Abstract
In mammals, Robertsonian (Rb) translocation (the joining of two telo/acrocentric chromosomes at their centromere to form a metacentric) is the most effective process in chromosomal evolution leading to speciation; its occurrence also affects human health (through the induction of trisomies) and the fertility of farm animals. To understand the mechanism of Rb translocation, we used the house mouse as a model system and studied the organization of pericentromeric satellite DNAs (satDNA) of telocentrics and Rb chromosomes, both minor and major satDNA. The chromosome-orientation fluorescence in situ hybridization (CO-FISH) technique was used to analyze the major satDNA. To detect the very small amount of minor satDNA, a procedure was developed that combines CO-FISH with primed in situ labeling and conventional FISH and is five times more sensitive than the CO-FISH procedure alone. It was found that both the major and the minor satDNA tandem repeats are oriented head-to-tail in telocentric and Rb chromosomes, and their polarity is always the same relative to the centromere. We suggest that all tandemly repetitive satDNAs in a species probably are locked into such a symmetry constraint as a universal consequence of chromosomal evolution. Rb translocation breakpoints were found localized within the minor satDNA of telocentrics, and these sequences contributed symmetrically to the formation of the centromeric region of the Rb chromosomes. These results are important for an understanding of the geometry of Rb translocations and suggest the study of DNA orientation as a new tool for investigating these rearrangements.
The Robertsonian (Rb) translocation is one of the most common chromosomal rearrangements in mammals (1, 2). In this translocation, two chromosomes with terminal or near-terminal centromeres (telocentrics or acrocentrics) fuse at or close to their centromeres to generate a bi-armed, metacentric chromosome, with a distinctly internal centromere. Rb chromosomes may occur as a polymorphism, as in humans, where their incidence is 1 in 813 newborn children (3, 4). They also may contribute to chromosomal differences between species, as within the lemurs (5) and mole rats (6). They also may define chromosome races within species, such as house mice (Mus musculus domesticus) for which more than 40 distinct local races have been described, characterized by about 100 types of Rb chromosomes with different arm combinations (7). The interest in Rb translocations derives not only from their high frequency in mammals, but also from their influence on fertility (8). Heterozygotes for Rb chromosomes have a tendency to be infertile or to produce offspring with birth defects. In humans, this has great medical consequences; in domesticated mammals of economic importance it may lead to loss of productivity. In wild mammals, Rb chromosomes may contribute to reproductive isolation and thus to speciation.
Because of its high frequency of Rb translocations, the house mouse constitutes a particularly amenable model to further our knowledge of this important mutational process (7). In this species, the pericentromeric regions of the standard telocentric chromosomes comprise a large block of major satellite DNA (satDNA, about 6 megabases) (9) flanked by a smaller block of minor satDNA (about 600 kb) (10) that is adjacent to the telomeric repeats (50–150 kb) present at the physical chromosome ends (11, 12). It is known that after Rb translocation, all telomeric and many minor satDNA sequences are lost, so that in the newly formed centromeric region of every Rb chromosome about 20–60 kb of minor satDNA are retained, sandwiched between two blocks of about 6 megabases each of major satDNA (13, 14).
Early studies (15–17) based on the quenching of Hoechst 33258-labeled chromosomes by BrdUrd determined the major satDNA polarity in the pericentromeric regions of mouse telocentrics and Rb chromosomes. The technique used at that time did not allow a rigorous analysis of the highly repetitive DNA families that constitute the pericentromeric region of mouse chromosomes, although the results obtained suggested that major satDNA polarity is maintained through the centromere in Rb chromosomes.
Here, we compare the molecular organization of pericentromeric regions of telocentrics and Rb chromosomes derived from wild mice of two chromosome races in northern Italy, the Poschiavo and Cremona races (18). We first re-examined the polarity of major satDNA by using the chromosome-orientation fluorescence in situ hybridization (CO-FISH) technique (19). Then, we used a method that combines CO-FISH with primed in situ labeling (PRINS) (20) and FISH for the detection of the very small amounts of the minor satDNA still harbored in the centromeric region of the Rb chromosomes. This method (CO-FISH/PRINS/FISH) allowed the localization of the breakpoints in telocentrics involved in Rb translocations. We showed that in Rb chromosomes the two telocentrics contribute symmetrically with 10–30 kb of minor satDNA each to the newly formed centromeric region. Moreover, this procedure revealed the organization of satDNA sequences in the centromeric regions of Rb chromosomes, thus defining the polarity of the minor satDNA at the fusion points.
