Abstract
The SNPWave marker system, based on SNPs between the reference accessions Colombia-0 and Landsberg erecta (Ler), was used to distinguish a set of 92 Arabidopsis accessions from various parts of the world. In addition, we used these markers to genotype three new recombinant inbred line populations for Arabidopsis, having Ler as a common parent that was crossed with the accessions Antwerp-1, Kashmir-2, and Kondara. The benefit of using multiple populations that contain many similar markers and the fact that all markers are linked to the physical map of Arabidopsis facilitates the quantitative comparison of maps. Flowering-time variation was analyzed in the three recombinant inbred line populations. Per population, four to eight quantitative trait loci (QTL) were detected. The comparison of the QTL positions related to the physical map allowed the estimate of 12 different QTL segregating for flowering time for which Ler has an allele different from one, two, or three of the other accessions.
FOR the genetic analysis of natural variation, so-called immortal mapping populations are very useful because they allow the localization of many traits that show allelic variation in the same mapping population (Koornneef et al. 2004). Furthermore, they allow replication of experiments and testing under various environmental conditions. Since genotypes differ in their genetic composition, the analysis of similar traits in different populations is required to get insight into the genetic variation of a specific trait within a species.
To allow a proper comparison of the locations of genes, it is important to use the same marker framework and markers should preferentially be anchored to the physical map of the species. Various marker systems have been used to genotype Arabidopsis recombinant inbred line (RIL) populations. In Arabidopsis, markers such as single sequence length polymorphisms (SSLPs) and cleaved amplified polymorphic sequences are anchored to the physical map. AFLP markers, as used to genotype the frequently analyzed Landsberg erecta (Ler) × Cape Verde Islands (Cvi) RILs (Alonso-Blanco et al. 1998b), can be anchored using bioinformatic tools only when it concerns AFLP bands that are characteristic for the sequenced genome of Columbia (Col-0) (Peters et al. 2001). On the basis of available sequence data of Arabidopsis accessions, many SNP markers have been detected [Schmid et al. 2003; The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org); Cereon database (http://www.arabidopsis.org/Cereon/index.jsp); and (http://walnut.usc.edu/2010.html)]. One of the various SNP detection systems (Cho et al. 1999) is the recently described SNPWave method (van Eijk et al. 2004).
In this article the usefulness of the SNPWave marker system was demonstrated on the basis of SNPs between the reference accessions Col-0 and Ler, to distinguish Arabidopsis accessions from various parts of the world. In addition, we used these markers to genotype three new sets of RILs derived from crosses between Ler and Antwerp (An-1), Kashmir (Kas-2), and Kondara (Kond), respectively. The populations studied were developed because their parents showed specific phenotypic differences and represent different geographical origins. To demonstrate their applicability for quantitative trait locus (QTL) mapping as well, we analyzed flowering time (FT) for all lines in the three populations as an example of a quantitative trait. Such analysis using three RIL populations having one common parent allows direct comparison of the loci segregating in these populations and facilitates identification of the different FT loci for which allelic variation is present among Arabidopsis accessions.
MATERIALS AND METHODS
Plant material and growth conditions:
Arabidopsis thaliana accessions were obtained from the Arabidopsis stock centers Arabidopsis Biological Resource Center (ABRC), Nottingham Arabidopsis Stock Center (NASC), and Sendai (http://www.arabidopsis.org), supplemented with accessions recently collected by members of the Laboratory of Genetics at Wageningen University and deposited at ABRC and NASC. Arabidopsis seeds were sown in petri dishes on water-saturated filter paper, followed by a 4-day cold treatment at 4°, and transferred to a climate room at 25° and 16 hr light for 2 days before planting in 7-cm pots with standard soil. In all descriptions of experiments, time is referred to as days after planting. The plants (12 plants/accession) were grown in an air-conditioned greenhouse with 70% relative humidity, supplemented with additional light (model SON-T plus 400W, Philips, Eindhoven, The Netherlands) providing a day length of at least 16 hr light (long day), with light intensity 125 μmol m−2 s−1, and maintained at a temperature between 22°–25° (day) and 18° (night).
New RIL populations were obtained from a cross between the accessions Landsberg erecta as a female (Ler, N20) and both Kashmir (Kas-2, N1264) and Kondara (Kond, CS6175) as a pollen parent, while Antwerp (An-1, N944) was reciprocally crossed with Ler. The F1 seeds of the different crosses were grown and allowed for self-fertilization to get F2 seeds. From the F2 seeds, sets of 120, 164, and 120 genotyped RILs for, respectively, An-1, Kas-2, and Kond, have been generated by a single-seed procedure until the F9 generation. To minimize any bias in the selection of plants taken to the next generation, six individuals per RIL were planted and plant number 3 was selected to go on for the next generation with the fifth one as a backup. Two plots containing six plants per RIL of the F9 generation were planted by the same procedures and under the same conditions mentioned before for growing the accessions. The flower heads of three individuals per RIL were harvested separately for DNA isolation and left for seed harvesting as well as for future use. The recombinant inbred lines with their marker data will be made available through the Arabidopsis stock centers.
