Abstract
The methylation of CpG dinucleotides has become a topic of great interest in cancer research, and the methylation of promoter regions of several tumor suppressor genes has been identified as a marker of tumorigenesis. Evaluation of DNA methylation markers in tumor tissue requires hundreds of samples, which must be analyzed quantitatively due to the heterogeneous composition of biological material. Therefore novel, fast and inexpensive methods for high throughput analysis are needed. Here we introduce a new assay based on peptide nucleic acid (PNA)-library hybridization and subsequent MALDI-TOF analysis. This method is multiplexable, allows the use of standard 384 well automated pipetting, and is more specific and flexible than established methods, such as microarrays and MS-SNuPE. The approach was used to evaluate three candidate colon cancer methylation markers previously identified in a microarray study. The methylation of the genes Ade-nomatous polyposis coli (APC), glycogen synthase kinase-β-3 (GSK3β) and eyes absent 4 (EYA4) was analyzed in 12 colon cancer and 12 normal tissues. APC and EYA4 were confirmed as being differentially methylated in colon cancer patients whereas GSK3β did not show differential methylation.
INTRODUCTION
The methylation of CpG dinucleotides is an epigenetic modification that has frequently been associated with control of gene expression (1). In particular, X-chromosomal silencing, inactivation of retrotransposon activity, imprinting, transcriptional regulation in embryogenesis and cell differentiation are influenced by DNA methylation (2–6). Aberrant DNA methylation patterns have been associated with a variety of genetic diseases and cancer (7). As such, DNA methylation markers hold great promise as research and diagnostic tools (8).
Due to the heterogenous composition of biological material from tissue samples, thorough investigations of methylation patterns necessitate the analysis of samples in a quantitative manner, in particular for applications, such as the analysis of allele-specific methylation (imprinting). In that respect there is a significant need for fast and cheap methods for quantitative DNA methylation analysis. Genome-wide discovery methods lead to a great number of candidate markers, which have to be evaluated on hundreds of samples (9–11). A final diagnostic application would also require an assay capable of handling many specimens.
Most techniques used to analyze methylation patterns depend on bisulfite treatment of the DNA. The bisulfite reaction converts all non-methylated cytosines to uracils, whereas methylated cytosines are not changed under the reaction conditions. The methylation level can then be determined by direct sequencing of subclones or PCR products (12,13), matrix-assisted laser desorption/ionization (MALDI) analysis of RNA transcripts from bisulfite treated DNA (14), fluorescence real-time PCR (MethyLight and QAMA) (15,16), microarray based systems (11), primer extension or pyrosequencing (17) and several others. Most of these techniques suffer from high analysis costs for kits and other consumables, in particular techniques applying fluorescence dyes (sequencing, real-time PCR and microarrays). Other methods are not capable of multiplex analysis (pyrosequencing and sequencing). In contrast MALDI is an excellent tool to analyze DNA methylation, due to its fast and accurate analysis power, its multichannel analysis capability and its ability to run quantitative analysis (18). The combination of MALDI detection with peptide nucleic acid (PNA) probe hybridization enables the complex analysis of DNA methylation (19). Compared to other MALDI based methods, such as the GOOD assay, no CpG-free extension primer sequences are required. Therefore, the method can also be used to study DNA methylation in CpG islands. CpG islands are often overlapping with promoters particularly in tumor suppressor genes. Their methylation status has been reported to inversely correlate with expression levels (20). The analysis of CpG island methylation status is therefore of great interest and PNA probe hybridization represents an excellent analysis tool. In addition, by design of suitable PNA probes the method allows the simultaneous detection of several CPGs (within a CPG island). Therefore patterns of CPG methylation can be analyzed in a single reaction.
PNAs are a class of DNA analogs in which the entire sugar-phosphate backbone is replaced by a pseudopeptide. Owing to its neutral character and the consequent lack of electrostatic repulsion, PNA exhibits very stable heteroduplex formation with complementary nucleic acid that is essentially ionic strength independent, enables hybridization under minimum salt conditions, and is more specific than DNA based hybridization methods. This feature, as well as its superior ion stability and easy ionization, compared to DNA, renders PNA very attractive for hybridization-based MALDI time-of-flight mass spectrometry (MALDI-TOF-MS) (21–23). Analytical sensitivity was further improved by introducing a ‘charge tag’ into the PNA molecule (24). In our work, a method has been established which enables the 384 well plate based analysis of immobilized amplificates from bisulfite treated DNA by PNA probes (Figure 1). Biotinylated PCR-amplificates were prepared from bisulfite treated DNA using 5′-biotinylated primers and immobilized in streptavidin coated microtiter plates without prior purification. Subsequently, a PNA-library hybridization was performed. After removing unspecifically bound probes, PNAs were analyzed with MALDI and the methylation level was estimated by dividing the signal intensity of a probe specific to the methylated allele by the sum of signal intensities of both probes (specific to the methylated and specific to the unmethylated allele).
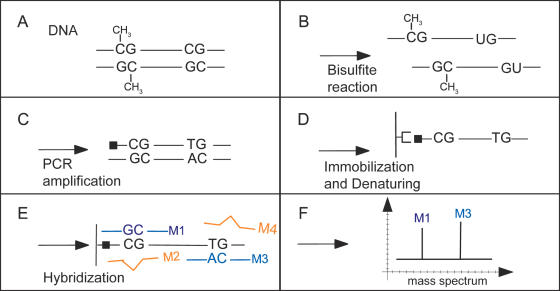
Figure 1.
