Abstract
The homothallic, filamentous ascomycete Sordaria macrospora is self-fertile and produces sexual fruiting bodies (perithecia) without a mating partner. Even so, S. macrospora transcriptionally expresses two pheromone-precursor genes (ppg1 and ppg2) and two pheromone-receptor genes (pre1 and pre2). The proteins encoded by these genes are similar to α-factor-like and a-factor-like pheromones and to G-protein-coupled pheromone receptors of the yeast Saccharomyces cerevisiae. It has been suggested that in S. macrospora, PPG1/PRE2 and PPG2/PRE1 form two cognate pheromone–receptor pairs. To investigate their function, we deleted (Δ) pheromone-precursor genes (Δppg1, Δppg2) and receptor genes (Δpre1, Δpre2) and generated single- as well as double-knockout strains. No effect on vegetative growth, fruiting-body, and ascospore development was seen in the single pheromone-mutant and receptor-mutant strains, respectively. However, double-knockout strains lacking any compatible pheromone-receptor pair (Δpre2/Δppg2, Δpre1/Δppg1) and the double-pheromone mutant (Δppg1/Δppg2) displayed a drastically reduced number of perithecia and sexual spores, whereas deletion of both receptor genes (Δpre1/Δpre2) completely eliminated fruiting-body and ascospore formation. The results suggest that pheromones and pheromone receptors are required for optimal sexual reproduction of the homothallic S. macrospora.
THE life cycle of ascomycetes can be either heterothallic or homothallic. Sexual reproduction in heterothallic fungi is possible only between cells of opposite mating type. However, homothallic ascomycetes are self-fertile and able to complete the sexual cycle without a mating partner. It has been shown that signaling through interaction of pheromone-receptor pairs plays an essential role during the mating of two cells of a heterothallic fungus (Bölker and Kahmann 1993; Kothe 1999; Casselton 2002; Souza et al. 2003). So far, the best-described model of pheromone involvement in the mating process is the pheromone-response system in the heterothallic ascomycetous yeast Saccharomyces cerevisiae. Here, pheromone-receptor interaction triggers a G-protein-linked signal-transduction pathway, which induces the expression of mating-specific genes (Bardwell 2005). Cells of one mating type (MATa) secrete a-factor lipopeptide pheromones, derived from precursors with C-terminal CaaX (C, cysteine; a, aliphatic; and X, any amino acid residue) motifs, whereas cells of the other mating type (MATα) secrete α-factor-peptide pheromones (Caldwell et al. 1995; Casselton 2002). Each kind of pheromone is able to bind only to cell-surface receptors present on cells of the opposite mating type. In S. cerevisiae, the pheromone receptor for the α-factor and the a-factor are Ste2p and Ste3p, respectively (Jenness et al. 1983; Hagen et al. 1986). Both are members of the large family of G-protein-coupled receptors (GPCRs), containing seven-transmembrane (7-TM) domains.
Interestingly, genes encoding two different pheromone precursors have been found not only in several heterothallic filamentous ascomycetes, such as Neurospora crassa, Podospora anserina, Magnaporthe grisea, and Cryphonectria parasitica, but also in the homothallic filamentous ascomycete Sordaria macrospora (Zhang et al. 1998; Shen et al. 1999; Pöggeler 2000; Bobrowicz et al. 2002; Coppin et al. 2005). Like the closely related N. crassa, the homothallic ascomycete S. macrospora has two pheromone-precursor genes, termed ppg1 and ppg2, and two pheromone-receptor genes, named pre1 and pre2. The pheromone-precursor gene ppg1 is predicted to encode an α-factor-like peptide pheromone and the ppg2 gene an a-factor-like lipopeptide pheromone (Pöggeler 2000). The deduced gene products of pre1 and pre2 show structural similarities to the yeast a-factor receptor Ste3p and the α-factor receptor Ste2p, respectively (Pöggeler and Kück 2001). Similar to yeast, the expression of pheromone genes and pheromone-receptor genes is supposed to be controlled directly by transcription factors encoded by the mating-type genes (Debuchy 1999). In heterothallic filamentous ascomycetes, either mat A or mat a mating-type specific transcription factors are present in mat A or mat a strains. Thus, pheromone-encoding genes of heterothallic ascomycetes are predominantly expressed in a mating-type-specific manner (Zhang et al. 1998; Shen et al. 1999; Bobrowicz et al. 2002; Kim et al. 2002; Turina et al. 2003; Coppin et al. 2005). However, both types of mating-type regulators are expressed in the same individual in the homothallic ascomycete S. macrospora (Pöggeler et al. 1997). For this reason, not surprisingly, both pheromone-precursor genes and pheromone-receptor genes of S. macrospora have been shown to be transcriptionally expressed during the entire sexual development (Pöggeler 2000; Pöggeler and Kück 2001).
Recently, it was demonstrated that male and female fertility of heterothallic mycelial ascomycetes depends on interactions of pheromones with their specific receptors. When pheromone genes were deleted, male spermatia were no longer able to fertilize the female partner, proving that pheromones are crucial for the fertility of male spermatia (Kim et al. 2002; Turina et al. 2003; Coppin et al. 2005). In P. anserina, the function of pheromones is restricted to fertilization, while the N. crassa lipopeptide-pheromone gene mfa-1 has also been shown to be involved in female sexual development, ascospore production, and vegetative growth of both mating types (Kim et al. 2002; Coppin et al. 2005). Kim and Borkovich (2004) demonstrated that deletion of the N. crassa pre-1 pheromone-receptor gene does not affect vegetative growth or male fertility. However, protoperithecia from Δpre-1 mat A mutants were shown to be female sterile because their trichogynes are unable to recognize and fuse with mat a spermatia.
It has recently been demonstrated that the disruption of the S. macrospora ppg1 gene, encoding the α-factor-like peptide pheromone, prevents production of the peptide pheromone. However, this does not affect vegetative growth, fruiting-body, and ascospore development (Mayrhofer and Pöggeler 2005). Using a heterologous yeast system, it has been shown that the S. macrospora receptor PRE2 facilitated all aspects of the S. cerevisiae pheromone response in S. cerevisiae MATa cells lacking the Ste2p receptor, when activated by the S. macrospora peptide pheromone. Therefore one may conclude that the receptor encoded by the pre2 gene functions as a GPCR in S. macrospora, too (Mayrhofer and Pöggeler 2005).
In contrast to the heterothallic N. crassa, fruiting-body development is an apandrous process in the homothallic S. macrospora and lacks the cooperative interaction of two opposite mating-type strains. In S. macrospora, fruiting-body development starts with the formation of female gametangia called ascogonia. The ascogenous cells are enveloped by sterile hyphae to form protoperithecia as fruiting-body precursors; conidiospores, spermatia, and trichogynes are lacking. After autogamous fertilization (i.e., pairwise fusion of nuclei present within the ascogonium, without cell fusion having taken place), the protoperithecia differentiate inner ascus initials, embedded in sterile paraphyses, and an outer-pigmented peridial tissue. The mature fruiting bodies, termed perithecia, harbor 200–300 asci. Meiosis and a postmeiotic division lead to eight meiotically derived ascospores in each ascus (Esser and Straub 1958; Esser 1982). The life cycle of S. macrospora suggests that pheromones are needed neither for attracting a mating partner nor for initializing fertilization events. Thus, S. macrospora is an ideal model system for analyzing the effects of pheromones on postfertilization events.