To genetically test the allele type for the flowering loci FRI and FLC in the accessions under study, F1 plants were made by crossing the five accessions An-1, Kas-2, Kond, Ler, and Sha with lines that carry either an active FLC allele (derived from Col) or an FRI allele (FRI–M73; derived from line M73) both in a predominantly Ler genetic background as described in Koornneef et al. (1994). The F1 progeny of the crosses as well as the control plants were grown in a randomized two-plot design with six plants per genotype and under the same conditions mentioned above.
DNA isolation and genotyping:
Genomic DNA of 92 accessions was isolated from leaf material of individual plants using a modified CTAB procedure (Stewart and Via 1993). Details of the protocol for genotyping these accessions, using SNP markers, were described previously (van Eijk et al. 2004). For the RIL populations, the flower buds of three F9 plants per genotype were harvested separately for DNA isolation. DNA extraction was performed as described above for the SNP markers. For the SSLP markers, DNA was extracted using the Wizard magnetic 96 (Promega, Madison, WI) DNA isolation kit. SSLP markers were described in Clerkx et al. (2004), the TAIR database, or the MSAT database (http://www.inra.fr/qtlat/msat/index.php). Primers used for novel markers that were developed are described in Table 1. In addition, T27K12-SP6 and F5I14-49495 are written as T27K12 and F5I14, respectively. For both markers, the physical position can be found in the TAIR database. All markers used have first been checked to determine if the parental accessions An-1, Kas-2, and Kond were polymorphic with Ler; thereafter, the polymorphic markers were used to genotype all individual RILs. For SSLP markers, a standard protocol of 30 sec at 94°, 30 sec at 50°, and 30 sec at 72° (35 cycles) was used except for FRI (54° annealing, 1-min extension) and FLC (52° annealing, 2-min extension). Marker data are presented in supplemental Table 2 at http://www.genetics.org/supplemental/.
TABLE 1.
New markers used to genotype the three populations
| Marker name | BAC | Primer 1 (5′ to 3′) | Primer 2 (5′ to 3′) |
|---|---|---|---|
| F12A24b | F12A24 | GGTGTGATGTCGACCGGTAAAG | TGCACAACGTGCTCTCCATG |
| F17A22 | F17A22 | ACACACGAATATTGATTGTCTAAGG | TCACTTGTCGGTTTGTGTGG |
| cF7M19 | F7M19 | AGCTTGTGTCGTTTCCGATAG | AGTTGCAGAAATAAGCAGTGGC |
| F8D20 | F8D21 | CTTAAATGCCGATCCAGTCGAGG | TTCATTCGCGCATTTATTGTTGC |
| K15I22 | K15I22 | TCGGTGGTTTACTTTCACTTT | GAATTGTAGCTTCTTCTGAACC |
TABLE 2.
Parental values, averages, and ranges of flowering time and heritabilities (h2) in the three populations
| RIL population | FT–Ler | FT non-Ler | Average FT RILs | Range RILs | h2 |
|---|---|---|---|---|---|
| Ler × An-1 | 23.4 | 22.3 | 23.2 | 19.1–28.3 | 0.88 |
| Ler × Kas-2 | 33.2 | 47.0 | 34.7 | 24.9–54.0 | 0.86 |
| Ler × Kond | 29.0 | 49.3 | 38.7 | 26.7–66.2 | 0.95 |
Measurement of flowering time:
F10 generation plants (12 plants/RIL) were grown in the greenhouse in a randomized two-block design to reduce environmental effects. FT for each plant was scored as the number of days from planting until opening of the first flower.
Map construction and QTL analysis:
Initially, the three linkage maps have been constructed using only the SNP markers; gaps between markers that were >13 cM were filled using SSLP markers to obtain uniformly distributed markers. The JoinMap program (version 3.0; http://www.kyazma.nl) was used to construct the genetic maps.
The software package MapQTL 5 was used to identify and locate QTL on the linkage map by using interval mapping and multiple-QTL model (MQM) mapping methods as described in its reference manual (http://www.kyazma.nl). In a first step, putative QTL were identified using interval mapping. Thereafter, the closest marker at each putative QTL was selected as a cofactor (van Ooijen and Maliepaard 1996; van Ooijen 2000) and the selected markers were used as genetic background controls in the approximate multiple QTL model of MapQTL. LOD threshold values applied to declare the presence of QTL were estimated by performing permutation tests implemented in MapQTL version 5.0 using at least 1000 permutations of the original data set, resulting in a 95% LOD threshold of 2.4. Two-LOD support intervals were established as 95% QTL confidence interval (van Ooijen 1999) using restricted MQM mapping implemented within MapQTL. The estimated additive genetic effect and the percentage of variance explained by each QTL and the total variance explained by all the QTL affecting a trait were obtained using MQM mapping.
Statistical analysis:
Using NTSYSpc version 2.10t. (Rohlf 2001), the tree plot of the 92 Arabidopsis accessions, based on UPGMA cluster analysis using the pattern of polymorphism between 79 SNP markers, was performed (data presented in supplemental Table 1 http://www.genetics.org/supplemental/).
Heritability (broad sense) was estimated as the proportion of variance explained by between-line differences using the general linear model module of the statistical package of SPSS version 11.0.1 (SPSS, Chicago) based on measurements of 6–12 plants per genotype.
Differences in recombination were tested using a chi-square test comparing the number of recombinant and parental lines for two identical markers in two populations where differences were observed.