Methylation analysis by PNA probe hybridization and MALDI-TOF analysis. (A) DNA extracted from tissue sample. (B) Conversion of unmethylated cytosine into uracil by bisulfite treatment. (C) PCR amplifing the upper strand with a 5′-biotinylated primer. (D) Amplicon immobilization on streptavidin coated microtiterplates; Denaturation using 200 mM NaOH. (E) PNA probe hybridization; Removal of excess probes (red) by stringent washing. (F) Release of specifically bound probes (green and blue) at elevated temperature (60°C); eluate transfer onto MALDI-TOF sample holder; MALDI analysis and matching of molecular mass to sequence database.
This enables the relative quantification for classification based on the methylation levels of samples. Altogether, twelve PNA hybridization assays were evaluated and multiplex analysis was demonstrated. Six assays were further characterized by analyzing mixtures of artificially methylated and unmethylated DNA. Design criteria were elucidated which will lead to an improved probe design for expanded DNA methylation research.
To demonstrate the viability of our method on patient samples we focused on colorectal cancer, the second most common cause of malignant death in industrialized countries. More accurate detection markers are needed to improve the effectiveness and efficiency of both screening and monitoring of colorectal neoplasia. DNA methylation in colorectal cancer has been well-studied in an effort to find new potential markers for early detection (25–27). We performed our analysis on a series of patient samples available from this neoplastic disorder. Two candidate DNA methylation cancer markers recently identified in our genome-wide discovery and microarray analysis (C. Lofton-Day, A. Sledziewski and K. Berlin, unpublished data) and one marker taken from literature were selected to evaluate the new PNA hybridization approach (36): Glycogen synthase kinase-β-3 (GSK3β), eyes absent 4 (EYA4) and Adenomatous polyposis coli (APC).
To evaluate the three candidate markers, 24 samples (12 cancer and 12 normal) were assayed and statistical analysis was performed. Subsequently, data were validated by pyrosequencing.
MATERIALS AND METHODS
PNA synthesis and modification
PNA design and synthesis has been performed according to the recommendations of Applied Biosystems (www.appliedbiosystems.com). All syntheses were performed in a 2 µmol-scale using a PerSeptive Biosystems Expedite 8909 synthesizer. PNAs were subsequently modified with one of the following trimethyl ammonium-alkyryl-NHS-ester: 5-Trimethyl ammonium- pentyryl- N-hydroxy- succinimidyl ester (CT 156), 4-Trimethyl ammonium- butyryl- N-hydroxy- succinimidyl ester (CT 142) and 3-Trimethyl ammonium- propyl- N-hydroxy- succinimidyl ester (CT 128). The reactions were performed according to methods described for oligonucleotide modification by Sauer et al. (28,29). Synthesized PNA probes and primers are listed in Table 1.
Table 1.
List of oligonucleotides and probes
| Assayname | Primer 1 | Primer 2 | M probea | U probeb |
|---|---|---|---|---|
| APC-1 | 5′Biotin-GGAGAGAGAAGTAGTTGTGTAATT | ACTACACCAATACAACCACATATC | CT128-OOc-ACTCCGCAATA | CT156-OOc-ACTCCACAATA |
| APC-2 | 5′Biotin-GGAGAGAGAAGTAGTTGTGTAATT | ACTACACCAATACAACCACATATC | CT128-OOc-AATCCGCATCC | CT156-OOc-AATCCACATCC |
| APC-3 | 5′Biotin-GGAGAGAGAAGTAGTTGTGTAATT | ACTACACCAATACAACCACATATC | CT156-OOc-ATCGACGAACT | CT156-Oc-ATCAACAAACT |
| EYA4-1 | 5′Biotin-TAGGTTTTTGGAAGAGGTGA | CCCAAAAATCAAACAACAAC | CT128-OOc-ATTTCGTATCT | CT142-Oc-ATTTCATATCT |
| EYA4-2 | 5′Biotin-TTTTAGTAATGTGTTTAGAGAAGTTTTG | CCCAAAAATCAAACAACAAC | CT128-OOc-ACGACGT | CT128-Oc-ACAACAT |
| GSKB-1 | 5′Biotin-AGTGATAAAGGAAGGAAGGAA | CAACCCAAAACCCTATCAAC | CT128-OOc-TCAACGACTAC | CT128-Oc-TCAACAACTAC |
| GSKB-2 | 5′Biotin-AGTGATAAAGGAAGGAAGGAA | CAACCCAAAACCCTATCAAC | CT156-OOc-CGAAACCGACT | CT156-Oc-CAAACCAACT |
| GSKB-3 | 5′Biotin-AGTGATAAAGGAAGGAAGGAA | CAACCCAAAACCCTATCAAC | CT128-OOc-TCCTCGCTTCC | CT128-Oc-TCCTCACTTCC |
| hMLH1-1 | 5′Biotin-TTTTTAGGAGTGAAGGAGGTT | TAAAACCCTATACCTAATCT | CT156-OOc-CGTCTACGTCA | CT142-OOc-CATCTACATCA |
| hMLH1-2 | 5′Biotin-TTTTTAGGAGTGAAGGAGGTT | TAAAACCCTATACCTAATCT | CT142-OOc-CGCCACTACGA | CT128-Oc-CACCACTACAA |
| hMLH1-3 | 5′Biotin-TGAAGGGTGGGGTTGGAT | TTCAACCAATCACCTCAATACC | CT156-Oc-CTCACGTTCTT | CT128-OOc-CTCACATTCTT |
| MDR1-1 | 5′Biotin-GAAGATTTATGTGAATTTTGAAAG | AAACATTTCAACTTATATAAACAC | CT156-OOc-AACACGTCTTT | CT156-OOc-AACACATCTTT |
aM probe: PNA probe reverse complementary to methylated sequence
bU probe: PNA probe reverse complementary to unmethylated sequence.
cChemical modifications: O = O-linker ({2-[2-(Fmoc-amino)ethoxy]acetic acid; CT156 = N-Propyl-5-Trimethylammoniumpentylamid; CT142 = N-Propyl-4-Trimethylammonium-butyrylamid and CT128 = N-Propyl-3-Trimethylammonium-propylamid.
