Abstract
For a newly arising mutation affecting a trait under selection, its degree of dominance relative to the preexisting allele(s) strongly influences its evolutionary impact. We have estimated dominance parameters for spontaneous mutations in a subset of lines derived from a highly inbred founder of Arabidopsis thaliana by at least 17 generations of mutation accumulation (MA). The labor-intensive nature of the crosses and the anticipated subtlety of effects limited the number of MA lines included in this study to 8. Each MA line was selfed and reciprocally crossed to plants representing the founder genotype, and progeny were assayed in the greenhouse. Significant mutational effects on reproductive fitness included a recessive fitness-enhancing effect in one line and fitness-reducing effects, one additive and the other slightly recessive. Mutations conferring earlier phenology or smaller leaves were significantly recessive. For effects increasing leaf number and reducing height at flowering, additive gene action accounted for the expression of the traits. The sole example of a significantly dominant mutational effect delayed phenology. Our findings of recessive action of a fitness-enhancing mutational effect and additive action of a deleterious effect counter a common expectation of (partial) dominance of alleles that increase fitness, but the frequency of occurrence of such mutations is unknown.
AS the ultimate source of genetic variation, which is the basis of any evolutionary change, spontaneous mutation is a fundamental evolutionary process. Theoretical investigations of the evolutionary consequences of mutation have addressed its role in numerous biological phenomena, including the maintenance of genetic variation (e.g., Lande 1976; Turelli 1984; most recently, Zhang et al. 2002, 2004a,b), the evolution of mating systems (e.g., Kondrashov 1988; Charlesworth et al. 1990), population extinction (e.g., Lande 1994; Lynch et al. 1995), and ecological specialization (Kawecki et al. 1997), among others. These studies have shown that predictions about the evolutionary implications of spontaneous mutation depend primarily on three of its properties, the rate of occurrence throughout the genome of mutations affecting fitness, the distribution of the effects of new mutations on fitness, and the gene action of new mutations.
Recent empirical studies of spontaneous mutation employing the mutation-accumulation (MA) approach have largely focused on quantifying the genomewide mutation rate, U (Drake et al. 1998; Lynch et al. 1999). Mutational properties have been studied in haploid organisms, as well as in diploids; we consider here studies of diploids. In studies where mutations are accumulated over generations in lines advanced by close inbreeding, phenotypic assays of the lines evaluate traits of highly inbred individuals (e.g., Keightley and Caballero 1997). Such studies yield information not only about U but also about the effects of new mutations on traits; given the extreme inbreeding of the lines, resulting estimates of mutational effect refer to the effect of mutations when expressed in homozygous state. Alternatively, MA lines may be propagated clonally, and, in this case, mutations are expressed in heterozygous state (e.g., Joseph and Hall 2004). There is increasing interest in the gene action of mutations in terms of the dominance or recessiveness of new mutations. This property can profoundly affect the evolutionary fate of new mutations (Haldane 1927; Caballero and Hill 1992; Charlesworth 1992; Caballero and Keightley 1994), because a fully dominant allele exerts its effect on phenotype in the first generation in which it appears as a single copy under heterozygous conditions, whereas a fully recessive allele affects the phenotype only when it occurs in homozygous state and may be lost by chance before this.
In an investigation of the effect of dominance on the evolutionary fate of new mutations, Caballero and Hill (1992) quantified the contribution to genetic variance for a new mutation, showing that a beneficial mutation of dominant effect makes the greatest contribution to genetic variance when the population mates randomly, whereas a recessive mutation contributes more to genetic variance in an inbred population. For a random mating population, Caballero and Keightley (1994) showed in a simulation study that the contribution to standing genetic variation of alleles generated by mutation can differ strikingly depending on the assumed distribution of mutational effects jointly on fitness and a correlated trait. In particular, when new mutations tend to reduce fitness severely and have more moderate and moderately correlated effects on the trait, as Mackay et al. (1992) found for P-element mutagenesis in Drosophila melanogaster, recessive mutations may contribute disproportionately to standing genetic variation in the trait. Alternatively, when mutational effects on both fitness and trait, as well as the correlation between them, tend to be weaker, as inferred from response to selection in highly inbred populations of D. melanogaster, partially dominant alleles contribute to standing genetic variance out of proportion to their mutational occurrence. They concluded that the degree of dominance of new mutations has little effect on the total genetic variance at equilibrium. Zhang et al. (2004a), however, have found that dominance can “dramatically alter the prediction of equilibrium genetic variance,” depending on the relative recessivity of mutational effect on fitness and of the pleiotropic effect on a trait.
As part of a study of evolutionarily important properties of spontaneous mutations in Arabidopsis thaliana (Shaw et al. 2000), we have studied gene action of mutations that originated during the course of up to 24 generations of MA. Specifically, to assess dominance relationships between new mutations and the alleles that characterize the highly inbred progenitor, we have conducted two sets of crosses between MA lines and the founder genotype, as well as selfs of both. We have grown progeny of these crosses in assays of individual fitness, as well as morphological and phenological traits. From these data, we have estimated homozygous effects of mutations on these traits, as well as the degree of dominance of mutations relative to progenitor alleles. Whereas, ultimately, the effect of individual mutations in homozygous and heterozygous state is of evolutionary importance, experiments of this kind (e.g., Lopez and Lopez-Fanjul 1993; Chavarrias et al. 2001; Peters et al. 2003) generally estimate the composite effect of the mutations in an MA line, rather than the effect of each mutation singly (for an exception, see Szafraniec et al. 2003). In this study, crosses were conducted considerably earlier in the course of line advancement than is often the case; it is therefore reasonable to expect that each line carries few mutations (0–5) affecting a given trait and estimates of mutational effect represent the composite effect of very few mutations.
MATERIALS AND METHODS
Experimental material:
Arabidopsis thaliana is a particularly tractable system for studies of spontaneous mutation in plants in part because of its high degree of autogamous selfing in natural populations (Abbot and Gomes 1989). As a result, the advancement of generations through selfing in mutation-accumulation lines does not constitute an unusual mode of reproduction for this species. Furthermore, its short generation time (8–10 weeks in our line advancement process) makes MA studies feasible within a relatively short period of time.
The MA lines used in this study were established from 120 progeny of a single founder individual from a Columbia accession of A. thaliana that had been advanced for several generations by selfing and single-seed descent. The founder individual obtained in this way was expected to be at mutation-drift equilibrium (Lynch and Hill 1986) and, hence, almost completely homozygous throughout its genome. Each MA line was propagated through selfing and single-seed descent through an individual chosen at random (see more details in Shaw et al. 2000). This protocol minimizes selection within lines such that, in advanced generations, phenotypic differences between MA lines and the founder and variation among MA lines reflect effects of mutations that have arisen spontaneously and subsequently fixed primarily by genetic drift.
