Abstract
Life-history theory and evolutionary theories of aging assume the existence of alleles with age-specific effects on fitness. While various studies have documented age-related changes in the genetic contribution to variation in fitness components, we know very little about the underlying genetic architecture of such changes. We used a set of recombinant inbred lines to map and characterize the effects of quantitative trait loci (QTL) affecting fecundity of Drosophila melanogaster females at 1 and 4 weeks of age. We identified one QTL on the second chromosome and one or two QTL affecting fecundity on the third chromosome, but these QTL affected fecundity only at 1 week of age. There was more genetic variation for fecundity at 4 weeks of age than at 1 week of age and there was no genetic correlation between early and late-age fecundity. These results suggest that different loci contribute to the variation in fecundity as the organism ages. Our data provide support for the mutation accumulation theory of aging as applied to reproductive senescence. Comparing the results from this study with our previous work on life-span QTL, we also find evidence that antagonistic pleiotropy may contribute to the genetic basis of senescence in these lines as well.
A major challenge in evolutionary genetics is to characterize the genetic architecture of natural variation in life-history traits, those components of fitness that directly influence age-specific survival and reproductive success. Life-history theory is founded on the idea that natural selection favors a particular strategy of age-specific allocation of energy to the competing demands of growth, development, reproduction, storage, maintenance, and repair in a given ecological setting (Stearns 1992). This suggests that understanding the genetic basis of life-history variation will require that we not only identify the genes that affect these traits but also characterize the age-specific effects of alleles at these loci. Knowledge of the genetic basis of life-history variation at the molecular genetic level not only would contribute to our understanding of the genetic architecture and evolution of quantitative traits in general but also would provide insight into mechanisms that maintain variation in fitness (Barton and Turelli 1989; Barton and Keightley 2002; Turelli and Barton 2004).
While numerous studies have documented the existence of genetically based variation in life-history traits in natural populations (Mousseau and Roff 1987; Hard et al. 1993; Shaw et al. 1995; Kruuk et al. 2000; Leips et al. 2000; Sommer and Pearman 2003; Drnevich et al. 2004; Fox et al. 2004; Windig et al. 2004), we know very little about the genes that underlie this variation. Further, although several studies have verified that mutations can have age-specific effects on fitness components (Houle et al. 1994; Hughes and Charlesworth 1994; Hughes 1995; Charlesworth and Hughes 1996; Promislow et al. 1996; Tatar et al. 1996; Pletcher et al. 1998, 1999; Mack et al. 2000; Yampolsky et al. 2000; Hughes et al. 2002), only a few have attempted to identify the actual loci that influence these age-related changes in natural populations (Curtsinger and Khazaeli 2002; Jackson et al. 2003; Khazaeli et al. 2005; Nuzhdin et al. 2005).
One of the more important life-history traits is fecundity, that is, the number of offspring, propagules, or eggs produced. The fecundity of an individual sets the baseline number of offspring that can be contributed to the next generation and so represents the maximum possible fitness of an organism. Further, the age-specific allocation of energy to reproduction is a more important determinant of fitness than life-time fecundity in populations with age structure (Charlesworth 1994). The age-specific allocation of energy to reproduction can also affect fitness by producing trade-offs with other traits such as adult survival and future reproduction (Rose 1984; Gustafsson et al. 1994; Tatar and Promislow 1997; Jouventin and Dobson 2002; Oksanen et al. 2002; Fedorka et al. 2004). Thus, loci that control the age-specific allocation of energy to reproduction should have pleiotropic effects on other fitness components and so may act as genetic constraints on the independent evolution of other fitness components.
Fecundity exhibits an age-related decline in many species (Aigaki and Ohba 1984; Rose 1991; Partridge and Barton 1993; Rauser et al. 2003; Ricklefs et al. 2003; Broekmans et al. 2004; Novoseltsev et al. 2004; Ottinger et al. 2004; Skoracka and Kuczynski 2004; Nichols et al. 2005) and so understanding the genetic basis of reproductive senescence should also be useful for understanding the genetics of aging. A large number of studies have identified genes through mutational analysis or genetic manipulation that influences rates of senescence as reflected in age-specific mortality rates (Kenyon et al. 1993; Lin et al. 1998; Hsin and Kenyon 1999; Tatar et al. 2001; Sun et al. 2002; Tu et al. 2002; Marden et al. 2003; Picard et al. 2004; Rogina and Helfand 2004; Halaschek-Wiener et al. 2005; Kaeberlein et al. 2005). However, genes that influence age-specific mortality may be different from those causing senescence in other traits, such as reproduction. Further, the above studies used artificially induced mutations to identify candidate genes for aging that may or may not contribute to natural variation in senescence. Thus, to understand how the genetic architecture of life-history traits influences senescence and the maintenance of variation in senescence it is important to characterize the influence of natural allelic variation (Knight et al. 2001).
Quantitative trait locus (QTL) mapping procedures provide a useful avenue to search for loci that contribute to variation in fitness components. Many QTL studies have identified chromosomal regions that affect some aspect of fitness (Nuzhdin et al. 1997, 2005; Fry et al. 1998; Shook and Johnson 1999; Lin 2000; Wayne et al. 2001; Slate et al. 2002; Steinmetz et al. 2002; Wayne and McIntyre 2002; Ayyadevara et al. 2003; Ungerer and Rieseberg 2003; Weinig et al. 2003; Moehring and Mackay 2004; Peripato et al. 2004; Valenzuela et al. 2004; Verhoeven et al. 2004; Colosimo et al. 2005; Fiumera et al. 2005; Khazaeli et al. 2005; Zhong et al. 2005). In Drosophila melanogaster fine mapping of these QTL has allowed refinement of the position of candidate genes within QTL that affect a number of traits, including life span (Pasyukova et al. 2000, 2004), male mating behavior (Moehring and Mackay 2004), ovariole number (Wayne and McIntyre 2002), starvation resistance (Harbison et al. 2004), and olfactory behavior (Fanara et al. 2002). Verification of the effects of natural genetic variation in candidate genes within QTL regions has also been initiated using association mapping (De Luca et al. 2003). While the “gold standard” for final confirmation of the effect of sequence variation on phenotypic variation is to functionally characterize the effects of alleles on phenotypes (Mackay 2001; Rong et al. 2002; Sun et al. 2004), the mapping process has proven to be a promising technique for identifying genes on which to focus our functional genetic efforts (Long et al. 1995; Gurganus et al. 1999; Geiger-Thornsberry and Mackay 2004; Moehring and Mackay 2004; Nguyen et al. 2004; Palsson and Gibson 2004; Pasyukova et al. 2004).
In this study we use a population of recombinant inbred lines (RIL) of D. melanogaster to address three issues related to the genetic architecture of age-specific fecundity. First, we map the location of QTL that produce variation in fecundity at two ages (1 week and 4 weeks) and assess their relative influences on fecundity at each age. Second, because the effects of allelic variation at QTL can be influenced by the genetic background (Mackay et al. 2005) we tested for epistatic effects on fecundity in a genome-wide analysis. Finally, because age-specific fecundity is often (Rose 1984, 1991) but not always (Harshman and Hoffmann 2000) negatively correlated with life span, we compare the fecundity results from this study to our previous study on life span to identify putative pleiotropic loci affecting these two traits. We discuss these results in light of the two leading evolutionary theories of aging, antagonistic pleiotropy (AP) (Williams 1957) and mutation accumulation (MA) (Medawar 1952). To explore the potential influence of AP, we compare the locations and age-specific allelic effects of fecundity QTL with the locations and allelic effects of QTL affecting mated life span, which were previously mapped using the same set of RIL under similar environmental conditions (Leips and Mackay 2002). Under the AP model, QTL affecting fecundity and life span should colocalize and the alleles at these loci should exhibit antagonistic effects on these traits. To examine our data for evidence in support of MA, we use data on the genetic components of variation in fecundity among lines at each age. A unique prediction of this theory applied to our mapping population of RIL (a population of homozygous lines) is that the genetic component of variation in fecundity should increase with age (Hughes and Charlesworth 1994; Hughes and Reynolds 2004).