DNA preparation
Fresh frozen samples were obtained from Integrated Laboratory Services (ILS, Chestertown, USA). A total of 24 patient samples were analyzed comprising 5 normals, 5 adenocarcinomas and 7 matched pairs (adenocarcinoma and normal adjacent tissue).
DNA was extracted using the QIAamp DNA purification kit (Qiagen, Hilden, Germany). Unmethylated DNA was prepared by MDA (multiple displacement amplification), a genome-wide amplification method described by Dean et al. (30). Methylated DNA was prepared by treating unmethylated MDA-DNA with SssI methyltransferase (New England Biolabs, Frankfurt, Germany) in the presence of S-adenosyl-methionine according to the manufacturer's instructions. Bisulfite treatment was performed as described previously (31).
PCR amplification and immobilization
Oligonucleotides listed in Table 1 were used for PCR amplification. Primers were designed for the following loci: APC: AC109473.2 (61 083–60 963); EYA4: AL121959.15, HSDJ78N10 (80 737–80 797); GSK3β: AC092910.9 (147 566–147 626); hMLH1: AC011816 (143 079–143 139); MDR1: AC002457.2 (36 792–36 839). PCR was performed in a total volume of 65 µl containing 10 ng template DNA, 2.6 U Hotstart Taq polymerase (Qiagen, Hilden, Germany), 32.5 pmol of each forward and reverse primer, 1× PCR buffer (Qiagen, Hilden, Germany) and 0.2 mM of each dNTP (MBI Fermentas, St. Leon-Rot, Germany). Cycling was done using a Mastercycler (Eppendorf, Hamburg, Germany) under the following conditions: 15 min at 95°C and 40 cycles at 95°C for 1 min, 55°C for 45 s and 72°C for 1:30 min with the following exceptions: for all GSK3β assays 53°C annealing temperature was used and primer 1 was used in a concentration of 1 µM. Assay hMLH1-1 and -2 were performed with annealing at 54°C, hMLH1-3 at 60°C and MDR1-1 at 51°C. For assay EYA4-2 1 µM of primer 1 was used. An aliquot of 5 µl of the PCR mix was loaded on a 2.5% agarose gel to check PCR performance. An aliquot of 60 µl was immobilized and denatured in a High Capacity Streptavidin (Sigma–Aldrich, Munich, Germany) 384 well plate according to the suppliers instructions. Wells were equilibrated for 10 min in the hybridization buffer containing 0.7 M ammonium chloride, 70 mM ammonium citrate and 3.6% (w/v) ammonium-laurylsarcosinate (pH 7). The buffer was replaced with fresh hybridization buffer containing 100 nM of each PNA probe. The plate was incubated while shaking (1000 U/min) using a Thermomixer 5355 Comfort (Eppendorf, Hamburg, Germany) for 1 h at 34°C. The EYA4-2 assay was incubated at 20°C. The plate was washed once with hybridization buffer heated to the hybridization temperature and twice with 4°C water. Subsequently the plate was filled with water and heated to 60°C for 10 min. Twice 0.5 µl of eluate was transfered to a ScoutTM384 stainless steel target plate precoated with 0.5 µl of 0.2% (w/v) α-cyano-4-methoxy-cinnamic acid, 1.8% (w/v) α-cyano-4-hydroxy-cinnamic acid methyl ester in acetone. After drying, 0.5 µl 0.01% ammonium hydroxide was added.
MALDI-TOF analysis
Automated analyses were carried out on a Biflex III mass spectrometer using the AutoXecute software (Bruker Daltonics, Bremen, Germany). Mass spectra were recorded in the positive ion linear time-of-flight mode. Acceleration potentials of IS/1 19 kV and 17.5 kV IS/2 were used. Detector potential was set on 1.7 kV. Ion extraction was delayed by 200 ns. The detector was gated to prevent saturation by molecules smaller than 1200 Da. Usually 16 × 25 shots were accumulated per sample spot, smoothed using a Golay-Savitzky filter and baseline-corrected. Spectra with a signal to noise range > 12 and a resolution > 310 were collected. All others were discarded. The Genotool SNP manager was used to automatically analyze the spectra and yield the relative intensities of the peaks. Each analysis was performed in duplicate and each eluate was measured twice resulting in four data points per analysis.
Data analysis
Methylation levels were calculated by dividing the signal intensity of the CG-peak by the sum of the signal intensities of both corresponding PNA probes [Methylation Level = CG/(CG + CA)]. Distributions of measured samples were visualized using a box-whisker-plot. A total of 75% of the data were represented by the box and 95% of the data by the whiskers. Data outside the 95th percentile were marked as outliers. Classifications of distributions were analyzed using a non-parametric (Wilcoxon) test.