Two-way interactions among the QTL identified for FT were tested by ANOVA employing the corresponding two markers as fixed factors and the trait as dependent variable and using the general linear model of the statistical package SPSS version 11.5.1. In the Ler × Kas-2 population, SNP32 was included in this analysis because a suggestive QTL with a LOD of 2.3 was found at that locus. A Bonferroni correction to adjust the 0.05 threshold of significance was applied if multiple tests were performed on the same data set. Only those interactions that were significant after the Bonferroni correction are presented.
RESULTS
Polymorphism between a set of Arabidopsis accessions using SNPWave markers:
A 100-plex SNPWave marker set of known SNPs between the two reference Arabidopsis accessions Col-0 and Ler (van Eijk et al. 2004) was used to genotype 92 Arabidopsis accessions. Among markers that could be amplified in most accessions, 37.6–62.4% of the markers were different from the Col-0 allele and 0–37.6% differed from the Ler allele. The polymorphism data indicated that, for many accessions, crosses made with one of the two reference accessions would yield reasonable numbers of polymorphic SNPWave markers.
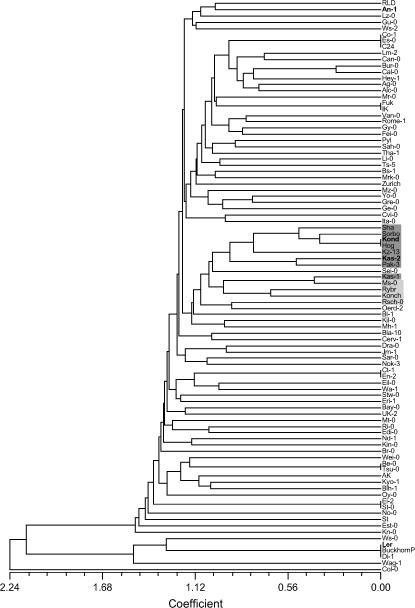
A few identical genotypes were detected, of which some have been described for other marker systems as well (e.g., Co-1, C24; Ler, Di-1; Buckhorn Pass) (Figure 1) (Torjek et al. 2003). For some other accessions (e.g., Co-1 and Es-0, Ct-1 and En-2, Be-0 and Tsu-1) this was not expected in view of their different geographical origins. As reported for many marker systems, no obvious structure related to the geographical origin was detected using UPGMA cluster analysis. However, a number of accessions from Central Asia and Russia (Figure 1) seem more related to each other than to accessions from other regions as was reported before (Schmuths et al. 2004; Nordborg et al. 2005). The data confirm that Kas-1 (N903) and Kas-2 (N1264) are genetically different (Levey and Wingler 2005), although both cluster in the Central Asian group.
Figure 1.
Tree plot of the 92 Arabidopsis accessions based on UPGMA cluster analysis using the pattern of polymorphism among 79 SNP markers. The dark-shaded block indicates accessions from Central Asia, while the light-shaded block refers to accessions from Russia.
Ler × An-1, Ler × Kas-2, and Ler × Kond linkage maps:
The accessions used to construct the RIL populations differed for 57.1, 55.8, and 54.5% of the markers from Ler for An-1, Kas-2, and Kond, respectively. These markers did not completely cover the genome. To construct genetic maps with equally spaced markers for the three different crosses (Ler × An-1, Ler × Kas-2, and Ler × Kond), either additional SNPWave markers had to be developed or publicly available markers (TAIR database) were used as mentioned in materials and methods.
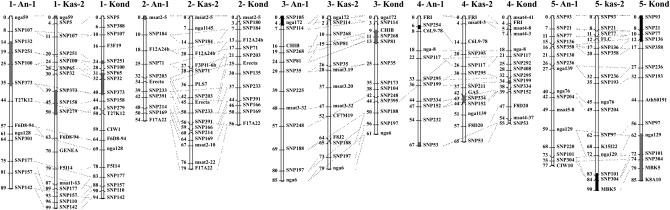
Linkage maps were obtained using 44, 45, and 51 SNP markers supplemented with 20, 31, and 23 SSLP markers for Ler × An-1, Ler × Kas-2, and Ler × Kond, respectively. In addition, the erecta mutation segregating in all three populations and the ga5-gibberellin-deficient mutation (Xu et al. 1995), segregating in the Ler × Kas-2 population (shown to be present in Kas-2 by the absence of complementation in the cross of the ga5 mutant and Kas-2), could be scored as morphological markers. This resulted in three genetic maps with 65, 78, and 75 markers for Ler × An-1, Ler × Kas-2, and Ler × Kond, respectively (Figure 2). The markers were assigned to five linkage groups for each population with a total genetic length of 371, 441, and 351 cM for Ler × An-1, Ler × Kas-2, and Ler × Kond, respectively. Most markers were located on the expected linkage groups predicted on the basis of the physical order of the markers in the sequenced Col-0 accession. Exceptions are SNP395, which was expected on chromosome 4 but mapped to chromosome 3 in Ler × Kond, and two pairs of markers that are inverted in the Ler × Kas-2 linkage map, viz., C6L9-78 and SNP395 on chromosome 4 and SNP77 and FLC on chromosome 5. This deviating order derived from normal recombination patterns (see supplemental Table 2 at http://www.genetics.org/supplemental/), which suggests that genotyping errors are not the reason for this changed order.
