Abstract
Interspecific chromosome substitution is among the most powerful means of introgression and steps toward quantitative trait locus (QTL) identification. By reducing the genetic “noise” from other chromosomes, it greatly empowers the detection of genetic effects by specific chromosomes on quantitative traits. Here, we report on such results for 14 cotton lines (CS-B) with specific chromosomes or chromosome arms from G. barbadense L. substituted into G. hirsutum and chromosome-specific F2 families. Boll size, lint percentage, micronaire, 2.5% span length, elongation, strength, and yield were measured by replicated field experiments in five diverse environments and analyzed under an additive–dominance (AD) genetic model with genotype and environment interaction. Additive effects were significant for all traits and dominance effects were significant for all traits except 2.5% span length. CS-B25 had additive effects increasing fiber strength and fiber length and decreasing micronaire. CS-B16 and CS-B18 had additive effects related to reduced yields. The results point toward specific chromosomes of G. barbadense 3-79 as the probable locations of the genes that significantly affect quantitative traits of importance. Our results provided a scope to analyze individual chromosomes of the genome in homozygous and heterozygous conditions and thus detected novel effects of alleles controlling important QTL.
UPLAND cotton (Gossypium hirsutum L., 2n = 52) is the most extensively cultivated of the four cultivated Gossypium species, and, as such, it has been the target of numerous genetic studies and breeding efforts. The level of genetic diversity is low in G. hirsutum, especially among agriculturally elite types, as revealed by all means of assessment (Gutiérrez et al. 2002; Ulloa and Meredith 2000; Wendel et al. 1989). Increasing diversity is therefore essential to genetic improvement efforts. Each of the three major approaches to increasing genetic diversity—mutagenesis, germplasm introgression, and transformation—has advantages and disadvantages. Interspecific germplasm introgression is particularly attractive in that it utilizes a broad germplasm base, can be targeted to one or more specific traits or genes or modulated to include thousands of genes or even entire genomes, and is readily coupled to marker-assisted genome analysis and selection. However, the biological and technical challenges of introgression increase as the phyletic distance between the donor and recipient genome increases. Not surprisingly, the amount of time typically needed for interspecific introgression considerably exceeds that for conventional varietal breeding. Long-term efforts for interspecific introgression are very challenging, especially when aimed at concomitant discovery and introgression of genetic variation that can be used to enhance multigenic traits of economic importance, for most of which cultivated species typically perform markedly better than their wild relatives, at least under conditions of cultivation. Although introgression of genes across species boundaries is difficult, it is quite desirable because the gene pools of cultivated species do not contain all of the desired alleles. Tanksley and McCouch (1997) have documented that beneficial alleles exist cryptically in wild germplasm. Such studies increase the awareness of the benefits from interspecific introgression projects that target essentially any multigenic trait, irrespective of the performance of the donor species.
The Gossypium germplasm most amenable to interspecific introgression into Upland cotton includes the four other 52-chromosome tetraploid species, all of which are thought to have arisen from a common ancient polyploidy event. Cytological observation of hybrids and comparative genome mapping indicate that synteny and colinearity are largely conserved among the five tetraploid species, suggesting that there are few cytostructural barriers to interspecific introgression among them, although genetic limitations exist, manifested by the significant level of F2 breakdown that is commonly observed in interspecific crosses between these species (Beasley and Brown 1942; Reinisch et al. 1994). Conventional breeding efforts aimed at interspecific introgression have transferred specific genes and useful traits, including stronger fibers, longer fibers, finer fibers, and resistance to drought (Mergeai 2003), but in general have culminated in an array of introgression products that is far less desirable relative to cultivars and more diverse than desired (Paterson and Smith 1999).
G. barbadense (L.) is the only 52-chromosome relative of Upland cotton (G. hirsutum, 2n = 52) that is cultivated. It is valued for its fiber length and quality, whereas Upland cotton is more valued for its high yield. While these species are hybridized easily, conventional backcrossing and/or inbreeding quickly lead to extensive germplasm loss (Stephens 1949). One of the challenges in interspecific introgression is to use valuable alien traits or genes of G. barbadense germplasm, such as fiber length, fineness, and strength for improvement of Upland cotton (Lacape et al. 2005). Traditional plant breeding approaches to improve fiber quality through interspecific introgression have been hindered by complex antagonistic genetic relationships (Culp et al. 1979). Attempts to incorporate genes from G. barbadense into Upland cotton have generally not achieved stable introgression of the G. barbadense fiber properties (Stephens 1949; McKenzie 1970). Associated with these attempts at introgression have been poor agronomic qualities of the progeny, distorted segregation, sterility, mote formation, and limited recombination due to incompatibility between the genomes (Reinisch et al. 1994).
Stagnant yield, declining fiber quality, and threats from biotic and abiotic stresses affect profitability of cotton production worldwide. The paucity of information about genes that control important traits and the need for more extensive usage of diverse germplasm impede the genetic improvement of cotton. The high value per hectare of cotton and global textile market demand for increased fiber uniformity, strength, extensibility, and quality clearly justify the importance of new and innovative approaches toward evaluating and understanding genetic mechanisms of fiber qualities. Recent molecular mapping studies associated G. barbadense chromosomes with many favorable QTL affecting fiber and agronomic traits (Rong et al. 2004; Lacape et al. 2005).
Chromosome substitution has been an indispensable method of interspecific germplasm introgression into bread wheat (Triticum aestivum, 2n = 42) for genetic analysis and breeding (Law 1966; Al-Quadhy et al. 1988; Berke et al. 1992a,b; Friebe et al. 2000). Prior to introgression, fertile or partially fertile isogenic hypoaneuploids were derived and identified in bread wheat. Germplasm introgression was then commenced by crossing each type of hypoaneuploid with the donor and then recurrently backcrossing the hypoaneuploid progeny to the respective bread wheat hypoaneuploid. Hemizygous hyppoaneuploid hybrids were recovered at each generation (F1, BC1F1, BC2F1,…, BCnF1), and self-progeny screened at the terminal backcross generation to identify disomic substitution lines. The process requires plant fertility and transmission of the hypoaneuploid condition via a nullisomic gamete, both of which are favored by the highly homozygous nature and disomic hexaploid constitution of bread wheat. This approach has been extended with various modifications to several species. Methods for development of interspecific chromosome substitution in G. hirsutum were outlined by Endrizzi (1963), and several of the initially discovered G. hirsutum monosomics were used to substitute G. barbadense chromosomes into G. hirsutum (White et al. 1967; Kohel et al. 1977; Endrizzi et al. 1984). To date, there have been no formal reports on long-term efforts to enhance productivity in Upland cultivars using interspecific chromosome substitution lines.
We have developed 14 disomic alien chromosome substitution (CS)-B lines through hypoaneuploid-based backcross chromosome substitution, using as recurrent parents previously developed monosomic or monotelodisomic near-isogenic backcross derivatives of TM-1. In each CS-B line, a pair of chromosomes (or chromosome arms) of G. hirsutum inbred TM-1 was replaced by the respective pair from G. barbadense doubled-haploid line 3-79 lines (Saha et al. 2004b). These substitution lines are nearly isogenic to the common parent TM-1 for 25 chromosome pairs, as well as to each other, for 24 chromosome pairs. Given n = 26, each replaces ∼4% of the genome with a G. barbadense homeolog, so they collectively enable dissection of quantitative traits of interest. They also can be used to determine additive or epistatic genetic effects by comparative analysis, because the parental lines are homozygous (Yen et al. 1997; Saha et al. 2004a,b). To detect dominant or epistatic effects in wheat, hybrids or their segregating progenies of chromosome substitution lines were studied (Yen et al. 1997). Comparable analyses in cotton using the CS-B lines would be of potential use in cotton breeding programs.
In this study, we used F2 hybrids to help search for chromosome-specific effects on key agronomic and fiber traits. We created a set of unique chromosome-specific F2 hybrids from each of the 14 CS-B lines crossed with TM-1 and evaluated the hybrids and their parents in replicated multilocation experiments. Due to heterozygosity specific to a chromosome, a somewhat expanded set of possible genetic effects arises when substitution lines are backcrossed to the donor type(s); these include single-locus (dominance and overdominance) as well as intrachromosomal interlocus (epistatic) effects. The additive–dominance (AD) genetic model helped us to dissect individual chromosomes and detect loci controlling agronomic and fiber traits.