Crossing design:
To examine the modes of gene action for the newly arisen mutations in our MA lines, we reciprocally crossed plants representing an MA line from an advanced generation to ones representing the founder generation (generation 0). For each line, plants representing the MA lines were grown from seeds sampled directly from our seed collection of the MA lines. Plants representing generation 0 were grown from seeds derived from the founder of the MA lines by two generations of selfing to increase numbers. Because of the minimal opportunity for mutations to accumulate during the propagation of these generation 0 sublines, the sublines are expected to be virtually identical genetically to the founder of the MA lines. Progeny of a cross between an MA line and generation 0 express the mutations of the MA line in heterozygous state, whereas progeny of the selfs of MA lines express the mutations carried by that line in homozygous state. Expression of traits in progeny of selfs of generation 0 provides a basis of reference. Comparison of heterozygous with homozygous expression of mutations reflects their dominance.
We conducted two sets of crosses, each of which included four MA lines from a single generation and two sublines from generation 0. The first crossing block (set A) included the following lines sampled at generation 17: lines 49, 69, 71, and 76 (designated as 17–49, 17–69, 17–71, and 17–76 hereafter) and 61 and 86 from generation 0 (0–61 and 0–86). The two generation 0 lines were chosen randomly, whereas the four generation 17 lines were chosen on the basis of preliminary assays of 20 lines in which they had the highest or lowest mean for some trait, although the differences were typically not statistically significant in these early assays. Both 17–49 and 17–71 produced larger leaves and tall inflorescences at flowering. Line 17–71 produced significantly more leaves than other lines. In contrast, lines 17–69 and 17–76 produced fewer leaves, and those of 17–76 were smaller than those of other lines (R. G. Shaw and D. L. Byers, unpublished results). Studies of gene action of mutations have typically focused on effects in lines chosen according to phenotype (e.g., Lopez and Lopez-Fanjul 1993; Peters et al. 2003). The second crossing block (set B) involved lines sampled at generation 24 of line advancement, including 24–3, 24–23, 24–39, and 24–102 and lines 0–7 and 0–18. All of the lines in set B were chosen randomly.
The following scheme was used consistently for the two sets of crosses both conducted in 2000. Plants from MA lines were reciprocally crossed to plants from generation 0 (Table 1). Each MA line was represented by five individuals as parents in the crosses, three serving as maternal and two as paternal parents. In addition to these crosses, self-pollination was also carried out manually to control for effects of floral manipulation and pollen transfer. Self-matings were conducted to yield at least three selfed fruits for each plant serving as a dam. Each of the crosses between pairs of lines was replicated six times (three dams × two sires). We used twice as many plants in the generation 0 sublines to ensure that there were enough flowers for the higher number of crosses involving these lines. Each individual plant in the generation 0 sublines was crossed to only two of the four MA lines.
TABLE 1.
One block of design of crosses between generation 17 MA lines and sublines sampled at generation 0
| Paternal lines
|
||||||
|---|---|---|---|---|---|---|
| Maternal lines | 1 | 2 | 3 | 4 | Y | Z |
| 1 | S | X | X | |||
| 2 | S | X | X | |||
| 3 | S | X | X | |||
| 4 | S | X | X | |||
| Y | X | X | X | X | S | |
| Z | X | X | X | X | S | |
Lines sampled in generation 17 for these crosses are designated 1–4. Sublines sampled at random from generation 0 are designated Y and Z. Lines were used as both paternal and maternal parents. X, crosses between generations; S, self-fertilization.
This design permits estimation of effects of mutations under homozygous and heterozygous condition and, hence, nonadditivity of gene action within loci. To the extent that multiple mutations have fixed in a given line, these estimates reflect the composite effect of all those mutations, rather than the effects of individual mutations. Inclusion of multiple maternal individuals per cross contributes to the accuracy of these estimates by permitting direct assessment of environmentally induced maternal effects. We implemented the crosses reciprocally to assess the role of the maternally inherited cytoplasm in transmission of mutational effects.
Because A. thaliana flowers normally self-pollinate in bud, crossing involves emasculating the miniscule flower buds before the anthers dehisce. To evaluate the possibility of contamination from self-pollen in our crosses, we carried out control emasculation of one flower on each maternal plant without subsequent pollination. None of the control emasculations resulted in any fruits, indicating that our emasculation was successful and that contamination by self or other pollen was unlikely. To accomplish cross-pollination, we plucked a dehiscent anther from an open flower on the paternal plant and brushed it directly against the stigmatic surface of an emasculated flower. Self-pollination was carried out in a similar fashion except the anther was obtained from another flower of the same plant. Fruits resulting from hand pollinations were collected individually when mature but before they dehisced.
Experimental assay:
The progeny from the two sets of crosses were grown in separate assays as follows. To reduce the number of plants to be measured at a time, we conducted each assay in two temporal blocks planted a week apart. Moreover, we grew plants from a subset of the available fullsibships; the progeny of two dams and one sire chosen at random from the parental individuals representing a line were grown in each temporal block. For the first assay, we grew 15 F1 offspring from each of the fruits produced from crosses between generations (15 × 8 line combinations × 2 for reciprocal crosses × 2 dams/combination = 480 plants). We also grew 60 and 30 progeny from the selfed fruits of generation 0 and MA lines, respectively (60 plants × 2 dams/line × 2 lines + 30 plants/dam ×2 dams/line × 4 lines = 480 plants). Each of the two temporal blocks comprised 15 trays of 32 plants each. To enhance precision of the comparisons of F1's to the respective parentals, each tray contained progeny derived from only 2 of the 4 MA lines. Within a tray, half of the plants represented cross-pollinations and the other half the corresponding self-pollinations. Plants were assigned to positions in a tray at random. We planted three seeds per pot and later thinned each to a single randomly chosen plant. To replace the pots in which all three seeds had failed to germinate, 27 additional pots were planted 2 weeks after the initial plantings. In four of the original pots that had been replanted, the seeds germinated later; these plants were included in the final analysis. In this assay, the number of plants totaled 964.
The assay of set B was grown like that of set A except for three main differences. First, this assay was conducted beginning in December and natural light was not supplemented; thus the day length was extremely short during most of the period of growth, such that development of plants differed considerably from development in the assay of set A, grown under long-day conditions. Second, rather than planting individuals in order according to the final randomization, as we did for the assay of the first set of crosses, we planted all at once the individuals representing a given cross and moved the pots into their randomized positions the same day when planting was complete. Third, the numbers of plants representing each cross were increased, with numbers of cross-progeny ranging from 157 to 205 and selfs of MA lines from 72 to 96 individuals, with the exception of the self of line 23. This line was represented in homozygous state by only 27 individuals, because three of the nine fruits for this cross produced no seed and seeds from three more fruits germinated poorly. Apart from this self-mating, only four other fruits were empty, no more than one for a particular cross combination. Including the 334 selfs of the founder, the number of plants in the second assay totaled 1363.
Plants were grown in the greenhouse at University of Minnesota using Sunshine Mix 5 (Sun Gro Horticulture, Seneca, IL) and 5-cm pots. All trays were subirrigated when necessary until plants senesced; i.e., all fruits had turned yellow or brown. For each individual, we recorded the phenological traits; days from planting to germination, bolting, and flowering; morphological traits expressed on the day it began to flower; height of inflorescence; leaf number; and length of the largest leaf. Once a plant began to flower, an Aracon (Lehle Seeds) was placed on it to keep it upright and also to collect material as fruits dehisced. At the end of the growing period after all fruits had matured, we recorded the number of fruits on each plant (set A only) and the dry biomass of reproductive structures. To do this, we cut off the infructescence and stored each plant separately in paper bags. These were dried at 60° for at least 24 hr before weighing on a digital balance (Mettler Toledo AT261) at 0.01 mg precision.