Methods
Chromosomes were prepared from splenocyte cultures of three male mice of each of the following types: laboratory strain C3H (Charles River Breeding Laboratories) (2n = 40, all-telocentric), Poschiavo race (POS, 2n = 26, seven pairs of Rb chromosomes), Cremona race (CRE, 2n = 22, nine pairs of Rb chromosomes), F1 POS×C3H (2n = 33, seven Rb chromosomes in a heterozygous state), and F1 CRE×3H (2n = 31, nine Rb chromosomes in a heterozygous state).
After 48-h culture using Con A (Sigma) stimulation (20 μg/ml), 10-5 M BrdUrd (Sigma) was added, and 17 h later cells were harvested. After 3-h incubation with colcemid (Sigma) (0.05 μg/ml), hypotonic treatment (75 mM KCl for 20 min) and fixation (methanol/acetic acid, 3:1, vol/vol), the cell suspensions were dropped onto slides and air-dried.
The synthetic oligonucleotides GGACCTGGAATATGGCGAGAAA (sat-1) (major satDNA) and TGATATACACTGTTCTACAAATCCCG (sat-2) (minor satDNA) were purchased from Perkin–Elmer and labeled with biotin by the end-labeling procedure (Roche Diagnostics) following the manufacturer's instructions.
Strand-specific hybridization was performed on two slides for each animal used. After digestion with a 0.005% pepsin, 10 mM HCl solution for 10 min at 37°C, the CO-FISH method described by Goodwin and Meyne (19) was followed. Chromosomes were hybridized with 0.4 μg/ml oligonucleotide probes at 37°C overnight. Posthybridization washes were in 2×SSC at 42°C. One round of staining with FITC-labeled avidin (4 μg/ml, fluorescein-avidin DCS, Vector Laboratories), biotinylated anti-avidin antibody (4 μg/ml, biotinylated anti-avidin D, Vector Laboratories) was used to increase the hybridization signal.
Two slides each from POS, F1 POS×C3H, and F1 CRE×C3H animals, hybridized with the sat-2 oligo probe, were subjected to target sequence amplification by the PRINS procedure. Amplification was performed by using the sat-2 oligonucleotide as forward primer and the oligonucleotide CAATGAGTTACAATGAGAAACATGG (sat-3) as reverse primer, 0.5 mM of each. The reaction was conducted at 72°C for 3 min followed by 60°C for 50 min. The signal then was detected with FISH. Metaphase chromosomes were hybridized with 50 ng of biotinylated minor satDNA probe (R198, kindly provided by D. Kipling, University of Wales College of Medicine, Cardiff, U.K.) at 37°C overnight. After conventional posthybridization washes, the probe was detected with FITC-labeled avidin (4 μg/ml, fluorescein-avidin DCS, Vector Laboratories).
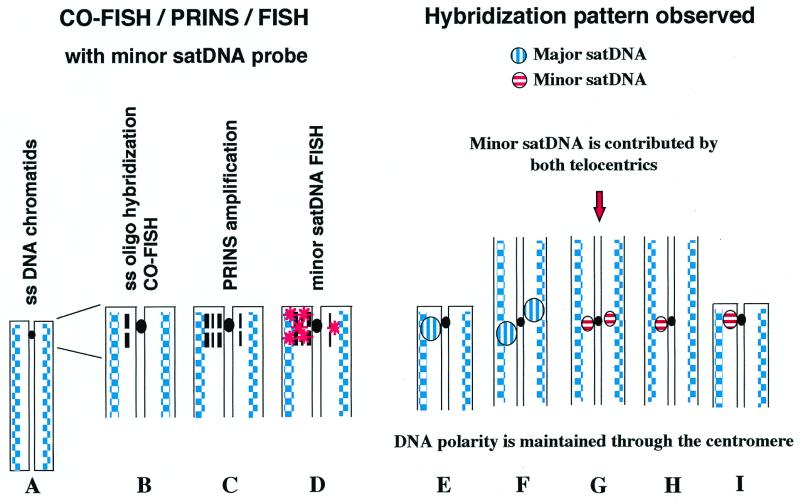
The experimental design used for the detection of the minor satDNA sequences on POS, F1 POS×C3H, and F1 CRE×C3H karyotypes entailed the sequential application of the CO-FISH/PRINS/FISH techniques and can be summarized as follows: (i) hybridization of the single-stranded chromosomes (Fig. 1A) with the sat-2 oligo (CO-FISH) (Fig. 1B); (ii) amplification of the double-stranded reconstituted region using the sat-2 and sat-3 oligos as primers (PRINS) (Fig. 1C); and (iii) in situ hybridization with the minor satDNA R198 probe (FISH) (Fig. 1D).