MATERIALS AND METHODS
Experimental materials:
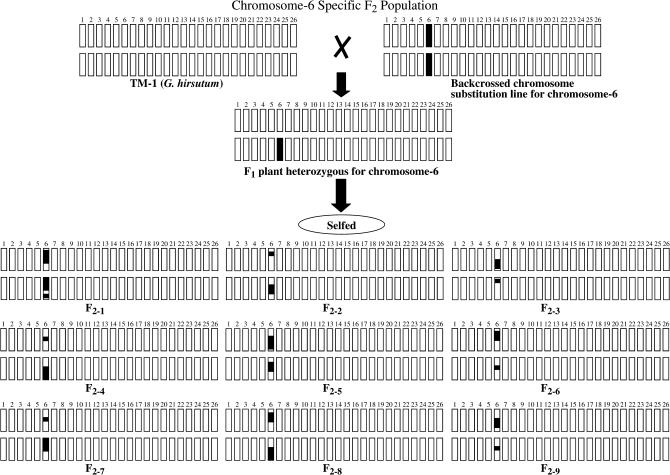
Fourteen near-isogenic BC5S1 chromosome substitution lines containing different pairs of G. barbadense chromosomes or segments (CS-B lines) were used as male parents and crossed to a common parent, TM-1 (G. hirsutum). In each CS-B line, a single chromosome pair of TM-1 had been replaced by the corresponding part of the 3-79 (G. barbadense) genome. BC5S1 seeds from euploid (2n = 52) CS-B lines specific to 13 different chromosomes were used in crossing with TM-1 to develop chromosome-specific F2 hybrids (Figure 1). These CS-B lines were listed with a number specific to the introgressed chromosome or chromosome arm of the alien species as follows: CS-B02, CS-B04, CS-B06, CS-B07, CS-B16, CS-B17, CS-B18, CSB-25, CS-B05sh (sh, short arm), CS-B14sh, CS-B15sh, CS-B22sh, CS-B22Lo (Lo, long arm), and CS-B26Lo. The development of these CS-B lines was described in previous studies (Saha et al. 2004a,b; Stelly et al. 2004, 2005). TM-1 is an inbred line derived from the commercial variety Deltapine 14 and maintained over 40 generations by selfing, as described by Kohel et al. (2001). Although line 3-79 is far from cultivar standards for productivity, it originated as a doubled haploid from G. barbadense germplasm and has higher fiber qualities than modern Pima germplasm. It is also distinctly different in agronomic and fiber traits from the other parent TM-1. These crosses were made at Mississippi State in the summer of 2000. F1 plants were grown at a winter nursery in Tecoman, Mexico to produce F2 hybrid seeds. Phenotypic effects of each substituted chromosome were determined by comparative analysis of TM-1 and all parental CS-B lines except CS-B26Lo and bulk analysis of all chromosome-specific F2 hybrids.
Figure 1.
Diagram of chromosome-specific F2 family, e.g., for CS-B06.
The 14 F2 hybrid families and TM-1 inbred were planted in a randomized complete block design with four replications at three locations, Mississippi (MS), New Mexico (NM), and Arizona (AZ) in 2001. The same 14 F2 hybrids, 13 different parental CS-B lines (except CS-B26Lo, due to shortage of sufficient seeds), and TM-1 were grown at MS and AZ with the same experimental design in 2002. Standard practices were followed in the growing season for all five environments.
A 25-boll hand-harvested seedcotton sample was collected from each plot prior to machine picking. These samples were weighed to determine boll size and ginned on a laboratory 10-saw gin to determine lint percentages and to provide lint samples for fiber analyses. Fiber samples were sent to STARLAB (Knoxville, TN) for determination of micronaire (MIC), 2.5% span length (SL2.5), elongation (E1), and fiber strength (T1) using single instrument measurements. Each plot was machine harvested, and the seedcotton was weighed. Lint yield was determined by multiplying seedcotton weight by lint percentage. Direct comparisons among the lines and TM-1 were used to detect effects of the substitutions on important characteristics.
Genetic models and statistical methods:
An AD genetic model with G × E interaction was used for our data analysis (Zhu 1994, 1998; Tang et al. 1996). This genetic model is based on the following two genetic assumptions: (1) normal disomic segregation and (2) dominance effects (interaction effects between alleles at each locus). The genetic model for parent i at environment h is expressed as follows: 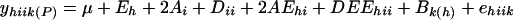 . The genetic model for an F2 between parents i and j at environment h is expressed as follows:
. The genetic model for an F2 between parents i and j at environment h is expressed as follows: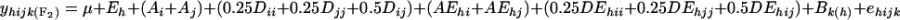 , where μ is the population mean; Eh is the environmental effect; Ai and Aj are the additive effects; Dii, Djj, and Dij are the dominance effects; AEhi and AEhj are the additive-by-environment interaction effects; DEhii, DEhjj, and DEhij are the dominance-by-environment interaction effects; Bk(h) is the block effect; and ehijk is the random error.
, where μ is the population mean; Eh is the environmental effect; Ai and Aj are the additive effects; Dii, Djj, and Dij are the dominance effects; AEhi and AEhj are the additive-by-environment interaction effects; DEhii, DEhjj, and DEhij are the dominance-by-environment interaction effects; Bk(h) is the block effect; and ehijk is the random error.
In this study, the data obtained in 2001 and 2002 were unbalanced and some coefficients for genetic effects were fractions rather than 0 and 1; thus, analysis of variance (ANOVA) and general linear model (GLM) approaches were not appropriate. The purposes of our study were to calculate the genetic variances and genetic effects for each genetic component; thus, we considered μ and Eh as fixed and the remaining effects as random. A mixed linear model, minimum norm quadratic unbiased estimation with an initial value of 1.0 [MINQUE (1)], was used to estimate the variance components (Zhu 1989). Genetic effects were predicted by the adjusted unbiased prediction (AUP) approach (Zhu 1993). Standard errors of variance components and genetic effects were estimated by jackknife resampling over one replication within each environment (Miller 1974). An approximate one-tailed t-test (d.f. = 19) was used to detect the significance of variance components and a two-tailed t-test was used to detect the significance of genetic effects. By this approach, the predicted genetic effects were deviations from the respective population mean μ, not from TM-1, so the significance of differences of chromosome-specific effects of 3-79 and TM-1 chromosomes was detected indirectly, not directly. Since each CS-B line is near isogenic to the recurrent parent TM-1 except for the substituted chromosome pair, the difference in additive effects between a CS-B line and TM-1 can be considered as the substituted chromosome additive effect deviations from TM-1. The differences of dominance effect between chromosomes of 3-79 × 3-79 (homozygous dominance effect, Dii) or of 3-79 × TM-1 (heterozygous dominance effect, Dij) and that of TM-1 × TM-1 can be considered the substituted chromosome dominance deviations from TM-1. A confidence-interval test (95%) was utilized to detect the significance of genetic effects between genotypes.
RESULTS
Mean comparisons for chromosome-specific F2 hybrids and parental lines:
The donor line, 3-79, produced less seedcotton (38%) and lint yield (39%), similar lint percentage, smaller bolls (60%), lower micronaire (3.65 vs. 4.85), similar elongation (101%), and fiber that was longer (121%) and stronger (140%) fiber than that of the recurrent parent TM-1 (Table 1). Eight of the parental CS-B lines yielded less seedcotton and lint than TM-1, but five of them yielded more seedcotton than TM-1 and one (CS-B06) exceeded that of TM-1 by 10%. Five CS-B lines had greater lint percentage than TM-1. No CS-B lines had greater boll size than TM-1. Seven CS-B lines had finer fibers than TM-1. Four CS-B lines had longer 2.5% span length than TM-1. Seven and eight CS-B lines had greater fiber elongation and strength than TM-1, respectively. All F2 hybrids (Table 2) produced less seed yield than TM-1 (Table 1). All F2 hybrids yielded less seedcotton and lint than their respective CS-B lines, except CS-B17, CS-B14sh, and CS-B22sh. The CS-B25 F2 yielded less seedcotton but more lint than CS-B25. Most F2 hybrids had greater boll size than their respective CS-B parents. Micronaire values of the CS-B F2 hybrids were close (±0.2) to that of TM-1 (4.85), except that of CS-B25 (4.40).
TABLE 1.