Statistical analyses:
The traits were analyzed according to the mixed model
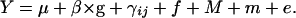 |
The trait mean expressed by progeny from selfing the founder, μ, is the basis against which we compare traits expressed by progeny from the remaining matings. The coefficient, β, accounts for the linear relationship between the trait and germination date, g. The parameter γij estimates the fixed effect of the ith MA line in self-matings (j = i) or in crosses to the founder (j = 0). Thus, in our crossing design, γij can be either γii, for homozygous effects, or γi0, for the heterozygous effects. The model for these effects is based on the model of Falconer and Mackay (1996, Figure 7.1). The genetic contribution to trait values expressed by progeny derived by selfing of the ith MA line is γii = 2ai, and that for progeny derived by crossing of the ith MA line to the founder is γi0 = ai + di (γ00 is necessarily zero). Thus, ai estimates half the homozygous effect of the ith mutation and di estimates the difference of heterozygote expression from additivity. Estimates of a and d were considered significantly different from zero at P < 0.05 if they exceeded 1.96 times their estimated standard errors, according to a t-test. No formal corrections for multiple tests were applied. We note that we detected far more effects than expected by chance, with nominal P-values ranging from 0.05 to 0.0001. Multiple effects within a line may reflect pleiotropic effects of the mutation(s) it carries, rather than distinct effects. The degree of dominance is given in two commonly used scalings: k = d/a (values of k = −1, 0, and 1 imply fully recessive, additive, and dominant gene action, respectively; Falconer and Mackay 1996) and h = (a+d)/2a (where h = 0, 0.5, and 1 have those respective interpretations; see, e.g., Caballero et al. 1997). They are related as h = (1 + k)/2. We considered as random the effects (1) of the flat in which the plant grew, f; (2) of maternal lineage, M (i.e., the effect of a line when used as maternal parent in excess of its effect as paternal parent); (3) of maternal individual, m, nested within maternal line; and (4) of the environment unique to individual progeny, e, and analyzed models that included components of variance attributable to these factors. This model was implemented in a version of the Quercus program, nf6.p, which conducts the analysis via restricted maximum likelihood (REML; Shaw and Shaw 1994). For each trait, residuals from the model were distributed approximately normally; no transformations were applied. In no case did the variance due to maternal lineage approach significance; accordingly, this factor was excluded from the analyses. We present estimates of the MA line effects obtained from models in which only significant variance components are retained. We note, however, that retention or exclusion of random factors affected the estimates very little and did not affect which were detected as significant.
Two aspects of the analysis of set B require further comment. The effect of the MA line was treated as fixed, as in the analysis of set A, because the motivation for the study was to estimate the line effects, rather than their variances. In addition, an estimate of a variance component based on four lines can be expected to be very unreliable. The difference in the planting of the assay of set B led to a difference in its analysis. Shortly after it was planted, we observed that the potting medium varied among lots in color and texture. To account for the possibility that this or some other factor influenced plants according to the planting order, we included in the analyses a further random factor, the flat into which an individual was planted, in addition to the flat in which it was grown. This factor contributed significantly to the variation for several traits, but, as with the other random effects, the estimates of the line effects differed little, depending on whether it was included in the model.
RESULTS
Set A:
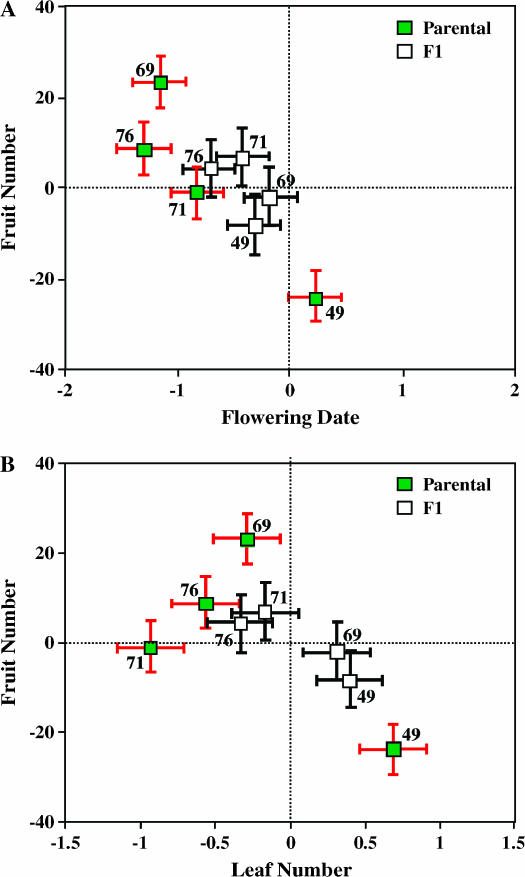
For all four lines, the composite effect of mutations was detected as significant for one or more traits (Table 2). In particular, for the fitness measure, number of fruits per plant, two parental lines expressed extreme values, reflecting effects on fitness of mutations in homozygous state. The fitness of line 49 was significantly lower than that of the plants representing the founder, whereas line 69 significantly exceeded the founder in its fitness (Table 2 and Figures 1A and 1B). The mutational effect on reproductive biomass was also significantly positive in the case of line 69. This positive effect jointly on fruit number and reproductive biomass is consistent with our earlier finding that mutational effects on these two traits are strongly positively correlated over lines (r = 0.7, Shaw et al. 2000), but line 49 showed no decline in reproductive biomass (Table 2).
TABLE 2.