In an attempt to understand the role of pheromones and pheromone receptors during postfertilization events, we deleted the pre1 a-factor-like and the pre2 α-factor-like receptor gene as well as the a-factor-like pheromone-precursor gene ppg2 of S. macrospora. Deletion (Δ) of the pheromone-precursor gene ppg2 or the pheromone-receptor genes resulted in no obvious phenotype. We therefore generated double-mutant strains to invalidate both pheromone systems. Double-mutant strains Δpre2/Δppg2, Δpre1/Δppg1, and Δppg1/Δppg2 displayed drastically reduced fruiting-body, ascus, and ascospore development, while the double-receptor mutant Δpre1/Δpre2 was not able to produce any fruiting bodies or ascospores. These results suggest that pheromone signaling is important for optimal sexual reproduction of homothallic ascomycetes.
MATERIALS AND METHODS
Strains and culture conditions:
Cloning and propagation of recombinant plasmids was done under standard conditions (Sambrook et al. 2001). The Escherichia coli strain SURE was used as the host for plasmid amplification (Greener 1990). All fungal strains used in this work are summarized in Table 1. The S. macrospora strain K (S17736) has a wild-type phenotype and was derived from our laboratory collection (Department of General and Molecular Botany, Bochum, Germany). All S. macrospora strains were cultivated on cornmeal medium or complete medium (Esser 1982; Nowrousian et al. 1999). S. macrospora strains used for the isolation of RNA were grown in Westergaard's synthetic medium (Westergaard and Mitchell 1947).
TABLE 1.
S. macrospora strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| S17736 | Wild type | Lab collectiona |
| S23443 | fus1-1 | Lab collectiona |
| S52063 | Δppg1∷hph | Mayrhofer and Pöggeler (2005) |
| S66385 | Δppg1∷hph/fus1-1 | S. Mayrhofer and S. Pöggeler (unpublished results) |
| S2.2-1 | Single-spore isolate Δpre1∷hph | This study |
| S60289 | Single-spore isolate Δpre2∷hph | This study |
| S54761 | Single-spore isolate Δppg2∷hph | This study |
| S59784 | Δppg2∷hph | This study |
| S59786 | Δppg2∷hph/fus1-1 | This study |
| S60446 | Δpre2∷hph | This study |
| S60447 | Δpre2∷hph/fus1-1 | This study |
| S66197 | Δppg1∷hph/Δppg2∷hph | This study |
| SA1-4 | Δpre1∷hph/Δpre2∷hph | This study |
| SD4-2 | Δpre1∷hph/Δppg1∷hph | This study |
| S61352 | Δpre2∷hph/Δppg2∷hph/fus1-1 | This study |
| S61357 | Δpre2∷hph/Δppg2∷hph | This study |
Department of General and Molecular Botany, Ruhr-University of Bochum, Germany.
Construction of knockout and rescue plasmids:
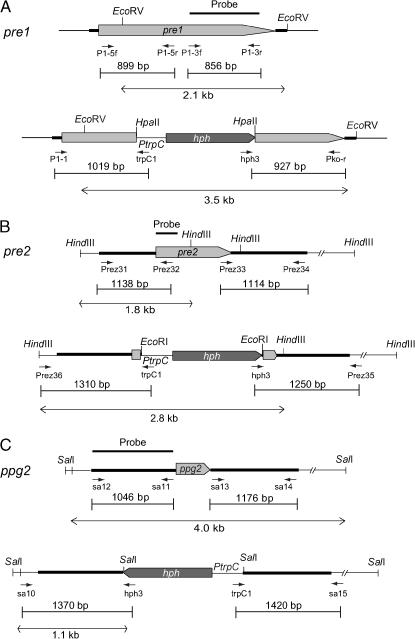
As described for knockouts of N. crassa genes, the deletion construct for the knockout of the S. macrospora pheromone-receptor gene pre1 was created utilizing homologous recombination in yeast (http://www.dartmouth.edu/∼neurosporagenome/protocols.html). The 5′- and 3′-regions of the pre1 gene were amplified from genomic DNA of the S. macrospora wild-type strain with primer pairs P1-5f/P1-5r and P1-3f/P1-3r, respectively. Within these PCR reactions specific 29-bp overhangs were added to the 5′- and 3′-pre1 fragments, homologous to plasmid pRS426 and the hygromycin-resistance cassette (hph), respectively. The sequences of all PCR primers used in this work are summarized in Table 2. The hph cassette was generated from plasmid pCB1003 (Carroll et al. 1994) by PCR using primers hph-f and hph-r. All three PCR fragments were cotransformed together with EcoRI–XhoI linearized plasmid pRS426 (Christianson et al. 1992) into yeast strain J69-4A (James et al. 1996), where homologous recombination took place (http://www.dartmouth.edu/∼neurosporagenome/protocols.html). The resulting plasmid pPRE1-KO was isolated from yeast strain J69-4A according to the method described at http://www.dartmouth.edu/∼neurosporagenome/protocols.html. Plasmid pPRE1-KO was used as a template to amplify the pre1-hph cassette with oligonucleotides P1-5f and P1-3r as primers. The 3222-bp PCR fragment obtained was transformed into a S. macrospora wild-type strain to achieve a knockout of the pre1 gene via homologous recombination (Figure 1).
TABLE 2.
PCR primers used in this work
| Oligo | Sequence (5′–3′) |
|---|---|
| P1-5f | GTAACGCCAGGGTTTTCCCAGTCACGACGCCAAGAACGAAATGGCCCTC |
| P1-5r | CCAAAAATGCTCCTTCAATATCAGTTAACTTCTGTCGTTGTGAACGAGAC |
| P1-3f | GAGTAGATGCCGACCGGGAACCAGTTAACCCAAGCTGTATCAAGAATACG |
| P1-3r | GCGGATAACAATTTCACACAGGAAACAGCTCCTAGAGTGGTGTTCTTGTG |
| hph-f | GTTAACTGATATTGAAGGAGCATTTTTGG |
| hph-r | GTTAACTGGTTCCCGGTCGGCATCTACTC |
| Prez31 | TCACATCCAGCGACCCGTGCT |
| Prez32 | GAATTCGGGGATGCTGAGCACAGTGG |
| Prez33 | GGGCCCGTGAGAATTGAGTGGACAATTG |
| Prez34 | GGCCCGTAGTTATAAGGGGTGTCAAGA |
| sa11 | CCATGGTGATGGTTTTAGGGTTGTTTGATA |
| sa12 | CCATGGTGCCGCGTGGGTGGTCATC |
| sa13 | GGGCCCTCACAGTTGTCTTGAAGGG |
| sa14 | GGGCCCGTTCAGTCTGGCATTTCTGG |
| trpC1 | GATCCGCCTGGACGACTAAACC |
| hph3 | ACTCGTCCGAGGGCAAAGGAATAG |
| P1-1 | ATGAACAACACTTTCTACACCACCG |
| Pko-r | TCCTTCTGGGAAGACCACACG |
| Prez36 | GACAATAAGACATTCTGGATGTCTC |
| Prez35 | GTCGATGCCTGCACGCGCGAA |
| sa10 | CGGAGTGAAGTTGCCGTTGCCG |
| sa15 | GATATCTGTTGATGTGGTTTCATG |
| MrezS2 | AGTATACGAACCGCGAAGACAA |
| Mrez32 | TTTCTGAATTCCTCATATGGAGAA |
| PrezS3 | CTCATGACGCCCCGCCGTCGGTT |
| PrezS4 | TGGTCCACATGTGCATGACCA |
| C-gpd | TTGGTGGTGCAAGAGGCGTTG |
| N-gpd | GGCATCAACGGTTTCGGCCG |
Figure 1.