MATERIALS AND METHODS
Fly stocks:
Our mapping population consisted of a set of 92 RIL (Nuzhdin et al. 1997) derived from a cross between two isogenic strains, the Oregon-R (Ore) (Lindsley and Zimm 1992) and the Russian 2b strain, an isogenic line derived from a population of flies selected for decreased male sexual activity (Pasyukova and Nuzhdin 1993). The procedure used to construct the RIL has been previously described (Nuzhdin et al. 1997) and is summarized here. F1 offspring of the cross between the Ore and 2b strains were backcrossed to the 2b strain and then randomly mated for four generations. After the last generation of random mating, 200 male-female pairs were used to create sublines from this population by carrying out brother-sister matings for 25 generations. Ninety-eight RIL were subsequently created from these sublines. Of the original 98 RIL, 92 were extant at the time this experiment was carried out.
Early and late-age fecundity assay:
The total number of eggs laid by single females over a 2-day period was used as an estimate of fecundity at 1 and 4 weeks of age. Two replicate sets of flies of each genotype were set up simultaneously so that early and late-age fecundity could be measured on different females from each RIL. We measured early and late-age fecundity on different females to minimize the effects of handling at early age on late-age fecundity and to decrease the influence of nongenetic phenotypic correlations between early and late-age fecundity. One limitation of this design, however, is that because fecundity was measured on different females at early and late age we could not calculate the phenotypic correlation between early and late-age fecundity.
To produce females for each fecundity estimate, 15–20 pairs of flies from the Ore and 2b strains and each of the 92 RIL were allowed to lay eggs over a 4-day period in egg-collecting chambers (described below). From these chambers, 50 first or second instar larvae of each genotype were collected and placed in vials containing 5 ml of standard cornmeal/agar/molasses medium to standardize larval density. This procedure was repeated with five replicate vials per line. Larval density was controlled in this manner to reduce the influence of variation in larval density on female size at eclosion, a trait that is positively correlated with fecundity (Robertson 1957; Tantawy and Rahka 1964; Partridge et al. 1986; Nunney 1996; Zwaan et al. 1995). From each replicate vial, three virgin females that emerged on the same day were collected and placed in a vial containing standard fly food. To provide mates, six young males (<1 week old) of the Samarkand (Sam) strain were added to each vial (two males:one female per vial). Sam (an unrelated isogenic laboratory stock) males were used as mates to standardize the potential effects of male genotype on female fecundity.
Age-specific fecundity of individual females was measured at young (4–5 days) and old (28 days) age in a fashion similar to that of Houle et al. (1994) and Service (2000). These dates were chosen because reproductive output typically peaks in D. melanogaster at ∼6–12 days posteclosion and declines beyond that point (Houle et al. 1994; Tatar et al. 1996; Gasser et al. 2000). At each age, single females were removed from each vial and placed in an egg-laying chamber with a single Sam male. Egg-laying chambers consisted of standard 10-ml fly vials, containing 1 ml of hardened 2% agar to provide a source of moisture and 1 ml of fly food to provide a site for oviposition. Food was placed on the flat side of a hardened foam plug (which was also used to cap the vial). The vial was then inverted so that the plug of food was on the bottom of the vial. Females were allowed to lay eggs on food plugs for 24 hr, after which each plug was removed to count eggs. A fresh food-containing plug was replaced in the vials and the above process was repeated for a second 24-hr period. After this second day of egg laying, all females were discarded.
At each age, fecundity estimates were made on 15 females/line (five replicate vials containing 3 females/vial/line) for a total of 2760 observations (1380 young flies and 1380 old flies).
Statistical analyses:
We tested for differences in age-specific fecundity between the inbred parental strains (Ore and 2b) and the RIL in separate analyses. Differences in fecundity between the parental strains were tested using a mixed-model ANOVA according to the model y = μ + A + S + (A × S) + R(A × S) + error, where μ is the overall mean, A is the fixed effect of age (1 or 4 weeks), S is the fixed effect of parental strain (Ore or 2b), and R is the random effect of the replicate nested within age and strain. Flies in a particular replicate were those that had shared a vial until the age that we measured fecundity.
To assess genetically based differences in fecundity among the RIL, we used three separate analyses. The first two analyses used a random-effects ANOVA to test for genetic differences among lines at 1 and 4 weeks of age and provided estimates for the among-line variance components for fecundity at each age. The model partitioned the random effects of line (L) and replicate (R) within line and residual error according to the model y = μ + L + R(L) + error for flies in each age group. In the third model we used a mixed-model ANOVA on the entire data set to examine the potential for a line-by-age interaction that would in essence tell us if the effect of age on fecundity was similar among lines. For this analysis we used the model y = μ + A + L + (A × L) + R(A × L) + error, where A is the fixed effect of age on fecundity and all other effects are random.
All statistical analyses were carried out using SAS V.9.1. The PROC GLM and VARCOMP procedures were used for the analyses of variance on RIL within each age and for estimating variance components within each age. The PROC MIXED procedure was used for both of the mixed-model analyses and the significance of random effects was determined using likelihood-ratio tests (Littell et al. 2002). Phenotypic data were ln-transformed to meet the assumptions of ANOVA.
Variance components from the random-effects analyses above were used to calculate the proportion of the total phenotypic variation in fecundity explained by genetic differences among lines at each age (also separately estimated as the coefficient of genetic variation) and the genetic correlation of fecundity at young and old ages. The coefficient of genetic variation (CVG) was calculated at each age as CVG = 100(VL)1/2/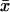 , where VL is the among-line variance component and
, where VL is the among-line variance component and 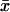 is the overall mean fecundity (Houle 1992). The genetic correlation across ages (rGA) was computed as cov12/(σL1σL2) (Robertson 1959), where cov12 is the covariance among-line means at 1 and 4 weeks of age and σL1 and σL2 are the square roots of the among-line variance components of fecundity at 1 and 4 weeks of age from the reduced model analyses.
is the overall mean fecundity (Houle 1992). The genetic correlation across ages (rGA) was computed as cov12/(σL1σL2) (Robertson 1959), where cov12 is the covariance among-line means at 1 and 4 weeks of age and σL1 and σL2 are the square roots of the among-line variance components of fecundity at 1 and 4 weeks of age from the reduced model analyses.
QTL mapping:
Molecular markers used to determine the genotype of the RIL were the cytological insertion sites of the roo transposable element (Nuzhdin et al. 1997). Eighty-one informative markers were used (Nuzhdin et al. 1997; Leips and Mackay 2000) with an average spacing between markers of 7.9 cM. Spacing between markers was estimated from the observed recombination (r) frequencies between pairs of markers using the Kosambi map function 100dM = 0.25 ln[(1 + 2r)/(1 − 2r)]. The distance between markers in this study is slightly greater than that of previous studies using these RIL (Nuzhdin et al. 1997; Wayne et al. 2001; Leips and Mackay 2002) because the loss of 6 of the original 98 lines reduced the number of observed recombination events in the mapping population.
