Abstract
Recombinant inbred (RI) mice are frequently used to identify QTL that underlie differences in measurable phenotypes between two inbred strains of mice. Here we show that one RI strain, C57BL/6J × DBA/2J (BXD29), does not develop an inflammatory response following inhalation of LPS. Approximately 25% of F2 mice [F1(BXD29 × DBA/2J) × F1] are also unresponsive to inhaled LPS, suggesting the presence of a recessive mutation in the BXD29 strain. A genomic scan of these F2 mice revealed that unresponsive animals, but not responsive animals, are homozygous for C57BL/6J DNA at a single locus on chromosome 4 close to the genomic location of Tlr4. All progeny between BXD29 and gene-targeted Tlr4-deficient mice are unresponsive to inhaled LPS, suggesting that the mutation in the BXD29 strain is allelic with Tlr4. Moreover, the intact Tlr4 receptor is not displayed on the cell surface of BXD29 macrophages. Finally, a molecular analysis of the Tlr4 gene in BXD29 mice revealed that it is interrupted by a large insertion of repetitive DNA. These findings explain the unresponsiveness of BXD29 mice to LPS and suggest that data from BXD29 mice should not be included when using BXD mice to study phenotypes affected by Tlr4 function. Our results also suggest that the frequency of such unidentified, spontaneously occurring mutations is an issue that should be considered when RI strains are used to identify QTL.
MOUSE models of human diseases have been used for several decades to explore the genetic basis of disease initiation and progression. These studies have greatly benefited from the recent sequencing of several inbred mouse strain genomes. Assays that detect single nucleotide polymorphisms (SNPs) distinguishing these strains are now used to expedite studies associating gene variants with disease phenotypes. However, the vast majority of SNPs are unrelated to the pathogenesis of the phenotype, and most candidate gene studies do not take into account modifying effects of genes situated at other loci throughout the genome. For this reason, genomic techniques are frequently used to identify discrete chromosomal intervals known as quantitative trait loci (QTL) that are associated with phenotype of interest. Comparative genomic analyses of these relatively small regions can be performed to yield a list of positional candidate genes that differ in sequence between phenotypically divergent strains.
Recombinant inbred (RI) strains are frequently used to identify QTL. These strains are created by breeding successive generations of sibling pairs from an initial cross between two different inbred strains until the mice are essentially homozygous at each locus for DNA derived from one of those two parental inbred strains. Because the RI lines differ from one another in the combinatorial distribution of parental strain DNA throughout the genome, the presence of a specific parental strain of DNA at one or more loci can sometimes be shown to correlate with the magnitude of a measurable phenotype. RI strains have several advantages over recombinant intercrossed F2 or backcrossed mice. First, RI strains have already been genotyped, so that little additional genotyping is necessary. Also, several individual mice can be analyzed for each strain, resulting in a more accurate estimation of the phenotype conferred by each combination of parental strain alleles. Finally, because RI strains have undergone more chromosomal crossover events than F2 or backcrossed mice, identified QTL can usually be localized to a smaller interval.
We previously used RI mice derived from the parental strains C57BL/6J and DBA/2J (BXD) to investigate genomic regions associated with the biologic response to inhaled lipopolysaccharide (LPS) (Cook et al. 2004). C57BL/6J and DBA/2J mice were chosen for this study because toll-like receptor 4 (Tlr4), the gene encoding the primary receptor for LPS, is identical in both strains of mice, even though they have very different biologic responses to inhaled LPS (Lorenz et al. 2001). While DBA/2J mice have a substantial inflammatory response following inhalation of LPS, C57BL/6J mice have a somewhat blunted response (Lorenz et al. 2001). In the process of screening BXD RI mice, we observed that one RI strain, BXD29, is essentially unresponsive to aerosolized LPS (Cook et al. 2004), as measured by concentrations in whole-lung lavage fluid of TNF-α and polymorphonuclear (PMN) cells. This unresponsiveness of BXD29 mice to LPS contrasted with the spectrum of responses seen in the other BXD RI strains. Here, we demonstrate that BXD29 mice have a spontaneously occurring disruption of the Tlr4 gene, which accounts for their unresponsiveness to inhaled LPS. This finding calls into question an assumption that underlies studies of RI strains, namely that their phenotypic variation results entirely from different combinations of parental-strain-derived loci, and not from unidentified, spontaneous mutations. It is likely that other unidentified mutations also exist in other genes of RI strains and that these mutations might impact the strength of QTL obtained from studies of these mice.