Construction of deletion strains. (A) Genomic pre1 region and construction of the Δpre1 gene replacement vector. Positions of primers used to amplify the disruption construct from plasmid pPRE1-KO and to verify the homologous recombination at the pre1 locus are indicated. (B) Genomic pre2 region and construction of the Δpre2 gene replacement vector. Positions of primers used to amplify the disruption construct from plasmid pPRE2-KO and to verify the homologous recombination at the pre2 locus are indicated. (C) The genomic ppg2 region and construction of the Δppg2 gene replacement vector. Positions of primers used to amplify the disruption construct from plasmid pPPG2-KO and to verify the homologous recombination at the ppg2 locus are indicated. PtrpC, A. nidulans trpC promoter.
Deletion of the S. macrospora pheromone-receptor gene pre2 was generated as follows. The 5′- and 3′-regions of the pre2 gene were amplified by PCR from genomic DNA of the wild-type S. macrospora strain. The oligonucleotide pairs Prez31/Prez32 and Prez33/Prez34 were used as primers (Figure 1; Table 2). Amplification with Prez33 and Prez34 generated ApaI sites at the ends. A pre2 fragment (1138 bp) containing the 5′-region of the pre2 gene was inserted into vector pGEM-T (Promega, Mannheim, Germany), resulting in plasmid p488.3. Plasmid p490.1 was generated by cloning the 1114-bp 3′-pre2 fragment into pGEM-T. The 1.1-kb ApaI fragment encoding the 3′-pre2 region was then inserted into the ApaI site of plasmid p488.3. The resulting plasmid p509.881 included the 5′-region of the pre2 gene adjacent to the 3′-region of the gene. The two sequences were separated by a single EcoRI restriction site. Next, the 1.4-kb EcoRI hph cassette of pCB1003 (Carroll et al. 1994) was inserted into the single EcoRI site of plasmid p509.881. The resulting plasmid pPRE2-KO was used as a template to amplify the pre2-hph cassette with oligonucleotides Prez31 and Prez34 as primers. The 3652-bp PCR fragment obtained was transformed into a S. macrospora wild-type strain to create a knockout of the pre2 gene by homologous recombination (Figure 1).
A similar strategy was followed to create the deletion of the ppg2 pheromone-precursor gene. The 1046-bp ppg2 5′-region was amplified from genomic DNA from S. macrospora wild type by PCR using oligonucleotides sa11 and sa12 as primers. Amplification with sa11 created a SalI restriction site. The PCR amplicon was ligated into PCR-cloning vector pDrive (QIAGEN, Hilden, Germany) to generate p433.14. Plasmid p428.4 contained the ppg2 3′-region. It was obtained by amplifying a 1176-bp fragment with the primer pair sa13/sa14, both generating ApaI ends, followed by cloning the amplicon into vector pGEM-T. The 1.2-kb ApaI fragment of plasmid p428.4 was afterward inserted into the ApaI site of p433.14. The resulting plasmid containing the promoter and terminator regions of the ppg2 gene, separated by a single SalI site, was termed p443.7. The ppg2 deletion construct pPPG2-KO was generated by cloning the 1.4-kb SalI hph cassette of pCB1003 (Carroll et al. 1994) into the single SalI site of plasmid p443.7. The 3.6-kb PCR fragment, which was amplified with oligonucleotides sa12/sa14 from plasmid pPPG2-KO, was used for transformation of S. macrospora wild type to achieve deletion of the ppg2 gene by means of homologous recombination (Figure 1).
Two different rescue vectors were designed to complement double-mutant strains with the coding sequences of the pre2 and the ppg2 gene, respectively. For these experiments, the nourseothricin-resistance gene nat1 was used as a selectable marker for transformation (Krugel et al. 1993). In plasmid pNat-1, the nat1 gene was put under the control of the gpd promotor and the trpC terminator of Aspergillus nidulans (U. Kück and S. Pöggeler, unpublished results). To complement the pre2 deficiency in the S. macrospora double-mutant strains Δpre2/Δppg2 and Δpre1/Δpre2, plasmid pNatpre2 was generated, which contained a wild-type 2.2-kb PstI–pre2 fragment. The lack of ppg2 in the double-mutant Δpre2/Δppg2 was complemented by pNatppg2. This plasmid carried a genomic 1.9-kb SspI–ppg2 fragment.
Single-mutant strains Δpre1, Δpre2, and Δppg2 were identified by Southern blot analysis (supplemental Figure S1 at http://www.genetics.org/supplemental/). Genomic DNA was isolated as described previously (Pöggeler et al. 1997). Successful homologous recombination was confirmed by PCR amplification. The 5′-flanking region of the pre1 gene was amplified with primers P1-1 and trpC1, whereas hph3 and Pko-r verified the 3′-flanking region. Primers Prez36 and trpC1 were used for the 5′-flanking region and primers Prez35 and hph3 for the 3′-flanking region of the pre2 gene. To confirm the deletion of the ppg2 gene, primers sa10 and hph3 were used for the 5′-flanking region and primers trpC1 and sa15 for the 3′-flanking region (Figure 1; supplemental Figure S1 at http://www.genetics.org/supplemental/).
Transformation of S. macrospora:
Transformation of S. macrospora was performed according to Nowrousian et al. (1999). Fungal protoplasts were transformed with linear PCR fragments amplified from plasmids pPRE1-KO, pPRE2-KO, and pPPG2-KO, respectively. Homologous recombination of the PCR fragments introduced into the S. macrospora genome resulted in disruption of the corresponding genes (Figure 1). Transformants were selected on hygromycin-B-containing medium (110 units/ml). For complementation of the double-mutant defects, strain Δpre2/Δppg2 (S61357) was transformed either with rescue plasmid pNatppg2, carrying the ppg2 gene, or with plasmid pNatpre2, carrying the pre2 gene. Additionally, we complemented the double-receptor mutant (SA1-4, Δpre1/Δpre2) with pNatpre2. Rescued transformants were selected on nourseothricin-containing medium (50 μg/ml).
Preparation of nucleic acids and hybridization protocols:
DNA isolation was performed as described by Pöggeler et al. (1997). Total RNA was isolated from S. macrospora at 3–7 days of growth using the method of Hoge et al. (1982). Southern and Northern blotting were performed according to Sambrook et al. (2001).
PCR analysis:
To obtain DNA fragments for further cloning or to confirm knockout transformants, PCR amplification was performed as described previously (Pöggeler et al. 1997). The different primers used are shown in Table 2. PCR products used for the transformation of S. macrospora strains were amplified with HotStarTaq DNA polymerase (QIAGEN) as described by the manufacturers.
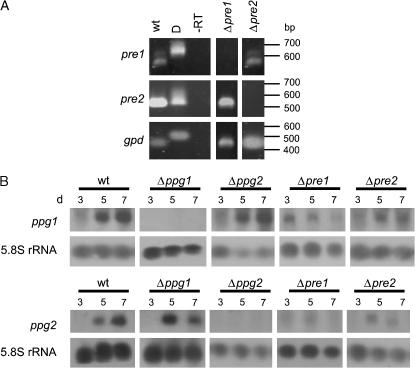
For RT–PCR, total RNA was treated with RNase-free DNase to remove any residual DNA and was then reverse transcribed. PCRs were performed with specific oligonucleotide-primer pairs MrezS2/Mrez32 and PrezS3/PrezS4 generated from the pre1 and pre2 sequence, respectively. In addition, gpd-specific primers (N-gpd/Cgpd) were used as a control (Table 2; Figure 2). Corresponding DNA fragments were amplified with each primer combination from S. macrospora wild type to serve as further controls, as well as PCRs without reverse-transcribed RNA as template (Figure 2).