QTL mapping was done using composite interval mapping (Zeng 1994) in QTL Cartographer (Version 1.14) and as outlined in Leips and Mackay (2002). This mapping procedure tests the hypothesis that an interval between adjacent markers contains a QTL affecting the quantitative trait, while controlling for the effects of linked QTL outside of the test interval. Markers on which the QTL analyses were conditioned were based on a forward-backward elimination stepwise regression analysis. Because the results of each analysis can be sensitive to the conditioning window used around each test interval, we tested a range of window sizes (5, 10, 15, and 20 cM) to evaluate the effect of window size on the likelihood ratios for each QTL. On the basis of the results from this set of analyses we used a window size of 10 cM because QTL identified with this window size were also determined to be significant in all analyses regardless of window size and so represent a conservative choice. The significance level for each QTL analysis was determined by randomly permuting the fecundity data 1000 times and calculating the maximum-likelihood ratio statistic across all test intervals for each permutation. LR statistics from the original data that were exceeded by the permutation maximum LR statistics <50 times were considered significant at ∝ = 0.05 (Churchill and Doerge 1994; Doerge and Churchill 1996).
We used ANOVA (PROC GLM in SAS V.9.1) to test for epistasis first by looking for significant pairwise interactions between QTL that had significant additive effects on fecundity on the basis of our mapping analysis. For each interaction, the genotype of each marker (homozygous for either the Ore or the 2b allele) closest to each significant QTL peak was used to evaluate the significance of marker interactions on fecundity. Because epistasis may also occur between QTL without main effects on the trait (Mackay et al. 2005), we performed a whole-genome screen for pairwise interactions between all possible pairs of markers using a two-way ANOVA where y = μ + Mi + Mj + (Mi × Mj) + error, where M is the genotype of each marker at positions i and j in each line. Using 81 markers means that we tested 3240 possible interactions. As such, we expect 162, 32.4, 3.24, and 0.324 significant interactions by chance alone at P < 0.05, < 0.01, < 0.001, and < 0.0001, respectively.
RESULTS
Genetic variation in age-specific fecundity (parental strains):
The average fecundity of the Ore strain (±1 standard error) at 1 and 4 weeks of age was 19.5 ± 3.0 and 10.06 ± 2.7 eggs, respectively. The average fecundity of the 2b strain at 1 and 4 weeks was 10.87 ± 2.6 and 8.53 ± 1.4 eggs, respectively. Despite these differences there was no significant difference in fecundity between the parental strains when averaged across both ages (F1,16 = 2.51, P = 0.13). There was also no significant effect of age (F1,16 = 3.36 P = 0.08) nor was there a significant age by parental strain interaction (F1,16 = 1.23, P = 0.28).
Genetic variation in age-specific fecundity (RIL):
Fecundity at 1 and 4 weeks averaged over all RIL was 14.8 and 15.2, respectively (Table 1) and the range of fecundity among the RIL at 1 and 4 weeks of age was similar (week 1: 5–23 eggs/female; week 4: 3–27 eggs/female). Thus, fecundity changed very little between weeks 1 and 4 when averaged over all lines.
TABLE 1.
Summary statistics for the fecundity data from the set of recombinant inbred lines
| Age | Fecundity | (VL/VL +VR)a | CVGb | P(GAI)c | rGAd (L1, L2)e |
|---|---|---|---|---|---|
| 1 wk | 14.8 ± 0.3 | 0.08 | 1.91 | **** | 0.05 (−0.11, 0.30) |
| 4 wk | 15.2 ± 0.1 | 0.13 | 2.46 |
Proportion of the total phenotypic variance explained by variation among RIL where VL is the variance component from ANOVA for testing differences among lines and VR is the variance component of the vial replicate plus the residual variance from ANOVA.
CVG = 100 (VL)1/2/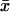 where
where 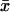 is the average life span among the RIL.
is the average life span among the RIL.
P(GAI) is from the test of significance for the line by age interaction term, ****P < 0.0001.
rGA is the genetic correlation of fecundity of the lines between each age.
Lower and upper confidence limits of rGA.
On the basis of the ANOVA at each age, however, we found significant differences in fecundity among the RIL at both 1 (F91,368 = 2.03, P < 0.0001) and 4 (F91,368 = 2.44, P < 0.0001) weeks of age. Notably, the genetic component of the total variation in fecundity at 4 weeks was almost twice what it was at 1 week (Table 1). This is not because there was less phenotypic variation in older aged individuals. In fact, the amount of residual variance in fecundity was 30% higher in the analysis of 4-week-old females compared to that of 1-week-old females. This increase in the environmental component of variance with age is consistent with other studies of fecundity (Rose and Charlesworth 1981) and longevity (Charlesworth and Hughes 1996) in Drosophila and suggests that older individuals may be more sensitive to environmental variation than younger flies (Charlesworth and Hughes 1996). Another possible explanation is that because older flies have experienced a greater range of environments the cumulative effect of this variation results in greater phenotypic variation among older aged individuals.
Although the RIL differed in fecundity at each age, the effect of age on fecundity differed dramatically among lines (Figure 1). The line-by-age interaction term was significant (χ2(1) = 23.4, P < 0.0001) and the genetic correlation of fecundity at 1 and 4 weeks of age was not significantly different from zero (Table 1). Together these results suggest that genes contributing to the variation in early age fecundity are distinct from those producing variation in fecundity at 4 weeks and/or that the allelic effects of loci that contribute to genetic variation in fecundity vary with age.
Figure 1.
The average fecundity of each line at weeks 1 and 4. Fecundity was measured as the number of eggs laid by a single female over a 2-day period. Fecundity counts within lines were measured for different females at each age.
Age-specific QTL for fecundity:
We identified two or three QTL affecting fecundity at 1 week of age, one on the second chromosome and another one or two on the third chromosome (Figure 2). Given the proximity of the two QTL on the third chromosome and the confidence intervals around each, it is not clear if these are distinct QTL. The most likely position of the QTL on the second chromosome is at cytological position 34E with the 2-LOD support interval (Lynch and Walsh 1998) extending from cytological positions 30D to 38A. Variation at this QTL explained 10% of the variation in fecundity among lines at 1 week of age. The additive effect of the Ore allele at this locus increased the fecundity of females by 0.78 eggs/day compared to the effects of the 2b allele. One of the QTL on the third chromosome has the highest likelihood of being at position at 85F (2-LOD support interval 73D–87E). Variation at this locus explained 14% of the genetic variation in fecundity, and the Ore allele at this locus also increased the fecundity by 0.74 eggs/day compared to the effect of the 2b allele. The third QTL appears at position 87B (2-LOD support interval also ranges from 73D to 87E) and explains an additional 10% of the genetic variation. At this locus, the Ore allele increases fecundity by 0.67 eggs/day relative to the 2b allele. Interestingly, the direction of effects of the 2b alleles is consistent at both sites and may reflect the fact that this line is derived from a population selected for decreased male mating activity. However, the 2b and Ore parental strains do not differ genetically for fitness (Wayne et al. 2001) so it is unlikely that the effects seen here are due to a general effect of 2b alleles in reducing overall fitness.
Figure 2.
Composite interval mapping results indicating the positions of QTL affecting fecundity at each age. The threshold value for significance at each age is given by the horizontal lines. Positions of informative markers are denoted by triangles on the x-axes. Lines above peaks indicate 2-LOD support intervals for the location of the QTL.