Phenotypic mean values for agronomic and fiber traits
| Parental lines | Seedcotton yield (kg/ha) | Lint yield (kg/ha) | Lint (%) | Boll weight (g) | Micronaire | 2.5% span length (mm) | Elongation (%) | Strength (g/tex) |
|---|---|---|---|---|---|---|---|---|
| CS-B02 | 2521 | 836 | 32.77 | 5.43 | 5.16 | 29.04 | 6.65 | 20.95 |
| CS-B04 | 2662 | 854 | 31.38 | 5.47 | 4.70 | 29.63 | 8.97 | 19.45 |
| CS-B06 | 2766 | 906 | 32.56 | 5.58 | 4.83 | 28.81 | 7.90 | 19.72 |
| CS-B07 | 2571 | 831 | 31.87 | 5.26 | 5.01 | 29.61 | 8.23 | 19.68 |
| CS-B16 | 1453 | 523 | 35.27 | 5.09 | 4.80 | 28.48 | 7.62 | 18.63 |
| CS-B17 | 2312 | 661 | 28.37 | 4.83 | 4.22 | 28.39 | 9.16 | 19.74 |
| CS-B18 | 1262 | 435 | 33.80 | 4.59 | 5.14 | 28.97 | 8.34 | 20.21 |
| CS-B25 | 2352 | 705 | 29.69 | 4.82 | 3.81 | 30.45 | 7.06 | 22.30 |
| CS-B05sh | 2387 | 811 | 33.88 | 4.79 | 5.12 | 28.52 | 7.73 | 18.49 |
| CS-B14sh | 1952 | 620 | 31.43 | 4.31 | 4.69 | 30.55 | 7.27 | 20.49 |
| CS-B15sh | 2699 | 848 | 31.24 | 5.34 | 4.75 | 29.97 | 7.95 | 20.42 |
| CS-B22sh | 1826 | 673 | 36.61 | 4.95 | 5.28 | 27.21 | 6.81 | 19.26 |
| CS-B22Lo | 2413 | 911 | 37.57 | 4.33 | 5.38 | 28.26 | 7.15 | 19.75 |
| TM-1 | 2507 | 824 | 33.00 | 5.79 | 4.85 | 29.35 | 7.70 | 19.69 |
| 3-79 | 963 | 320 | 32.88 | 3.48 | 3.65 | 35.44 | 7.78 | 27.61 |
| LSD (P = 0.05) | 267.7 | 87.8 | 0.54 | 0.21 | 0.15 | 0.52 | 0.42 | 0.71 |
CS-B02, chromosome 2 substituted from 3-79 in TM-1; CS-B05sh, substituted chromosome 5 short arm from 3-79 in TM-1; CS-B22Lo, substituted chromosome 22 long arm from 3-79 in TM-1.
Means of CS-B lines were based on two environments in 2002; Means of F2 and TM-1 were based on five environments in 2001 and 2002.
TABLE 2.
Phenotypic mean values for agronomic and fiber traits in F2 lines developed from crosses of the common parent (TM-1) with the other parental lines (column 1)
| Parent of F2 lines | Seedcotton yield (kg/ha) | Lint yield (kg/ha) | Lint (%) | Boll weight (g) | Micronaire | 2.5% span length (mm) | Elongation (%) | Strength (g/tex) |
|---|---|---|---|---|---|---|---|---|
| CS-B02 | 2194 | 740 | 33.78 | 5.34 | 4.88 | 29.66 | 7.37 | 20.26 |
| CS-B04 | 2293 | 751 | 32.92 | 5.46 | 4.77 | 29.85 | 8.21 | 19.46 |
| CS-B06 | 2271 | 730 | 32.26 | 5.67 | 4.79 | 29.57 | 7.15 | 19.80 |
| CS-B07 | 2176 | 713 | 32.82 | 5.56 | 5.02 | 29.24 | 7.50 | 19.70 |
| CS-B16 | 2046 | 668 | 32.57 | 5.66 | 4.72 | 29.16 | 7.67 | 20.13 |
| CS-B17 | 2465 | 787 | 32.09 | 5.80 | 4.69 | 29.01 | 8.19 | 19.98 |
| CS-B18 | 1878 | 644 | 34.22 | 5.25 | 4.73 | 29.54 | 7.89 | 19.58 |
| CS-B25 | 2410 | 777 | 32.48 | 5.31 | 4.40 | 30.11 | 7.39 | 21.20 |
| CS-B05sh | 2165 | 692 | 31.96 | 5.31 | 4.87 | 29.28 | 8.31 | 19.49 |
| CS-B14sh | 1978 | 659 | 33.21 | 4.84 | 4.80 | 30.08 | 7.41 | 19.73 |
| CS-B15sh | 2333 | 764 | 32.85 | 5.73 | 4.80 | 29.77 | 7.99 | 19.74 |
| CS-B22sh | 2153 | 744 | 34.52 | 5.24 | 4.94 | 28.68 | 8.16 | 19.42 |
| CS-B22Lo | 2143 | 743 | 34.65 | 4.97 | 4.99 | 28.90 | 7.11 | 19.76 |
| CS-B26Lo | 2112 | 689 | 32.69 | 5.43 | 4.75 | 29.92 | 7.70 | 19.64 |
| 3-79a | 728 | 220 | 29.96 | 3.15 | 3.51 | 32.21 | 8.05 | 24.29 |
| LSD (P = 0.05) | 267.7 | 87.8 | 0.54 | 0.21 | 0.15 | 0.52 | 0.42 | 0.71 |
Means of CS-B lines were based on two environments in 2002; Means of F2 and TM-1 were based on five environments in 2001 and 2002.
Conventional F2 hybrid from the cross of TM-1 and 3-79.
Variance components:
Residual effects accounted for just 7.8% (lint percentage, LP) to 34.9% (T1) of the phenotypic variance for the traits measured, indicating that genotypic effects and genotype-by-environment interaction effects were accurately detected (Table 3). All agronomic and fiber traits were significantly affected by additive effects ranging from 7.7 to 33%, and, except length, by dominance effects ranging from 8.9 to 61.0%. The relative importance of simple additive vs. dominance effects varied widely. Fiber length was subject to no dominance effects. Most simple genetic effects were due to additive effects for lint percentage and elongation, ∼4:1 additive:dominance ratio. Additive effects moderately exceeded dominance effects for boll weight (1.38:1) and fiber strength (1.18:1). In contrast, simple dominance effects strongly exceeded simple additive effects by ∼4-fold for lint percentage and fiber elongation and were moderately stronger (∼1.25- to 1.5-fold) for lint yield and boll weight. Additive G × E interaction effects ranged from 16.3% (lint percentage) to 55.3% (seedcotton yield) of overall phenotypic variance, indicating that the additive genetic effects were also environmentally dependent. Large dominance-by-environment interaction effects (D × E) were detected for seedcotton and lint yields, lint percentage, and all fiber traits, but not for boll weight.
TABLE 3.
Estimated proportions of variance components to the phenotypic variance for agronomic and fiber traits
| Parameter | Seedcotton yield | Lint yield | Lint (%) | Boll weight | Micronaire | 2.5% span length | Elongation | Strength |
|---|---|---|---|---|---|---|---|---|
| VA/VP | 0.145** | 0.106** | 0.149** | 0.237** | 0.289** | 0.330** | 0.077* | 0.230** |
| VD/VP | 0.089* | 0.135** | 0.610** | 0.325** | 0.209** | 0.000 | 0.341** | 0.195** |
| VAE/VP | 0.000 | 0.000 | 0.048* | 0.205** | 0.000 | 0.000 | 0.000 | 0.000 |
| VDE/VP | 0.553** | 0.548** | 0.115** | 0.000 | 0.299** | 0.444** | 0.340** | 0.226** |
| Ve/VP | 0.214** | 0.210** | 0.078** | 0.233** | 0.203** | 0.226** | 0.241** | 0.349** |
Variance components are significant at *P ≤ 0.05 and **P ≤ 0.01, respectively.
Chromosome effect:
Additive effects were calculated for each CS-B line and TM-1 (Table 4). Interaction effects between the same allele vs. between different alleles at a locus were calculated as homozygous and heterozygous dominance effects, respectively, by comparative analysis of TM-1 and parental CS-B lines and bulk analysis of the CS-B × TM-1 F2 population. Differences among the lines were attributed to the substituted chromosome according to the simplifying assumption of isogenicity of nonsubstituted chromosomes (Tables 5 and 6). The deviation of homozygous dominance effects of a CS-B line from TM-1 measures the difference of interaction effects between the substituted chromosome pair and the interaction effects between the respective pair of TM-1 chromosomes. The deviation of heterozygous dominance effects of the substituted chromosome (chromosome interaction effects in bulk F2) from TM-1 measures the difference of the interaction effects between the 3-79 and TM-1 alleles, from the interaction effects of the TM1 alleles on the same chromosome.