Figure 2.
Integrated genetic maps linked through anchoring markers scored in the three populations Ler × An-1, Ler × Kas-2, and Ler × Kond. Distorted regions [regions deviating from the 1:1 ratio (P < 0.05)] are indicated by shaded boxes (regions with a significantly higher number of RILs with the Ler allele) or solid boxes (regions with significantly higher numbers of lines in favor of An-1, Kas-2, or Kond alleles).
Since in each generation heterozygosity per locus is reduced by half after selfing, the probability that a specific locus is heterozygous is 0.39% for the F9 generation. The average frequency of heterozygosity for all loci is 0.28, 0.25, and 0.25% for Ler × An-1, Ler × Kas-2, and Ler × Kond, respectively, with no locus having a significantly higher value than predicted.
For each marker, the expected segregation ratio would be 1:1 for each parental allele in the case of no bias in the selection of individual plants during the maintenance of the populations. Figure 2 indicates regions with significantly distorted segregation (at 0.0005 < P < 0.05) for the three populations. Markers showing significant segregation distortion clustered in a certain region of the genome in the three maps, with ratios ranging from 1.4:1 to 2.1:1. These regions either partially overlap in the different populations as on chromosome 1 or are population specific in other chromosome regions. The distortion in most regions favored Ler alleles, although in four different regions of the three populations, the non-Ler alleles were in excess (Figure 2).
Comparison of the Ler × An-1, Ler × Kas-2, and Ler × Kond genetic maps:
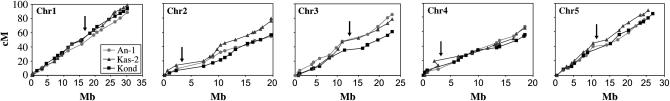
Figure 2 shows the Ler × An-1, Ler × Kas-2, and Ler × Kond genetic maps linked to each other by 41 anchoring markers scored in the three populations. Comparison between the physical map of Col-0 and the three newly generated linkage maps indicates that the overall recombination rates are similar over chromosomes with suppression of recombination observed around the centromeres of chromosomes 2, 3, 4, and 5 (Figure 3). The maps are colinear with the exception of the two inverted pairs of markers in the Ler × Kas-2 population described above. When comparing recombination frequencies in regions where maps appeared different (Figure 2) among the three populations, recombination was found to be significantly higher (P = 0.001) in the Kas-2 cross between SNP71 and SNP203 on chromosome 2 compared to the Kond cross and recombination was not significantly different when compared to the An-1 cross (P = 0.2). In other regions differences were not statistically significant.
Figure 3.
The relationship between the genetic maps of Ler × An-1, Ler × Kas-2, and Ler × Kond and the physical map along the five chromosomes. The arrows indicate the position of the centromeres.
The pattern of similarity in recombination described above results in similar genetic lengths of the five chromosomes in the three crosses. The largest differences were observed for chromosome 2 where the genetic map of Ler × Kas-2 is longer than the other two crosses by >20 cM and for chromosome 3 where the Ler × Kond map is shorter than the other two maps.
QTL mapping of flowering time:
Flowering-time QTL have been mapped in several Arabidopsis RIL populations (Kowalski et al. 1994; Clarke et al. 1995; Jansen et al. 1995; Kuittinen et al. 1997; Alonso-Blanco et al. 1998a; Loudet et al. 2002; El-Lithy et al. 2004; Koornneef et al. 2004 for review). Since different populations may segregate for different loci depending on the genetic composition of their parental lines, a comparison between multiple crosses is needed to obtain information about the variation present among Arabidopsis accessions. This allows the description of the so-called global genetic architecture (Symonds et al. 2005) of a trait within a species. Comparison between different crosses can be done accurately only when the same markers are used and/or when these markers are anchored to the Arabidopsis physical map, which acts as a reference map.
To illustrate this approach, we analyzed FT in the three RIL populations, which were grown in the same greenhouse under long-day conditions but in independent experiments. For all populations, heritabilities were high and transgression beyond the parental values was observed toward both earliness and lateness (Table 2). In total four, six, and eight QTL were identified per population (Figure 4). However, because the populations were not grown in the same experiment, some of the differences among the populations may be due to genotype × environment interactions in cases where specific QTL are expressed only under specific environmental conditions.
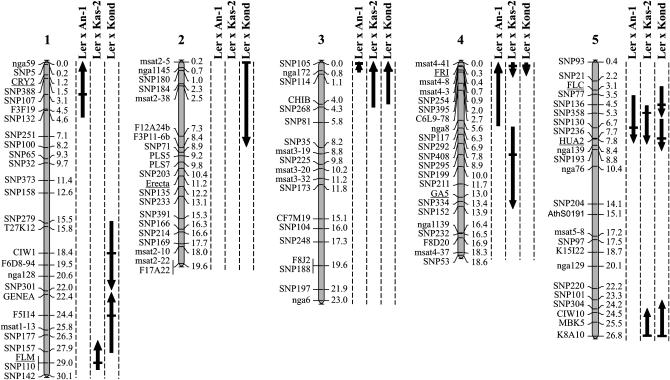
Figure 4.