Unexpectedly, we found no QTL that influenced fecundity at 4 weeks of age, despite the fact that the ANOVA indicated a higher degree of genetically based variation in late-age fecundity than in early age fecundity. This suggests that genetic variation in fecundity among lines at the older age is due to alleles with small late-age specific effects. Our observation that there was no genetic correlation of fecundity between young and old ages is thus explained by the diminution of the effects of QTL affecting early age fecundity.
Effects of epistasis among marker loci on fecundity:
None of the pairwise tests for epistasis among the QTL with main effects on fecundity were significant. In the global test for pairwise interactions among loci at early age, a total of 97 markers exhibited significant epistatic effects on fecundity (P < 0.05). Of these, 83 interactions had P-values between 0.01 and ≤0.05, 13 had P-values between 0.001 and <0.01, and only one interaction had a P-value of <0.001. In a comparable test using late-age fecundity a total of 98 markers exhibited significant epistatic effects on fecundity (P < 0.05). Of these, 92 interactions had P-values between 0.01 and ≤0.05, and 6 had P-values between 0.001 and <0.01. Given 3240 possible pairwise interactions, the number of significant interactions that we found were well within the numbers of interactions expected by chance alone (expected: 162 P ≤ 0.05, 32.4 P < 0.01, and 3.2 P < 0.001).
DISCUSSION
Age-specific effects of QTL:
Our results show that the relative influence of genes regulating the age schedule of reproduction changes with age. This interpretation is supported by the high degree of variation among lines in the effect of age on fecundity and the lack of a genetic correlation between early and late-age fecundity in the RIL. Combining the QTL results with those from the analyses of genetic components of variation leads to the interpretation that variation in fecundity at the younger age was determined at least in part by a few genes of moderate effect but fecundity at old age was determined by many more loci of smaller effect. While it is unclear how many actual genes within the QTL regions contribute to the variation in fecundity at the early age, it is clear that their relative effect on fecundity diminishes greatly with age.
Genes with age-specific effects on other fitness components have been implicated in many other studies of D. melanogaster (Kosuda 1985; Engstrom et al. 1989; Hughes and Charlesworth 1994; Hughes 1995; Charlesworth and Hughes 1996; Promislow et al. 1996; Tatar et al. 1996; Pletcher et al. 1998, 1999; Mack et al. 2000; Yampolsky et al. 2000; Curtsinger and Khazaeli 2002; Hughes et al. 2002; Snoke and Promislow 2003) and a few studies have begun to map the QTL underlying these age-specific effects on mortality rates (Curtsinger and Khazaeli 2002; Nuzhdin et al. 2005) and metabolic rates (Khazaeli et al. 2005). Our findings of the age-specific effects of QTL are similar to those seen by Khazaeli et al. (2005). In their study, metabolic rates at days 16 and 29 posteclosion and life span appeared to be affected by the same QTL, but these QTL did not affect metabolic rates at the youngest and oldest ages examined in their study. The results from Nuzhdin et al. (2005) are also consistent with ours in that no QTL identified in their study had a significant influence on mortality rate at all ages studied. Even more intriguing, their results suggested that at two of the QTL, the allelic effects on mortality rate were negatively correlated across different ages, with the same allele having either a positive or a negative effect on mortality, depending on age.
Given the transient nature of the effects of QTL on senescent phenotypes, an important goal for future studies will be to identify the factors that give rise to age-specific genetic effects. One possibility is that age-specific mutational effects result from genotype-by-environment interactions. Under this scenario, genetic influences on the phenotype depend on the internal physiological conditions; age-related changes in the physiology of the organism modulate the effects of these loci such that they have a notable phenotypic effect only within a certain range of conditions. In addition, age-specific changes in gene expression may give rise to age-specific effects. Numerous studies have demonstrated age-dependent changes in gene expression (Rogina et al. 1998; Jin et al. 2001; Weindruch et al. 2001; Pletcher et al. 2002; Seroude et al. 2002; McCarroll et al. 2004; Kim et al. 2005) and it may be that the age at which the effects of allelic variation are notable coincides with the ages of peak expression of these loci. Along these same lines, a growing body of evidence implicates age-related changes in chromatin structure, which directly regulates gene expression, as a mechanism for regulating aging (Chang and Min 2002; Rogina et al. 2002; Tissenbaum and Guarente 2002; Issa 2003; Jaenisch and Bird 2003). It is possible that genetic variation in the enzymes controlling age-related changes in chromatin remodeling produces the age-specific effects of particular loci on senescence.
Gene action:
We found little compelling evidence for the influence of epistasis on fecundity among our RIL at either age. Admittedly, the method used to correct for the number of expected false positives is conservative and so does not preclude the potential importance of epistasis on fecundity in our mapping population. A more sophisticated statistical approach that has greater power to detect epistasis might have been more useful in this regard (Kao et al. 1999) but such methods require much larger sample sizes than we had in this study. If our interpretation about epistasis is correct—that there are few if any significant epistatic interactions among loci that affect fecundity—then this strengthens our interpretation of the QTL results. This is because the composite interval mapping method used produces biased estimates of the position and marginal effects of QTL, given any amount of epistasis and linkage between epistatic QTL (Kao and Zeng 2002).
The lack of epistatic interactions affecting fecundity is in stark contrast with previous work on these lines, which found extensive epistasis among QTL affecting virgin life span (Leips and Mackay 2000; Mackay et al. 2005). Interestingly, when these lines were measured for mated longevity, only a single pair of markers appeared to interact epistatically. Thus, differences in the mating status appeared to influence the degree to which epistasis affected longevity. More work is necessary to evaluate the extent to which epistasis is dependent on the environmental/physiological condition and whether different traits are more or less affected by epistatic interactions.
Implications for evolutionary theories of aging:
Many studies on the genetic basis of senescence have used age-specific variance components to test the predictions of the MA and AP theories of aging. These studies typically use breeding designs that allow estimation of the additive and dominance genetic components of variation in a trait with age that can then be evaluated in light of the predictions of each theory. While increases in the additive component of variation (VA) with age can be expected from either model of aging (Charlesworth and Hughes 1996), a unique prediction of the MA theory is that the genetic component of variation among homozygous lines will increase with age. Using this metric, the results of our study lend support for MA producing variation among lines in reproductive senescence. Within the age span covered in this experiment (ages 1 to 4 weeks) our results are in agreement with most studies of age-specific genetic effects on a number of traits, including fecundity (Engstrom et al. 1989; Tatar et al. 1996), age-specific mortality (Hughes and Charlesworth 1994; Charlesworth and Hughes 1996; Promislow et al. 1996; Tatar et al. 1996; Hughes et al. 2002; Snoke and Promislow 2003), and aspects of male mating success (Kosuda 1985; Hughes 1995; Charlesworth and Hughes 1996; Hughes et al. 2002; Snoke and Promislow 2003). The lack of a significant genetic correlation between early and late-age fecundity adds additional support for the MA theory because this model assumes that the effects of alleles on fitness early in life are uncorrelated with allelic effects on fitness later in life (Partridge and Barton 1993). Under the AP model we would expect to see a negative genetic correlation between early and late-age fecundity. On the basis of these summary statistics we can conclude that MA contributes to the variation in age-specific fecundity in our mapping population.