Figure 1.
Schematic representation of the CO-FISH/PRINS/FISH method as applied to mouse chromosomes. Single-stranded (ss) chromatids (A) were hybridized with a ss minor satDNA oligo (thick dash) (B). Sat-2 (thick dash) and sat-3 (thin dash) oligos then were used as primers for PRINS amplification (C) followed by conventional FISH with the R198 probe (stars) (D). Although the amplification steps involved both chromatids at a hybridization site, the efficiency of amplification was higher for the double-stranded chromatid reconstituted by ss oligo hybridization, so that a bright signal was detected only for that chromatid. For telocentric chromosomes, hybridization signals were always present on only one chromatid (I), whereas for Rb chromosomes hybridization signals were present either on both chromatids each side of the centromere (i.e., in contra-lateral disposition: G) or on only one of the two chromatids (H). A single signal on telocentrics (E) and two contra-lateral signals on Rb chromosomes (F) also were detected after CO-FISH with the major satDNA probe. These patterns of hybridization indicate that the tandem repeats of both major and minor satDNAs are oriented head-to-tail along the DNA strand. The contra-lateral disposition relative to the centromere of the hybridization signals in Rb chromosomes shows that the DNA polarity is maintained through the centromere. Both telocentrics contributed minor satDNA to the newly formed centromeric regions of Rb chromosomes. The centromeres are represented by black dots.
Chromosomes were counterstained with propidium iodide (0.3 μg/ml) or 4′-6-diamidino-2-phenylindole (DAPI, 0.05 μg/ml), mounted with an antifade and observed under a fluorescent microscope (Olympus, Provis) equipped with appropriate filters.
Results
In the CO-FISH procedure, cells are grown in the presence of BrdUrd for one round of replication so that sister chromatids are singly substituted. After culture, routine chromosome preparations are exposed to UV light and exonuclease III treatment to remove the newly synthesized strands. At the end of the procedure, chromatids are single-stranded, i.e., contain only a complementary prereplication DNA strand (Fig. 1A). Making use of a single-stranded oligonucleotide probe, only one chromatid will show a hybridization signal (Fig. 1B) if the tandem repeats are oriented head-to-tail along the DNA strand (19).
Visualization of the major satDNA hybridization was achieved both for telocentric and Rb chromosomes. For each animal, 10 metaphases on each of two slides were examined. All of the telocentrics and Rb chromosomes showed positive hybridization signals. Confirming previous results (21), telocentric chromosomes had a single spot located in the pericentromeric region of only one of the two chromatids (Figs. 1E and 2A), indicating that the tandem repeats are arranged with the same orientation along the DNA strand. Rb chromosomes showed two fluorescent bright spots in the pericentromeric regions, one on each chromosome arm but on different chromatids, i.e., the signals had a contra-lateral disposition relative to the centromere (Figs. 1F and 2A). This hybridization pattern suggests that the orientation of the tandem repeats is head-to-tail along the DNA strand in both arms of the Rb chromosomes. The orientation is such that the heads (an arbitrary designation) always point to the centromere. All of the different karyotypes examined, i.e., C3H, POS, CRE, F1 POS×C3H, and F1 CRE×C3H, showed the same hybridization results, with a single spot detected on the telocentric chromosomes and the two contra-lateral spots on the Rb chromosomes. We never detected two spots with lateral symmetry, i.e., on the same chromatid for both arms of an Rb chromosome.