TABLE 4.
Additive effects for agronomic and fiber traits
| CS-B line | Seedcotton yield (kg/ha) | Lint yield (kg/ha) | Lint (%) | Boll weight (g) | Micronaire | 2.5% span length (nm) | Elongation | Strength (g/tex) |
|---|---|---|---|---|---|---|---|---|
| CS-B02 | 74.89 ± 34.29* | 31.48 ± 10.10* | 0.21 ± 0.08* | 0.05 ± 0.04 | 0.12 ± 0.04* | 0.13 ± 0.10 | −0.26 ± 0.13 | 0.54 ± 0.17* |
| CS-B04 | 93.90 ± 48.29 | 25.55 ± 14.42 | −0.12 ± 0.12 | 0.10 ± 0.04* | −0.05 ± 0.04 | 0.29 ± 0.12* | 0.25 ± 0.13 | −0.28 ± 0.13* |
| CS-B06 | 209.09 ± 58.29* | 56.04 ± 19.00* | −0.18 ± 0.05* | 0.21 ± 0.05* | −0.02 ± 0.03 | −0.06 ± 0.13 | −0.17 ± 0.08* | −0.04 ± 0.13 |
| CS-B07 | 82.43 ± 55.96 | 15.76 ± 16.14 | −0.22 ± 0.07* | 0.09 ± 0.04* | 0.14 ± 0.03* | −0.05 ± 0.07 | −0.04 ± 0.04 | −0.22 ± 0.17 |
| CS-B16 | −296.73 ± 73.87* | −80.08 ± 26.02* | 0.22 ± 0.08* | 0.20 ± 0.05 | −0.04 ± 0.03 | −0.43 ± 0.07* | −0.08 ± 0.06 | −0.23 ± 0.17 |
| CS-B17 | 65.46 ± 58.41 | −8.00 ± 16.13 | −0.80 ± 0.14* | 0.17 ± 0.02* | −0.18 ± 0.02* | −0.47 ± 0.09* | 0.29 ± 0.15 | −0.13 ± 0.16 |
| CS-B18 | −395.57 ± 60.37* | −102.93 ± 23.52* | 0.52 ± 0.09* | −0.13 ± 0.04* | 0.06 ± 0.03 | 0.05 ± 0.09 | 0.13 ± 0.08 | −0.20 ± 0.12 |
| CS-B25 | 57.28 ± 69.56 | −1.59 ± 18.86 | −0.52 ± 0.13* | −0.10 ± 0.03* | −0.41 ± 0.03* | 0.61 ± 0.08* | −0.20 ± 0.10 | 1.37 ± 0.47* |
| CS-B05sh | −21.48 ± 61.65 | −10.29 ± 16.96 | −0.11 ± 0.09 | −0.13 ± 0.03* | 0.06 ± 0.05 | −0.30 ± 0.09* | 0.10 ± 0.07 | −0.45 ± 0.20* |
| CS-B14sh | −188.59 ± 57.80* | −59.27 ± 19.37* | −0.15 ± 0.08 | −0.37 ± 0.05* | −0.01 ± 0.03 | 0.72 ± 0.11* | −0.16 ± 0.08 | 0.01 ± 0.12 |
| CS-B15sh | 171.87 ± 61.11* | 41.98 ± 18.58* | −0.22 ± 0.06* | 0.14 ± 0.04* | −0.02 ± 0.03 | 0.36 ± 0.14* | 0.12 ± 0.08 | −0.07 ± 0.16 |
| CS-B22sh | −125.97 ± 54.90* | −4.70 ± 15.20 | 1.03 ± 0.16* | −0.08 ± 0.04 | 0.16 ± 0.04* | −0.91 ± 0.13* | 0.05 ± 0.04 | −0.33 ± 0.17 |
| CS-B22Lo | 26.86 ± 55.23 | 51.06 ± 22.62* | 1.21 ± 0.19* | −0.33 ± 0.06* | 0.21 ± 0.03* | −0.55 ± 0.09* | −0.25 ± 0.15 | −0.15 ± 0.11 |
| CS-B26Lo | −89.14 ± 66.33 | −33.08 ± 21.36 | −0.22 ± 0.07* | 0.00 ± 0.04 | −0.05 ± 0.03 | 0.44 ± 0.14* | 0.00 ± 0.04 | −0.18 ± 0.12 |
| TM1 | 336.07 ± 84.27* | 78.16 ± 27.53* | −0.65 ± 0.08* | 0.18 ± 0.04* | 0.02 ± 0.03 | 0.18 ± 0.14 | 0.22 ± 0.09* | 0.37 ± 0.17* |
Significant from zero at P = 0.05.
TABLE 5.
Homozygous dominance effects (Dii) for agronomic and fiber traits
| CS-B line | Seedcotton yield (kg/ha) | Lint yield (kg/ha) | Lint (%) | Boll weight (g) | Micronaire | Elongation | Strength (g/tex) |
|---|---|---|---|---|---|---|---|
| CS-B02 | 88 ± 126 | 34 ± 42 | −0.60 ± 0.25* | 0.36 ± 0.07* | 0.15 ± 0.04* | −0.43 ± 0.22 | 0.54 ± 0.43 |
| CS-B04 | 48 ± 62 | 16 ± 26 | −0.22 ± 0.32 | 0.25 ± 0.11* | −0.05 ± 0.06 | 0.04 ± 0.26 | −0.09 ± 0.10 |
| CS-B06 | 248 ± 207 | 102 ± 47* | 1.07 ± 0.20* | 0.16 ± 0.07* | −0.03 ± 0.05 | 0.47 ± 0.09* | 0.01 ± 0.15 |
| CS-B07 | 175 ± 153 | 53 ± 30 | −0.44 ± 0.15* | 0.00 ± 0.13 | −0.06 ± 0.05 | 0.31 ± 0.15* | −0.49 ± 0.20* |
| CS-B16 | −311 ± 232 | −71 ± 31* | 2.24 ± 0.26* | 0.13 ± 0.07 | 0.06 ± 0.03 | −0.30 ± 0.21 | −1.34 ± 1.13 |
| CS-B17 | −270 ± 193 | −107 ± 41* | −1.55 ± 0.18* | −0.25 ± 0.07* | −0.26 ± 0.09* | 0.27 ± 0.14 | −0.30 ± 0.70 |
| CS-B18 | −177 ± 199 | −70 ± 48 | −0.15 ± 0.18 | −0.06 ± 0.04 | 0.27 ± 0.09* | 0.20 ± 0.10 | 0.22 ± 0.15 |
| CS-B25 | −216 ± 151 | −81 ± 32* | −1.10 ± 0.31* | −0.10 ± 0.10 | −0.27 ± 0.08* | −0.17 ± 0.14 | 0.95 ± 0.59 |
| CS-B05sh | 1 ± 51 | 30 ± 29 | 2.07 ± 0.30* | −0.19 ± 0.10 | 0.03 ± 0.05 | −0.83 ± 0.18* | −0.71 ± 0.42 |
| CS-B14sh | 1 ± 34 | −11 ± 20 | −0.99 ± 0.11* | 0.00 ± 0.08 | −0.02 ± 0.03 | −0.08 ± 0.21 | 0.06 ± 0.36 |
| CS-B15sh | 150 ± 99 | 31 ± 26 | −0.49 ± 0.18* | −0.21 ± 0.06* | −0.05 ± 0.03 | −0.02 ± 0.12 | −0.07 ± 0.11 |
| CS-B22sh | −144 ± 132 | −33 ± 29 | 1.61 ± 0.18* | 0.14 ± 0.07 | 0.13 ± 0.07 | −0.70 ± 0.19* | −0.12 ± 0.10 |
| CS-B22Lo | 163 ± 100 | 79 ± 32* | 2.16 ± 0.19* | −0.13 ± 0.07 | 0.16 ± 0.04* | 0.11 ± 0.20 | −0.18 ± 0.18 |
| CS-B26Lo | −27 ± 19 | −9 ± 5 | −0.13 ± 0.05 | 0.00 ± 0.02 | −0.01 ± 0.01 | 0.00 ± 0.04 | −0.08 ± 0.06 |
| TM-1 | 72 ± 99 | 42 ± 42 | 1.93 ± 0.24* | 0.40 ± 0.12* | 0.07 ± 0.06 | −0.66 ± 0.27* | −1.03 ± 0.80 |
Significant from zero at P = 0.05.