To examine evidence supporting the AP theory, we compared the results of this study with our earlier study mapping QTL affecting the life span of mated males and females in the same population of RIL (Leips and Mackay 2002). The earlier experiment differed from the current one in that the life-span measurements were made on the offspring of the cross between each RIL and the inbred parental strains, Ore and 2b. Thus, the allelic effects of QTL on life span were estimated in different genetic backgrounds. However, there was only one instance in which life-span QTL were shown to interact epistatically in that earlier study and neither of the two QTL involved were those identified as fecundity QTL. Therefore, comparison of the additive allelic effects of fecundity QTL on life span using these two studies is appropriate in looking for evidence supporting the AP theory. While the antagonistic relationships assumed by AP could presumably exist between any number of traits that influence fitness at early age and that are negatively correlated with longevity (Leroi et al. 2005), one of the most commonly observed trade-offs that support the AP theory is the trade-off between early age fecundity and life span (Rose and Charlesworth 1981; Rose 1991; Marden et al. 2003; Leroi et al. 2005). Application of this theory to our QTL mapping studies would predict a negative correlation between early age fecundity and life span among the RIL. At the QTL level we would expect that QTL affecting life span and early age fecundity should colocalize and that the allelic effects at these loci should have opposite effects on these two traits. To investigate these possibilities, we first calculated the correlation between the average mated life span of males and females from our earlier study with early and late-age fecundity. As predicted by AP, we did find a significant negative correlation (r = −0.43, P < 0.0001) but interestingly this correlation was between early female fecundity and the life span of males from the RIL × Ore cross. At the QTL level, both of the early age fecundity QTL colocalize with QTL affecting life span and alleles at these loci do indeed have antagonistic effects. Oddly enough and in accordance with the correlation analysis described above, it is the life span of males and not females that exhibits antagonistic allelic effects with female fecundity at early age. At the QTL on the second chromosome the Ore allele increases fecundity by 0.78 eggs/day compared to the 2b allele but decreases male life span by 4 days. The allelic effects at the QTL on the third chromosome are similar to those at the QTL on the second; the Ore allele increases early age fecundity by 0.74 eggs/day but decreases male life span by 5 days. Thus, it appears that the allelic effects of these QTL may exhibit sexual antagonism with alleles having an advantageous effect on one sex but a deleterious effect on the other. Sexual antagonism in QTL studies is not unusual (Nuzhdin et al. 1997; Leips and Mackay 2000; Vieira et al. 2000; Wayne et al. 2001) although neither is it universal (Curtsinger and Khazaeli 2002). Another possible explanation is that male longevity is genetically correlated with male fecundity (which was not measured) and so would indicate a positive correlation between male and female fitness. It should be noted here that the allelic effects at these QTL potentially represent the combined effects of many genes within the QTL region. Therefore, the actual genes that contribute to the variation in fecundity may be in the same region as, but distinct from, those affecting male life span. Only by identifying the actual loci underlying the variation in these traits can this issue be resolved. Also, genotype-by-environment interactions can influence the sign and magnitude of the allelic effects of QTL (Leips and Mackay 2000; Vieira et al. 2000) and it may well be that under different environmental conditions a trade-off between fecundity and female life span would be evident. Marden et al. (2003) found just such a situation in studying the combined influence of the Indy mutation on life span and fecundity. A trade-off between these traits was evident only when flies were reared on a calorically restricted diet. Confirmation of the degree to which particular alleles contribute to trade-offs among traits will require that we not only identify the loci that affect these traits, but also observe the allelic effects on all traits affected in a range of ecologically relevant environments.
From QTL to gene:
Identification of QTL with age-specific effects on life-history traits represents the first of many steps toward understanding the complexities of the genetic basis of variation in these traits. Identification of the actual genes that contribute to the variation in fecundity identified in this study will require fine-scaled mapping of the QTL regions using crosses to deficiency strains and complementation tests to candidate genes within refined QTL regions (e.g., Pasyukova et al. 2000). Once candidates are identified, testing for the effects of naturally segregating variation on fecundity can be accomplished by association mapping studies (e.g., De Luca et al. 2003). The QTL regions identified in this study contain many candidate genes that are involved in some aspect of reproduction/oogenesis and might contribute to the variation in this study. These genes include daughterless, rho-6, zucchini, kekkon-1, vasa, cactus, and kelch on the second chromosome and maelstrom, jim, rpk, bicoid, poached, and squid on the third chromosome. As there are hundreds of genes within each of these QTL regions, most of unknown function, resolving the actual loci contributing to variation in fecundity will require additional mapping efforts.
One limitation of this study, which is indeed a limitation of QTL studies in general, is that only a limited sample of genetic diversity is represented in our lines. Also, the mapping population used was derived from two inbred laboratory strains and not isolates from a natural population. As such, the generality of our results needs to be tested by repeating this study with independent lines ideally constructed from a natural population. The fact that the parental strains did not differ in fecundity but we were still able to map QTL in the RIL derived from them is not unusual (e.g., Leips and Mackay 2000; Vieira et al. 2000). These results suggest that QTL with positive and negative effects on fecundity that were fixed for each parental strain were revealed when they appeared in different combinations in the RIL.
Assuming that we can make use of the rapidly developing technological tools to identify and characterize the genetic architecture of life-history traits at the molecular genetic level, many questions will remain after we have the loci in hand. For example, given genetic variation in the age-specific expression of a trait, does variation result from the action of a different subset of genes acting on the trait at different ages or does variation arise from differences in the influence of particular alleles at the same genes with age? Do physiological changes with age modulate allelic effects on traits (in a fashion similar to genotype-by-environment interactions)? And if so, what are the relevant physiological changes that alter these allelic effects? Such questions represent a few of the many challenges that remain in understanding the genetic basis of life-history variation.
Acknowledgments
We thank Stephanie Rollmann for thoughtful discussions on many aspects of the project and Kimberly Hughes, Tashauna Felix, and two anonymous reviewers for their constructive comments on the manuscript. This work was supported by National Science Foundation grant DEB-0349856 and National Research Service Award National Institutes of Health (NIH) grant GM18818-03 to J.L. and NIH grant GM45344 to T.F.C.M.
References
- Aigaki, T., and S. Ohba, 1984. Individual analysis of age-associated changes in reproductive activity and lifespan of Drosophila virilis. Exp. Gerontol. 19: 13–23. [DOI] [PubMed] [Google Scholar]
- Ayyadevara, S., R. Ayyadevara, A. Vertino, A. Galecki, J. J. Thaden et al., 2003. Genetic loci modulating fitness and life span in Caenorhabditis elegans: categorical trait interval mapping in CL2a × Bergerac-BO recombinant-inbred worms. Genetics 163: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., and P. D. Keightley, 2002. Understanding quantitative genetic variation. Nat. Rev. Genet. 3: 11–21. [DOI] [PubMed] [Google Scholar]
- Barton, N. H., and M. Turelli, 1989. Evolutionary quantitative genetics: How little do we know? Annu. Rev. Genet. 23: 337–370. [DOI] [PubMed] [Google Scholar]
- Broekmans, F. J., M. J. Faddy, G. Scheffer and E. R. te Velde, 2004. Antral follicle counts are related to age at natural fertility loss and age at menopause. Menopause 11: 607–614. [DOI] [PubMed] [Google Scholar]
- Chang, K. T., and K. T. Min, 2002. Regulation of lifespan by histone deacetylase. Ageing Res. Rev. 1: 313–326. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., 1994. Evolution in Age-Structured Populations. Cambridge University Press, Cambridge, UK.