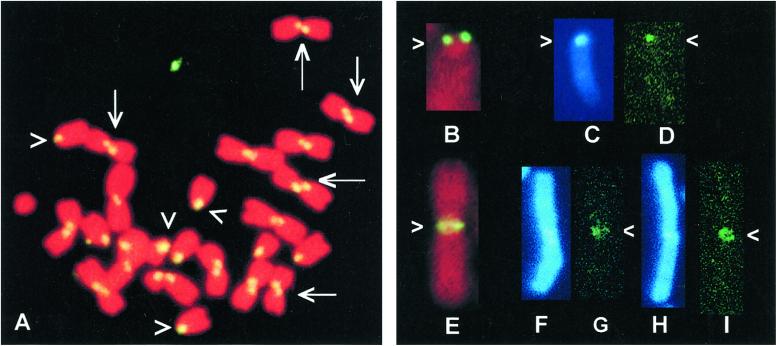
Figure 2.
(A) CO-FISH staining of a chromosome spread from a POS mouse using a major satellite oligonucleotide (sat-1). A contra-lateral signal was obtained for Rb chromosomes (examples: arrows) and a single lateral signal on telocentrics (examples: arrowheads). (B and E) Conventional FISH with the minor satDNA probe (R 198) on a telocentric (B) and an Rb chromosome (E): two bright spots are visible in the centromeric regions of both chromosomes. (D, G, and I) CO-FISH/PRINS/FISH staining of the minor satDNA in the centromeric regions of telocentric and Rb chromosomes. In telocentric chromosomes, hybridization signals were present on one chromatid only (D); in Rb chromosomes, hybridization signals were present either on one (G) or on both chromatids (I). (C, F, and H) DAPI (4′-6-diamidino-2-phenylindole) staining of chromosomes after the CO-FISH/PRINS/FISH procedure. The AT-rich heterochromatin located in the pericentromeric region of mouse chromosomes fluoresces brightly (arrow).
In contrast to conventional FISH (Fig. 2 B and E), CO-FISH with a minor satDNA probe gave negative results. Such a difference between the FISH and CO-FISH hybridization patterns are probably caused by the lower sensitivity of the CO-FISH procedure, which does not allow the detection of hybridization sites smaller than 50 kb (22). Both conventional amplification of hybridization signals through the addition of layers of antibodies and in situ PCR gave unsatisfactory results. For this reason, to detect the small amount of minor satDNA sequences at the centromeric regions of the Rb chromosomes (only 20–60 kb), we applied the PRINS technique followed by FISH for further amplification of the signal.
After the CO-FISH/PRINS/FISH procedure, the minor satDNA hybridization patterns were evaluated on at least 10 metaphases per slide. Two hybridization patterns were detected: (i) hybridization signals were present on one chromatid at the centromeres of telocentric (Figs. 1I and 2D) and Rb (Figs. 1H and 2G) chromosomes; (ii) hybridization signals were present on both chromatids in the centromeric regions of Rb chromosomes (Figs. 1G and 2I), the latter being the most frequent pattern of hybridization for Rb chromosomes. From over 5,300 chromosomes examined, for all of the karyotypes considered, only 5% of telocentrics (corresponding to 1.75 ± 0.34, mean ± standard deviation, chromosomes per metaphase) and 4% of Rb chromosomes (1.16 ± 0.26 chromosomes per metaphase) completely lacked a signal, whereas only 3.8% of the Rb chromosomes (1.08 ± 0.44 chromosomes per metaphase) showed a hybridization signal on only one chromatid at the centromere.
The visualization of a single lateral spot on telocentric chromosomes suggests the presence of a single minor satDNA block composed of tandem repeats oriented head-to-tail along the DNA strand, i.e., a similar organization within the satellite block to that recorded for the major satDNA.
For all of the different karyotypes examined, the presence of minor satDNA hybridization signals on both chromatids in Rb chromosomes (Fig. 2I) indicates that two blocks of minor satDNA are present in the centromeric regions and that they have a contra-lateral orientation relative to the centromere. The two hybridization signals were not as clearly visible in a contra-lateral orientation as was the case for the CO-FISH analysis of major satDNA, but appear on the same plane. This finding suggests that the two minor satDNA blocks are adjacent (Fig. 1G). In support of this interpretation, it is well known that in metaphase chromosomes two distinct signals can be optically resolved only when they are 1–3 megabases apart (23). The occurrence of two contra-lateral blocks of minor satDNA at the centromere of Rb chromosomes is expected if the breakpoints are in the minor satDNA of both telocentrics, with each contributing approximately equally to the newly formed Rb centromeric region (Fig. 1G).