Pyrosequencing
Pyrosequencing was performed as described by Tost et al. (17) using a PSQ™ station and a PSQ 96MA system (Pyrosequencing AB). For the amplification, the following primers were used: Biotin-AGTGAtAAAGGAAGGAAGGAA and AAACAACTCCCTTCAAACC (GSK3β), Biotin-GGTTAGGGTTAGGTAGGTTGT and ACTACACCAATACAACCACATATC (APC), and for EYA4 Biotin-TAGGTTTTTGGAAGAGGTGA and CCCAAAAATCAAACAACAAC (EYA4). PCR was performed in a total volume of 25 µl containing 10 ng template DNA, 1 U Hotstar Taq polymerase (Qiagen, Hilden, Germany), 12.5 pmol of forward and reverse primers, 1× PCR buffer (Qiagen, Hilden, Germany), 0.2 mM of each dNTP (MBI Fermentas, St. Leon-Rot, Germany). Cycling was done using a Mastercycler (Eppendorf, Hamburg, Germany) with the following conditions: 15 min at 95°C and 40 cycles at 95°C for 1 min, 55°C for 45 s and 72°C for 1:30 min. The following biotinylated oligonucleotides were used as extension primers for the pyrosequencing reaction: APC CCACACCCAACCAA; EYA4 ACTTACTTCCAAAC and GSK3β CCCAAAACCCTATCAA.
RESULTS AND DISCUSSION
MALDI-TOF represents a sensitive and easily automatable method for the detection and unambigious identification of biomolecules and fragments thereof. Recently, quantitative analysis using MALDI-TOF has been described by standardizing the signal to an internal control (14,18). To study the suitability of the technique for quantitative methylation analysis, bisulfite specific primers with a 5′-biotin label on the forward primer were used to amplify the promoter region of the multidrug resistance protein 1 (MDR1). Amplification was performed with mixtures of bisulfite treated methylated and unmethylated template DNA. Amplificates were immobilized in different wells of a 384 well streptavidin coated microtiter plate and subjected to a NaOH washing step to remove the unlabeled reverse strand (Figure 1D). A set of two methylation-specific PNA probes was hybridized (Figure 1E). After removal of the supernatant and stringent washing the wells were incubated with elution buffer at elevated temperature. Microliter volumes of the elution buffer were then directly transferred to a MALDI steel target, mixed with organic matrix and analyzed in the positive ion mode using a Bruker Biflex III instrument (Figure 1F).
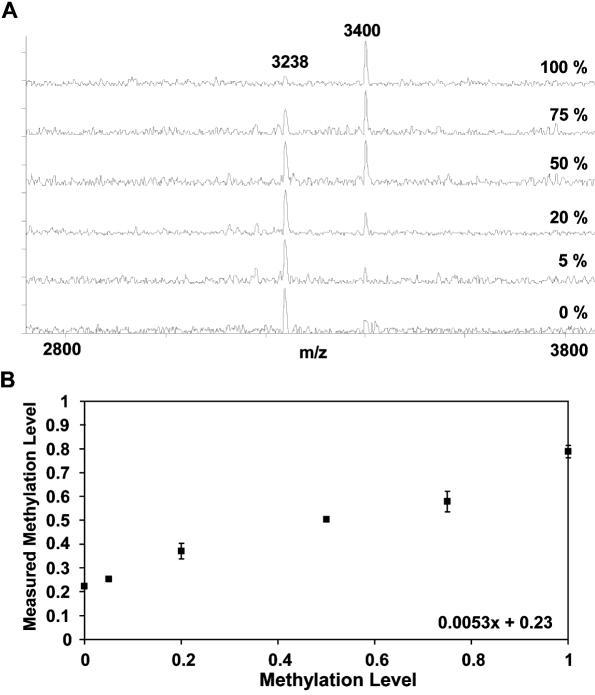
MALDI spectra for DNA mixtures representing different methylation states of a specific CpG position and PNA probe sequences are shown in Figure 2. Different numbers of 5′ linker residues were incorporated during PNA probe synthesis to guarantee optimum peak separation of the two probe signals. A quarternary ammonium moiety was added to the linker to facilitate ionization (24). Comparison of the spectra obtained for 100% methylated and 0% methylated DNA demonstrates that both PNA probes hybridize with high, although not complete specificity to the respective immobilized templates (Figure 2A). In addition, methylation levels were calculated and plotted against the methylation state represented by the hybridization template. The resulting response curve is linear with a correlation coefficient of 0.98. Only minor deviations were detectable between two independent experiments indicating the feasibility of MALDI-TOF analysis for quantitative methylation analysis via PNA probe hybridization. Due to a low level of unspecific hybridization a slope <1 is observed.
Figure 2.
Quantitative analysis of single CpG methylation within the MDR1 gene. (A) MALDI spectra of calibration experiment: PCR on mixtures of bisulfite treated DNA representing different methylation levels; Hybridization using MDR1-U1 AACACATCTTT (3238.98 Da) and MDR1-M1 AACACGTCTTT (3399.64 Da). (B) Response curve: ratios of the signal intensity of the probe reverse complementary to the methylated DNA sequence to the sum of both probes plotted against the methylation levels present in the mixtures. Squares represent median of two independent experiments with two replicates each; R2 = 0.98.
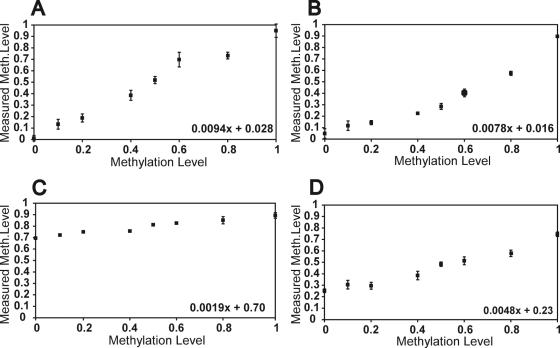
To further characterize the scope of the method and to pinpoint important design parameters five additional assays were designed and characterized as described before. For the analysis of EYA4 (Figure 3A) and APC (3B) PNA probes covering two adjacent CpG postions were synthesized. In addition, probes addressing single CpG positions were used in hMLH1 (3C) and in GSK3β (3D).