- Charlesworth, B., and K. A. Hughes, 1996. Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc. Natl. Acad. Sci. USA 93: 6140–6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo, P. F., K. E. Hosemann, S. Balabhadra, G. Villarreal, Jr., M. Dickson et al., 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307: 1928–1933. [DOI] [PubMed] [Google Scholar]
- Curtsinger, J. W., and A. A. Khazaeli, 2002. Lifespan, QTLs, age-specificity, and pleiotropy in Drosophila. Mech. Ageing Dev. 123: 81–93. [DOI] [PubMed] [Google Scholar]
- De Luca, M., N. V. Roshina, G. L. Geiger-Thornsberry, R. F. Lyman, E. G. Pasyukova et al., 2003. Dopa decarboxylase (Ddc) affects variation in Drosophila longevity. Nat. Genet. 34: 429–433. [DOI] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drnevich, J. M., M. M. Reedy, E. A. Ruedi, S. Rodriguez-Zas and K. A. Hughes, 2004. Quantitative evolutionary genomics: differential gene expression and male reproductive success in Drosophila melanogaster. Proc. Biol. Sci. 271: 2267–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom, G., L.-E. Liljedahl, M. Rasmuson and T. Bjorklund, 1989. Expression of genetic and environmental variation during ageing. Theor. Appl. Genet. 77: 119–122. [DOI] [PubMed] [Google Scholar]
- Fanara, J. J., K. O. Robinson, S. M. Rollmann, R. R. Anholt and T. F. Mackay, 2002. Vanaso is a candidate quantitative trait gene for Drosophila olfactory behavior. Genetics 162: 1321–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorka, K. M., M. Zuk and T. A. Mousseau, 2004. Immune suppression and the cost of reproduction in the ground cricket, Allonemobius socius. Evolution 58: 2478–2485. [DOI] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169: 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, C. W., M. L. Bush, D. A. Roff and W. G. Wallin, 2004. Evolutionary genetics of lifespan and mortality rates in two populations of the seed beetle, Callosobruchus maculatus. Heredity 92: 170–181. [DOI] [PubMed] [Google Scholar]
- Fry, J. D., S. V. Nuzhdin, E. G. Pasyukova and T. F. Mackay, 1998. QTL mapping of genotype-environment interaction for fitness in Drosophila melanogaster. Genet. Res. 71: 133–141. [DOI] [PubMed] [Google Scholar]
- Gasser, M., M. Kaiser, D. Berrigan and S. C. Stearns, 2000. Life-history correlates of evolution under high and low adult mortality. Evolution 54: 1260–1272. [DOI] [PubMed] [Google Scholar]
- Geiger-Thornsberry, G. L., and T. F. Mackay, 2004. Quantitative trait loci affecting natural variation in Drosophila longevity. Mech. Ageing Dev. 125: 179–189. [DOI] [PubMed] [Google Scholar]
- Gurganus, M. C., S. V. Nuzhdin, J. W. Leips and T. F. Mackay, 1999. High-resolution mapping of quantitative trait loci for sternopleural bristle number in Drosophila melanogaster. Genetics 152: 1585–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson, L., D. Nordling, M. S. Andersson, B. C. Sheldon and A. Qvarnstrom, 1994. Infectious diseases, reproductive effort and the cost of reproduction in birds. Philos. Trans. R. Soc. Lond. B Biol. Sci. 346: 323–331. [DOI] [PubMed] [Google Scholar]
- Halaschek-Wiener, J., J. S. Khattra, S. McKay, A. Pouzyrev, J. M. Stott et al., 2005. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 15: 603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison, S. T., A. H. Yamamoto, J. J. Fanara, K. K. Norga and T. F. Mackay, 2004. Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166: 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hard, J. J., W. E. Bradshaw and C. M. Holzapfel, 1993. The genetic basis of photoperiodism and its evolutionary divergence among populations of the pitcher-plant mosquito, Wyeomyia smithii. Am. Nat. 142: 457–473. [DOI] [PubMed] [Google Scholar]
- Harshman, L. G., and A. A. Hoffmann, 2000. Laboratory selection experiments using Drosophila: What do they really tell us? Trends Ecol. Evol. 15: 32–36. [DOI] [PubMed] [Google Scholar]
- Houle, D., 1992. Comparing evolvability and variability of quantitative traits. Genetics 130: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle, D., K. A. Hughes, D. K. Hoffmaster, J. Ihara, S. Assimacopoulos et al., 1994. The effects of spontaneous mutation on quantitative traits. I. Variances and covariances of life history traits. Genetics 138: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin, H., and C. Kenyon, 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., 1995. The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution 49: 521–537. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., and B. Charlesworth, 1994. A genetic analysis of senescence in Drosophila. Nature 367: 64–66. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., and R. M. Reynolds, 2004. Evolutionary and mechanistic theories of aging. Annu. Rev. Entomol. 50: 421–445. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., J. A. Alipaz, J. M. Drnevich and R. M. Reynolds, 2002. A test of evolutionary theories of aging. Proc. Natl. Acad. Sci. USA 99: 14286–14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa, J. P., 2003. Age-related epigenetic changes and the immune system. Clin. Immunol. 109: 103–108. [DOI] [PubMed] [Google Scholar]
- Jackson, A. U., A. T. Galecki, D. T. Burke and R. A. Miller, 2003. Genetic polymorphisms in mouse genes regulating age-sensitive and age-stable T cell subsets. Genes Immun. 4: 30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch, R., and A. Bird, 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl): 245–254. [DOI] [PubMed] [Google Scholar]
- Jin, W., R. M. Riley, R. D. Wolfinger, K. P. White, G. Passador-Gurgel et al., 2001. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat. Genet. 29: 389–395. [DOI] [PubMed] [Google Scholar]
- Jouventin, P., and F. S. Dobson, 2002. Why breed every other year? The case of albatrosses. Proc. R. Soc. Lond. B Biol. Sci. 269: 1955–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein, M., K. T. Kirkland, S. Fields and B. K. Kennedy, 2005. Genes determining yeast replicative life span in a long-lived genetic background. Mech. Ageing Dev. 126: 491–504. [DOI] [PubMed] [Google Scholar]
- Kao, C. H., and Z. B. Zeng, 2002. Modeling epistasis of quantitative trait loci using Cockerham's model. Genetics 160: 1243–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, C. H., Z. B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C., J. Chang, E. Gensch, A. Rudner and R. Tabtiang, 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464. [DOI] [PubMed] [Google Scholar]
- Khazaeli, A. A., W. Van Voorhies and J. W. Curtsinger, 2005. Longevity and metabolism in Drosophila melanogaster: genetic correlations between life span and age-specific metabolic rate in populations artificially selected for long life. Genetics 169: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. N., J. H. Rhee, Y. H. Song, D. Y. Park, M. Hwang et al., 2005. Age-dependent changes of gene expression in the Drosophila head. Neurobiol. Aging 26: 1083–1091. [DOI] [PubMed] [Google Scholar]
- Knight, C. G., R. B. Azevedo and A. M. Leroi, 2001. Testing life-history pleiotropy in Caenorhabditis elegans. Evolution 55: 1795–1804. [DOI] [PubMed] [Google Scholar]
- Kosuda, K., 1985. The aging effect on male mating activity in Drosophila melanogaster. Behav. Genet. 15: 297–303. [DOI] [PubMed] [Google Scholar]
- Kruuk, L. E., T. H. Clutton-Brock, J. Slate, J. M. Pemberton, S. Brotherstone et al., 2000. Heritability of fitness in a wild mammal population. Proc. Natl. Acad. Sci. USA 97: 698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips, J., and T. F. Mackay, 2000. Quantitative trait loci for life span in Drosophila melanogaster: interactions with genetic background and larval density. Genetics 155: 1773–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips, J., and T. F. Mackay, 2002. The complex genetic architecture of Drosophila life span. Exp. Aging Res. 28: 361–390. [DOI] [PubMed] [Google Scholar]
- Leips, J., J. Travis and F. H. Rodd, 2000. Genetic influences on experimental population dynamics of the least killifish. Ecol. Monogr. 70: 289–309. [Google Scholar]
- Leroi, A. M., A. Bartke, G. De Benedictis, C. Franceschi, A. Gartner et al., 2005. What evidence is there for the existence of individual genes with antagonistic pleiotropic effects? Mech. Ageing Dev. 126: 421–429. [DOI] [PubMed] [Google Scholar]
- Lin, J. Z., 2000. The relationship between loci for mating system and fitness-related traits in Mimulus (Scrophulariaceae): a test for deleterious pleiotropy of QTLs with large effects. Genome 43: 628–633. [DOI] [PubMed] [Google Scholar]
- Lin, Y. J., L. Seroude and S. Benzer, 1998. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science 282: 943–946. [DOI] [PubMed] [Google Scholar]
- Lindsley, D. L., and G. G. Zimm, 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego.