Discussion
Pericentromeric regions involved in Rb translocations are composed of highly repetitive DNA families (satDNA). Illegitimate recombination between homologous sequences, such as satDNA, on nonhomologous chromosomes has been suggested as a possible mode of formation of Rb chromosomes in both house mice and humans (3, 9, 24, 25). In fact, more than 100 different Rb chromosomes have been found in the house mouse (7) involving all 19 autosomes and the X chromosome, which share the same pericentromeric satDNA. The Y chromosome, which does not share that satDNA, has never been found involved in Rb translocation. In humans, the most frequent Rb translocations occur between chromosomes 13 and 14 and chromosomes 14 and 21, which share homologous pericentromeric satDNA sequences (3, 25). Also, in the less-frequent Rb translocations, e.g., those between chromosomes 13 and 15 or chromosomes 14 and 22, interspersed homologous sequences may be involved (25, 26).
We determined the orientation of the two families of satDNA located in the pericentromeric regions of both telocentric and Rb metacentric chromosomes in the house mouse. Both satDNA families were found to be organized head-to-tail, because all of the blocks of major and minor satDNA showed the same orientation pointing to the centromere. When Rb translocations occur, the overall DNA polarity is maintained through the centromere in Rb chromosomes, i.e., from one telocentric through the joining point of the other telocentric, as shown by the contra-lateral symmetry of the hybridization signals (Fig. 2). Previous results obtained by BrdUrd quenching of Hoechst 33258 fluorescence in Rb chromosomes present in the mouse L-cell line (17) and in the POS wild-derived mice (15) indicated that the major satDNA maintained consistently the same polarity across the centromeres for all Rb chromosomes. Our findings obtained from POS and CRE-wild-derived mice, directly visualizing the two families of satDNA, demonstrate that in mouse chromosomes both major and minor satDNA sequences have exactly the same polarity relative to centromeres and telomeres. These results are strongly supported by the use of CRE mice, which possess the highest number of Rb chromosomes found in natural populations. Therefore, both families of satDNAs apparently are maintained with exactly the same polarity relative to the centromere and telomeres, a disposition that precludes U-type exchanges between homologous satellite sequences that lead to losses of large chromosome segments (28). If terminal satellite blocks had opposite centric orientations, exchange would have produced acentric chromosomes. The finding that both satDNA families present in the pericentromeric regions of mouse chromosomes share this polarity constraint corroborates the existence of a base-pairing dependent mechanism of interchromosomal exchange between satellite blocks (27). The implication is that all tandemly repetitive DNAs within a species probably are locked into this symmetry constraint. Our results on satDNA orientation suggest that Rb translocation occurs between DNA strands ending 3′ of one telocentric and 5′ of another telocentric to generate an Rb metacentric chromosome with an antiparallel symmetry at the fusion point. The maintenance of this symmetry across the centromere in Rb chromosomes might permit whole arm reciprocal translocations (WARTs) between nonhomologous Rb chromosomes, accelerating chromosomal evolution in the house mouse. In this regard, WART chromosomes have been described in a number of feral house mouse populations (29–32).
The CO-FISH/PRINS/FISH results indicate that the two telocentrics contribute minor satDNA symmetrically to the centromeric region of the newly formed Rb chromosome. The capability to detect the two minor satDNA blocks, contributed by both telocentrics, shows that the CO-FISH/PRINS/FISH method has a sensitivity of 10–30 kb, 2–5 times higher than the previously reported limit of 50 kb for the CO-FISH technique alone (22).
In the house mouse, the very high frequency of Rb chromosomes, and the random involvement of the telocentrics in the translocation process, could be caused by “inherent genomic traits” (33) such as the clustering of heterochromatic regions (34–36), the homology of satDNA sequences shared by the telocentric chromosomes (37), and the nicking activity of the CENP-B protein. CENP-B protein binds the CENP-B box present within the minor satDNA in mice as well as within centromeric satDNA of other species (10, 38–40) and could play an important role in recombination events leading to Rb translocation. Kipling and Warburton (41) suggested that the dimerization ability of the CENP-B protein could not only promote misalignment between higher-order repeat units of alpha satDNA on nonhomologous chromosomes in humans, but also could facilitate recombination, mediated by its nicking activity. In this context, the minor satDNA–CENP-B protein complex appears to be the precise molecular substrate for Rb translocations in house mice. In support of this view all of the breakpoints were found within minor satDNA.