TABLE 6.
Heterozygous dominance effects (Dij) for agronomic and fiber traits
| Agronomic traits
|
Fiber traits
|
||||||
|---|---|---|---|---|---|---|---|
| CS-B line | Seedcotton yield (kg/ha) | Lint yield (kg/ha) | Lint (%) | Boll weight (g) | Micronaire | Elongation | Strength (g/tex) |
| CS-B02 | −112 ± 195 | −38 ± 70 | 1.69 ± 0.45* | −0.63 ± 0.16* | −0.15 ± 0.07* | 0.30 ± 0.42 | −0.12 ± 0.31 |
| CS-B04 | 4 ± 97 | −8 ± 51 | 0.15 ± 0.60 | −0.33 ± 0.22 | 0.05 ± 0.11 | 0.44 ± 0.50 | −0.41 ± 0.51 |
| CS-B06 | −278 ± 259 | −147 ± 74 | −2.55 ± 0.41* | 0.03 ± 0.15 | 0.04 ± 0.11 | −1.29 ± 0.17* | −0.07 ± 0.28 |
| CS-B07 | −255 ± 251 | −88 ± 59 | 0.36 ± 0.31 | 0.16 ± 0.25 | 0.29 ± 0.13* | −0.68 ± 0.27* | 0.54 ± 0.25 |
| CS-B16 | 320 ± 223 | 61 ± 38 | −3.95 ± 0.46* | 0.07 ± 0.15 | −0.17 ± 0.06* | 0.41 ± 0.36 | 2.28 ± 1.99 |
| CS-B17 | 598 ± 462 | 204 ± 84* | 1.20 ± 0.34* | 0.78 ± 0.19* | 0.28 ± 0.13* | 0.08 ± 0.29 | 0.43 ± 1.26 |
| CS-B18 | −44 ± 100 | 39 ± 60 | 1.52 ± 0.33* | −0.10 ± 0.10 | −0.47 ± 0.17* | −0.11 ± 0.21 | −0.72 ± 0.47 |
| CS-B25 | 481 ± 363 | 158 ± 66* | 0.97 ± 0.59 | 0.04 ± 0.20 | 0.02 ± 0.10 | −0.07 ± 0.25 | 0.76 ± 0.69 |
| CS-B05sh | −29 ± 104 | −72 ± 57 | −4.39 ± 0.49* | 0.17 ± 0.16 | 0.02 ± 0.10 | 1.84 ± 0.37* | 0.53 ± 0.36 |
| CS-B14sh | −194 ± 171 | −36 ± 43 | 1.64 ± 0.21* | −0.63 ± 0.18* | 0.04 ± 0.06 | −0.19 ± 0.41 | −0.23 ± 0.63 |
| CS-B15sh | −114 ± 86 | −19 ± 44 | 0.47 ± 0.35 | 0.64 ± 0.14* | 0.07 ± 0.06 | 0.31 ± 0.23 | 0.01 ± 0.27 |
| CS-B22sh | 158 ± 171 | 63 ± 58 | −0.79 ± 0.30* | −0.41 ± 0.16* | −0.05 ± 0.11 | 1.51 ± 0.35* | −0.40 ± 0.49 |
| CS-B22Lo | −284 ± 186 | −104 ± 46* | −1.47 ± 0.42* | −0.30 ± 0.11* | −0.05 ± 0.05 | −0.77 ± 0.35* | 0.19 ± 0.23 |
| CS-B26Lo | −53 ± 38 | −19 ± 10 | −0.26 ± 0.10* | 0.01 ± 0.04 | −0.03 ± 0.02 | 0.00 ± 0.07 | −0.17 ± 0.13 |
Significant from zero at P = 0.05.
Seedcotton yield:
The average seedcotton yields for TM-1 and 3-79 and conventional F2 hybrids from the cross of TM-1 and 3-79 were 2507, 963, and 728 kg/ha, respectively (Tables 1 and 2). CS-B lines carrying 3-79 chromosomes 2, 6, or 15sh resulted in significant positive additive effects, while substituted 3-79 chromosomes 16, 18, 14sh, and 22sh had significant negative additive effects on seedcotton yield, suggesting that genes associated with seedcotton yield might be located on these chromosomes (Table 4). However, none of the CS-B lines had a greater additive effect on seedcotton yield compared to TM-1. There were no homozygous or heterozygous dominance effects for seedcotton yield (Tables 5 and 6).
Lint yield:
The average lint yields for TM-1 and 3-79 and conventional F2 hybrids from the cross of TM-1 and 3-79 were 824, 320, and 220 kg/ha, respectively (Tables 1 and 2). Several CS-B lines including lines for chromosomes 2, 6, 15sh, and 22 Lo had significant positive additive effects on lint yield (Table 4). CS-B lines carrying alien chromosomes 16, 18, and 14sh had negative additive effects for lint yield. CS-B lines for chromosomes 6 and 22Lo had positive homozygous dominance effects on lint yield. The heterozygous dominance effects on lint yield for substituted chromosome 17 and 25 were greater than their homozygous chromosomal dominance effects (Tables 5 and 6). CS-B16 had significant negative heterozygous dominance effects on lint yield. Results suggested that these substituted chromosomes were more likely carrying genes that had an effect on lint yield. However, none of the CS-B lines had greater additive effect on lint yield compared to TM-1.
Lint percentage:
The average lint percentage values for TM-1 and 3-79 and conventional F2 hybrids from the cross of TM-1 and 3-79 were 33, 32.88, and 29.96%, respectively (Tables 1 and 2). However, several CS-B lines (02, 06, 16, 18, 22sh, and 22Lo) had additive effects on lint percentage greater than that of TM-1. This suggested that epistatic interactions may occur between the individual 3-79 chromosomes and nonhomologous chromosomes. Several CS-B lines including lines for chromosomes 2, 16, 18, 22sh, and 22Lo had positive additive effects on lint percentage (Table 4). CS-B lines with alien chromosomes 6, 7, 17, 25, 15sh, and 26Lo had negative additive effects on lint percentage. CS-B lines for chromosomes 2, 6, 17, 5sh, 22sh, and 22Lo had opposing dominance effects on lint percentage in heterozygous vs. homozygous condition, suggesting that the difference is due to interactions of different alleles in the same chromosome pair or with alleles in other chromosome pairs (Tables 5 and 6). Several other CS-B lines also had dominance effects on lint percentage (Tables 5 and 6).
Boll weight:
The average boll weights (g) for TM-1 and 3-79 and conventional F2 hybrids from the cross of TM-1 and 3-79 were 5.79, 3.48, and 3.15 g, respectively. CS-B lines involving chromosomes 4, 6, 7, and 17 had positive additive effects on boll weight, while five CS-B lines (18, 25, 5sh, 14sh, and 22 Lo) had negative additive effects for boll weight, suggesting the association of these chromosomes with boll weight (Table 4). CS-B lines carrying alien species chromosomes 2, 4, and 6 had positive homozygous dominance effects for boll weight, showing that genes for boll size might be located on these chromosomes. There was no heterozygous dominance effect for boll weight with any of the 3-79 chromosomes.
Micronaire:
The average MIC values for TM-1 and 3-79 and conventional F2 hybrids from the cross of TM-1 and 3-79 were 4.85, 3.65, and 3.51, respectively (Tables 1 and 2). Upland cotton prices are discounted when micronaire values exceed 5.0 or fall below 3.0 (Calhoun and Bowman 1999). CS-B lines with alien chromosomes 2, 7, 22sh, and 22Lo had positive additive effects on micronaire. CS-B lines carrying 3-79 chromosomes 17 and 25 had negative additive and homozygous dominance effects on micronaire, suggesting that these chromosomes might carry genes that can help in reducing the micronaire. CS-B02, CS-B18, and CS-B22Lo had positive homozygous dominance effects on micronaire, indicating that the respective 3-79 chromosomes might carry genes that increase micronaire (Table 5).
2.5% span length:
The average 2.5% span lengths (mm) for TM-1 and 3-79 and normal F2 hybrids from the cross of TM-1 and 3-79 were 29.35, 35.44, and 32.21, respectively (Tables 1 and 2). CS-B04, CS-B25, CS-B14sh, CS-B15sh, and CS-B26Lo had positive additive effects, while CS-B16, CS-B17, CS-B05sh, CS-B22sh, and CS-B22Lo had negative additive effects on this trait, suggesting that genes affecting 2.5% span length are likely associated with these chromosomes.