- Littell, R. C., W. W. Stroup and R. J. Freund, 2002. SAS for Linear Models. SAS Institute, Cary, NC.
- Long, A. D., S. L. Mullaney, L. A. Reid, J. D. Fry, C. H. Langley et al., 1995. High resolution mapping of genetic factors affecting abdominal bristle number in Drosophila melanogaster. Genetics 139: 1273–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mack, P. D., V. K. Lester and D. E. Promislow, 2000. Age-specific effects of novel mutations in Drosophila melanogaster. II. Fecundity and male mating ability. Genetica 110: 31–41. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., N. V. Roshina, J. W. Leips and E. G. Pasyukova, 2005. Complex genetic architecture of Drosophila longevity, pp. 181–216 in Handbook of the Biology of Aging, edited by E. J. Masoro and S. N. Austad. Elsevier, Oxford.
- Marden, J. H., B. Rogina, K. L. Montooth and S. L. Helfand, 2003. Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc. Natl. Acad. Sci. USA 100: 3369–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll, S. A., C. T. Murphy, S. Zou, S. D. Pletcher, C. S. Chin et al., 2004. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 36: 197–204. [DOI] [PubMed] [Google Scholar]
- Medawar, P. B., 1952. An Unsolved Problem of Biology. H. K. Lewis, London.
- Moehring, A. J., and T. F. Mackay, 2004. The quantitative genetic basis of male mating behavior in Drosophila melanogaster. Genetics 167: 1249–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousseau, T. A., and D. A. Roff, 1987. Natural selection and the heritability of fitness components. Heredity 59 (Pt. 2): 181–197. [DOI] [PubMed] [Google Scholar]
- Nguyen, T. T., N. Klueva, V. Chamareck, A. Aarti, G. Magpantay et al., 2004. Saturation mapping of QTL regions and identification of putative candidate genes for drought tolerance in rice. Mol. Genet. Genomics 272: 35–46. [DOI] [PubMed] [Google Scholar]
- Nichols, S. M., B. D. Bavister, C. A. Brenner, P. J. Didier, R. M. Harrison et al., 2005. Ovarian senescence in the rhesus monkey (Macaca mulatta). Hum. Reprod. 20: 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoseltsev, V. N., R. Arking, J. R. Carey, J. A. Novoseltseva and A. I. Yashin, 2004. How an individual fecundity pattern looks in Drosophila and medflies. Ann. NY Acad. Sci. 1019: 577–580. [DOI] [PubMed] [Google Scholar]
- Nunney, L., 1996. The response to selection for fast larval development in Drosophila melanogaster and its effect on adult weight: an example of a fitness trade-off. Evolution 50: 1193–1204. [DOI] [PubMed] [Google Scholar]
- Nuzhdin, S. V., E. G. Pasyukova, C. L. Dilda, Z-B. Zeng and T. F. Mackay, 1997. Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 94: 9734–9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin, S. V., A. Khazaeli and J. Curtsinger, 2005. Survival analysis of life span quantitative trait loci in Drosophila melanogaster. Genetics 170: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, T. A., E. Koskela and T. Mappes, 2002. Hormonal manipulation of offspring number: maternal effort and reproductive costs. Evolution Int. J. Org. Evolution 56: 1530–1537. [DOI] [PubMed] [Google Scholar]
- Ottinger, M. A., M. Abdelnabi, Q. Li, K. Chen, N. Thompson et al., 2004. The Japanese quail: a model for studying reproductive aging of hypothalamic systems. Exp. Gerontol. 39: 1679–1693. [DOI] [PubMed] [Google Scholar]
- Palsson, A., and G. Gibson, 2004. Association between nucleotide variation in Egfr and wing shape in Drosophila melanogaster. Genetics 167: 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge, L., and N. H. Barton, 1993. Optimality, mutation and the evolution of ageing. Nature 362: 305–311. [DOI] [PubMed] [Google Scholar]
- Partridge, L., K. Fowler, S. Trevitt and W. Sharp, 1986. An examination of the effects of males on the survival and egg production rates of female Drosophila melanogaster. J. Insect Physiol. 32: 925–929. [Google Scholar]
- Pasyukova, E. G., and S. V. Nuzhdin, 1993. Doc and copia instability in an isogenic Drosophila melanogaster stock. Mol. Gen. Genet. 240: 302–306. [DOI] [PubMed] [Google Scholar]
- Pasyukova, E. G., C. Vieira and T. F. Mackay, 2000. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics 156: 1129–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasyukova, E. G., N. V. Roshina and T. F. Mackay, 2004. Shuttle craft: a candidate quantitative trait gene for Drosophila lifespan. Aging Cell 3: 297–307. [DOI] [PubMed] [Google Scholar]
- Peripato, A. C., R. A. De Brito, S. R. Matioli, L. S. Pletscher, T. T. Vaughn et al., 2004. Epistasis affecting litter size in mice. J. Evol. Biol. 17: 593–602. [DOI] [PubMed] [Google Scholar]
- Picard, F., M. Kurtev, N. Chung, A. Topark-Ngarm, T. Senawong et al., 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429: 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., D. Houle and J. W. Curtsinger, 1998. Age-specific properties of spontaneous mutations affecting mortality in Drosophila melanogaster. Genetics 148: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., D. Houle and J. W. Curtsinger, 1999. The evolution of age-specific mortality rates in Drosophila melanogaster: genetic divergence among unselected lines. Genetics 153: 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletcher, S. D., S. J. Macdonald, R. Marguerie, U. Certa, S. C. Stearns et al., 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr. Biol. 12: 712–723. [DOI] [PubMed] [Google Scholar]
- Promislow, D. E., M. Tatar, A. A. Khazaeli and J. W. Curtsinger, 1996. Age-specific patterns of genetic variance in Drosophila melanogaster. I. Mortality. Genetics 143: 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauser, C. L., L. D. Mueller and M. R. Rose, 2003. Aging, fertility, and immortality. Exp. Gerontol. 38: 27–33. [DOI] [PubMed] [Google Scholar]
- Ricklefs, R. E., A. Scheuerlein and A. Cohen, 2003. Age-related patterns of fertility in captive populations of birds and mammals. Exp. Gerontol. 38: 741–745. [DOI] [PubMed] [Google Scholar]
- Robertson, A., 1959. The sampling variance of the genetic correlation coefficient. Biometrics 15: 469–485. [Google Scholar]
- Robertson, F. W., 1957. Studies in quantitative inheritance. XI. Genetic and environmental correlation between body size and egg production in Drosophila melanogaster. J. Genet. 55: 428–443. [DOI] [PubMed] [Google Scholar]
- Rogina, B., and S. L. Helfand, 2004. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. USA 101: 15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina, B., J. W. Vaupel, L. Partridge and S. L. Helfand, 1998. Regulation of gene expression is preserved in aging Drosophila melanogaster. Curr. Biol. 8: 475–478. [DOI] [PubMed] [Google Scholar]
- Rogina, B., S. L. Helfand and S. Frankel, 2002. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science 298: 1745. [DOI] [PubMed]
- Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. R., 1984. Laboratory evolution of postponed senescence in Drosophila melanogaster. Evolution 38: 1004–1010. [DOI] [PubMed] [Google Scholar]
- Rose, M. R., 1991. Evolutionary Biology of Aging. Oxford University Press, New York.