The re-examination with the CO-FISH and CO-FISH/PRINS/FISH techniques of Rb chromosomes in species other than the house mouse would be most desirable to establish the generality of our findings.
Acknowledgments
We are deeply indebted to Drs. Gerald Holmquist, Helmut Zacharias, and Mels Van der Ploeg for their valuable suggestions and Dr. Ton Raap for comments on the technique. The work was supported by The Italian MURST (Progetti di Ricerca Scientifica di Rilevante Interesse Nazionale), Fondazione Cassa di Risparmio delle Provincie Lombarde per la Ricerca Scientifica, Olympus Foundation Science for Life, and Telethon Fondazione Onlus (Grant A132).
Abbreviations
- Rb
Robertsonian
- satDNA
satellite DNA
- FISH
fluorescent in situ hybridization
- CO-FISH
chromosome orientation FISH
- PRINS
primed in situ labeling
- POS
Poschiavo race
- CRE
Cremona race
References
- 1.King M. Species Evolution: The Role of Chromosome Change. Cambridge, U.K.: Cambridge Univ. Press; 1993. [Google Scholar]
- 2.Searle J B. In: Hybrid Zones and the Evolutionary Process. Harrison R G, editor. New York: Oxford Univ. Press; 1993. pp. 309–353. [Google Scholar]
- 3.Therman E, Susman B, Denniston C. Ann Hum Genet. 1989;53:49–65. doi: 10.1111/j.1469-1809.1989.tb01121.x. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen J, Wohlert M. Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 5.Dutrillaux B. Hum Genet. 1979;48:251–314. doi: 10.1007/BF00272830. [DOI] [PubMed] [Google Scholar]
- 6.Nevo E, Filippucci M G, Redi C A, Korol A, Beilis A. Proc Natl Acad Sci USA. 1994;91:8160–8164. doi: 10.1073/pnas.91.17.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nachman M W, Searle J B. Trends Ecol Evol. 1995;10:397–402. doi: 10.1016/s0169-5347(00)89155-7. [DOI] [PubMed] [Google Scholar]
- 8.Daniel A. The Cytogenetics of Mammalian Autosomal Rearrangements. New York: Liss; 1988. [Google Scholar]
- 9.Vissel B, Choo K H A. Genomics. 1989;5:407–414. doi: 10.1016/0888-7543(89)90003-7. [DOI] [PubMed] [Google Scholar]
- 10.Joseph A, Mitchell A R, Miller O J. Exp Cell Res. 1989;183:494–500. doi: 10.1016/0014-4827(89)90408-4. [DOI] [PubMed] [Google Scholar]
- 11.Narayanswami S, Dogget N A, Clark L M, Hildebrand C E, Weier H U, Hamkalo B A. Mamm Genome. 1992;2:186–194. doi: 10.1007/BF00302876. [DOI] [PubMed] [Google Scholar]
- 12.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 13.Garagna S, Broccoli D, Redi C A, Searle J B, Cooke H J, Capanna E. Chromosoma. 1995;103:685–692. doi: 10.1007/BF00344229. [DOI] [PubMed] [Google Scholar]
- 14.Nanda I, Schneider-Rasp S, Winking H, Schmid M. Chromosome Res. 1995;3:399–409. doi: 10.1007/BF00713889. [DOI] [PubMed] [Google Scholar]
- 15.Lin M S, Davidson R L. Science. 1974;185:1179–1181. doi: 10.1126/science.185.4157.1179. [DOI] [PubMed] [Google Scholar]
- 16.Lin M S, Davidson R L. Nature (London) 1975;254:354–356. doi: 10.1038/254354a0. [DOI] [PubMed] [Google Scholar]
- 17.Holmquist G P, Comings D E. Chromosoma. 1975;52:245–259. doi: 10.1007/BF00332114. [DOI] [PubMed] [Google Scholar]