Estimates of the genetic parameters with respect to each trait for the MA lines included in set A
| Line 49
|
Line 69
|
Line 71
|
Line 76
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k = d/a | k = d/a | k = d/a | k = d/a | |||||||||
| Set A | a (SE) | d (SE) | h | a (SE) | d (SE) | h | a (SE) | d (SE) | h | a (SE) | d (SE) | h |
| Bolting date | −0.20 | −0.17 | 0.83 | −0.42* | 0.35 | −0.84 | −0.43* | 0.10 | −0.24 | −0.65* | −0.03 | 0.05 |
| (0.16) | (0.21) | 0.92 | (0.16) | (0.21) | 0.08 | (0.16) | (0.21) | 0.38 | (0.16) | (0.20) | 0.52 | |
| Flowering date | 0.11 | −0.42* | −3.73 | −0.58* | 0.39 | −0.67 | −0.41* | −0.01 | 0.02 | −0.65* | −0.06 | 0.09 |
| (0.17) | (0.21) | −1.36 | (0.17) | (0.21) | 0.16 | (0.17) | (0.22) | 0.51 | (0.17) | (0.21) | 0.54 | |
| Height at flowering | 3.17 | 0.24 | 0.08 | 0.84 | 1.05 | 1.25 | 1.65 | −0.55 | −0.33 | 0.67 | −3.07 | −4.61 |
| (1.01) | (1.62) | 0.54 | (1.00) | (1.61) | 1.12 | (1.00) | (1.62) | 0.33 | (1.01) | (1.60) | −1.80 | |
| Leaf no. | 0.34* | 0.06 | 0.17 | −0.15 | 0.46* | −3.11 | −0.47* | 0.30 | −0.63 | −0.28 | −0.05 | 0.17 |
| (0.16) | (0.19) | 0.58 | (0.16) | (0.19) | −1.05 | (0.16) | (0.19) | 0.18 | (0.16) | (0.19) | 0.58 | |
| Leaf length | 0.47 | 0.47 | 1.00 | −0.94* | 1.05 | −1.11 | −1.20* | 1.20* | −0.99 | −0.29 | −0.13 | 0.44 |
| (0.34) | (0.55) | 1.00 | (0.34) | (0.54) | −0.05 | (0.34) | (0.55) | 0.005 | (0.35) | (0.54) | 0.72 | |
| Rep. biomass | 0.01 | −0.0001 | −0.16 | 0.01* | −0.01 | −1.16 | −0.004 | 0.02* | −3.89 | 0.01 | −0.0002 | −0.04 |
| (0.01) | (0.01) | 0.42 | (0.01) | (0.01) | −0.08 | (0.01) | (0.01) | −1.44 | (0.01) | (0.01) | 0.48 | |
| Fruit no. | −11.92* | 3.56 | −0.30 | 11.63* | −13.64* | −1.17 | −0.53 | 7.40 | −13.88 | 4.39 | −0.08 | −0.02 |
| (4.03) | (6.42) | 0.35 | (4.03) | (6.40) | −0.08 | (4.03) | (6.41) | −6.44 | (4.07) | (6.38) | 0.49 | |
Definitions of a and d are similar to those given in Falconer and Mackay (1996; see materials and methods). The degree of dominance is given in two commonly used scalings: k = d/a and h = (a + d)/2a. They are related as h = 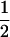 (1 + k). Both k and h were calculated using the original values of a and d before rounding. Standard errors are given in parentheses. *P < 0.05; **P < 0.01.
(1 + k). Both k and h were calculated using the original values of a and d before rounding. Standard errors are given in parentheses. *P < 0.05; **P < 0.01.
Figure 1.
Predicted values of traits expressed by the progeny of crosses of MA lines sampled at generation 17 (set A). Shaded symbols refer to progeny obtained by selfing an MA line; predicted values, which are expected to equal those of the parental MA lines, are obtained as 2ai. Open symbols refer to progeny obtained by crossing the MA line to a plant representing the founder of the MA lines; in this case, predicted values are obtained as ai + di. The values are given as deviations from the trait value of progeny obtained by self-mating the founder (indicated by lines at 0). (A) Fruit number vs. flowering date; (B) fruit number vs. leaf number. Bars, 1 SE.
In addition to effects on fitness, we also detected significant mutational effects on each of the phenological and morphological traits. Plants representing three of the four MA lines, 69, 71, and 76, developed significantly more rapidly than those from the founder. This is true for both the time to bolting and the time to anthesis of the first flower (Table 2, Figure 1A). In the case of leaf number at flowering, line 49 tended to produce more leaves by the date of first flowering, whereas line 71 produced fewer (Figure 1B). Concerning leaf length, two lines, 69 and 71, produced shorter leaves than did the founder. Estimates of mutational effect on height at flowering were positive for each of the lines, and this effect was significant in the case of line 49 (Table 2). We note that three of the significant cases of homozygous effect were opposite to the expected direction (leaf length and number for line 71 and flowering date for line 76). These discrepancies may merely reflect the unreliability of the preliminary results but could be due to differences in genotypic expression in the environments of the assays (G × E interaction).
Estimates of dominance parameters scaled to the homozygous effect, k = d/a (Table 2), ranged widely (−13.9 < k < 1.2), but almost 2/3 of them (18/28) suggested partially or fully recessive gene action, regardless of the direction of the homozygous effect. For MA line 69, for example, sizable, positive estimates of d were obtained for both bolting and flowering time; thus, the earliness of the homozygous effect of this line, evidenced by the significant negative estimates of a, is largely recessive. Despite this wide range of estimates, the model of additive gene action cannot be rejected for many of the traits expressed in each line. Nevertheless, several cases of significant nonadditivity of gene action were found (Table 2).
The enhanced fitness of line 69 was estimated to be fully recessive, as were the reductions in leaf length of lines 69 and 71 (Table 2). The reduction in leaf number of line 71 appeared to be at least partially recessive, whereas the gene action conferring increased leaf number of line 49 cannot be distinguished from additive. The estimate of d for leaf number in line 69 suggests the possibility of overdominance with respect to leaf number (i.e., the mean of the F1 exceeds that of both parents), but, given the sampling variance for a, the data are also consistent with recessive action of alleles reducing leaf number in line 69. In three cases, the point estimates of k and h are near 1, suggesting dominance of the mutational effects (height at flowering, line 69; leaf length and bolting date, line 49). However, in none of these cases did the estimate of either d or a approach significance (Table 2).
Beyond these mutational effects on traits, environmental differences among flats and environmental variation within flats accounted for almost all the variation in traits (Table 3). For three traits, length of longest leaf, reproductive biomass and the number of fruits per plant, the variance attributable to differences among flats was considerable and significant, contributing 68%–75% of the variance due to random effects. In no case did maternal contributions account for a sizable portion of the variance. For the phenological traits, variance attributable to the maternal individual contributed significantly, accounting for 2.6% of the total; for leaf number, this component accounted for 5% of the total and was marginally significant. The estimate of maternal variance accounted for <1% of the total variance of the remaining traits.
TABLE 3.
Estimates of the mean for plants representing generation 0 and variance components for each trait in set A
| Set A | Mean | Ve | Vmaternal individual | Vgrowing flat |
|---|---|---|---|---|
| Bolting date | 22.91 | 2.98 | 0.10 | 0.61 |
| (0.14) | (0.06) | (0.19) | ||
| Flowering date | 29.30 | 3.24 | 0.11 | 0.66 |
| (0.15) | (0.07) | (0.20) | ||
| Height at flowering | 47.63 | 189.80 | 0.00 | 5.95 |
| (8.83) | (0.00) | (3.21) | ||
| Leaf no. | 13.17 | 2.54 | 0.16 | 0.31 |
| (0.12) | (0.08) | (0.10) | ||
| Leaf length | 43.00 | 21.45 | 0.00 | 56.91 |
| (1.00) | (0.00) | (14.99) | ||
| Rep. biomass | 0.53 | 0.01 | 0.00 | 0.03 |
| (0.00) | (0.00) | (0.00) | ||
| Fruit no. | 332.21 | 2964.9 | 0.00 | 6406 |
| (137.8) | (0.00) | (1701) |
Standard errors are given in parentheses.