MATERIALS AND METHODS
Mice:
Genetically engineered C57BL/6J–Tlr4 mice were provided by Osamu Takeuchi, Osaka University. C57BL/6J, DBA/2J, and all BXD RI mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in the vivarium at Duke University. All procedures were carried out according to the Duke University guidelines for animal use and care.
Endotoxin exposures and phenotypic analyses:
Mice were exposed to a monitored, 4-hr exposure to an aerosol of LPS as described previously (Cook et al. 2004). The target concentration of LPS in this aerosol was 6 μg/m3. Actual concentrations of LPS in the chamber were determined by periodically placing filters in the exhaust tubing and measuring levels of LPS on them by a Limulus amebocyte lysate assay (QCL-1000; Whittaker Bioproducts, Walkersville, MD). These LPS concentrations ranged from 4.7 to 6.16 μg/m3. Following exposure to LPS, mice were sacrificed by CO2, their lungs lavaged, and differential cell counts performed. TNF-α levels were assayed using a commercial ELISA kit (R&D Systems, Minneapolis) according to the manufacturer's instructions.
Quantitative PCR-based genotyping assays:
Real-time PCR assays were performed using a 7900 HT thermocycler (Applied Biosystems, Foster City, CA), using standard conditions recommended by the manufacturer. Sequences corresponding to parental strain allele-specific primers were obtained from the Roche mouse SNP website (http://mousesnp.roche.com/). Percentages of parental strain DNAs were calculated using the formula, 100 × 1/(1 + 2ΔCt), where Ct is the PCR cycle number (C) at which the intensity of fluorescence crosses a set threshold (t). For genomewide analysis of F2 [F1(BXD29 × DBA/2J) × F1] populations, parental allele frequencies at 305 different loci were determined. High- and low-responder DNA pools were created by combining equal quantities of DNA from the 25 mice having the highest and lowest biologic responses, respectively (Germer et al. 2000).
Flow cytometric analysis:
Antibodies against F4/80 and murine Tlr4/MD-2 were purchased from Bioscience (San Diego). Stained cells were analyzed using a BD LSRII cytometer (Becton Dickinson Biosciences, San Jose, CA) and FlowJo software (Tree Star, Ashland, OR).
Subcloning of BXD29 Tlr4 gene segments by inverse PCR:
BXD29 genomic DNA was purchased from the Jackson Laboratories. To subclone the junction between the intact 5′ Tlr4 DNA and the presumptive insertion BXD29, genomic DNA was cleaved to completion with the restriction enzyme EcoRI. Circular DNA was generated from this DNA by self-ligating it at a concentration of 20 μg/ml. This circular DNA was then used in an inverse PCR (Ochman et al. 1988) together with two divergent primers. One primer (5′-ACATTCCTGTAAGTTACCTGCATATTT-3′), in the sense orientation, corresponded to a region in exon 3 of Tlr4 immediately upstream of the presumptive insertion. The other primer (5′-GCTAAACGTTGGAGAAAGCACA-3′), in the antisense orientation, corresponded to a region of Tlr4 5′ of the sense primer and immediately 3′ of an EcoRI restriction site. Following this first PCR reaction, a portion of the reaction mixture was used as a template for a second, nested PCR reaction using a second set of divergent primers (5′-CATATTTTTCCAATCTGACGAACC-3′ and 5′-CCAACAGAGCTGTGGTCCTC-3′) corresponding to regions predicted to be present at the ends of the first PCR product. Following agarose gel electorphoresis of this second PCR reaction mixture, a single band of DNA was seen. It was excised from the gel and subcloned into a pTA plasmid (Invitrogen, Carlsbad, CA). Recombinant plasmids containing the PCR product were subjected to DNA sequencing using the SP6 and T7 primers.
To subclone the 3′ junction of the BXD29 Tlr4 gene with the presumptive insertion, a similar inverse PCR approach was used, except that HindIII-cleaved BXD29 DNA was circularized and used as a template in the first PCR reaction. The antisense primer for this reaction corresponded to Tlr4 DNA immediately downstream of the insertion. A divergent sense orientation primer corresponded to a region in Tlr4 immediately upstream of the HindIII site following the insertion.
Statistics:
P-values corresponding to differences between means of experimental groups were calculated using the two-tailed t-test; values <0.05 were considered significant.