Combined physical map of Ler × An-1, Ler × Kas-2, and Ler × Kond containing markers that are used in this study in addition to several candidate genes (CRY2, FLM, HUA2). Flowering-time QTL are indicated by arrows along the chromosomes. Thick horizontal dashes in the arrows indicate the marker fixed during the MQM mapping analysis. The lengths of the arrows indicate the 2-LOD support intervals. The direction of the arrows indicates the allelic effect: upward, Ler increasing the FT and the other allele decreasing; downward, the non-Ler allele increasing and the Ler decreasing.
Although the FT differences between Ler and An-1 were very small (Table 2), variation among the RILs is considerable and is explained by four QTL, of which for three the An-1 allele causes early flowering (Figure 4; Table 3). In the Kas-2 and Kond populations, the parents differed much more and the genetic differences could be explained by six and eight QTL, for which in three and five cases the Ler alleles accelerate flowering, respectively (Figure 4; Table 3). The detected QTL explained 68.3, 78.8, and 84.8% of the phenotypic variance for the Ler × An-1, Ler × Kas-2, and Ler × Kond populations, respectively. Within the three populations significant interactions among several QTL were detected (Table 3).
TABLE 3.
Characteristics of flowering-time QTL detected in the three populations
| RILs | QTL at nearest marker | Map positiona | LOD score | % of variance | Additive allele effect (days)d |
|---|---|---|---|---|---|
| Ler × An-1 | 68.3c | ||||
| SNP107 | 1, 7.9 | 2.8 | 4.0 | 0.6 | |
| SNP105 | 3, 0.0 | 16.4 | 29.4 | 1.8 | |
| SNP254 | 4, 6.2 | 3.3 | 4.9 | 0.8 | |
| SNP130 | 5, 21.0 | 14.2 | 24.7 | −1.6 | |
| SNP105 × SNP130 | P = 0.000045b | 6.1 | |||
| Ler × Kas-2 | 78.8c | ||||
| SNP110 | 1, 95.5 | 7.8 | 6.5 | 3.0 | |
| nga172 | 3, 0.0 | 3.5 | 2.4 | 2.0 | |
| FRI | 4, 0.0 | 23.8 | 24.0 | −6.0 | |
| SNP295 | 4, 30.0 | 3.8 | 2.8 | −2.6 | |
| SNP358 | 5, 20.2 | 8.8 | 7.2 | −3.4 | |
| MBK5 | 5, 89.6 | 16.8 | 15.0 | 4.8 | |
| SNP110 × MBK5 | P = 0.006b | 4.7 | |||
| SNP32 × SNP295 | P = 0.008b | 4.0 | |||
| Ler × Kond | 84.8c | ||||
| CIW1 | 1, 59.2 | 4.2 | 2.6 | −3.0 | |
| F5I14 | 1, 78.4 | 6.2 | 4.2 | 3.6 | |
| msat2-5 | 2, 0.0 | 5.1 | 3.4 | −3.0 | |
| nga172 | 3, 0.0 | 5.0 | 3.5 | 3.0 | |
| FRI | 4, 4.1 | 36.5 | 46.6 | −11.4 | |
| SNP136 | 5, 12.4 | 3.2 | 2.0 | −2.8 | |
| SNP236 | 5, 27.4 | 8.9 | 6.4 | −5.0 | |
| K8A10 | 5, 84.8 | 5.4 | 3.6 | 3.2 | |
| FRI × SNP136 | P = 0.008b | 2.3 | |||
| FRI × SNP236 | P = 0.009b | 2.2 |
Chromosome number is given, followed by the marker position in centimorgans.
For interactions, the P-values are given instead of the LOD scores.
Values in italics refer to the total variance explained by the additive effects of significant QTL and significant two-way interactions.
Positive values indicate that Ler alleles increase the trait value and negative values indicate that the non-Ler alleles increase the trait value.
Relating the map positions and 2-LOD intervals to the physical map on the basis of the Col-0 sequence allows a comparison among the QTL in the three populations. In most cases the comparison was relatively straightforward. However, for QTL with relatively large 2-LOD intervals (top chromosome 1 and 2 for Ler × An-1 and Ler × Kond crosses, respectively) and for regions where two linked QTL were detected in the same population, interpretation is more complex.
Two FT QTL are in common among the three populations: the QTL on top of chromosome 3 (around nga172) for which the Ler allele delays flowering and the QTL located around SNP130 (chromosome 5) for which the Ler allele accelerates flowering. In addition, there are several QTL common between two of the three populations. These are the QTL located at the FRI locus for Ler × Kas-2 and Ler × Kond and around SNP136 (chromosome 5) for Ler × An-1 and Ler × Kond and around K8A10 (chromosome 5) for Ler × Kas-2 and Ler × Kond. For the Ler × An-1 population the QTL around SNP136 could not be separated from another QTL around SNP236. However, the presence of two distinct QTL in this region was clear for the Ler × Kond population.
