. Rev. Genet. 37: 31–66. [DOI] [PubMed] [Google Scholar]
- Ravanat, J. L., T. Douki and J. Cadet, 2001. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B 63: 88–102. [DOI] [PubMed] [Google Scholar]
- Roush, A. A., M. Suarez, E. C. Friedberg, M. Radman and W. Siede, 1998. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 257: 686–692. [DOI] [PubMed] [Google Scholar]
- Sabbioneda, S., B. K. Minesinger, M. Giannattasio, P. Plevani, M. Muzi-Falconi et al., 2005. The 9–1-1 checkpoint clamp physically interacts with Polζ and is partially required for spontaneous Polζ-dependent mutagenesis in Saccharomyces cerevisiae. J. Biol. Chem. 280: 38657–38665. [DOI] [PubMed] [Google Scholar]
- Sancar, G. B., 2006. Damage recognition by DNA photolyases, pp. 95–110 in DNA Damage Recognition, edited by W. Siede, Y. W. Kow and P. W. Doetsch. Taylor & Francis, New York.
- Schaaper, R. M., R. L. Dunn and B. W. Glickman, 1987. Mechanisms of ultraviolet-induced mutation. Mutational spectra in the Escherichia coli lacI gene for a wild-type and an excision-repair-deficient strain. J. Mol. Biol. 198: 187–202. [DOI] [PubMed] [Google Scholar]
- Sherman, F., 1991. Getting started with yeast. Methods Enzymol. 194: 3–20. [DOI] [PubMed] [Google Scholar]
- Stelter, P., and H. D. Ulrich, 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191. [DOI] [PubMed] [Google Scholar]
- Swanson, R. L., N. J. Morey, P. W. Doetsch and S. Jinks-Robertson, 1999. Overlapping specificities of base excision repair, nucleotide excision repair, recombination, and translesion synthesis pathways for DNA base damage in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 2929–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J.-S., 2006. Structure and properties of DNA photoproducts, pp. 67–94 in DNA Damage Recognition, edited by W. Siede, Y. W. Kow and P. W. Doetsch. Taylor & Francis, New York.
- Thoma, F., 1999. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18: 6585–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, H. T., J. D. Keen, M. Kricker, M. A. Resnick and D. A. Gordenin, 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol. Cell. Biol. 17: 2859–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington, M. T., R. E. Johnson, S. Prakash and L. Prakash, 1999. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J. Biol. Chem. 274: 36835–36838. [DOI] [PubMed] [Google Scholar]
- Yu, S.-L., R. E. Johnson, S. Prakash and L. Prakash, 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21: 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., and W. Siede, 2002. UV-induced T→C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase η in vivo. Nucleic Acids Res. 30: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]