Figure 3.
Comparison of PNA probes covering one or two CpG positions. (A and B) Assays with PNA probes containing two CG or CA-sequences each. (C and D) Assays with PNA probes covering one CpG. Analyzed genes: (A) EYA4-2, (B) APC-3, (C) hMLH1-3 and (D) GSK3β-1.
When comparing the different response curves, one can conclude that the slope is high on average when a PNA probe pair addresses two CpG positions simultaneously but is lower when a single CpG is analyzed (3C). Since in this experiment both positions are either methylated or unmethylated this observation is easily explained by the increased hybridization temperature difference between match and mismatch. Evaluation of a total of 12 PNA hybridization assays led to the observation that the assay performed better with short amplificates and probes with a low C-content (Table 2). This was indicated by a higher difference in the measured methylation level between artificially methylated and unmethylated DNA, respectively, which results in an increased measurement range of the method. The negative influence of longer amplificates (>100 bp) can be explained by the higher tendency of long fragments to form secondary structures (32,33). The finding that neighbored C:G pairing stabilizes mismatch pairing has been already described by Ratilainen et al. (34). In summary the number of analyzed CpGs per PNA probe, the length of the PCR product and the C-content all influence assay performance.
Table 2.
Characteristic values of evaluated assays
| Assay | PCR length(bp) | Number of CpGs in probe | C-content(%) in probe | Measurement rangea |
|---|---|---|---|---|
| APC-2 | 123 | 1 | 45 | 0.02 |
| APC-1 | 123 | 1 | 36 | 0.16 |
| EYA4-1 | 227 | 1 | 18 | 0.17 |
| GSK3β-3 | 66 | 1 | 55 | 0.18 |
| hMLH1-2 | 180 | 2 | 45 | 0.19 |
| hMLH1-3 | 77 | 1 | 36 | 0.26 |
| hMLH1-1 | 180 | 2 | 36 | 0.4 |
| GSK3β-1 | 66 | 1 | 36 | 0.54 |
| APC-3 | 123 | 2 | 27 | 0.57 |
| GSK3β-2 | 66 | 2 | 36 | 0.68 |
| MDR1-1 | 53 | 1 | 27 | 0.71 |
| EYA4-2 | 69 | 2 | 29 | 0.88 |
aDifference of measured methylation levels for methylated and unmethylated DNA = (CG/(CG+CA)meth − (CG/(CG+CA)unmeth) = Maximum range of measurable values. High values allow a higher resolution of methylation measurement.
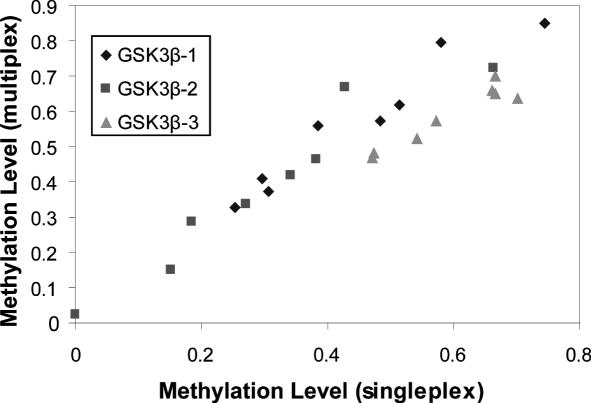
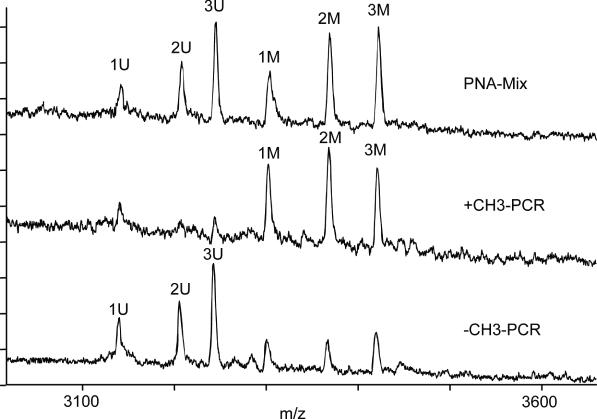
Multiplex analysis was performed to analyze several CpG positions simultaneously, and was compared to the singleplex analysis (Figure 4). Mixtures of different methylation levels were analyzed and a CG / (CG + CA) ratio was calculated. As shown in Figure 4 the results obtained by singleplex and multiplex analysis are highly correlated. In Figure 5 representative spectra from the corresponding multiplex analysis are shown. The methylation level of the analyzed sample can be clearly identified when comparing the peak patterns. Lower masses 1 U to 3 U identified PNA probes with reverse complementarity to unmethylated DNA. High mass probes 1 M to 3 M were specific for the methylated allele.
Figure 4.
Comparison of singleplex and multiplex analysis. GSK3β promoter methylation analysis on DNA methylation mixtures using three PNA probe pairs both in singleplex and multiplex hybridizations. Correlation coefficients: GSK3β-1 (0.935); GSK3β-2 (0.921), GSK3β-3 (0.896).
Figure 5.
Analysis using 6 PNA probes in parallel. Used PNAs: 1 U and 1 M. (GSK3β-U3: 3138.64 Da and GSK3β-M3: 3299.64 Da); 2 U and 2 M (GSK3β-U1: 3204.48 Da and GSK3β-M1: 3365.64 Da); 3 U and 3 M (GSK3β-U2: 3241.64 and GSK3β-M2: 3418.64 Da). Top spectrum: used hybridization mixture. Bottom spectrum: analysis of eluates from methylated and unmethylated template.