Set B:
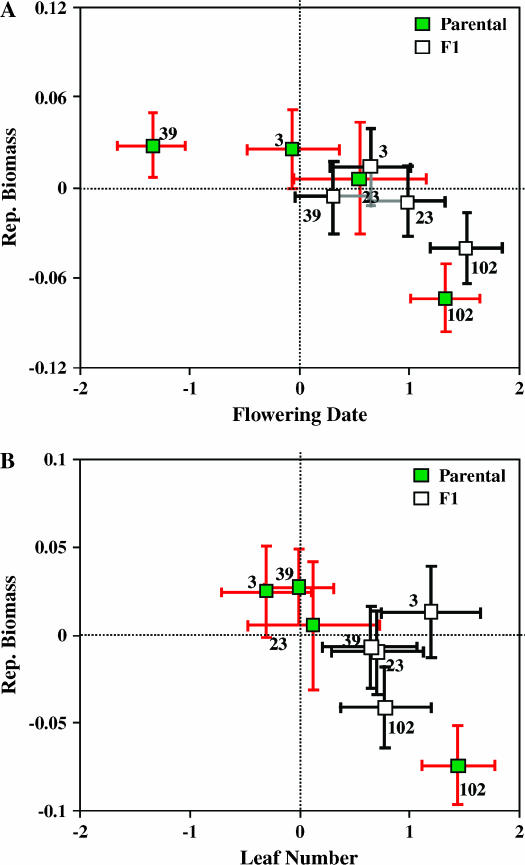
Of this set of four lines randomly chosen from generation 24, three of the lines exhibited significant effects of mutations (Table 4). Line 23, for which no effects were detected as significant, was the line having poor representation as progeny of self-matings (see materials and methods). With respect to reproductive biomass, the homozygous effect of line 102 was significantly lower than that of the founder (Figure 2). This line was also extreme in several other traits; relative to the founder, it was delayed in its time of bolting and flowering, bore more rosette leaves and was shorter at the time of flowering, and carried its fruits more densely along the stem. The homozygous effect of line 39 differed detectably from the founder only in its flowering time, which was earlier by 2/3 of a day, on average (Figure 2).
TABLE 4.
Estimates of the genetic parameters with respect to each trait for the MA lines included in set B
| Line 3
|
Line 23
|
Line 39
|
Line 102
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| k = d/a | k = d/a | k = d/a | k = d/a | |||||||||
| Set B | a (SE) | d (SE) | h | a (SE) | d (SE) | h | a (SE) | d (SE) | h | a (SE) | d (SE) | h |
| Bolting date | −0.13 | 0.74* | −5.91 | 0.46 | 0.4 | 0.88 | −0.34 | 0.57* | −1.69 | 0.56* | 0.64* | 1.16 |
| (0.29) | (0.36) | −2.45 | (0.39) | (0.43) | 0.94 | (0.24) | (0.29) | −0.34 | (0.24) | (0.29) | 1.08 | |
| Flowering date | −0.03 | 0.68 | −21.12 | 0.28 | 0.71 | 2.59 | −0.67* | 0.97* | −1.44 | 0.66* | 0.85* | 1.28 |
| (0.30) | (0.41) | −10.06 | (0.43) | (0.49) | 1.79 | (0.22) | (0.31) | −0.22 | (0.23) | (0.32) | 1.14 | |
| Ht at flowering | 0.02 | 0.87 | 49.9 | −1.25 | 0.16 | −0.13 | 0.30 | −1.24 | −4.10 | −6.85* | 0.04 | −0.01 |
| (1.08) | (1.63) | 25.45 | (1.56) | (1.90) | 0.43 | (0.93) | (1.39) | −1.55 | (0.96) | (1.43) | 0.49 | |
| Leaf no. | −0.15 | 1.35* | −8.93 | 0.06 | 0.65 | 10.44 | −0.0009 | 0.65 | −731.0 | 0.72* | 0.06 | 0.08 |
| (0.31) | (0.43) | −3.96 | (0.44) | (0.51) | 5.72 | (0.29) | (0.38) | −365 | (0.30) | (0.39) | 0.54 | |
| Leaf length | 0.46 | 0.71 | 1.55 | 0.27 | 0.15 | 0.54 | 0.83 | −0.31 | −0.37 | 0.05 | 0.88 | 16.23 |
| (0.72) | (1.08) | 1.27 | (1.04) | (1.27) | 0.77 | (0.61) | (0.92) | 0.31 | (0.63) | (0.95) | 8.61 | |
| Rep. biomass | 0.01 | 0.0006 | 0.05 | 0.003 | −0.01 | −4.25 | 0.01 | −0.02 | −1.48 | −0.04* | −0.0035 | 0.09 |
| (0.02) | (0.03) | 0.52 | (0.03) | (0.03) | −1.62 | (0.02) | (0.02) | −0.24 | (0.02) | (0.02) | 0.54 | |
| Final height | 3.76 | 2.87 | 0.76 | 0.25 | 4.02 | 16.05 | −2.54 | 3.75 | −1.48 | −10.86* | 2.99 | −0.27 |
| (2.91) | (4.34) | 0.88 | (4.21) | (5.09) | 8.52 | (2.58) | (3.79) | −0.24 | (2.68) | (3.93) | 0.36 | |
Notation is as in Table 2.
Figure 2.
Predicted values of traits expressed by the progeny of crosses of MA lines sampled at generation 24 (set B). Symbols are as in Figure 1. (A) Reproductive biomass vs. flowering date; (B) reproductive biomass vs. leaf number.
In conjunction with more limited detection of homozygous effects for this set of lines, we also detected fewer instances of significant allelic interaction (Table 4). In particular, for reproductive biomass we cannot reject the model of additive gene action, although the point estimate of k suggests that the increasing mutational effect in line 39 is recessive. Concerning the phenological traits, the delay in bolting and flowering found for line 102 was significantly dominant, and line 39's earliness in flowering was significantly recessive. As in set A, we found an instance suggestive of overdominance of the founder allele with respect to leaf number; the dominance estimate for line 3 was significantly positive, whereas its estimate of a was slightly negative (not statistically significant).
In this assay, as in the previous one, variation among the flats in which plants were grown contributed significantly to the overall variance for every trait (Table 5). Beyond this, the flat into which individuals were initially planted also contributed significantly to the variance of all but one trait (leaf length), but estimates of this component were in no case as much as a third of those accounting for variation among flats in which plants were grown. Estimates of the variance due to maternal effect, whether the maternal lineage or the maternal individual, were generally not significant. The earliest expressed trait, date of bolting, is an exception, as is leaf number. For these traits, variance due to maternal individual made a small (2%) but statistically significant contribution to the overall variance (Table 5).
TABLE 5.