Figure 2.
Transcript analysis of S. macrospora wild-type and mutant strains. (A) RT–PCR. pre1 (MrezS2, Mrez32)-, pre2 (PrezS3, PrezS4)-, and gpd (C-gpd, N-gpd)-specific oligonucleotide pairs were employed in an RT–PCR of S. macrospora total RNA. D, DNA template (S. macrospora wild type); -RT, RNA template (S. macrospora wild type), no reverse transcription prior to PCR. (B) Northern analysis. Twenty micrograms of total RNA, isolated at different time points during the life cycle of wild-type and single-mutant strains (at 3, 5, and 7 days) was loaded per lane. Northern blots were probed using a ppg1 and ppg2 gene-specific probe, respectively. As a control, the blots were striped and reprobed with a 5.8S rRNA-specific probe.
Conventional genetic analysis:
Fungal transformants are often heterokaryotic and mycelia carry transformed and nontransformed nuclei. Therefore, homokaryotic single-spore isolates of primary transformants carrying the single knockouts were created by selfing the primary transformant and selecting the hygromycin-resistant progeny. The single-spore isolates were then crossed with the nontransformed S. macrospora spore-color mutant fus1-1. The latter produces brown ascospores. Conventional genetic analysis of >10 ordered tetrads from the cross of S2.2-1 × fus1-1, S60289 × fus1-1, and S54761 × fus1-1 revealed a Mendelian 1:1 segregation of hygromycin-resistant and nonresistant progeny. Southern analysis indicated that in the hygromycin-resistant progeny the hph gene was integrated only into the pre1, pre2, and ppg2 locus, respectively (supplemental Figure S1 at http://www.genetics.org/supplemental/). In addition, we isolated the Δpre2/fus1-1 mutant (S60447) and the Δppg2/fus1-1 mutant (S59786) from these crosses (Table 1).
Double-knockout strains were generated by crossing the single-knockout strains. We crossed S52063 [Δppg1 (Mayrhofer and Pöggeler 2005)] × S59786 (Δppg2/fus1-1) to obtain a double-pheromone mutant (Δppg1/Δppg2) and S2-2.1 (Δpre1) × S60447 (Δpre2/fus1-1) to produce the double-receptor knockout (Δpre1/Δpre2). Double-knockout strains without compatible pheromone-receptor pairs Δpre2/Δppg2 and Δpre1/Δppg1 were generated by crossing S60446 (Δpre2) × S59786 (Δppg2/fus1-1), S59784 (Δppg2) × S60447 (Δpre2/fus1-1), or S2.2-1 (Δpre1) × S66385 (Δppg1/fus1-1). From the progeny of these crosses, we chose for further analyses the single-spore isolates S66197 (Δppg1/Δppg2), S61357 (Δpre2/Δppg2), S61352 (Δpre2/Δppg2/fus1-1), SD4-2 (Δpre1/Δppg1), and SA1-4 (Δpre1/Δpre2) (Table 1).
Microscopic investigation:
The Zeiss Axiophot microscope was used for light microscopy. Pictures were captured with an AxioCam camera. Recorded images were edited using Adobe Photoshop 6.0.
Quantitative analysis of fruiting-body production was done at ×50 magnification under the microscope. The number of protoperithecia and perithecia from wild-type and mutant strains produced per 2.4 mm2 were counted at eight different sections of two agar plates each. The diameters of all protoperithecia and perithecia found in these sections were measured under the microscope using the Zeiss Axiovision digital imaging system. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; 0.5 μg/μl).
RESULTS
Construction of the S. macrospora single- and double-knockout strains:
Our aim was to determine the role of the pheromones and pheromone receptors during the vegetative and sexual development of the homothallic ascomycete S. macrospora. To investigate this, we first generated Δpre1, Δpre2, and Δppg2 single-mutant strains. The disruption of the ppg1 gene was recently described (Mayrhofer and Pöggeler 2005).
Both S. macrospora pheromone-receptor genes are predicted to encode G-protein-coupled 7-TMD receptors with an extracellular N-terminal tail, three outer and three cytoplasmic loops, and an inner C-terminal tail. The pre1 gene codes for a 717-amino-acid protein, which shares homologies to the yeast Ste3p a-factor receptor, whereas the 554-amino-acid α-factor receptor PRE2 is homologous to Ste2p of S. cerevisiae (Pöggeler and Kück 2001). In a recent study, we reported the pre1 ORF much shorter (1232 bp interrupted by one intron of 56 bp). RT–PCR analysis performed during this study revealed the presence of a second intron in the pre1 ORF. This intron is 70 bp in length with conserved 5′ donor and 3′ acceptor sequences (Pöggeler 1997). When the second intron is spliced from the pre1 transcript, 326 amino acids are added to the N-terminal 376 amino acids of the previously defined 391-amino-acid PRE1 protein (Pöggeler and Kück 2001). The pre1 cDNA encoding the 391-amino-acid polypeptide was amplified by RT–PCR using a primer located within the second intron (Pöggeler and Kück 2001), indicating that the second intron is not removed in a portion of the pre1 transcripts.
The pre1 and pre2 knockouts were generated as described in materials and methods and confirmed by Southern analysis and PCR (Figure 1; supplemental Figure S1 at http://www.genetics.org/supplemental/). After homologous recombination, a 141-bp fragment (47 amino acids, corresponding to TMD VII of the receptor) within the pre1 coding sequence was replaced by the hygromycin-resistance cassette. The desired pre1 knockout strain (single-spore isolate S2.2-1, Table 1) was selected as described in materials and methods. Successful knockout of the pre1 gene was confirmed by Southern blot and PCR amplification (Figure 1; Table 2; supplemental Figure S1, A and B, at http://www.genetics.org/supplemental/).
Replacement of a 1087-bp fragment within the pre2 coding sequence deleted 362 amino acids of the PRE2 receptor corresponding to TMD III–TMD VII and the largest part of the C terminus. The selection procedure (see materials and methods) of the pre2 knockout identified the homokaryotic Δpre2 single-spore isolate S60289. After crossing S60289 × fus1-1, we isolated the Δpre2 mutant (S60446) and the Δpre2/fus1-1 mutant (S60447) from the progeny (Table 1). The pre2 knockout was confirmed by Southern blot and PCR analysis (Figure 1; Table 2; supplemental Figure S1, C and D, at http://www.genetics.org/supplemental/).
The S. macrospora pheromone-precursor gene ppg2 encodes a 24-amino-acid polypeptide. The precursor is thought to be N- and C-terminally processed and further modified by farnesylation and methylation at the C-terminal cysteine residue (Pöggeler 2000). Deletion of the S. macrospora ppg2 gene was achieved by a strategy similar to the one used for the knockout of the pre2 gene and is described in materials and methods (Figure 1). However, in this case the entire coding region was replaced by the hph cassette. The hygromycin-resistant single-spore isolate S54761 was selected after selfing of a primary transformant carrying the Δppg2 knockout (Table 1). We then isolated the Δppg2 mutant (S59784) and the Δppg2/fus1-1 mutant (S59786) from the resulting progeny of the cross S54761 × fus1-1 (Table 1). The correct deletion in knockout strains was confirmed by Southern blot and PCR analysis (Figure 1; Table 2; supplemental Figure S1, E and F, at http://www.genetics.org/supplemental/).