To demonstrate that the method is not limited to the analysis of artificial template material, the methylation state of a set of colon tumor and normal adjacent colon tissue samples was analyzed using three assays (Figure 6). The promoter regions of two of these genes were previous reported to be differentially methylated in colon cancer by a genome-wide discovery method (methylation-specific arbitrarily primed PCR) (9) and microarray analysis (F. Model, C. Lofton-Day and K. Berlin, unpublished data). The gene APC has been described (26). DNA was extracted, bisulfite treated (31) and assayed as described previously (Figure 1). Valid data were obtained for all samples and are presented in Figure 6.
Figure 6.
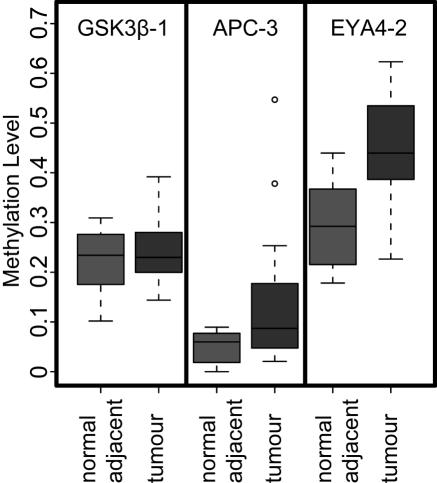
Methylation analysis of candidate colon cancer markers on patient DNA samples. Results of PNA hybridizations 24 DNA samples from colon cancer patients. (samples: 5 normals, 5 adenocarcinoma and 7 matched normal adjacent/adenocarcinoma; data shown in box-whisker plots. Wilcoxon test P-values for class separation of normal versus cancer samples: GSK3β-1 (0.932), APC-3 (0.0478), EYA4-2 (0,00143).
APC promoter
The product of the APC tumor suppressor gene was reported to regulate the Wnt signaling pathway through beta-catenin (35,36). APC promoter methylation has been frequently described (37–40). [Using methylation specifc PCR (MSP)] Esteller et al. (26) reported that 18% of patients with colorectal colon carcinoma without inherited APC mutations to exhibit hypermethylation of the APC promoter in the tumor tissue. The hybridization assay was designed to fit the promoter region analyzed by these authors. In most samples, negligible methylation was measured which cannot be separated from the background control (methylation level ≤0.1). However, 3 out of 12 tumor samples exhibited significantly elevated methylation levels, similar to the proportion reported by Esteller et al. (26). Methylation indices from cancer samples differed significantly from normals (non-parametric Wilcoxon test P-value 0.04783). Using pyrosequencing as an independent method the mean methylation level of the two CpG positions of interest in these tumor samples was verified to be indeed significantly higher with 0.08 (0.25), 0.54 (0.38) and 0.79 (0.55), respectively, than for all other samples, for which values close to 0 (<0.1) were obtained (PNA hybridization data given in parenthesis).
GSK3β
GSK3β has been identified by methylation-specific AP-PCR as a candidate methylation marker in colorectal cancer (C. Haefliger, R. Lesche, S. Maier and K. Berlin, unpublished data). Like APC, Glycogen-Synthase-Kinase-3 (GSK3β) has been implicated with the regulation of the β-Catenin/Tcf signaltransduction and is regulated by the Wnt-pathway (41). When analyzing the GSK3β promoter region similar methylation indices were found for all analyzed samples, i.e. no differential methylation in colon cancer could be demonstrated. As indicated by Figure 3D a methylation level of approximately 0.2 was obtained on unmethylated DNA for this assay. Accordingly, the elevated methylation indices for all samples may be attributed to non-specific binding of the CG specific probe regardless of methylation state. The lack of classification by PNA hybridization was again confirmed by pyrosequencing analysis of the same samples.
EYA4
EYA4 was first identified by microarray analysis as being differentially methylated in tumors (42). EYA4 is one of four members of the EYA gene family that is homologous to the eyes absent gene in Drosophila melanogaster (43). It is a non-thiol protein tyrosine phosphatase with an intrinsic ability to regulate transcriptional effects in mammalian organogenesis (44). Mutations in EYA4 are correlated with defects in ocular development in Drosophila flies and dilated cardiomyopathy and deafness in humans. It has been shown that aberrant expression in the steady state level of EYA4 triggers distinct apoptotic mechanisms, implicating these genes as possible regulators of programmed cell death (45). EYA4 hypermethylation has been shown in esophageal adenocarcinomas and colorectal cancer (46). EYA4 amplificates have been selected according to Haefliger et al. (44). PNA hybridization demonstrates methylation in all analyzed samples, but significantly higher methylation levels were found in tumors. A median methylation level of 0.451 for the tumor and 0.294 for the normal samples has been determined (Figure 6A). Wilcoxon analysis identified a significant class separation with a P-value of 0.00143. A detailed analysis of cancer samples compared with their corresponding normal adjacent tissue demonstrated an increased methylation level of the tumors in all cases. Pyrosequencing confirmed the PNA hybridization data.