RESULTS
BXD29 mice are unresponsive to inhaled LPS:
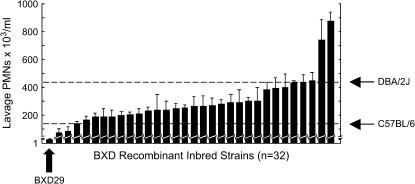
We challenged 32 strains of BXD RI mice to inhaled LPS and assessed their biologic responses by measuring levels of TNF-α and PMNs in whole-lung lavage fluid. Although the intensity of their responses varied, all but one of these strains displayed significant and marked increases in TNF-α and PMNs compared to their unchallenged counterparts (Figure 1). The one unresponsive strain, BXD29, had a lung inflammatory response that was indistinguishable from unchallenged mice.
Figure 1.
PMN response of BXD RI mice. Mice were exposed for 4 hr to aerosolized LPS and harvested immediately thereafter. Whole-lung lavage was performed and the fluid assayed for the number of PMNs per milliliter. Mean neutrophil counts for parental strains are indicated by open rectangles. Error bars represent standard deviation (n ≥ 5 mice per strain). BXD29 mice are essentially unresponsive to inhaled LPS.
BXD29 mice have a recessive mutation:
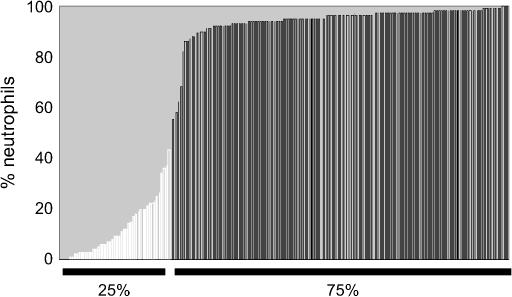
The virtual absence of a biologic response to inhaled LPS in the BXD29 RI mice suggested that during the generation of this strain it might have sustained a mutation in a gene controlling responses to LPS. To test this possibility and to determine whether such a mutation was dominant or recessive, we backcrossed BXD29 mice to DBA/2J mice, interbred their F1 progeny, and exposed the resulting F2 mice to aerosols of LPS. Analysis of TNF-α and PMNs in the lavage fluid revealed that 43 of 191 (73%) challenged F2 mice had >80% neutrophils, while 23% had <25% neutrophils (Figure 2). These values are close to the 1:3 ratio expected for a recessive mutation in a single gene.
Figure 2.
The unresponsiveness of BXD29 mice results from a recessive mutation. F2 [F1(BXD29 × DBA/2J) × F1] mice were exposed to aerosolized LPS and lavaged, and the percentage of neutrophils in this fluid was determined. Mice having the highest 75% and lowest 25% of neutrophils are indicated.
The BXD29 mutation resides in chromosome 4:
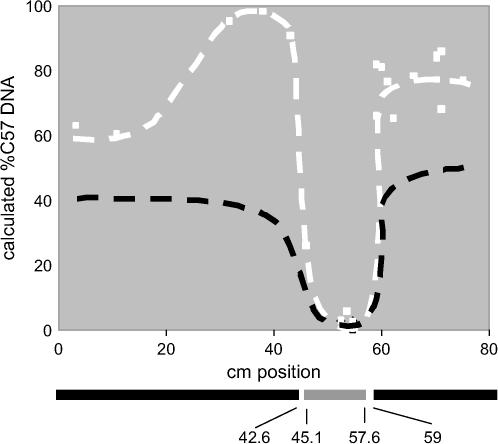
The mutation underlying the unresponsiveness to LPS in BXD29 mice might have occurred in an uncharacterized gene or in one of several genes known to be required for this response. We therefore undertook a mapping experiment to identify the chromosomal location of the mutation in BXD29 RI mice. This experiment was based on the following line of reasoning. Analysis of the F2 [F1(BXD29 × DBA/2J) × F1] mice suggested that the mutation is recessive. If the mutation arose on C57BL/6J-derived DNA, all unresponsive F2 mice should be homozygous at this locus for C57BL/6J DNA. By contrast, all other loci in this unresponsive group should be represented either equally by both parental strains or entirely by DBA/2J DNA, depending on whether that locus in BXD29 mice is derived from the C57BL/6J or the DBA/2J strain. Conversely, if the mutation resides on DBA/2J DNA, then all unresponsive F2 mice should be homozygous for DBA/2J DNA at this locus. To determine the chromosomal location of the BXD29 mutation, we prepared separate pools of DNA from 25 responsive F2 mice and 25 unresponsive F2 mice from these crosses. Parental allele frequencies in each of these pools were determined for ∼300 loci using a quantitative, PCR-based, genomewide scan (Germer et al. 2000). This experiment revealed that, as expected, all loci but one were represented either by equal amounts of each parental strain or entirely by DBA/2J DNA. However, in the low-responder pool of F2 [F1(BXD29 × DBA/2J) × F1] mice, a single region on chromosome 4 near cM 40 was represented entirely by C57BL/6J DNA (Figure 3). The percentage of C57BL/6J DNA in the low-responder pool declined with distance from this locus, presumably as the numbers of mice bearing chromosomal crossovers between the mutation and the assayed locus increased. An exception to this finding was the region between cM 46 and 60. Within this region, both the high- and low-responder F2 [F1(BXD29 × DBA/2J) × F1] pools were represented entirely by DBA/2J DNA. Inspection of the known crossover points for C57BL/6J and DBA/2J DNA in BXD29 mice revealed that this region of the chromosome was derived from DBA/2J (http://www.informatics.jax.org/searches/riset.cgi?sort=cM&format=html&set=BXD&chromosome). Therefore, a cross between BXD29 and DBA/2J mice must necessarily yield DBA/2J DNA at this interval. Taken together, these findings indicate that the mutation in BXD29 mice resides in C57BL/6J-derived DNA on chromosome 4 near cM 40.