To confirm the successful knockout of pre1, pre2, and ppg2 in the single-knockout strains on transcript level, we performed Northern and RT–PCR analysis. Previous expression studies of the pheromone-receptor genes pre1 and pre2 revealed only weak pheromone-receptor-specific signals in Northern hybridizations with enriched poly(A) mRNA (Pöggeler and Kück 2001). Thus, we performed RT–PCR with total RNA of wild-type, Δpre1 (S2.2-1), and Δpre2 (S60446) strains (Figure 2A). The RT–PCR analysis indicated that Δpre1 and Δpre2 are knockout mutants with no detectable pre1 and pre2 transcript, respectively. As expected, the pre2 transcript was present in the Δpre1 single-knockout strain and the pre1 transcript in the Δpre2 mutant. As a positive control, we amplified the gpd transcript (Figure 2A). Corresponding DNA fragments were amplified to serve as controls. RT–PCR of the pre1 and gpd transcript revealed differently sized amplicons when DNA and cDNA was used as a template. This result can be explained by the presence of an intron within the amplified sequence. Primer pair MrezS2 and Mrez32 used for amplification of the pre1 cDNA framed the second intron of the pre1 ORF. As mentioned above, the second intron of the pre1 transcript seems to be optionally spliced. Thus, the two bands present after amplification of the pre1 cDNA in wild type and the Δpre2 mutant can be attributed to completely and partially spliced forms of the pre1 transcript.
To confirm the knockout of ppg1 and ppg2 in the Δppg1 and Δppg2 mutants on transcript level, respectively, and to investigate whether the absence of receptor genes altered the expression of pheromone-precursor genes, Northern blot analyses were performed (Figure 2B). Total RNA was isolated from the wild-type strain and from mutant strains Δppg1 (S52063), Δppg2 (S59784), Δpre1 (S2.2-1), and Δpre2 (S60446) at day 3, 5, and 7 of sexual development. In our experiments, sexual development started 2 days after inoculation with the appearance of ascogonia and ended with the discharge of mature ascospores from perithecia 7 days after inoculation. Figure 2B shows that in the wild-type strain both pheromone-precursor genes were transcribed during the entire sexual phase and that until day 7 the ppg1 and ppg2 mRNA levels increased. As expected, no ppg1 and ppg2 mRNA was detected in the Δppg1 and Δppg2 mutant strains, respectively. As can be seen in Figure 2B, the transcript level of both pheromone-precursor genes seemed to be downregulated in the Δpre1 and Δpre2 mutant strains, suggesting that loss of the pheromone receptors indirectly had a negative effect on the expression of both pheromone genes. The Northern blots shown are representatives of Northerns from three independent samples.
To analyze strains lacking any compatible pheromone-receptor pair, we generated double-knockout strains by crossing the single mutants in different combinations. As described in materials and methods, we obtained a double-pheromone knockout (S66197, Δppg1/Δppg2), a double-receptor knockout (SA1-4, Δpre1/Δpre2), as well as strains containing no cognate pheromone-receptor pair: Δpre1/Δppg1 (SD4-2), Δpre2/Δppg2 (S61357), and Δpre2/Δppg2/fus1-1 (S61352).
Phenotype of single- and double-knockout strains:
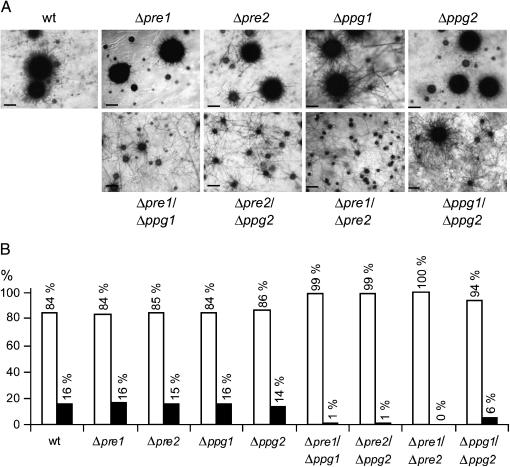
All mutant strains generated were examined for defects in vegetative and sexual development. No abnormality in vegetative growth and mycelial morphology was seen when single-mutant strains and double-knockout strains were compared to S. macrospora wild type. The development of sexual reproductive structures was analyzed on cornmeal medium. After 7 days on this fructification medium, the single-mutant strains Δpre1, Δpre2, Δppg1, and Δppg2 produced protoperithecia (<200 μm) and perithecia (>200 μm), which were different in size and developmental stage. This was very similar to the development of the wild-type strain (Figure 3).
Figure 3.
Distribution of protoperithecia and perithecia produced by the S. macrospora wild-type, single-, and double-mutant strains. (A) Microscopic analysis of fruiting-body development on solid cornmeal medium after 7 days. Wild type, single-, and double-mutant strains are given in the margin. Bars, 200 μm. (B) Percentage of protoperithecia (<200 μm, open bar) and perithecia (>200 μm, solid bar) produced by wild-type, single-, and double-mutant strains on agar plates, calculated from eight measurements from two different plates.
However, all double mutants were impaired in fruiting-body development and predominately formed pigmented protoperithecia with a diameter of ∼100 μm. Formation of fully differentiated perithecia (>200 μm, with necks) was completely prevented in the double-receptor mutant Δpre1/Δpre2, while perithecia production was only 4% and 8% of wild-type level in the Δpre2/Δppg2 and Δpre1/Δppg1 double-mutant strains, respectively (Figure 3B). The least impairment in respect to fruiting-body development was observed in the double-pheromone mutant Δppg1/Δppg2. In this mutant strain, perithecia development was 36% of wild-type level (Figure 3B). Prolonged incubation did not increase the number of perithecia in the double-knockout strains.
To confirm that the defects in sexual development observed in the double-mutant strains were due to the deletion of the pheromone- and pheromone-receptor genes, complementation assays were performed. Strain Δpre2/Δppg2 (S61357) was transformed either with rescue-plasmid pNatppg2, carrying a wild-type copy of the ppg2 gene (Δpre2/Δppg2 + ppg2), or with plasmid pNatpre2, containing a wild-type copy of the pre2 gene (Δpre2/Δppg2 + pre2). In addition, strain Δpre1/Δpre2 was transformed with pNatpre2 (Δpre1/Δpre2 + pre2). The introduced plasmids complemented the double-knockout strains and restored normal fruiting-body development (data not shown). This phenotype is in agreement with the observations made in the Δpre1, Δpre2, and Δppg2 single mutants lacking any phenotypic defects.