CONCLUSIONS
A novel DNA methylation analysis method for the validation of candidate markers in tissue samples has been introduced. Reliable relative quantitation of methylation levels in heterogeneous tissues is achieved by PNA hybridization and subsequent MALDI-TOF-MS analysis. This method enables the rapid and simultaneous evaluation of DNA methylation markers in a single measurement and can therefore be performed in high throughput. In contrast to primer extension and sequencing based methods, our approach uses hybridization for the sequence identification. This enables the investigation of adjacent methylated CpGs (co-methylated). Due to their more stable nature, co-methylated sequences have a higher biological impact than single methylated CpGs, which can be the result of a non-specific random process (47). PNA hybridization therefore represents a significant advantage over the direct PCR product sequencing or primer extension based methods. These techniques do not have the ability to distinguish specific methylation patterns. In contrast they measure an average methylation level for each position. Identified co-methylated CpGs are suitable for the subsequent development of methylation detection assays, such as MethyLight, QAMA (quantitative analysis of methylated alleles), HM (HeavyMethyl) or MSP analysis (15,16,48,49).
The method can be used to perform multiplex analysis which allows the usage of small amounts of sample material and enables the parallel analysis of several methylation positions. The method is relatively inexpensive when large numbers of samples have to be analyzed.
Acknowledgments
This work was co-funded by the German Ministry of Education and Research (BMBF) KW01101. The authors thank Ina Schwope, Janine Notzke and Anne Hentschel (Epigenomics AG) for their productive support Theo deVos, Debjani Roy and Sue Cottrell (Epigenomics AG) for reading the manuscript and Jörn Walter (Saarland University) for helpful discussions. Funding to pay the Open Access publication charges for this article was provided by the Epigenomics AG.
Conflict of interest statement. None declared.
REFERENCES
- 1.Holliday R., Pugh J.E. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Wutz A., Rasmussen T.P., Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nature Genet. 2002;30:167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 3.Martin C., Prescott A., Lister C., MacKay S. Activity of the transposon Tam3 in Antirrhinum and tobacco: possible role of DNA methylation. EMBO. J. 1989;8:997–1004. doi: 10.1002/j.1460-2075.1989.tb03466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reik W., Collick A., Norris M.L., Barton S.C., Surani M.A. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 5.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 6.Lunyak V.V., Prefontaine G.G., Rosenfeld M.G. REST and peace for the neuronal-specific transcriptional program. Ann. NY Acad. Sci. 2004;1014:110–120. doi: 10.1196/annals.1294.011. [DOI] [PubMed] [Google Scholar]
- 7.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 8.Widschwendter M., Jones P.A. The potential prognostic, predictive, and therapeutic values of DNA methylation in cancer. Clin. Cancer Res. 2002;8:17–21. [PubMed] [Google Scholar]
- 9.Gonzalgo M.L., Liang G., Spruck C.H., Zingg J.M., Rideout W.M., Jones P.A. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 1997;57:594–599. [PubMed] [Google Scholar]
- 10.Huang T.H., Perry M.R., Laux D.E. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 1999;8:459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 11.Adorjan P., Distler J., Lipscher E., Model F., Muller J., Pelet C., Braun A., Florl A.R., Gutig D., Grabs G., et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002;30:e21. doi: 10.1093/nar/30.5.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakyan V.K., Hildmann T., Novik K.L., Lewin J., Tost J., Cox A.V., Andrews T.D., Howe K.L., Otto T., Olek A., et al. DNA methylation profiling of the human major histocompatibility complex: a pilot study for the human epigenome project. PLoS Biol. 2004;2:e405. doi: 10.1371/journal.pbio.0020405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewin J., Schmitt A.O., Adorjan P., Hildmann T., Piepenbrock C. Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics. 2004;20:3005–3012. doi: 10.1093/bioinformatics/bth346. [DOI] [PubMed] [Google Scholar]
- 14.Schatz P., Dietrich D., Schuster M. Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF. Nucleic Acids Res. 2004;32:e167. doi: 10.1093/nar/gnh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeschnigk M., Bohringer S., Price E.A., Onadim Z., Masshofer L., Lohmann D.R. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tost J., Dunker J., Gut I.G. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–156. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- 18.Tost J., Schatz P., Schuster M., Berlin K., Gut I.G. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31:e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehrach H., Berlin K., Gut I.G. Method for identifying nucleic acids by means of matrix-assisted laser desorption/ionization mass spectrometry. 2000. Patent EP-1036202; PCT/WO/99/29898.
- 20.Jaenisch R., Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 21.Ross P.L., Lee K., Belgrader P. Discrimination of single-nucleotide polymorphisms in human DNA using peptide nucleic acid probes detected by MALDI-TOF mass spectrometry. Anal. Chem. 1997;69:4197–4202. doi: 10.1021/ac9703966. [DOI] [PubMed] [Google Scholar]
- 22.Griffin T.J., Tang W., Smith L.M. Genetic analysis by peptide nucleic acid affinity MALDI-TOF mass spectrometry. Nat. Biotechnol. 1997;15:1368–1372. doi: 10.1038/nbt1297-1368. [DOI] [PubMed] [Google Scholar]
- 23.Bauer O., Guerasimova A., Sauer S., Thamm S., Steinfath M., Herwig R., Janitz M., Lehrach H., Radelof U. Multiplexed hybridizations of positively charge-tagged peptide nucleic acids detected by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:1821–1829. doi: 10.1002/rcm.1554. [DOI] [PubMed] [Google Scholar]
- 24.Gut I.G., Jeffery W.A., Pappin D.J.C., Beck S. Analysis of DNA by ‘charge tagging’ and matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1997;11:43–50. [Google Scholar]
- 25.Toyota M., Ahuja N., Ohe-Toyota M., Herman J.G., Baylin S.B., Issa J.P. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esteller M., Sparks A., Toyota M., Sanchez-Cespedes M., Capella G., Peinado M.A., Gonzalez S., Tarafa G., Sidransky D., Meltzer S.J., et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- 27.Lofton-Day C., Lesche R. DNA methylation markers in patients with gastrointestinal cancers. Current understanding, potential applications for disease management and development of diagnostic tools. Dig. Dis. 2003;21:299–308. doi: 10.1159/000075352. [DOI] [PubMed] [Google Scholar]
- 28.Sauer S., Lechner D., Berlin K., Lehrach H., Escary J.L., Fox N., Gut I.G. A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:E13. doi: 10.1093/nar/28.5.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gut I.G., Berlin K., Kucharzak R., Wiesner S., Guerasimova A., Lehrach H. Characterisation of Nucleic Acids with PNA Probes. Proceedings of the 46th ASMS Conference on Mass Spectrometry and Allied Topics; Orlando, FL, USA. 1998. [Google Scholar]
- 30.Dean F.B., Hosono S., Fang L., Wu X., Faruqi A.F., Bray-Ward P., Sun Z., Zong Q., Du Y., Du J., et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA. 2002;99:5261–5266. doi: 10.1073/pnas.082089499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berlin K., Ballhause M., Cardon K. Improved bisulfite conversion of DNA. 2005. Patent PCT/WO/2005/038051.