Figure 3.
Identification of a QTL for PMN recruitment on chromosome 4. The percentage of C57BL/6J DNA in the high-responder pool (solid dashed line) and low-responder pool (open dashed line) are shown for various loci on this chromosome. Maximum divergence between these two pools occurs at a region between cM 30 and 40. Note that in BXD29 mice the region between 45.1 and 57.6 is represented by DBA/2J DNA. As a result, both pools must have 100% DBA/2J DNA (0% C57BL/6J DNA).
The BXD29 mutation is allelic with Tlr4:
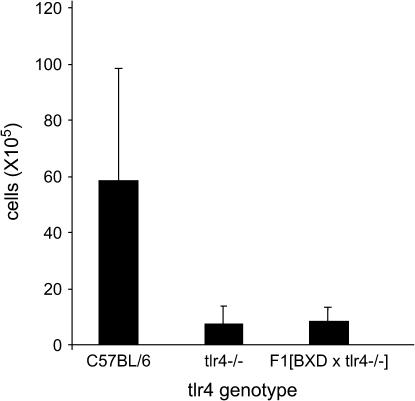
Tlr4 is required for the response to LPS and its gene is known to reside on chromosome 4 at cM 34. Although the sequence of this gene is identical in C57BL/6J and DBA/2J mice, it was nonetheless an obvious candidate for having undergone a recessive mutation in BXD29 mice. We therefore performed an experiment to determine if the mutation in BXD29 mice is allelic with Tlr4. BXD29 mice were crossed to mice bearing a targeted disruption of the Tlr4 gene (C57BL/6J–Tlr4), and the F1 progeny were challenged with aerosolized LPS. The responses of these F1 mice were indistinguishable from those of Tlr4-deficient mice (Figure 4), indicating that the mutation in BXD29 mice is allelic with Tlr4.
Figure 4.
Cellular response of F1 (BXD29 × C57BL/6J–Tlr4) mice to inhaled LPS. Wild-type C57BL/6J mice have a robust cellular infiltration to the lung airspace, whereas the response of F1 (BXD29 × C57BL/6J–Tlr4) mice is extremely low and indistinguishable from that of C57BL/6J–Tlr4 mice.
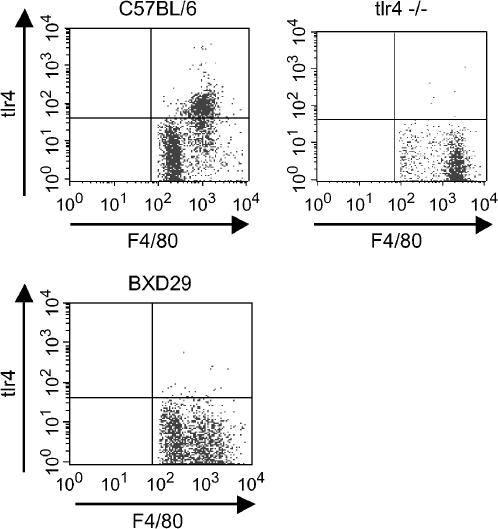
Tlr4 is not displayed on the cell surface of BXD29 macrophages:
Quantitative PCR analyses did not reveal differences between wild-type and BXD29 mice in levels of Tlr4-specific mRNA (data not shown). We therefore investigated whether Tlr4 is expressed on the surface of cells from BXD29 RI mice. Macrophages were elicited from the peritoneum of C57BL/6J, BXD29, and Tlr4-deficient mice and stained with antibodies against Tlr4 and F4/80, a molecule on the cell surface of macrophages. Flow cytometric analysis revealed that cells from C57BL/6J mice were stained with antibodies against Tlr4 and F4/80, whereas cells from the BXD29 and C57BL/6J–Tlr4 mice were stained with antibodies against F4/80, but not with antibodies against Tlr4 (Figure 5). This result demonstrates that macrophages from BXD29 mice either lack Tlr4 or display a form of the protein that is not recognized by the antibody used.