Elongation:
The average fiber elongation values for TM-1, 3-79, and normal F2 hybrids from the cross of TM-1 and 3-79 were 7.70, 7.78, and 8.05%, respectively (Tables 1 and 2). CS-B06 had a significant negative additive effect on elongation. CS-B06 and CS-B07 had significant positive homozygous dominant effects on elongation. However, both chromosome 6 and chromosome 7 showed significant negative dominant heterozygous effects on elongation. CS-B05sh and CS-B22sh had negative homozygous dominant effects on elongation. CS-B05sh and CS-B22sh showed positive heterozygous dominant effects on elongation.
Fiber strength:
Fiber strength is measured in grams per tex. It is the force in grams required to break a bundle of fiber 1 tex unit in size. A tex unit is equal to the weight in grams of 1000 m of fiber. The average fiber strengths (g/tex) for TM-1, 3-79, and normal F2 hybrids from the cross of TM-1 and 3-79 were 19.69, 27.61, and 24.29, respectively (Table 1). CS-B02 and CS-B25 had significant positive additive effects on fiber strength, suggesting that these chromosomes carry genes for improving fiber strength. CS-B04 and CS-B05sh had negative additive effects on fiber strength, showing that the respective 3-79 chromosomes might have some genes or transcription factors repressing fiber strength. CS-B07 had negative homozygous dominant effects on fiber strength.
DISCUSSION
One of the challenges in cotton improvement is the paucity of information about genes controlling quantitative traits, including agronomic performance and fiber qualities. Studies that dissect genetic effects into additive effects, dominance effects (Cockerham 1980), and their G × E interaction effects (Zhu 1994) are conventionally applied in a whole-genome manner and thus detect cumulative genetic effects from all genes in the genome. In this study, the genetic dissection is applied to CS-B lines, each of which is divergent from TM-1 for essentially just one pair of chromosomes introgressed from G. barbadense, well known for superior fiber quality. Due to their quasi-isogenic nature, CS-B lines enable a much more detailed and precise quantitative genetic dissection of agronomic and fiber traits. Theoretically, the reduction in genetic complexity for single-locus effects would be approximately inversely proportional to the haploid chromosome number, 1/(n − 1) for whole-chromosome substitutions. For multilocus interactions, the reductions would be geometric and thus much more extreme. In each CS-B line, the alien alleles of one 3-79 chromosome can interact with genes of one 3-79 chromosome and the other 25 TM-1 chromosome pairs. In CS-B F2's, segregation is largely to completely eliminated for 25 of the 26 chromosomes (∼96% of genome), rendering CS-B F2 analyses relatively free of the extensive numbers and kinds of interlocus interactions that arise in a conventional interspecific F2 population (Figure 1). The CS-B F2's thus provided an opportunity to discern effects of alleles in homozygous vs. heterozygous conditions, on a chromosome-by-chromosome or arm-by-arm basis (Figure 1). Multiple comparisons such as this can be used to determine if a substituted chromosome(s) or a chromosome arm(s) is associated with a quantitative trait of interest.
Previous studies have effectively utilized lines similar to CS-B lines in other crops to determine additive or epistatic genetic effects by comparative analysis of homozygous lines (Yen et al. 1997). Actually, additive and epistatic effects are confounded in homozygous CS-B lines. To detect dominant or epistatic effects, hybrids or the segregating progeny have also been used (Yen et al. 1997).
Chromosome additive effects are equivalent to general combining ability (GCA) in this study. The deviation of the additive effect for a CS-B line from TM-1 can be considered as a QTL additive effect associated with the specific chromosome or chromosome arm of 3-79. In our previous analysis of cotton CS-B lines (Saha et al. 2004a,b), the chromosomal effects on important fiber and agronomic traits could not be partitioned into different components because only the CS-B lines and their recurrent parent line were analyzed. Chromosome and interaction effects were thus confounded. An important advantage in this study was to separate additive effects from other genetic effects using the crosses between the CS-B lines and the recurrent parent. Our results showed that chromosome additive effects were important for all traits. All of the alien chromosomes affected seedcotton and lint yields negatively relative to TM-1, suggesting that the overall effect of each alien chromosome in these CS-B lines is reduced yield. This is not surprising given that line 3-79 had very low seedcotton and lint yield compared to TM-1. Given the marked inferiority of the donor parent relative to the recipient, the likelihood of discovering a positive whole-chromosome effect for any trait is intuitively inversely related to the number of polymorphic loci that govern the trait. On the presumption that some 3-79 loci affect seedcotton and lint yield positively relative to TM-1 alleles, we accordingly predict on empirical grounds that recombinational dissection of these CS-B lines will reveal beneficial 3-79 QTL for these traits.
The data indicate that CS-B25 overall is a good general combiner for achieving finer fibers, longer 2.5% span length, and stronger fibers, but not for improving yield. Dissection of this 3-79 chromosome seems especially likely to yield important QTL for these traits, if separated from yield-depressing QTL.
In this study, dominance effects significantly affected most traits. Comparative analysis of the F2 hybrids from the crosses of TM-1 with 3-79 and with the CS-B lines showed that some undesirable agronomic and fiber traits of G. barbadense could be removed using some CS-B lines in the crosses (Table 2). Line 3-79 had low lint percentage compared to TM-1. However, several CS-B lines also had increased lint percentage relative to TM-1. This could be due to major dominance and additive effects by relatively few loci (at least some being separated at the chromosome level). The variance for lint percentage arose mostly from simple dominance, not additive or interaction effects. Thus, the CS-B chromosome effects on lint percentage for several lines may arise largely from interactions within loci (dominance, overdominance, and underdominance). This also could be due to interaction between genes in chromosome 25, per se, and/or interactions with the TM-1 alleles elsewhere in the genome. Similar genetic phenomena were also found for other traits.
Study of several early versions of CS-B lines indicated that chromosome 6 of 3-79 confers a higher lint percentage, finer fiber, and later flowering relative to TM-1, and that chromosome 17 confers short fiber length (Kohel et al. 1977). Ren et al. (2002) mapped QTL for boll size, lint percentage, fiber length, and fiber elongation on chromosome 16 using 178 families from the cross of a CS-B line for chromosome 16 and TM-1. Recent analyses of an expanded set of new and resynthesized CS-B lines per se showed that chromosomes 16 and 18 of 3-79 were associated with reductions in yield and that chromosome 25 was associated with reduced micronaire and increased fiber length and strength compared to TM-1 (Saha et al. 2004a,b).
The genetic effect of any CS-B line that differs from TM-1 is likely due to genes in the alien substituted chromosome and/or due to interactions between TM-1 genes on the other chromosomes and genes on the alien chromosome (Saha et al. 2004a,b). The 3-79 lines produced superior fiber but inferior agronomic qualities compared to TM-1. In situations where a donor is inferior to the recurrent parent for a multigenic trait, chromosome substitutions lines are generally inferior to the recurrent parent. However, when individual substitution lines are superior for the trait, then it is likely attributable to two causes: (1) a limited number of loci having major influence over variation for the trait for the parental combination and/or (2) there are strong interactions and epistatic effects between the respective donor and recipient alleles, on other (nonsubstituted) chromosomes.
The results here revealed the presence of some new QTL and QTL–chromosome associations that were not detected earlier by Saha et al. (2004b). The difference in analyses is that here we compared homozygous CS-B parental lines and the chromosome-specific heterozygous F2 lines while in the earlier study only the homozygous parental CS-B lines were compared. Given the 25-fold differences in the numbers of genes present on a single substituted chromosome pair vs. the nonsubstituted chromosome pairs, it is likely that most of the inferred interlocus interactions, i.e., epistatic effects, involve interactions between genes in the alien chromosome and genes in the remainder of the recipient genome.