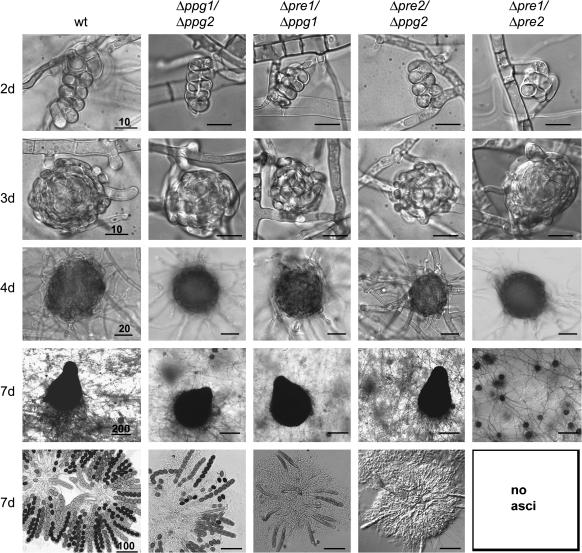
In addition, the morphology of all mutant strains and the wild-type strain as a control was examined under the microscope in more detail. We looked at different time points during their sexual development when grown under homothallic conditions (Figure 4; supplemental Figure S2 at http://www.genetics.org/supplemental/). Similar to wild type, the single-knockout strains Δpre1, Δpre2, Δppg1, and Δppg2 developed ascogonia, young and pigmented protoperithecia, as well as perithecia all containing 200–300 asci with eight ascospores (supplemental Figure S2 at http://www.genetics.org/supplemental/). No formation of perithecia with asci and ascospores was observed in the double-receptor strain Δpre1/Δpre2. The Δpre1/Δppg1 and Δpre2/Δppg2 double-knockout mutants rarely produced perithecia (8% and 4% of wild-type level, respectively). Most of these (60%) harbored undifferentiated asci and no viable ascospores (Figure 4). Protoperithecia of ∼100 μm did not contain any croziers or ascus initials (data not shown). To analyze the defect within the reproductive structures of the Δpre2/Δppg2 double mutant, DAPI staining was performed. This staining of nuclei revealed that the development of fruiting bodies was blocked in most cases at the stage of hook-cell formation or young ascus initials (Figure 5, A and B). In 30% of analyzed perithecia, ascus rosettes were found to be composed of only 3–20 asci (Figure 5C). Normal developed ascus rosettes were observed in only 10% of the rarely produced perithecia (Figure 5D). With respect to ascospore production, the phenotype of the Δpre1/Δppg1 mutant was very similar to that of the Δpre2/Δppg2 mutant. However, the double-pheromone mutant Δppg1/Δpgg1 showed less impairment in ascospore production. In the Δppg1/Δppg2 mutant, 20% of the analyzed fruiting bodies contained wild-type ascus rosettes with 200–300 asci, 50% of the fruiting bodies were composed of only a few asci, and 30% of the perithecia were blocked at the stage of hook-cell formation or young ascus initials (data not shown).
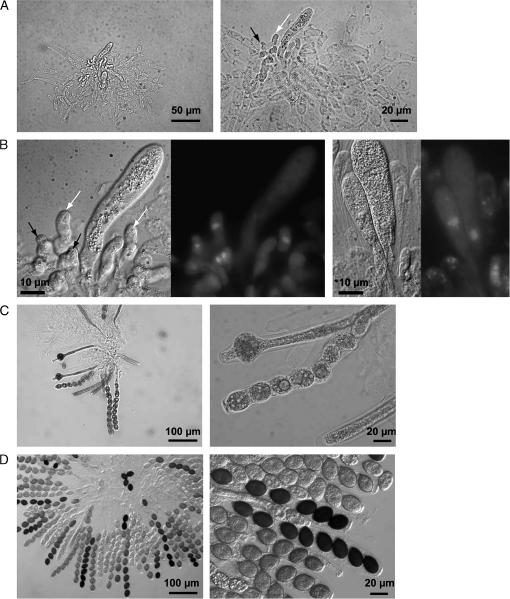
Figure 4.
Microscopic investigation of sexual developmental stages of the wild-type and double-knockout strains. Differential interference microscopy identified ascogonia (at 2 days), protoperithecia (at 3 days), pigmented protoperithecia (at 4 days), and perithecia (at 7 days) in the wild-type and mutant strains. Strains were grown on fructification medium and analyzed after growth at 25° for the number of days indicated. For examination of ascospore development, the content of perithecia was dissected. The perithecia of the Δppg1/Δppg2 mutant contained fewer asci and ascospores than the wild type. The sparsely produced perithecia of the Δpre1/Δppg1 and Δpre2/Δppg2 double mutants contain either ascus rosettes with viable ascospores or, in most cases, undifferentiated asci without ascospores. No perithecia, and thus no asci and ascospores, are produced in the Δpre1/Δpre2 mutant. Bars represent sizes in micrometers as indicated.
Figure 5.
Microscopic analysis of ascus development in the rarely produced perithecia of the Δpre2/Δppg2 double-mutant strain. (A) Sixty percent of all squeezed perithecia contain few hook cells (solid arrow) and undifferentiated asci (ascus initial, open arrow). (B) DAPI staining identified nuclei during crozier (solid arrows) and ascus formation (ascus initial, open arrow). (C) Thirty percent of ascus rosettes carry 3–20 more or less differentiated asci with ascospores. (D) Ten percent of perithecia contents represent normally developed asci with ascospores.
In summary, double-mutant strains lacking a functioning pheromone-receptor system showed drastically reduced fruiting-body, ascus, and ascospore production.
Outcrossing of double mutants:
Recently, Seo et al. (2004) demonstrated that in the homothallic ascomycete A. nidulans strains lacking both pheromone-receptor genes are not able to form cleistothecia and ascospores under homothallic (selfing) conditions. However, double-knockout strains could be forced to form heterokaryons. Under these heterothallic conditions, they produced normal cleistothecia containing ascospores with wild-type-level viability. It was therefore suggested that heterokaryon formation may obviate the need for pheromone receptors during sexual development (Seo et al. 2004).
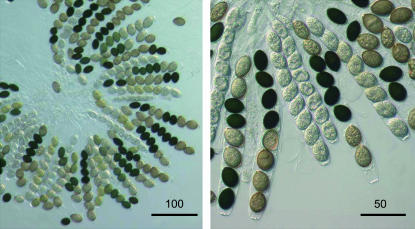
We outcrossed double-knockout strains to investigate whether pheromones and receptors are required for sexual development under heterothallic conditions in S. macrospora. As mentioned above, even Δpre2/Δppg2 strains produced perithecia, although less numerously than in wild-type strains. Since S. macrospora is a homothallic fungus that produces perithecia by selfing, we used the spore-color mutant to distinguish between self-fertile and hybrid perithecia in crosses between two Δpre2/Δppg2 strains (Δpre2/Δppg2 × Δpre2/Δppg2/fus1-1). As under the homothallic condition, only very few hybrid perithecia developed. However, on opening, 10% of these perithecia contained ascus rosettes with eight viable ascospores. In these asci, the spore-color marker (fus1-1) was segregated as expected in a 4:4 Mendelian ratio (Figure 6). Similarly, fruiting-body and ascospore production was not restored or increased when we outcrossed Δpre1/Δpre2 or Δppg1/Δppg2 and Δpre1/Δppg1 double-knockout strains (data not shown). In conclusion, we were unable to demonstrate any change in fruiting-body and ascospore production under homothallic and heterothallic conditions in the double-knockout strains.
Figure 6.
Light microscopic investigation of ascus rosettes resulting from crosses between Δpre2/Δppg2/fus1-1 and Δpre2/Δppg2. Bars represent sizes in micrometers as indicated.
DISCUSSION
Despite being self-fertile, the filamentous ascomycete S. macrospora carries and expresses two pheromone-precursor genes and two pheromone-receptor genes. These genes encode two pheromone-receptor pairs (PPG1/PRE2 and PPG2/PRE1), which upon interaction are supposed to trigger pheromone-induced responses (Pöggeler 2000; Pöggeler and Kück 2001; Mayrhofer and Pöggeler 2005). To determine the function of pheromone and receptor genes in S. macrospora, we replaced them with an hph resistance-marker cassette (Figure 1) and generated double-mutant strains that did not retain any compatible pheromone-receptor pair. No differences in vegetative growth were observed in either single- or double-knockout strains compared to the wild type. The fact that all mutant strains displayed normal vegetative growth means that pheromone-receptor interactions are not required during filamentous growth in S. macrospora. Similarly, the previously described knockout of ppg1 in S. macrospora or deletion of both pheromone genes in P. anserina, and deletion of the pre-1 receptor gene in N. crassa, as well as deletion of both pheromone-receptor genes in A. nidulans, caused no changes in vegetative growth (Kim and Borkovich 2004; Seo et al. 2004; Coppin et al. 2005; Mayrhofer and Pöggeler 2005). Vegetative growth was delayed and reduced only in N. crassa mfa-1 pheromone mutants (Kim et al. 2002).