- Rose, M. R., and B. Charlesworth, 1981. Genetics of life history in Drosophila melanogaster. II. Exploratory selection experiments. Genetics 97: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seroude, L., T. Brummel, P. Kapahi and S. Benzer, 2002. Spatio-temporal analysis of gene expression during aging in Drosophila melanogaster. Aging Cell 1: 47–56. [DOI] [PubMed] [Google Scholar]
- Service, P. M., 2000. The genetic structure of female life history in D. melanogaster: comparisons among populations. Genet. Res. 75: 153–166. [DOI] [PubMed] [Google Scholar]
- Shaw, R. G., G. A. Platenkamp, F. H. Shaw and R. H. Podolsky, 1995. Quantitative genetics of response to competitors in Nemophila menziesii: a field experiment. Genetics 139: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook, D. R., and T. E. Johnson, 1999. Quantitative trait loci affecting survival and fertility-related traits in Caenorhabditis elegans show genotype-environment interactions, pleiotropy and epistasis. Genetics 153: 1233–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoracka, A., and L. Kuczynski, 2004. Demography of the cereal rust mite Abacarus hystrix (Acari: Eriophyoidea) on quack grass. Exp. Appl. Acarol. 32: 231–242. [DOI] [PubMed] [Google Scholar]
- Slate, J., P. M. Visscher, S. MacGregor, D. Stevens, M. L. Tate et al., 2002. A genome scan for quantitative trait loci in a wild population of red deer (Cervus elaphus). Genetics 162: 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoke, M. S., and D. E. L. Promislow, 2003. Quantitative genetic tests of recent senescence theory: age-specific mortality and male fertility in Drosophila melanogaster. Heredity 91: 546–556. [DOI] [PubMed] [Google Scholar]
- Sommer, S., and P. B. Pearman, 2003. Quantitative genetic analysis of larval life history traits in two alpine populations of Rana temporaria. Genetica 118: 1–10. [DOI] [PubMed] [Google Scholar]
- Stearns, S. C., 1992. The Evolution of Life Histories. Oxford University Press, Oxford.
- Steinmetz, L. M., H. Sinha, D. R. Richards, J. I. Spiegelman, P. J. Oefner et al., 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Sun, J., D. Folk, T. J. Bradley and J. Tower, 2002. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics 161: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S., C. T. Ting and C.-I Wu, 2004. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science 305: 81–83. [DOI] [PubMed] [Google Scholar]
- Tantawy, A. O., and F. A. Rahka, 1964. Studies on natural populations of Drosophila. IV. Genetic variances and correlations between four characters in D. melanogaster and D. simulans. Genetics 50: 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar, M., and D. E. Promislow, 1997. Fitness costs of female reproduction. Evolution 51: 1323–1326. [DOI] [PubMed] [Google Scholar]
- Tatar, M., D. E. Promislow, A. A. Khazaeli and J. W. Curtsinger, 1996. Age-specific patterns of genetic variance in Drosophila melanogaster. II. Fecundity and its genetic covariance with age-specific mortality. Genetics 143: 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar, M., A. Kopelman, D. Epstein, M. P. Tu, C. M. Yin et al., 2001. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292: 107–110. [DOI] [PubMed] [Google Scholar]
- Tissenbaum, H. A., and L. Guarente, 2002. Model organisms as a guide to mammalian aging. Dev. Cell 2: 9–19. [DOI] [PubMed] [Google Scholar]
- Tu, M. P., D. Epstein and M. Tatar, 2002. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell 1: 75–80. [DOI] [PubMed] [Google Scholar]
- Turelli, M., and N. H. Barton, 2004. Polygenic variation maintained by balancing selection: pleiotropy, sex-dependent allelic effects and G × E interactions. Genetics 166: 1053–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M. C., and L. H. Rieseberg, 2003. Genetic architecture of a selection response in Arabidopsis thaliana. Evolution Int. J. Org. Evolution 57: 2531–2539. [DOI] [PubMed] [Google Scholar]
- Valenzuela, R. K., S. N. Forbes, P. Keim and P. M. Service, 2004. Quantitative trait loci affecting life span in replicated populations of Drosophila melanogaster. II. Response to selection. Genetics 168: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven, K. J., T. K. Vanhala, A. Biere, E. Nevo and J. M. van Damme, 2004. The genetic basis of adaptive population differentiation: a quantitative trait locus analysis of fitness traits in two wild barley populations from contrasting habitats. Evolution Int. J. Org. Evolution 58: 270–283. [PubMed] [Google Scholar]
- Vieira, C., E. G. Pasyukova, Z. B. Zeng, J. B. Hackett, R. F. Lyman et al., 2000. Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154: 213–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., and L. M. McIntyre, 2002. Combining mapping and arraying: an approach to candidate gene identification. Proc. Natl. Acad. Sci. USA 99: 14903–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne, M. L., J. B. Hackett, C. L. Dilda, S. V. Nuzhdin, E. G. Pasyukova et al., 2001. Quantitative trait locus mapping of fitness-related traits in Drosophila melanogaster. Genet. Res. 77: 107–116. [DOI] [PubMed] [Google Scholar]
- Weindruch, R., T. Kayo, C. K. Lee and T. A. Prolla, 2001. Microarray profiling of gene expression in aging and its alteration by caloric restriction in mice. J. Nutr. 131: 918S–923S. [DOI] [PubMed] [Google Scholar]
- Weinig, C., L. A. Dorn, N. C. Kane, Z. M. German, S. S. Halldorsdottir et al., 2003. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics 165: 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, G. C., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11: 398–411. [Google Scholar]
- Windig, J. J., R. F. Veerkamp and S. Nylin, 2004. Quantitative genetic variation in an island population of the speckled wood butterfly (Pararge aegeria). Heredity 93: 450–454. [DOI] [PubMed] [Google Scholar]
- Yampolsky, L., L. E. Pearse and D. E. Promislow, 2000. Age-specific effects of novel mutations in Drosophila melanogaster. I. Mortality. Genetica 110: 11–29. [DOI] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, D., A. Pai and G. Yan, 2005. Costly resistance to parasitism: evidence from simultaneous quantitative trait loci mapping for resistance and fitness in Tribolium castaneum. Genetics 169: 2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaan, B. J., R. Bijlsma and R. F. Hoekstra, 1995. Artificial selection for development time in Drosophila melanogaster in relation to aging: direct and correlated responses. Evolution 49: 635–648. [DOI] [PubMed] [Google Scholar]