G. hirsutum and G. barbadense contain 52 chromosomes and are disomic polyploids. The 26-chromosome genomes of G. hirsutum and G. barbadense are known as (AD)1 and (AD)2, respectively. On the basis of preferential meiotic pairing with their 13-chromosome A and D genomic relatives, the chromosomes of G. hirsutum were numbered 1–13 [A subgenome of (AD)1] and 14–26 [D subgenome of (AD)1], respectively. For some chromosomes, earlier studies with molecular markers have provided detailed information on the genetics and homeologous relationships (Reinisch et al. 1994; Lacape et al. 2003). Our results on chromosomal association with the QTL affecting the same traits also supported the homeologous relationship (Endrizzi et al. 1984). For example, chromosome 2 had significant positive additive effect and its homeologous chromosome 14sh had significant negative additive effect on seedcotton yield (Table 4). Results showed that chromosome 6 had additive negative effect on lint percentage and positive effect on boll weight, whereas its homeologous chromosome 25 also showed additive negative effect on lint percentage and boll weight, supporting their homeologue relationship. The average distribution of the QTL per chromosome was almost the same (8.2 QTL/A genome-specific chromosome vs. 8.3 QTL/D genome-specific chromosome) in the A and D genomes, respectively. Our results based on individual chromosomes are slightly different from those of earlier molecular mapping studies that reported slight overrepresentation of QTL in the D genome (58%) compared to A genome-specific chromosomes (Lacape et al. 2005) . However, association of several fiber-related QTL with the D genome is surprising considering that the “D” subgenome derived from an ancestor that did not produce spinnable fibers. Jiang et al. (1998) also concluded that the merger of A and D genomes with different evolutionary histories in a common nucleus of the tetraploid appeared to offer unique avenues for phenotypic selection during polyploidy formation. The domestication and breeding of tetraploid cottons (AD) resulted in superior fiber quality and yield in the D subgenome compared to its ancestor diploid species.
Some of the aneuploid lines used in developing these CS-B lines originated from sources other than TM-1 and were backcrossed with TM-1 to recover the hypoaneuploid TM-1 isogenic parents used to breed the CS-B lines. It is conceivable that some remnants of the genome, in whose genetic background the original hypoaneuploid was discovered, persisted in the TM-1 isogenic stock, in spite of several backcrossings. If so, it may have confounded our results and contributed to the variation among CS-B lines and thus to the trait–chromosome associations detected. Similarly, there is also a possibility that the observed genetic effects could have been due to some unlinked residual effect of barbadense chromatin in other chromosomes that is independent of the homozygous condition of the substituted chromosome or arm from G. barbadense (Saha et al. 2004a,b). Since this amount of chromatin could be significant relative to that borne by the alien CS-B, and highly genetically divergent from TM-1, we consider that this may be an experimentally significant source of possible confounding. Other, more remote possibilities are that mutation or contaminants confounded results.
Traditional plant breeding approaches that strive to improve yield and fiber quality in parallel have been hindered by complex antagonistic genetic relationships between important fiber and agronomic traits (Culp et al. 1979; Green and Culp 1990). The results here on CS-B lines allowed the effects of whole chromosomes or chromosome arms to be studied. The findings indicate that pursuit of additional genetic resolution is desirable for scientific and breeding purposes and in some cases suggest which chromosomes are most likely to yield the desired sorts of information and genetic products. To help extend the genetic definitions, CS-B lines are being intermated and the progeny will be used to define interchromosomal interlocus interactions. Moreover, chromosome-specific recombinant inbred lines (RILs) or recombinant substituted (RS) lines can be used to separate, identify, and map gene(s) controlling agronomic traits and fiber traits by linkage with molecular markers (Kaeppler 1997; Shah et al. 1999). Studies have demonstrated the advantages of QTL mapping using chromosome substitution and chromosome-specific recombinant lines in wheat, tomato, and mice (Zemetra et al. 1986; Zemetra and Morris 1988; Yen and Baenziger 1992; Weide et al. 1993; Shah et al. 1999; Nadeau et al. 2000; McDonald et al. 2001). Indeed, we are developing RS populations to be used for high-resolution dissection and mapping of the QTL governing cotton yield and fiber qualities.
Cotton is one of the most important economic crops in some developed and developing countries of the world. Production of U.S. cotton results in revenues exceeding $6 billion annually, with an additional $120 billion being generated by industries using cotton as raw materials. Two major economic forces driving the global cotton market are competition from synthetic fibers and technological changes in the textile industry that require improved fiber quality. The exceptional fiber length, strength, and fineness of Pima and Sea Island cotton (G. barbadense L.) give it a 30–50% price advantage over the more widely grown Upland cotton (G. hirsutum L.) because of its superior spinning and manufacturing performance. However, Upland cotton occupies >90% of the cotton growing area because of superior agronomic properties and yield potential. The genetic base that cotton breeders use in applied Upland cotton breeding programs to develop new varieties is narrow. The need to improve fiber quality while maintaining the superior agronomic and yield properties of Upland cottons requires innovative research approaches and a greater understanding of the genetics of fiber quality. The quality of cotton fibers comes from several traits including length, fineness, and strength. Our results showed that controlled interspecific introgression of G. hirsutum with G. barbadense through chromosome substitution provided a scope to demonstrate that fiber qualities in G. hirsutum can be improved without genetic drag effect of poor agronomic qualities of G. barbadense. It is the only method that allows introgression of a complete chromosome or chromosome segment, and it complements other methods of introgression.
The strength of this article derived from several perspectives including: (1) the effect of each chromosome was dissected on the basis of its individual homozygous or heterozygous nature using a novel genetic strategy; (2) some new QTL effects associated with some chromosomes were discovered, which exist cryptically without showing such effects in the parental line; and (3) this is the first detailed report of some of the new chromosome substitution lines in cotton. One of the overriding long-term goals of this research is to enhance germplasm for agronomic and fiber qualities that will benefit the cotton producers. To complement this work, CS-B line intermatings are in progress. These lines will be used to define interchromosomal interlocus interactions; and chromosome-specific RILs will be developed to empower the QTL mapping process with both high sensitivity and high resolution. Moreover, while early generation data have limited resolution and discrimination among sources of genetic effects, we anticipate that extension of the CS-B chromosome substitution approach will be extremely powerful as both an analytical and a breeding tool for cotton improvement.
Acknowledgments
Alan Zipf provided valuable suggestions regarding this research and also with improving the manuscript. We also appreciate the help of Russell Hayes in collection of experimental data and Ramesh Katam, Lillie Hendrix, Douglas Dollar, and Parvati Ynturi for assisting in the field experiments. We also thank Mauricio Ulloa, U.S. Department of Agriculture/Agricultural Research Service (USDA/ARS), Shafter, California; T. Brooks of USDA/ARS, Mississippi State, Mississippi; and Ramesh Kantety, Alabama A&M University, Normal, Alabama, for their critical evaluations of the manuscript and helpful comments. Mention of a trademark or a proprietary product does not constitute a guarantee or warranty of the product by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may also be suitable.
We dedicate this article to the memory of the late Allan Zipf (9/6/1953–1/25/2004).
References
- Al-Quadhy, R., W. R. Morris and R. F. Mumm, 1988. Chromosomal locations of genes for traits associated with lodging in winter wheat. Crop Sci. 28: 631–635. [Google Scholar]
- Beasley, J. O., and M. S. Brown, 1942. Asynaptic Gossypium plants and their polyploids. J. Agric. Res. 65: 421–427. [Google Scholar]
- Berke, T. G., P. S. Baenziger and W. R. Morris, 1992. a Chromosomal location of wheat quantitative trait loci affecting agronomic performance, using reciprocal chromosome substitutions. Crop Sci. 32: 621–627. [Google Scholar]
- Berke, T. G., P. S. Baenziger and W. R. Morris, 1992. b Chromosomal location of wheat quantitative trait loci affecting stability of six traits, using reciprocal chromosome substitutions. Crop Sci. 32: 628–633. [Google Scholar]
- Calhoun, D. S., and D. T. Bowman, 1999. Techniques for development of new cultivars, pp. 361–414 in Cotton Origin, History, Technology and Production, edited by C. W. Smith and J. T. Cothren. John Wiley & Sons, New York.
- Cockerham, C. C., 1980. Random and fixed effect in plant genetics. Theor. Appl. Genet. 56: 119–131. [DOI] [PubMed] [Google Scholar]
- Culp, T. W., D. C. Harrell and T. Kerr, 1979. Some genetic implications in the transfer of high fiber strength genes to upland cotton. Crop Sci. 19: 481–484. [Google Scholar]
- Endrizzi, J. E., 1963. Genetic analysis of six primary monosomes and one tertiary monosome in Gossypium hirsutum. Genetics 48: 1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi, J. E., E. L. Turcotte and R. J. Kohel, 1984. Quantitative genetics, cytology and cytogenetics, pp. 81–129 in Cotton, edited by R. J. Kohel and C. F. Lewis. American Society of Agronomy, Madison, WI.