Throughout this work no rigorous phenotypic changes regarding sexual development were observed in the S. macrospora pheromone or receptor single-mutant strains. They were able to develop fruiting-body precursors, fruiting bodies, asci, and ascospores (supplemental Figure S2 at http://www.genetics.org/supplemental/). These results are in agreement with our earlier findings that a ppg1 mutant strain showed no impairment in sexual reproduction (Mayrhofer and Pöggeler 2005). Since S. macrospora does not produce conidia, it was impossible to investigate whether there were any defects in male fertility. In the heterothallic ascomycete P. anserina, the deletion of pheromone genes affects only male fertility without impairing postfertilization events (Coppin et al. 2005). However, deletion of one of the two copies of the C. parasitica gene encoding the a-factor-like pheromone resulted in a pleiotropic phenotype. These Mf2-2 mutants displayed reduced asexual reproduction. Crossing of a Mf2-2 mutant (as the female) with a wild-type strain (as the male) produced only barren perithecia (Zhang et al. 1993). It has therefore been postulated that Mf2-2 is required during a developmental phase after fertilization in C. parasitica and that it acts in a dose-dependent manner in postfertilization events (Turina et al. 2003). In additon to this, introduction of mutations into the mfa-1 gene of N. crassa had several interesting effects, including delayed conidial germination, drastically reduced protoperithecial formation, and highly abnormal perithecial development during homozygous crosses (Kim et al. 2002). Deletion of one of the two putative pheromone-receptor genes, gprA or gprB, in the homothallic filamentous ascomycete A. nidulans resulted in the production of fewer and smaller fruiting bodies with a reduced number of ascospores (Seo et al. 2004).
In contrast to the above observations, our experiments revealed no phenotypic defects with respect to development of perithecia and ascospores as well as timing of sexual development in S. macrospora single-pheromone or single-receptor mutants (supplemental Figure S2 at http://www.genetics.org/supplemental/). This suggests that, in the absence of one of the two expressed pheromone-receptor pairs, the remaining pheromone-receptor pair can compensate for the loss of the other.
To investigate whether pheromone-receptor interactions are involved in fertilization and/or postfertilization events in the homothallic S. macrospora, we constructed double-knockout strains and invalidated both pheromone systems. The resulting double mutants had no compatible pheromone-receptor pair or no pheromones or no pheromone receptors (Table 1). With respect to sexual development, all double mutants displayed a clear phenotype: fruiting bodies either were completely lacking or were less numerous than in the wild-type strain (Figure 3). In the double-knockout strains, the impairment of sexual reproduction was independent of the choice of medium. Even so, the transfer of strains on fresh medium did not allevite the fertility defect. Thus, the defect in sexual development was not due to energy limitation.
We observed the strongest effect on fruiting-body formation in the double-receptor mutant Δpre1/Δpre2. Similar to our results, a double-knockout of two receptor genes in the homothallic A. nidulans (strain ΔgprA/ΔgprB) completely abolished fruiting-body and ascospore formation (Seo et al. 2004). In those cases where perithecia were produced in S. macrospora double-knockout strains, ascospore production was predominantly impaired and only very few hook cells were formed (Figure 5). Normal sexual development was restored in the double-knockout strains by introducing a gene encoding either the missing pheromone or the receptor. This suggests that at least one pheromone-receptor system is necessary for proper formation of dikaryotic ascogenous hyphae and ascus initials. In filamentous ascomycetes, the nuclear sorting occurring in the dikaryotic ascogenous hyphae is reminiscent of the synchronous division of two nuclei in the dikaryotic hyphae of basidiomycetes (Casselton 2002). In contrast to ascomycetes, pheromones and receptors are encoded by one of two mating-type loci in mushroom fungi and are not needed for the initial fusion of mating partners. However, they are required to promote nuclear migration and clamp cell fusion after the dikaryon is established (Wendland et al. 1995; Vaillancourt et al. 1997; O'Shea et al. 1998). Pheromones secreted from the clamp cell are believed to activate receptors on the surface of the subterminal cell and vice versa (Brown and Casselton 2001). Similarly, one may speculate that the pheromone-receptor system of filamentous ascomycetes is also involved in the nuclear migration of heterogenic nuclei in ascogenous hyphae. It has also been suggested that, in dikaryotic hyphae, recognition between nuclei is mediated by mating-type-specific pheromones and receptors and that the expression of pheromone and receptor genes is nucleus limited. All signaling components are thus restricted to the plasma region close to the individual nucleus, a feature that is thought to facilitate recognition between two nuclei (Schuurs et al. 1998; Debuchy 1999; Shiu and Glass 2000). In the S. macrospora double-knockout strain, the disturbance of this putative pheromone-triggered nuclear recognition system might result in the defects in crozier and ascus formation, which in turn might lead to the reduction in perithecia production. All fruiting-body structures produced in the Δpre1/Δpre2 mutant and the vast majority of fruiting bodies in the other double-mutant strains contained no ascospores. However, the pheromone-driven nuclear-recognition system seemed to be leaky in some cases. Then, probably by chance or due to nonspecific activation of the pheromone receptors, two nuclei come together in the apical cell of the crozier and undergo regular karyogamy, meiosis, and ascosporogenesis.
With respect to fruiting-body and ascospore development, the least effect was observed in the double-pheromone mutant Δppg1/Δppg2, which displayed only a threefold reduction in perithecia formation in comparison to the wild type. Approximately 70% of these perithecia produced asci and ascospores, although in most cases to a lesser extent than in the wild type. Thus, we can conclude from our experiments that pheromones seem to be less important than pheromone receptors for fruiting-body and ascospore formation. It might be possible that pheromone receptors can be activated in a nonspecific manner and that this activation can in turn lead to the formation of ascogenous hyphae in the double-pheromone mutant. Since the Δpre1/Δppg1 and Δpre2/Δppg2 mutants retain only one intact receptor gene, a nonspecific activation might occur to a lesser extent. Previously, we have expressed the pre2 receptor gene of S. macrospora in S. cerevisiae and observed a low nonspecific activation of PRE2 after incubation of yeast cells with 2% dimethyl sulfoxide (Mayrhofer and Pöggeler 2005).
Interestingly, outcrossing of A. nidulans receptor-mutant strains (ΔgprA/ΔgprB × ΔgprA/ΔgprB) resulted in fruiting-body and ascospore formation at wild-type level, suggesting that, in A. nidulans pheromone receptors, GprA and GprB are required only for self-fertilization and not for sexual development per se (Seo et al. 2004). However, in contrast to the differences observed under homothallic and outcross conditions in A. nidulans, our analysis of S. macrospora mutants revealed no differences between the two conditions (Figure 6).
In summary, the phenotype of the S. macrospora double-mutant strains suggests that pheromones and receptors play an important role in fruiting-body development and ascosporogenesis. Similarly to the function in basidiomycetes, where pheromones regulate only nuclear migration and clamp-cell fusion in the dikaryotic mycelium, the pheromone system of a homothallic ascomycete seems to promote crozier formation and ensure the stability of the dikaryon.