- 18.Gropp A, Winking H, Redi C A, Capanna E, Britton-Davidian J, Noack G. Cytogenet Cell Genet. 1982;3:67–77. doi: 10.1159/000131794. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin E, Meyne J. Cytogenet Cell Genet. 1993;63:126–127. doi: 10.1159/000133516. [DOI] [PubMed] [Google Scholar]
- 20.Koch J E, Kolvraa S, Petersen K B, Gregersen N, Bolund L. Chromosoma. 1989;98:259–265. doi: 10.1007/BF00327311. [DOI] [PubMed] [Google Scholar]
- 21.Goodwin E, Meyne J, Bailey S M, Quigley D. Chromosoma. 1996;104:345–347. doi: 10.1007/BF00337223. [DOI] [PubMed] [Google Scholar]
- 22.Meyne J, Goodwin E H, Moyzis R K. Chromosoma. 1994;103:99–103. doi: 10.1007/BF00352318. [DOI] [PubMed] [Google Scholar]
- 23.Wiegant J, Kalle W, Mullenders L, Brookes S, Hoovers J M N, Dauwerse J G, van Ommen G J, Raap A K. Hum Mol Genet. 1992;1:587–591. doi: 10.1093/hmg/1.8.587. [DOI] [PubMed] [Google Scholar]
- 24.Redi C A, Garagna S, Capanna E. J Mol Evol. 1990;3:133–137. [Google Scholar]
- 25.Page S L, Shin J C, Han J Y, Choo K H A, Shaffer L G. Hum Mol Genet. 1996;5:1279–1288. doi: 10.1093/hmg/5.9.1279. [DOI] [PubMed] [Google Scholar]
- 26.Gravholt C H, Friedrich U, Caprani M, Jorgensen A L. Genomics. 1992;14:924–930. doi: 10.1016/s0888-7543(05)80113-2. [DOI] [PubMed] [Google Scholar]
- 27.Smith G P. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 28.Holmquist G P, Dancis B. Proc Natl Acad Sci USA. 1979;76:4566–4570. doi: 10.1073/pnas.76.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capanna E, Redi C A. Chromosome Res. 1995;3:135–137. doi: 10.1007/BF00710676. [DOI] [PubMed] [Google Scholar]
- 30.Garagna S, Zuccotti M, Redi C A, Capanna E. Nature (London) 1997;390:241–242. doi: 10.1038/36760. [DOI] [PubMed] [Google Scholar]
- 31.Hauffe H C, Piálek J. Biol J Linn Soc. 1997;62:255–278. [Google Scholar]
- 32.Castiglia R, Capanna E. Chromosome Res. 1999;7:493–495. doi: 10.1023/a:1009206131364. [DOI] [PubMed] [Google Scholar]
- 33.Redi C A, Garagna S, Zuccotti M. Biol J Linn Soc. 1990;41:235–255. [Google Scholar]
- 34.Manuelidis L. In: Genome Evolution. Dover G A, Flavell R B, editors. London: Academic; 1982. pp. 263–285. [Google Scholar]
- 35.Rattner J B, Lin C C. Chromosoma. 1985;92:325–329. doi: 10.1007/BF00327462. [DOI] [PubMed] [Google Scholar]
- 36.Cerda M C, Berrios S, Fernandez-Donoso R, Garagna S, Redi C A. Biol Cell. 1999;91:55–65. [PubMed] [Google Scholar]
- 37.Redi C A, Garagna S, Della Valle G, Bottiroli G, Dell'Orto P, Viale G, Peverali F, Raimondi E, Forejt J. Chromosoma. 1990;99:11–17. doi: 10.1007/BF01737284. [DOI] [PubMed] [Google Scholar]
- 38.Masumoto H, Masukata H, Muro Y, Nosaki N, Okasaki T. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volobouev V, Vogt N, Viegas-Pequignot E, Malfoy B, Dutrillaux B. Chromosoma. 1995;104:252–259. doi: 10.1007/BF00352256. [DOI] [PubMed] [Google Scholar]
- 40.Haff T, Mater A G, Wienberg J, Ward D C. J Mol Evol. 1995;41:487–491. doi: 10.1007/BF00160320. [DOI] [PubMed] [Google Scholar]
- 41.Kipling D, Warburton P E. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]