- Friebe, B., L. L. Qi, S. Nasuda, P. Zhang, N. A. Tuleen et al., 2000. Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theor. Appl. Genet. 101: 51–58. [Google Scholar]
- Green, C., and T. W. Culp, 1990. Simultaneous improvement of yield, fiber quality, and yarn strength in upland cotton. Crop Sci. 30: 66–69. [Google Scholar]
- Gutiérrez, O. A., S. Basu, S. Saha, J. N. Jenkins, D. B. Shoemaker et al., 2002. Genetic distance of cotton cultivars and germplasm lines based on SSR markers and its association with agronomic and fiber traits of their F2 hybrids. Crop Sci. 42: 1841–1847. [Google Scholar]
- Jiang, C. R., C. R. Wright, K. M. El-Zik and A. H. Paterson, 1998. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 95: 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler, S. M., 1997. Quantitative trait locus mapping using set of near-isogenic lines: relative power comparisons and technical considerations. Theor. Appl. Genet. 95: 384–392. [Google Scholar]
- Kohel, R. J., J. E. Endrizzi and T. G. White, 1977. An evaluation of Gossypium barbadense L. chromosome 6 and 17 in the G. hirsutum L. genome. Crop Sci. 17: 404–406. [Google Scholar]
- Kohel, R. J., J. Yu, Y.-H. Park and G. R. Lazo, 2001. Molecular mapping and characterization of traits controlling fiber quality in cotton. Euphytica 121: 163–172. [Google Scholar]
- Lacape, J. M., T. B. Nguyen, S. Thibivilliers, B. Courtois, B. M. Bojinov et al., 2003. A combined RFLP-SSR-AFLP map of tetraploid cotton based on a Gossypium hirsutum x Gossypium barbadense backcross population. Genome 46: 612–626. [DOI] [PubMed] [Google Scholar]
- Lacape, J. M., T. B. Nguyen, B. Courtois, J. L. Belot, M. Giband et al., 2005. QTL analysis of cotton fiber quality using multiple Gossypium hirsutum X Gossypium barbadense backcross generations. Crop Sci. 45: 123–140. [Google Scholar]
- Law, C. N., 1966. The location of genetic factors affecting a quantitative character in wheat. Genetics 53: 487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald, M. P., N. W. Galway, P. Ellneskog-Stamm and T. D. Colmer, 2001. Effects on growth and development of individual chromosome from slow-growing Lophopyrum elongatum Love when incorporated into bread wheat (Triticum aestivum L.). Ann. Bot. 88: 215–223. [Google Scholar]
- Mckenzie, W. H., 1970. Fertility relationships among interspecific hybrid progenies of Gossypium. Crop Sci. 10: 571–574. [Google Scholar]
- Mergeai, G., 2003. Forty years of genetic improvement of cotton through interspecific hybridization at Gembloux Agricultural University: achievement and prospects, pp. 119–133 in Cotton Production for the New Millennium: World Cotton Research Conference–3 Proceedings, edited by A. Swanepoel. Agricultural Research Council: Institute for Industrial Crops, Pretoria, South Africa.
- Miller, R. G., 1974. The jackknife: a review. Biometrika 61: 1–15. [Google Scholar]
- Nadeau, J. H., J. B. Singer, A. Martin and E. S. Lander, 2000. Analyzing complex genetic traits with chromosome substitution strains. Nat. Genet. 24: 221–225. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., and R. Smith, 1999. Future horizons: biotechnology for cotton improvement, pp. 415–432 in Cotton Origin, History, Technology and Production, edited by C. W. Smith and J. T. Cothren. John Wiley & Sons, New York.
- Reinisch, M. J., J. Dong, C. L. Brubaker, D. M. Stelly, J. F. Wendel et al., 1994. A detailed RFLP map of cotton, Gossypium hirsutum x Gossypium barbadense: chromosome organization and evolution in a disomic polyploidy genome. Genetics 138: 829–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, L., W. Guo and T. Zhang, 2002. Identification of quantitative trait loci (QTLs) affecting yield and fiber properties in chromosome 16 in cotton using substitution line. Acta Bot. Sin. 44: 815–820. [Google Scholar]
- Rong, J., C. Abbey, J. E. Bowers, C. L. Brubaker, C. Chang et al., 2004. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166: 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., D. A. Raska, J. N. Jenkins, J. C. Mccarty, Jr., O. A. Gutiérrez et al., 2004. a Effect of chromosome on important quantitative traits of agronomic and fiber traits using Gossypium barbadense chromosome-specific recombinant lines of of G. hirsutum, pp. 170–174 in Cotton Production for the New Millennium: World Cotton Research Conference-3 Proceedings, edited by A. Swanepoel. Agricultural Research Council: Institute for Industrial Crops, Pretoria, South Africa.
- Saha, S., J. Wu, J. N. Jenkins, J. C. Mccarty, Jr., O. A. Gutiérrez et al., 2004. b Effect of chromosome substitutions from Gossypium barbadense L. 3–79 into G. hirsutum L. TM-1 on agronomic and fiber traits. J. Cot. Sci. 8: 162–169. [Google Scholar]
- Shah, M. M., K. S. Gill, P. S. Baenziger, Y. Yen, S. M. Kaeooler et al., 1999. Molecular mapping of loci for agronomic traits on chromosome 3A of bread wheat. Crop Sci. 39: 1728–1732. [Google Scholar]
- Stelly, D. M., S. Saha, D. A. Raska, J. Wu, J. N. Jenkins et al., 2004. Chromosome-specific resources for germplasm introgression and genomics in Gossypium. W187. Plant and Animal Genome XII, The International Conference, San Diego.
- Stelly, D. M., S. Saha, D. A. Raska, J. N. Jenkins, J. C. McCarty et al., 2005. Registration of 17 Upland (Gossypium hirsutum) germplasm lines disomic for different G. barbadense chromosome or arm substitutions. Crop Sci. 45: 2663–2665. [Google Scholar]
- Stephens, S. G., 1949. The cytogenetics of speciation in Gossypium. I. Selective elimination of the donor parent genotype in interspecific backcrosses. Genetics 34: 627–637. [DOI] [PubMed] [Google Scholar]
- Tang, B., J. N. Jenkins, C. E. Watson, J. C. Mccarty and R. G. Creech, 1996. Evaluation of genetic variances, heritabilities, and correlations for yield and fiber traits among cotton F2 hybrid populations. Euphytica 91: 315–322. [Google Scholar]
- Tanksley, S. D., and S. R. Mccouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Ulloa, M., and W. R. Meredith, Jr., 2000. Genetic linkage map and QTL analysis of agronomic and fiber quality traits in an interspecific population. J. Cot. Sci. 4: 161–170. [Google Scholar]
- Weide, M., F. V. Wordragen, R. K. Lankhorst, R. Verkrek, C. Hanhart et al., 1993. Integration of the classical and molecular linkage map of tomato chromosome 6. Genetics 135: 1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., P. D. Olson and J. Mcd Stewart, 1989. Genetic diversity, introgression and independent domestication of Old World cultivated cottons. Am. J. Bot. 76: 1795–1806. [Google Scholar]
- White, T. G., T. R. Richmond and C. F. Lewis, 1967. Use of cotton monosomes in developing interspecific substitution lines. Crops Research USDA Report ARS 34-91, Washington, DC.
- Yen, Y., and P. S. Baenziger, 1992. A better way to construct recombinant chromosome lines and their controls. Genome 35: 827–830. [Google Scholar]
- Yen, Y., P. S. Baenziger, R. Bruns, J. Reeder, B. Moreno-Sevilla et al., 1997. Agronomic performance of hybrids between cultivars and chromosome substitution lines. Crop Sci. 37: 396–399. [Google Scholar]
- Zemetra, R. S., and W. R. Morris, 1988. Effects of an intercultivaral chromosome substitution on winterhardiness and vernalization in wheat. Genetics 119: 453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemetra, R. S., W. R. Morris and J. W. Schmidt, 1986. Gene locations for heading date using reciprocal chromosome substitutions in winter wheat. Crop Sci. 26: 531–533. [Google Scholar]
- Zhu, J., 1989. Estimation of genetic variance components in the general mixed model. Ph.D. Dissertation, North Carolina State University, Raleigh, NC.
- Zhu, J., 1993. Methods of predicting genotype value and heterosis for offspring of hybrids (Chinese). J. Biomath. 8(1): 32–44. [Google Scholar]
- Zhu, J., 1994. General genetic models and new analysis methods for quantitative traits. J. Zhejianiang Agric. Univ. 20(6): 551–559. [Google Scholar]
- Zhu, J., 1998. Analytical Methods for Genetic Models. Press of China Agriculture, Bejing, China.