Acknowledgments
The authors thank Silke Nimtz, Gisela Isowitz-Seidel, and Susanne Schlewinski for excellent technical assistance; Michael Karus for help with some experiments; and Ulrich Kück (Ruhr-University Bochum) for laboratory resources. This work was funded by the Deutsche Forschungsgemeinschaft PO523/3-1 (Bonn, Germany) and the Ruhr-University of Bochum.
References
- Bardwell, L., 2005. A walk-through of the yeast mating pheromone response pathway. Peptides 26: 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowicz, P., R. Pawlak, A. Correa, D. Bell-Pedersen and D. J. Ebbole, 2002. The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol. Microbiol. 45: 759–804. [DOI] [PubMed] [Google Scholar]
- Bölker, M., and R. Kahmann, 1993. Sexual pheromones and mating responses in fungi. Plant Cell 5: 1461–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A. J., and L. Casselton, 2001. Mating in mushrooms: increasing the chances but prolonging the affair. Trends Genet. 17: 393–400. [DOI] [PubMed] [Google Scholar]
- Caldwell, G. A., F. Naider and J. M. Becker, 1995. Fungal lipopeptide mating pheromones: a model system for the study of protein prenylation. Microbiol. Rev. 59: 406–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, A. M., J. A. Sweigard and B. Valent, 1994. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41: 22. [Google Scholar]
- Casselton, L. A., 2002. Mate recognition in fungi. Heredity 88: 142–147. [DOI] [PubMed] [Google Scholar]
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero and P. Hieter, 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Coppin, E., C. de Renty and R. Debuchy, 2005. The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryot. Cell 4: 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy, R., 1999. Internuclear recognition: a possible connection between euascomycetes and homobasidiomycetes. Fungal Genet. Biol. 27: 218–223. [DOI] [PubMed] [Google Scholar]
- Esser, K., 1982. Cryptogams—Cyanobacteria, Algae, Fungi, Lichens. Cambridge University Press, London.
- Esser, K., and J. Straub, 1958. Genetische Untersuchungen an Sordaria macrospora Auersw.: Kompensation und Induktion bei genbedingten Entwicklungsdefekten. Z. Vererbungslehre 89: 729–746. [PubMed] [Google Scholar]
- Greener, A., 1990. New competent cells for highest transformation efficiencies. Strategies 3: 5–6. [Google Scholar]
- Hagen, D. C., G. McCaffrey and G. F. Sprague, Jr., 1986. Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a-factor: gene sequence and implications for the structure of the presumed receptor. Proc. Natl. Acad. Sci. USA 83: 1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoge, J. H. C., J. Springer, B. Zantige and J. G. H. Wessels, 1982. Absence of differences in polysomal RNA from vegetative monokaryotic and dikaryotic cells of the fungus Schizophyllum commune. Exp. Mycol. 6: 225–232. [Google Scholar]
- James, P., J. Halladay and E. A. Craig, 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness, D. D., A. C. Burkholder and L. H. Hartwell, 1983. Binding of α-factor pheromone to yeast a cells: chemical and genetic evidence for an α-factor receptor. Cell 35: 521–529. [DOI] [PubMed] [Google Scholar]
- Kim, H., and K. A. Borkovich, 2004. A pheromone receptor gene, pre1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52: 1781–1798. [DOI] [PubMed] [Google Scholar]
- Kim, H., R. L. Metzenberg and M. A. Nelson, 2002. Multiple functions of mfa-1, a putative pheromone precursor gene of Neurospora crassa. Eukaryot. Cell 1: 987–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe, E., 1999. Mating types and pheromone recognition in the Homobasidiomycete Schizophyllum commune. Fungal Genet. Biol. 27: 146–152. [DOI] [PubMed] [Google Scholar]
- Krugel, H., G. Fiedler, C. Smith and S. Baumberg, 1993. Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 127: 127–131. [DOI] [PubMed] [Google Scholar]
- Mayrhofer, S., and S. Pöggeler, 2005. Functional characterization of an α−factor-like Sordaria macrospora peptide pheromone and analysis of its interaction with its cognate receptor in Saccharomyces cerevisiae. Eukaryot. Cell 4: 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowrousian, M., S. Masloff, S. Pöggeler and U. Kück, 1999. Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP citrate lyase activity. Mol. Cell. Biol. 19: 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea, S. F., P. T. Chaure, J. R. Halsall, N. S. Olesnicky, A. Leibbrandt et al., 1998. A large pheromone and receptor gene complex determines multiple B mating type specificities in Coprinus cinereus. Genetics 148: 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöggeler, S., 1997. Sequence characteristics within nuclear genes from Sordaria macrospora. Fungal Genet. Newsl. 44: 41–44. [Google Scholar]
- Pöggeler, S., 2000. Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr. Genet. 37: 403–411. [DOI] [PubMed] [Google Scholar]
- Pöggeler, S., and U. Kück, 2001. Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280: 9–17. [DOI] [PubMed] [Google Scholar]
- Pöggeler, S., S. Risch, U. Kück and H. D. Osiewacz, 1997. Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147: 567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 2001. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schuurs, T. A., H. J. Dalstra, J. M. Scheer and J. G. Wessels, 1998. Positioning of nuclei in the secondary mycelium of Schizophyllum commune in relation to differential gene expression. Fungal Genet. Biol. 23: 150–161. [DOI] [PubMed] [Google Scholar]
- Seo, J. A., K. H. Han and J. H. Yu, 2004. The gprA and gprB genes encode putative G protein-coupled receptors required for self-fertilization in Aspergillus nidulans. Mol. Microbiol. 53: 1611–1623. [DOI] [PubMed] [Google Scholar]
- Shen, W. C., P. Bobrowicz and D. J. Ebbole, 1999. Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet. Biol. 27: 253–263. [DOI] [PubMed] [Google Scholar]
- Shiu, P. K., and N. L. Glass, 2000. Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr. Opin. Microbiol. 3: 183–188. [DOI] [PubMed] [Google Scholar]
- Souza, C. A., C. C. Silva and A. V. Ferreira, 2003. Sex in fungi: lessons of gene regulation. Genet. Mol. Res. 2: 136–147. [PubMed] [Google Scholar]
- Turina, M., A. Prodi and N. K. Alfen, 2003. Role of the Mf1–1 pheromone precursor gene of the filamentous ascomycete Cryphonectria parasitica. Fungal Genet. Biol. 40: 242–251. [DOI] [PubMed] [Google Scholar]
- Vaillancourt, L. J., M. Raudaskoski, C. A. Specht and C. A. Raper, 1997. Multiple genes encoding pheromones and a pheromone receptor define the Bβ1 mating-type specifity in Schizophyllum commune. Genetics 146: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland, J., L. J. Vaillancourt, J. Hegner, K. B. Lengeler, K. J. Laddison et al., 1995. The mating-type locus Bα1 of Schizophyllum commune contains a pheromone receptor gene and putative pheromone genes. EMBO J. 14: 5271–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard, M., and H. K. Mitchell, 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. [Google Scholar]
- Zhang, L., A. C. Churchill, P. Kazmeirczak, D. H. Kim and N. K. van Alfen, 1993. Hypovirulence-associated traits induced by a mycovirus of Cryphonectria parasitica are mimicked by targeted inactivation of a host gene. Mol. Cell. Biol. 13: 7782–7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., R. A. Baasiri and N. K. van Alfen, 1998. Viral repression of the fungal pheromone precursor gene expression. Mol. Cell. Biol. 18: 953–959. [DOI] [PMC free article] [PubMed] [Google Scholar]