Estimates of the mean for plants representing generation 0 and variance components for each trait in set B
| Set B | Mean | Ve | Vmaternal individual | Vgrowing flat | Vplanting flat |
|---|---|---|---|---|---|
| Bolting date | 60 | 4.01 | 0.20 | 2.10 | 0.88 |
| (0.16) | (0.10) | (0.47) | (0.21) | ||
| Flowering date | 66.3 | 4.50 | 0.00 | 4.70 | 1.36 |
| (0.18) | (0.00) | (1.02) | (0.30) | ||
| Height at flowering | 34.11 | 145.32 | 0.00 | 9.43 | 7.81 |
| (5.81) | (0.00) | (3.09) | (2.84) | ||
| Leaf no. | 47.2 | 14.03 | 0.26 | 1.65 | 0.16 |
| (0.56) | (0.18) | (0.45) | (0.16) | ||
| Leaf length | 50.2 | 62.08 | 0.00 | 0.00 | 3.92 |
| (2.44) | (0.00) | (0.00) | (1.31) | ||
| Rep. biomass | 0.18 | 0.04 | 0.00 | 0.02 | 0.00 |
| (0.00) | (0.00) | (0.00) | (0.00) | ||
| Final height | 348.6 | 1315.5 | 0.00 | 247.07 | 29.19 |
| (52.47) | (0.00) | (61.62) | (17.46) |
DISCUSSION
In this study, we have documented interactions between new mutations and alleles characterizing the founder in a subset of MA lines of A. thaliana. The point estimates of the parameters of gene action suggest that mutational effects can range from recessive through additive to dominant in their effects, and, indeed, over- and underdominant gene action was also estimated. In view of considerable sampling variance of these estimates, however, the hypothesis of additive gene action cannot be rejected for the expression of most of the traits in these eight lines. In cases where significant allelic interactions were detected, the mutational effect tended to be recessive or partially recessive, regardless of the direction of the effect. The sole exception was a clearly dominant mutational effect delaying bolting and flowering (line 102).
Before considering these results further, we emphasize two limitations on our inferences. First, of primary interest from an evolutionary perspective is the effect of individual mutations on fitness and other traits. Studies of this kind do not permit inference of dominance of single mutations because each MA line may differ from the founder by mutations at multiple loci. Thus, our estimates of the parameters of gene action, like those from most other studies of this kind (e.g., Peters et al. 2003) reflect the composite effects of new mutations at all these loci, rather than the effects of individual mutations. Moreover, to the extent that an MA line differs from the founder by multiple mutations, we could fail to detect a homozygous mutational effect if mutations having opposite effects on the phenotype have accumulated in a single line (i.e., association in repulsion phase). More important for the purposes of this study of gene action, we could fail to detect allelic interaction if mutations at different loci have homozygous effects in the same direction, but one is recessive and the other dominant. Alternatively, the appearance of overdominance could result if two mutations, one of dominant, positive effect and the other of recessive, negative effect, are fixed in an MA line. This phenomenon, termed associative overdominance, could account for our two cases of overdominance with respect to leaf number. We emphasize that in our MA lines, relative to other studies of the gene action of new mutations, it is reasonable to expect that few mutations contribute to the observed mutational effects, because relatively few generations separate each MA line from the founder (17 generations for set A and 24 for set B).
Second, we have assayed a small subset of the 117 MA lines that are available, because considerable replication of each cross is required to achieve acceptable statistical precision. Our choice to limit the number of lines studied allowed us to employ a crossing design and degree of replication that together provided sufficient power to detect effects that are relatively small (e.g., <2% for earlier bolting and 3.5% for enhanced fruit production), although of an evolutionarily considerable magnitude. Moreover, only 4 of the 8 lines were chosen at random; the remaining 4 evaluated in set A were chosen on the basis of suggestive (not statistically significant) evidence that they might be extreme with respect to one or more morphological traits, a common practice in studies of this kind (e.g., Lopez and Lopez-Fanjul 1993; Peters et al. 2003). Both because the number of lines is small and because some were included on the basis of preliminary findings, our study, like others of this kind, cannot yield generalizations about the dominance of single mutations. In particular, we cannot infer a distribution of dominance of individual mutations, as we have for homozygous effects of mutations for this set of MA lines (Shaw et al. 2002), nor can we infer a general relationship between a and d for new mutations. Nonetheless, to our knowledge, these are the first estimates of gene action of spontaneous mutations for a vascular plant.
Having noted these caveats, we consider the key findings of this study. Of particular evolutionary interest in MA studies is the effect of new mutations on the traits that most directly mediate individual fitness, because mutations affecting these traits can be expected to influence the evolutionary dynamics of adaptation most directly. More specifically, models accouning for the maintenance of genetic variation, the evolution of inbreeding avoidance, the extinction of small populations, and ecological specialization focus on the role of new mutations affecting individual fitness in making evolutionary predictions. Of the eight lines examined in this study, for three we detected significant homozygous effects with respect to the primary component of fitness (number of fruits per plant in set A and reproductive biomass in set B). In addition to two lines expressing fitness inferior to the founder, one line significantly exceeded the founder in fitness. This finding confirms our previous inference that fitness-enhancing mutations have occurred in this set of MA lines (Shaw et al. 2002, 2003). We emphasize the importance of substantial replication in detecting these effects conclusively; in previous studies in which fitness-enhancing homozygous effects were not detected as significant, replication was considerably less (e.g., Peters et al. 2003). In the case of three additional lines, our evidence suggests an enhancement of fitness relative to the founder but was not conclusive (Tables 2 and 4). Still greater replication would be necessary to detect subtler effects if they exist. Even though set B was more highly replicated than set A for most crosses, however, we detected fewer mutational effects for those lines. This may have resulted, in part, because the lines in set B were sampled at random, rather than chosen as phenotypically extreme, as in set A. In addition, it appears that the assay of set B reflected effects less precisely than did that of set A. For example, mutational effects on biomass on the order of 5% were not detected as significant in the more highly replicated set B, whereas effects on biomass of 2.4% were detected in set A.
Considering the mutational effects that we have documented collectively over all the traits, we focus on cases for which evidence of a mutational effect is most conclusive, noting the difficulty of inference about mutations of more slight effect because they are less likely to be detected with statistical support. For the 21 cases of significant homozygous mutational effect on a trait, we have found that mutations tend to range in gene action from recessive to additive, regardless of the direction of the effect. We obtained conclusive evidence for a dominant mutational effect only for the delay in bolting and flowering of line 102 (Table 4, Figure 2). In this case of effects on phenology, our evidence is suggestive of gene action that is associated with the direction of the effect, given that, for three lines manifesting early development, the mutational effect ranged from recessive (e.g., line 69) to additive (e.g., line 76, Tables 2 and 4, and Figures 1 and 2). However, among known Arabidopsis mutations, those delaying flowering tend toward recessive action (the Arabidopsis Information Resource, http://www.arabidopsis.org).
In six cases a mutational effect was detected in the progeny of the crosses but not in the corresponding pure lines (i.e., significant d but not a). In three of these, the heterozygote was larger with respect to some traits than either homozygote, including one case of greater reproductive biomass (line 71). In the remaining cases, the heterozygote was more extreme in phenology than either parent (line 49, earlier; lines 3 and 39, later). Such findings are suggestive of overdominant gene action but could result if two (or more) alleles that affect the trait in the opposite direction occur in a single MA line (i.e., associative overdominance; see Fry 2004 for interpretation of apparent overdominance in the context of MA studies).