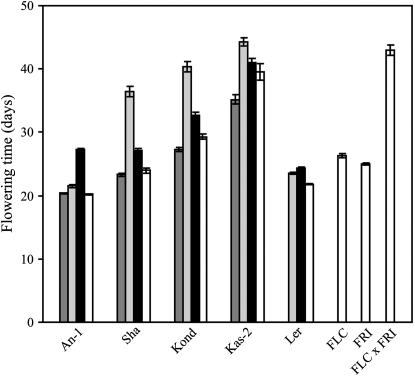
It is known that Ler carries a weak FLC allele in contrast to many other accessions that contain active FLC alleles that, together with active FRI alleles, confer late flowering, which can be overcome by vernalization (Koornneef et al. 1994; Michaels et al. 2003; Shindo et al. 2005). Various authors (Koornneef et al. 1994; Gazzani et al. 2003; Michaels et al. 2003) used hybrids of accessions with lines containing only either an active FRI allele or an active FLC allele, expecting that when both active copies are present in the hybrids the plants will be late flowering. To test whether active alleles of FRI and FLC are present, we also applied this procedure in the accessions that we have used to construct our populations. In addition, we included the accession Shakdara (Sha) that is assumed to contain a weak FLC allele (Gazzani et al. 2003; Michaels et al. 2003). This is in agreement with the absence of a late-flowering Sha allele at the FLC locus in populations made from crosses with Sha (Loudet et al. 2002; El-Lithy et al. 2004). Flowering-time data of the hybrids and the parents are shown in Figure 5 and confirm that all accessions except An-1 contain active FRI alleles resulting in late flowering, as compared to the parents, when combined with a line containing an active FLC allele. Hybrids from crosses made with FRI–M73 are less late than the FLC hybrids but slightly later than the hybrids with Ler or the accession parents. This observation suggests that all accessions have weak FLC alleles (compared to the Col alleles) but stronger than Ler, except An-1, where the FRI hybrid is later flowering than the hybrid with FLC.
Figure 5.
Flowering time in hybrids of accessions with near-isogenic lines containing, respectively, FLC–Col and FRI–M73 in a predominantly Ler genetic background. Dark-shaded bars represent the cross with Ler, light-shaded bars represent the cross with FLC–Col-0, solid bars represent the cross with FRI–M73, and open bars represent the accessions and pure lines.
In addition, population-specific QTL (Table 3) were detected around markers SNP107 and SNP254 for Ler × An-1, where Ler alleles delayed flowering. For Ler × Kas-2, two QTL with different allele effects were identified around the markers SNP110 and SNP295. In the Ler × Kond population-specific QTL could be identified around markers CIW1, F5I14, and msat2-5 with different allele effects. The number of colocating QTL might be higher since in a few cases suggestive QTL (LOD between 1.5 and 2.4) were detected in one population at a position where significant QTL were detected in another population (data not shown). This was true for two suggestive QTL at SNP301 and GENEA for Ler × An-1 and Ler × Kas-2, respectively, which colocate with a significant QTL in that region in Ler × Kond (chromosome 1). When taking these suggestive QTL into account, the total number of QTL did not increase. In total we identified 12 different QTL for FT for which Ler has alleles different from the alleles in one, two, or three of the other accessions.
DISCUSSION
In this study we describe three new recombinant inbred line populations for Arabidopsis having Ler as a common parent. The SNPWave technique was applied to genotype these RIL populations on the basis of SNPs between Col-0 and Ler. Of the SNPWave markers that are polymorphic between Ler and Col-0, ∼50% (37.6–62.5%) could be used to genotype populations made from crosses with Ler. Some regions are not covered by the SNPWave markers, such as the upper part of the lower arm of chromosome 1 for all three populations, the middle part of chromosome 3 for Ler × Kas-2, the top of chromosome 4 for all populations, and the middle part of chromosome 5 for Ler × An-1. As far as this was due to insufficient coverage of the SNPWave markers in the three crosses, it implied that additional markers, such as the common PCR markers used here, were required to obtain genetic maps with equally distributed markers.
Since the maps contain many similar markers and, more importantly, all markers are linked to the physical map of Arabidopsis, a quantitative comparison of maps could be performed. This analysis showed that map lengths are quite similar and also in the same range as those published for other populations (Lister and Dean 1993; Alonso-Blanco et al. 1998b; Loudet et al. 2002; Clerkx et al. 2004). The two inverted pairs of markers that were detected in the Kas-2 population and the reduced recombination between the two markers on chromosome 2 might also be explained by structural chromosomal inversions between accessions. However, differences in local recombination rate as such may exist, as suggested by cytogenetic data (Sanchez-Moran et al. 2002). Structural chromosome variants between accessions have not been studied frequently but are not uncommon (reviewed in Koornneef et al. 2003). Suppression of recombination in specific regions makes map-based cloning in such regions difficult. In general, a solution for this problem is to perform mapping in a cross with another accession that does not show suppression of recombination.
RIL populations allow the identification of natural genetic variants for which the parents differ. Such populations facilitate the mapping of many traits in the same population (Koornneef et al. 2004). However, since the parents might not be different for a specific QTL for which variation is present within the germ plasm pool, additional mapping populations are being developed (http://www.inra.fr/qtlat/naturalvar/rilsummary.htm). The power of using multiple populations was recently demonstrated by Symonds et al. (2005), who identified nine QTL for trichome density in a total of four RIL populations, whereas individual population segregated for three to five QTL.