Figure 5.
Flow cytometric analysis of macrophages in BXD29 mice. Macrophages elicited by thioglycollate from mice were stained with anti-F4/80 and anti-Tlr4 antibodies. Note that F4/80-staining cells of C57BL/6J mice also stain for Tlr4. By contrast, no Tlr4 staining is seen in cells from C57BL/6J–Tlr4 mice or from BXD29 mice.
Analysis of the Tlr4 locus in BXD29 DNA:
To identify the molecular lesion in the Tlr4 gene of BXD29 mice, we first used PCR to generate a series of overlapping amplified products that collectively encompassed the entire Tlr4 coding region. For most of these PCR reactions, both C57BL/6J DNA and BXD29 DNA gave PCR products of identical size and DNA sequence. However, for a region in exon 3 of Tlr4 bounded by nucleotides 279 and 708 of the cDNA sequence, we were able to generate a product of the expected size from C57BL/6J DNA, but not from BXD29 DNA. This finding suggested that this region of the BXD29 Tlr4 gene was mutated and likely contained an insertion of sufficient size to preclude amplification of a PCR product containing it. To characterize junctions between the intact Tlr4 sequence and this presumptive DNA insertion, we employed a technique termed inverse PCR. In this procedure, genomic DNA is first cleaved with a restriction enzyme that cleaves once within the known (Tlr4) DNA and also within the DNA of unknown sequence. These DNAs are then ligated under dilute conditions to generate circular molecules, some of which contain the unknown DNA of interest, flanked by regions of known sequence. Using divergent primers corresponding to regions in Tlr4, we used PCR to separately amplify the 5′ and 3′ junctions between Tlr4 and the unknown DNA. Sequence analysis of the ends of the 1.3-kb product encompassing the 5′ junction revealed that Tlr4 was interrupted by repetitive DNA whose sequence was at least 98% identical to >200 sites throughout the mouse genome. The other end of this PCR product was also composed of this same repeat sequence. Analysis of the PCR product encompassing the 3′ junctions revealed that repetitive DNA was situated immediately upstream of the 3′ portion of Tlr4. In addition to repetitive DNA, this PCR product also contained DNA that was identical to a 239-bp region on chromosome 10 between the nucleotides 51,365,897 and 51,366,135. Inspection of the Tlr4 sequence adjacent to each end of the repetitive DNA revealed that no Tlr4 DNA had been deleted, but that an eight-nucleotide stretch (TCTTTCTT) of Tlr4 DNA was repeated at each end of the DNA insertion.
DISCUSSION
We have shown here that BXD29 RI mice have a mutation in the Tlr4 gene, causing them to be unresponsive to LPS. The evidence supporting this conclusion is as follows. First, the hyporesponsive phenotype of F2 mice [F1(BXD29 × DBA/2J) × F1] maps to a locus on chromosome 4 that contains Tlr4. Second, all F1(BXD29 × C57BL/6J–Tlr4) mice fail to respond to inhaled LPS. Third, anti-Tlr4 antibodies do not recognize Tlr4 on BXD29 macrophages. Finally, exon 3 of the Tlr4 gene in BXD29 mice is interrupted by a large stretch of repetitive DNA. Together, these findings conclusively identify another mouse strain that contains a defective Tlr4 gene and at the same time raise questions related to the study of RI strains.
The disruption of Tlr4 in BXD29 mice provides a ready explanation for their lack of an innate immune response to inhaled LPS, but the actions of this receptor are not limited to its function in innate immune responses. Tlr4 also triggers adaptive immune responses to pathogens by upregulating costimulatory molecules in dendritic cells (Reis e Sousa and Kapsenberg 2004), and its actions have been associated with a variety of inflammatory diseases, including atherosclerosis (Kiechl et al. 2002). Therefore, BXD29 mice are likely to have defects in multiple aspects of immune and inflammatory responses.