The relatively few studies of gene action based on MA lines derived in other organisms support a generalization of recessivity of spontaneous mutations. Lopez and Lopez-Fanjul (1993) found allelic interaction that was additive or recessive, in approximately equal proportions, for mutational effects on abdominal bristle number in 18 of 22 MA lines of D. melanogaster, whereas they found fewer instances of recessive gene action for lines derived by selection on a highly inbred base population. Only 3 of 22 MA lines and none of the lines obtained by selection exhibited unambiguous dominance of the phenotype expressed by the MA line (2 of increasing effect in homozygous state and 1 of decreasing effect).
Focusing on mutations affecting fitness, we found evidence of differences among them in their gene action. The deleterious effect of line 49 appears to be partially recessive, whereas that of line 102 appears to be additive. The fitness enhancement of line 69 is fully recessive. For lines 76, 3, and 39, whose fitness enhancement is less conclusive, point estimates of d suggest a range of gene action for their effects, with line 76 apparently additive and lines 3 and 39 showing recessive action, but these inferences must remain tentative, because sampling variance obscures the gene action for these small genetic effects.
If our findings concerning gene action of mutations directly affecting fitness under the conditions of this study apply beyond this species, they do not support the view that plant populations steadily incur partially recessive, deleterious mutations. This view is a central component of theory to account for inbreeding depression (e.g., Charlesworth et al. 1990). Among the few lines we studied, line 102 in set B appears to be an exception to the expectation that mutations of deleterious effect are generally recessive or partially so, although we cannot definitively rule out weak recessivity in this case. Of the eight lines we studied, only line 71 arguably corresponds to the expectation of a weakly deleterious mutation whose effect is masked in the heterozygote. Moreover, under the hypothesis, we might have expected that a fitness-enhancing mutation would tend to be dominant (e.g., Fry 2004), although evidence for recessive fitness-enhancing mutations was given by Charlesworth (1992). In fact, our clearest case of a fitness-enhancing effect was found to be fully recessive. In sum, our findings suggest that the influx of deleterious mutations does not, by itself, account for the ubiquity of inbreeding depression. Further studies of the distribution of mutational effects and the relationship between homozygous and heterozygous effect in other plants are needed to assess the generality of our findings and the role in inbreeding depression of ongoing mutation generating deleterious alleles.
For mutations affecting fitness, estimates of the average dominance of new mutations have been obtained from a small, but growing number of experiments, primarily in D. melanogaster. Inferences have been clouded by doubts about experimental procedures (see, e.g., Garcia-Dorado and Caballero 2000) and biases of estimation (Caballero et al. 1997). Garcia-Dorado and Caballero (2000) have reviewed early studies by Mukai et al. (1972) that approximated the average dominance, h, of mildly deleterious mutations at 0.4, i.e., slightly recessive (see also Fry and Nuzhdin 2003). Their reanalysis of experiments by Ohnishi (1977) suggests that effects were much more nearly recessive, h ∼ 0.1. From newly conducted experiments on MA lines of D. melanogaster advanced to generation 250, Chavarrias et al. (2001) inferred an intermediate degree of recessiveness of effects of mutations on competitive viability (h = 0.3). Fry and Nuzhdin (2003) inferred still more recessive gene action (h = 0.17) for a set of MA lines at generation 33. This contrasts with their estimate for additivity of effects of insertions of the transposable element, copia, (h = 0.51). This finding, with their own reanalysis of separate data from Ohnishi, supported their hypothesis that mutations due to transposable elements have considerably greater effect in heterozygotes, compared to point mutations, which tend toward more nearly recessive action.
Szafraniec et al. (2003) have estimated average dominance, h, of single EMS-induced mutations that slightly reduce colony size in the yeast, Saccharomyces cerevisiae, as 0.197, reflecting the largely recessive action of most of the mutations. Of the 74 mutations studied, however, three acted as partial dominants, exceeding h = 0.5. Peters et al. (2003), using a set of 19 lines derived following EMS mutagenesis of the N2 strain of Caenorhabditis elegans, found that mutations were, on average, largely recessive (h ∼ 0.1). As in the study of Szafraniec et al. (2003), they observed considerable variability in the dominance of deleterious effects, which ranged from recessive to dominant for different lines. Moreover, for several lines whose homozygous fitness was not significantly inferior to that of the progenitor, they detected fitness enhancement in the heterozygote, suggesting overdominance. The authors acknowledged that occurrence of numerous fitness-enhancing mutations could produce these results via associative overdominance but suggested that this is unlikely. Fry (2004), however, has pointed out that even a low fraction of fitness-enhancing mutations could account for the apparent overdominance. We further note that there is suggestive, although not significant, evidence of homozygous lines having fitness greater than that of the progenitor (Peters et al. 2003, Figure 2B, lines E13, E46; see also their Figure 1).
We have graphically examined associations between mutational effects on fitness, on the one hand, and on a phenological and a morphological trait, on the other (Figures 1 and 2). We note that, in the parental MA lines, reproductive fitness tends to increase as development time and number of leaves decrease, trends that Pigliucci et al. (2003) also documented in trait correlations among eight Scandinavian accessions of A. thaliana. Because each MA line may harbor multiple mutations distinguishing it from the founder and other MA lines, such trait associations among MA lines could appear even if different traits are influenced by distinct mutations that arose in the same line. Alternatively, particularly given that we expect few mutations per line in 24 generations of MA, they may reflect pleiotropic effects of mutations.
We conducted the crosses in a design that would allow us to distinguish effects of nuclear mutations on phenotype from contributions due to maternal effect. Further, our design avoids confounding of effects due to maternal environment (e.g., variation among replicate, genotypically identical mothers within lines) with those due to mutations in either nuclear or organelle genomes. Mutations in organelle genomes, which are maternally inherited in A. thaliana, would be evidenced by differences in progeny phenotype between reciprocally crossed MA lines. Male-biased transmission of mutations, previously inferred in A. thaliana for mutations induced by UV in a single generation (Whittle and Johnston 2003), is not expected as a source of reciprocal differences in our study because, under our MA protocol, most mutations that are retained are expected to become fixed within few generations.
In our analyses of the full models, variances attributable to both environmental and cytoplasmic maternal effects tended to be estimated as slightly negative. In the few cases of positive estimates, they were very small. For no trait did we obtain even suggestive evidence of cytoplasmic (i.e., maternal lineage) transmission of a mutational effect. Concerning environmental maternal effects, these were statistically significant in the case of few traits, but even in these cases they contributed no more than 2% of the variance. Thus, neither source of maternal effect makes a strong contribution to phenotypic differences in this study. We acknowledge that the design likely did not have adequate power to reveal subtle effects of these kinds. Nevertheless, the nuclear mutational effects we estimated were detected as significant regardless of whether either maternal component was included in the statistical model.
In conclusion, we have found that the composite effects of spontaneous mutations generally range from additive to recessive in gene action, regardless of the direction of the mutational effect in homozygous state. We obtained conclusive evidence for a single exception, a dominant mutational effect delaying bolting and flowering. This study has confirmed mutational enhancement of reproductive fitness of one line and provided further suggestive evidence of fitness increases in other lines. Further studies of gene action of these lines are under way.