- 32.Atha D.H., Kasprzak W., O'Connell C.D., Shapiro B.A. Prediction of DNA single-strand conformation polymorphism: analysis by capillary electrophoresis and computerized DNA modeling. Nucleic Acids Res. 2001;29:4643–4653. doi: 10.1093/nar/29.22.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen D.A., Novoradovsky A., Goldman D. SSCP primer design based on single-strand DNA structure predicted by a DNA folding program. Nucleic Acids Res. 1995;23:2287–2291. doi: 10.1093/nar/23.12.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratilainen T., Holmen A., Tuite E., Nielsen P.E., Norden B. Thermodynamics of sequence-specific binding of PNA to DNA. Biochemistry. 2000;39:7781–7791. doi: 10.1021/bi000039g. [DOI] [PubMed] [Google Scholar]
- 35.Behrens J., Jerchow B.A., Wurtele M., Grimm J., Asbrand C., Wirtz R., Kuhl M., Wedlich D., Birchmeier W. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 36.Brunner E., Peter O., Schweizer L., Basler K. Pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya T., Tamura G., Sato K., Endoh Y., Sakata K., Jin Z., Motoyama T., Usuba O., Kimura W., Nishizuka S., et al. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642–3646. doi: 10.1038/sj.onc.1203704. [DOI] [PubMed] [Google Scholar]
- 38.Usadel H., Brabender J., Danenberg K.D., Jeronimo C., Harden S., Engles J., Danenberg P.V., Yang S., Sidransky D. Quantitative adenomatous polyposis coli promoter methylation analysis in tumor tissue, serum, and plasma DNA of patients with lung cancer. Cancer Res. 2002;62:371–375. [PubMed] [Google Scholar]
- 39.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Danenberg P.V., Laird P.W. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 40.Chen J., Rocken C., Lofton-Day C., Schulz H.U., Muller O., Kutzner N., Malfertheiner P., Ebert M.P. Molecular analysis of APC promoter methylation and protein expression in colorectal cancer metastasis. Carcinogenesis. 2005;26:37–43. doi: 10.1093/carcin/bgh280. [DOI] [PubMed] [Google Scholar]
- 41.Smalley M.J., Sara E., Paterson H., Naylor S., Cook D., Jayatilake H., Fryer L.G., Hutchinson L., Fry M.J., Dale T.C. Interaction of axin and Dvl-2 proteins regulates Dvl-2-stimulated TCF-dependent transcription. EMBO. J. 1999;18:2823–2835. doi: 10.1093/emboj/18.10.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haefliger C., Lesche R., Maier S., Nimmrich I., Burger M., Becker E., Adorjan P., Lofton-Day C., Sledziewski A., Berlin K., et al. Methylation profiling to identify colon cancer markers for early detection of disease. Proc. Am. Assoc. Cancer Res. 2003;44:R1131. [Google Scholar]
- 43.Borsani G., DeGrandi A., Ballabio A., Bulfone A., Bernard L., Banfi S., Gattuso C., Mariani M., Dixon M., Donnai D., et al. EYA4, a novel vertebrate gene related to Drosophila eyes absent. Hum. Mol. Genet. 1999;8:11–23. doi: 10.1093/hmg/8.1.11. [DOI] [PubMed] [Google Scholar]
- 44.Tootle T.L., Silver S.J., Davies E.L., Newman V., Latek R.R., Mills I.A., Selengut J.D., Parlikar B.E., Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 45.Clark S.W., Fee B.E., Cleveland J.L. Misexpression of the eyes absent family triggers the apoptotic program. J. Biol. Chem. 2002;277:3560–3567. doi: 10.1074/jbc.M108410200. [DOI] [PubMed] [Google Scholar]
- 46.Zou H., Osborn N.K., Harrington J.J., Klatt K.K., Molina J.R., Burgart L.J., Ahlquist D.A. Frequent methylation of eyes absent 4 gene in Barrett's esophagus and esophageal adenocarcinoma. Cancer Epidemiol. Biomarkers Prev. 2005;14:830–834. doi: 10.1158/1055-9965.EPI-04-0506. [DOI] [PubMed] [Google Scholar]
- 47.Song J.Z., Stirzaker C., Harrison J., Melki J.R., Clark S.J. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- 48.Cottrell S.E., Distler J., Goodman N.S., Mooney S.H., Kluth A., Olek A., Schwope I., Tetzner R., Ziebarth H., Berlin K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herman J.G., Graff J.R., Myöhänen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]