Although spontaneous mutations have not been previously demonstrated in RI strains, a spontaneous deletion in a stock of C57BL/6 mice was recently shown to impact electroconvulsive responses (Yang et al. 2003). That spontaneous mutations should occur in mice is not surprising. Indeed, genetic variation between mouse inbred strains is the basis for most gene mapping techniques in the mouse. However, our demonstration of a spontaneous Tlr4 mutation in BXD29 mice suggests that this type of mutation might be more common in RI mice than previously appreciated. Because there are more BXD RI strains than any other set of RI strains (http://www.jax.org/jaxmice), BXD RI mice have been extensively used for QTL mapping. The impact of BXD29 Tlr4 mutation will likely vary, depending on the location of the mapped QTL, and be greatest for QTL residing between 0 and 46 cM on chromosome 4 as this region is derived from C57BL/6J in BXD29 mice (http://www.informatics.jax.org/searches/riset.cgi?sort=cM&format=html&set=BXD&chromosome).
RI strains are extremely useful for identifying QTL in part because they provide a source of multiple, genetically identical animals whose genotypes and phenotypes can be readily compared. However, the discovery of the Tlr4 mutation in BXD29 raises a more general issue relating to the frequency of such mutations. We were able to identify this Tlr4 mutation in BXD29 mice because of its dramatic effect on the biologic response to inhaled LPS. If the phenotype of the BXD29 mice had been less marked, we would likely have assumed that it resulted from the inheritance of multiple QTL negatively affecting the response to LPS. The effect of such erroneous assumptions on QTL analyses and genetic correlation studies relating the genotype of RI strains to immune and inflammation-related phenotypes will depend on the impact of the mutation of the phenotype of interest. Although minor phenotypic effects of such mutations on QTL analyses are likely minimal, steps should be taken to minimize the impact of larger phenotypic effects. Such steps could include statistical approaches such as Jacknife resampling to minimize the impact of single outlier strains. Additional steps include introducing fresh breeding stock at regular intervals to reduce fixation of spontaneous mutations and increasing the number of RI strains in the study, as was done recently for BXD crosses (Peirce et al. 2004). Together, these steps should enhance the utility of RI strains in uncovering genetic loci affecting phenotypes of interest.
Acknowledgments
This study was supported by grants from the Department of Veterans' Affairs (Merit Review), the National Institute of Environmental Health Sciences (ES11375, ES07498, and ES09607), and the National Heart, Lung, and Blood Institute (HL66604, HL66611, and HL66580).
References
- Cook, D. N., S. Wang, Y. Wang, G. P. Howles, G. S. Whitehead et al., 2004. Genetic regulation of endotoxin-induced airway disease. Genomics 83: 961–969. [DOI] [PubMed] [Google Scholar]
- Germer, S., M. J. Holland and R. Higuchi, 2000. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 10: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiechl, S., E. Lorenz, M. Reindl, C. J. Wiedermann, F. Oberhollenzer et al., 2002. Toll-like receptor 4 polymorphisms and atherogenesis. N. Engl. J. Med. 347: 185–192. [DOI] [PubMed] [Google Scholar]
- Lorenz, E., M. Jones, C. Wohlford-Lenane, N. Meyer, K. L. Frees et al., 2001. Genes other than TLR4 are involved in the response to inhaled LPS. Am. J. Physiol. Lung Cell. Mol. Physiol. 281: L1106–L1114. [DOI] [PubMed] [Google Scholar]
- Ochman, H., A. S. Gerber and D. L. Hartl, 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirce, J. L., L. Lu, J. Gu, L. M. Silver and R. W. Williams, 2004. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis e Sousa, C., and M. L. Kapsenberg, 2004. Activation of dendritic cells: translating innate into adaptive immunity: dendritic-cell control of pathogen-driven T-cell polarization. Curr. Opin. Immunol. 16: 21–25. [DOI] [PubMed] [Google Scholar]
- Yang, Y., B. J. Beyer, J. F. Otto, T. P. O'Brien, V. A. Letts et al., 2003. Spontaneous deletion of epilepsy gene orthologs in a mutant mouse with a low electroconvulsive threshold. Hum. Mol. Genet. 12: 975–984. [DOI] [PubMed] [Google Scholar]