Acknowledgments
We thank Lorelle Berkeley, Chris Kavanaugh, and Jason Hill for assistance with the crosses. Frank Shaw provided key support for analysis of the data. Peter Tiffin and Matt Rutter made valuable suggestions for improving the manuscript, as did John Kelly, Joy Bergelson, and an anonymous reviewer. We gratefully acknowledge funding from Pioneer Hi-Bred International for establishment of the MA lines and from the U.S. National Science Foundation (DEB-9629457 and DEB-9981891).
References
- Abbott, R. J., and M. F. Gomes, 1989. Population genetic structure and outcrossing rate of Arabidopsis thaliana. Heredity 62: 411–418. [Google Scholar]
- Caballero, A. M., and W. G. Hill, 1992. Effects of partial inbreeding on fixation rates and variation of mutant genes. Genetics 131: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero, A. M., and P. D. Keightley, 1994. A pleiotropic nonadditive model of variation in quantitative traits. Genetics 138: 883–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero, A., P. D. Keightley and M. Turelli, 1997. Average dominance for polygenes: drawbacks of regression estimates. Genetics 147: 1487–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B, 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140: 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., M. T. Morgan and B. Charlesworth, 1990. Inbreeding depression, genetic load, and the evolution of outcrossing rates in a multilocus system with no linkage. Evolution 44: 1469–1489. [DOI] [PubMed] [Google Scholar]
- Chavarrias, D., C. Lopez-Fanjul and A. Garcia-Dorado, 2001. The rate of mutation and the homozygous and heterozygous mutational effects for competitive viability: a long-term experiment with Drosophila melanogaster. Genetics 158: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake, J. W., B. Charlesworth, D. Charlesworth and J. F. Crow, 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D.S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Ed. 4. Prentice Hall, Harlow, UK.
- Fry, J. D., and S. V. Nuzhdin, 2003. Dominance of mutations affecting viability in Drosophila melanogaster. Genetics 163: 1357–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, J. D., 2004. How common are overdominant mutations? Genetics 167: 1031–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dorado, A., and A. Caballero, 2000. On the average coefficient of dominance of deleterious spontaneous mutations. Genetics 155: 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1927. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc. Cambridge Phil. Soc. 23: 838–844. [Google Scholar]
- Joseph, S. B., and D. W. Hall, 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae: more beneficial than expected. Genetics 168: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki, T. J., N. H. Barton and J. D. Fry, 1997. Mutational collapse of fitness in marginal habitats and the evolution of ecological specialization. J. Evol. Biol. 10: 407–429. [Google Scholar]
- Keightley, P.D., and A. Caballero, 1997. Genomic mutation rates for lifetime reproductive output and lifespan in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 94: 3823–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A. S., 1988. Deleterious mutation and the evolution of sexual reproduction. Nature 336: 435–440. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1976. The maintenance of genetic variability by mutation in a polygenic character with linked loci. Genet. Res. 26: 221–235. [DOI] [PubMed] [Google Scholar]
- Lande, R., 1994. Risk of population extinction from fixation of new deleterious mutations. Evolution 48: 1460–1469. [DOI] [PubMed] [Google Scholar]
- Lopez, M. A., and C. Lopez-Fanjul, 1993. Spontaneous mutation for a quantitative trait in Drosophila melanogaster. II. Distribution of mutant effects on the trait and fitness. Genet. Res. 61: 117–126. [DOI] [PubMed] [Google Scholar]
- Lynch, M., J. Blanchard, D. Houle, T. Kibota, S. Schultz et al., 1999. Perspective: spontaneous deleterious mutation. Evolution 53: 1–19. [DOI] [PubMed] [Google Scholar]
- Lynch, M., J. Conery and R. Bürger, 1995. Mutation accumulation and the extinction of small populations. Am. Nat. 146: 489–518. [Google Scholar]
- Lynch, M., and W. G. Hill, 1986. Phenotypic evolution by neutral mutation. Evolution 40: 915–935. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., R. F. Lyman, M. S. Jackson, C. Terzian and W. G. Hill, 1992. Polygenic mutation in Drosophila melanogaster: estimates from divergence among inbred strains. Evolution 46: 300–316. [DOI] [PubMed] [Google Scholar]
- Mukai, T., S. I. Chigusa, L. E. Mettler and J. F. Crow, 1972. Mutation rate and dominance of genes affecting viability in Drosophila melanogaster. Genetics 72: 333–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi, O., 1977. Spontaneous and ethyl methane-sulfonate induced mutations controlling viability in Drosophila melanogaster. III. Heterozygous effect of polygenic mutations. Genetics 87: 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A. D., D. L. Halligan, M. C. Whitlock and P. D. Keightley, 2003. Dominance and overdominance of mildly deleterious induced mutations for fitness traits in Caenorhabditis elegans. Genetics 165: 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci, M., H. Pollard and M. B. Cruzan, 2003. Comparative studies of evolutionary responses to light environments in Arabidopsis. Am. Nat. 161: 68–82. [DOI] [PubMed] [Google Scholar]
- Shaw, F. H., C. J. Geyer and R. G. Shaw, 2002. A comprehensive model of mutations affecting fitness and inferences for Arabidopsis thaliana. Evolution 56: 453–463. [DOI] [PubMed] [Google Scholar]
- Shaw, R. G., D. L. Byers and E. Darmo, 2000. Spontaneous mutational effects on reproductive traits of Arabidopsis thaliana. Genetics 155: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, R. G., and F. H. Shaw, 1994. Quercus: programs for quantitative genetic analysis using maximum likelihood (http://www.cbs.umn.edu/eeb/events/quercus.shtml).
- Shaw, R. G., F. H. Shaw and C. J. Geyer, 2003. What fraction of mutations reduces fitness?: a reply to Keightley and Lynch. Evolution 57: 686–689. [Google Scholar]
- Szafraniec, K., D. M. Wloch, P. Sliwa, R. H. Borts and R. Korona, 2003. Small fitness effects and weak genetic interactions between deleterious mutations in heterozygous loci of the yeast Saccharomyces cerevisiae. Genet. Res. 82: 19–31. [DOI] [PubMed] [Google Scholar]
- Turelli, M., 1984. Heritable genetic variation via mutation-selection balance: Lerch's zeta meets the abdominal bristle. Theor. Popul. Biol. 25: 138–193. [DOI] [PubMed] [Google Scholar]
- Whittle, C., and M. O. Johnston, 2003. Male-biased transmission of deleterious mutations to the progeny in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 100: 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.-S., J. Wang and W. G. Hill, 2002. Pleiotropic model of maintenance of quantitative genetic variance at mutation–selection balance. Genetics 161: 419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.-S., J. Wang and W. G. Hill, 2004. a Influence of dominance, leptokurtosis and pleiotropy of deleterious mutations on quantitative genetic variation at mutation-selection balance. Genetics 166: 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.-S., J. Wang and W. G. Hill, 2004. b Redistribution of gene frequency and changes of genetic variation following a bottleneck in population size. Genetics 167: 1475–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]