In this study similar results were obtained for FT, a frequently studied trait showing large natural variation in Arabidopsis. In this species extreme lateness is mainly due to the presence of dominant alleles at the FRI and FLC loci (Caicedo et al. 2004; Hagenblad et al. 2004; Koornneef et al. 2004). These large-effect loci mask the segregation of other minor-effect loci, which are easier to detect in mapping populations where these large-effect alleles do not segregate. This is well illustrated in the present example, where early and middle late accessions are combined and where, in total, 12 QTL could be detected. One of these is probably FRI, detected in the Ler × Kas-2 and Ler × Kond populations. Kas-2 and Kond have late alleles at the FRI locus, which is at least functional in Kond (Gazzani et al. 2003; Michaels et al. 2003; Hagenblad et al. 2004). Active FRI alleles may confer lateness without FLC, as was also found in Sha-derived populations (Loudet et al. 2002; El-Lithy et al. 2004). In the Ler × An-1 population, a novel locus was also identified at the top of chromosome 4, for which An-1 accelerates FT.
The situation at the top of chromosome 5 is complex. As in the Ler × Sha population (El-Lithy et al. 2004), we could not identify a putative FLC QTL in either the Ler × Kas-2 or the Ler × Kond population. This was confirmed by our hybrid data (Figure 5) and that of Michaels et al. (2003), which suggest that Kond has a weak FLC allele. This is in agreement with the fact that Kas-2, Kond, as well Sha all have a 1.2-kb insertion within the FLC gene, which might reduce FLC function (Hagenblad et al. 2004). However, we could identify a QTL at marker SNP136 close to the position of FLC, which was also used as a marker in the Ler × Kond population. As explained above, we assume that this is not FLC, although the significant interaction between the QTL at the FRI locus and SNP136 (Table 3) is in agreement with the epistatic interaction described previously for FRI and FLC. However, its effect is relatively small and it cannot be excluded that other loci located below FLC are responsible for this interaction. In the Ler × Cvi population (Alonso-Blanco et al. 1998a), two linked loci on chromosome 5 were found, both conferring lateness, but only when both alleles were derived from the non-Ler parent. It has been suggested that the upper locus is FLC and that the lower locus (named FLG by Alonso-Blanco et al. 1998a) might encode HUA2 (Doyle et al. 2005). For the An-1 cross, the 2-LOD interval includes the FLC locus but we could not separate this SNP130 QTL into two QTL. On the contrary, for total leaf number and rosette leaf number, traits that are known to be correlated to FT (Koornneef et al. 1991; Alonso-Blanco et al. 1998a), we could detect two distinct QTL (data not shown). Thus, probably An-1 contains an active, although not very strong, FLC allele that was confirmed by our hybrid data (Figure 5).
It appears that this region below FLC on chromosome 5 contains at least one and maybe two other QTL for FT for which Ler has alleles different from the other parents. The observation that one or two loci are present in this region, depending on the cross, most likely can be explained by the difficulty to statistically separate linked QTL.
The Ler × Kas-2 and Ler × Kond populations have common QTL with the same allelic effects at nga172, at FRI, and at K8A10. At these positions also the Sha accession from Tadjikistan carries similar alleles (Loudet et al. 2002; El-Lithy et al. 2004). Kas-2 and Sha may have similar alleles for the QTL at the bottom of chromosome 1 for which the FLM locus is a candidate gene (Werner et al. 2005).
This analysis shows that for a single trait additional genetic variation is detected when different populations are analyzed for the same trait. However, the accuracy of QTL mapping is such that colocation can also be due to two different closely linked QTL. Having available different sources for the same type of allelic variation allows the selection of the populations with the strongest alleles for future fine mapping and cloning. In addition, these genetic studies provide the basis of the correlation between function and molecular haplotype as has been described for the FRI and FLC loci (Gazzani et al. 2003; Michaels et al. 2003; Caicedo et al. 2004; Hagenblad et al. 2004). The new RIL populations show segregation for various other traits such as seed dormancy and plant performance (our unpublished data) that are currently being analyzed.
Acknowledgments
M. El-Lithy was supported by a grant from the Ministry of Higher Education of the Egyptian government. Leónie Bentsink and Gerda Ruys were supported by the European Union program NATURAL (contract QLG2-CT-2001-01097). This project was carried out within the research program of the Centre of BioSystems Genomics, which is part of The Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. The SNPWave technology is covered by patent applications owned by Keygene N. V. An application for trademark registration for SNPWave has been filed by Keygene N. V.
References
- Alonso-Blanco, C., S. E. El-Assal, G. Coupland and M. Koornneef, 1998. a Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco, C., A. J. M. Peeters, M. Koornneef, C. Lister, C. Dean et al., 1998. b Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14: 259–271. [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., J. R. Stinchcombe, K. M. Olsen, J. Schmitt and M. D. Purugganan, 2004. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc. Natl. Acad. Sci. USA 101: 15670–15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, R. J., M. Mindrinos, D. R. Richards, R. J. Sapolsky, M. Anderson et al., 1999. Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat. Genet. 23: 203–207. [DOI] [PubMed] [Google Scholar]
- Clarke, J. H., R. Mithen, J. K. Brown and C. Dean, 1995. QTL analysis of flowering time in Arabidopsis thaliana. Mol. Gen. Genet. 248: 278–286. [DOI] [PubMed] [Google Scholar]
- Clerkx, E. J., M. E. El-Lithy, E. Vierling, G. J. Ruys, H. Blankestijn-De Vries et al., 2004. Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiol. 135: 432–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, M. R., C. M. Bizzell, M. R. Keller, S. D. Michaels, J. Song et al., 2005. HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 41: 376–385. [DOI] [PubMed] [Google Scholar]
- El-Lithy, M. E., E. J. Clerkx, G. J. Ruys, M. Koornneef and D. Vreugdenhil, 2004. Quantitative trait locus analysis of growth-related traits in a new Arabidopsis recombinant inbred population. Plant Physiol. 135: 444–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzani, S., A. R. Gendall, C. Lister and C. Dean, 2003. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 132: 1107–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenblad, J., C. Tang, J. Molitor, J. Werner, K. Zhao et al., 2004. Haplotype structure and phenotypic associations in the chromosomal regions surrounding two Arabidopsis thaliana flowering time loci. Genetics 168: 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, R. C., J. W. van Ooijen, P. Stam, C. Lister and C. Dean, 1995. Genotype by environment interaction in genetic mapping of multiple quantitative trait loci. Theor. Appl. Genet. 91: 33–37. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., C. J. Hanhart and J. H. van der Veen, 1991. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229: 57–66. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., H. Blankesijn-de Vries, C. Hanhart, W. Soppe and T. Peeters, 1994. The phenotype of some late flowering time mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J. 6: 911–919. [Google Scholar]
- Koornneef, M., P. Fransz and H. de Jong, 2003. Cytogenetic tools for Arabidopsis thaliana. Chromosome Res. 11: 183–194. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., C. Alonso-Blanco and D. Vreugdenhil, 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Kowalski, S. P., T. H. Lan, K. A. Feldmann and A. H. Paterson, 1994. QTL mapping of naturally-occurring variation in flowering time of Arabidopsis thaliana. Mol. Gen. Genet. 245: 548–555. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., M. J. Sillanpää and O. Savolainen, 1997. Genetic basis of adaptation: flowering time in Arabidopsis thaliana. Theor. Appl. Genet. 95: 573–583. [Google Scholar]
- Levey, S., and A. Wingler, 2005. Natural variation in the regulation of leaf senescence and relation to other traits in Arabidopsis. Plant Cell Environ. 28: 223–231. [Google Scholar]
- Lister, C., and C. Dean, 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745–750. [DOI] [PubMed] [Google Scholar]
- Loudet, O., S. Chaillou, C. Camilleri, D. Bouchez and F. Daniel-Vedele, 2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor. Appl. Genet. 104: 1173–1184. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D., Y. He, K. C. Scortecci and R. M. Amasino, 2003. Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., T. T. Hu, Y. Ishino, J. Jhaveri, C. Toomajian et al., 2005. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J. L., H. Constandt, P. Neyt, G. Cnops, J. Zethof et al., 2001. A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol. 127: 1579–1589. [PMC free article] [PubMed] [Google Scholar]
- Rohlf, F. J., 2001. NTSYSpc: numerical taxonomy and multivariate analysis system, version 2.10x. Exeter Software, Setauket, NY.
- Sanchez-Moran, E., S. J. Armstrong, J. L. Santos, F. C. Franklin and G. H. Jones, 2002. Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., T. R. Sorensen, R. Stracke, O. Torjek, T. Altmann et al., 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths, H., M. H. Hoffmann and K. Bachmann, 2004. Geographic distribution and recombination of genomic fragments on the short arm of chromosome 2 of Arabidopsis thaliana. Plant Biol. 6: 128–139. [DOI] [PubMed] [Google Scholar]
- Shindo, C., M. J. Aranzana, C. Lister, C. Baxter, C. Nicholls et al., 2005. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 138: 1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, C. N., Jr., and L. E. Via, 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14: 748–750. [PubMed] [Google Scholar]
- Symonds, V. V., A. V. Godoy, T. Alconada, J. F. Botto, T. E. Juenger et al., 2005. Mapping quantitative trait loci in multiple populations of Arabidopsis thaliana identifies natural allelic variation for trichome density. Genetics 169: 1649–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torjek, O., D. Berger, R. C. Meyer, C. Mussig, K. J. Schmid et al., 2003. Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J. 36: 122–140. [DOI] [PubMed] [Google Scholar]
- van Eijk, M. J., J. L. Broekhof, H. J. van der Poel, R. C. Hogers, H. Schneiders et al., 2004. SNPWave: a flexible multiplexed SNP genotyping technology. Nucleic Acids Res. 32: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 1999. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 2000. MapQTL (R) Version 4.0: User Friendly Power in QTL Mapping—Addendum to the Manual of Version 3.0. Plant Research International, Wageningen, The Netherlands.
- Van Ooijen, J. W., and C. Maliepaard, 1996. MapQTLTM Version 4.0: Software for the Calculation of QTL Positions on Genetic Maps. Plant Research International, Wageningen, The Netherlands.
- Werner, J. D., J. O. Borevitz, N. Warthmann, G. T. Trainer, J. R. Ecker et al., 2005. Quantitaive trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 102: 2460–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. L., L. Li, K. Wu, A. J. Peeters, D. A. Gage et al., 1995. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: molecular cloning and functional expression. Proc. Natl. Acad. Sci. USA 92: 6640–6644. [DOI] [PMC free article] [PubMed] [Google Scholar]