Abstract
To be effective, antisense molecules should be stable in biological fluids, non-toxic, form stable and specific duplexes with target RNAs and readily penetrate through cell membranes without non-specific effects on cell function. We report herein that negatively charged DNA mimics representing chiral analogues of peptide nucleic acids with a constrained trans-4-hydroxy-N-acetylpyrrolidine-2-phosphonate backbone (pHypNAs) meet these criteria. To demonstrate this, we compared silencing potency of these compounds with that of previously evaluated as efficient gene knockdown molecules hetero-oligomers consisting of alternating phosphono-PNA monomers and PNA-like monomers based on trans-4-hydroxy-L-proline (HypNA-pPNAs). Antisense potential of pHypNA mimics was confirmed in a cell-free translation assay with firefly luciferase as well as in a living cell assay with green fluorescent protein. In both cases, the pHypNA antisense oligomers provided a specific knockdown of a target protein production. Confocal microscopy showed that pHypNAs, when transfected into living cells, demonstrated efficient cellular uptake with distribution in the cytosol and nucleus. Also, the high potency of pHypNAs for down-regulation of Ras-like GTPase Ras-dva in Xenopus embryos was demonstrated in comparison with phosphorodiamidate morpholino oligomers. Therefore, our data suggest that pHypNAs are novel antisense agents with potential widespread in vitro and in vivo applications in basic research involving live cells and intact organisms.
INTRODUCTION
Over the last years, antisense approaches to the specific in vivo suppression of gene expression received an increased attention. Significant progress in this direction was achieved by the design of serum-stable nucleic acid analogues and mimics with improved target affinity and low toxicity and by the development of highly efficient antisense strategies using the cellular RNA interference (RNAi) pathway. The use of RNAi has been found to be an efficient silencing mechanism based on double-stranded RNA molecules, particularly small interfering RNAs (siRNAs), and the high potency of siRNAs to suppress gene expression was demonstrated [for recent reviews see (1–3)]. However, it has become clear that effectively delivered and rationally targeted traditional gene silencing molecules such as oligodeoxynucleotide analogues and mimics can silence gene expression as efficiently as the best siRNA molecules (4,5). Among earlier developed antisense agents, locked nucleic acids (LNAs) (6,7), phosphorodiamidate morpholino oligomers (MOs) (8) and peptide nucleic acids (PNAs) (9) can be evaluated as the most promising molecules. Charge-neutral DNA mimics, PNAs and MOs have been developed as compounds, which are unaffected by cellular nucleases and have strong RNA binding. In spite of high affinity towards DNA and RNA and stringent mismatch discrimination, the antisense application of unmodified PNAs is restricted by their inefficient cell membrane translocation, poor water-solubility and tendency to self-aggregation (10,11). Nevertheless, promising results have been reported in a range of systems exploited cell-penetrating peptides as carriers for PNAs delivery (12). At their turn, MOs exhibit high solubility in water despite their lack of charge, owing to their strong polarity, but they have lower affinity to nucleic acids and mismatch discrimination ability than PNAs (8,13,14).
For some years, we have studied PNA analogues, which contain monomer units connected with phosphonate ester bonds. A set of negatively charged PNA-like DNA mimics was designed, and their properties were investigated (15–17). Much of our interest has been focused on the preparation of oligomers containing conformationally restricted elements. Thus, hetero-oligomers consisting of alternating phosphono-PNA monomers and PNA-like monomers based on trans-4-hydroxy-L-proline (HypNA-pPNAs) were designed (17,18). The evaluation of physico-chemical properties of this chimeric DNA mimic revealed that HypNA-pPNA oligomers have excellent solubility in water and at the same time preserve biological stability and high hybridization characteristics of classical PNAs demonstrating strong and specific binding to DNA and RNA targets (19,20). The direct comparison of gene silencing efficiency of HypNA-pPNAs with the efficiency of PNAs and MOs validate this type of negatively charged mimics as an effective alternative to PNAs and MOs for gene knockdown experiments in vivo (21,22).
More recently, conformationally constrained chiral phosphono-PNA analogues representing derivatives of trans-4-hydroxy-N-acetyl-pyrrolidine-2-phosphonic acid (pHypNAs) were prepared (23,24), and examination of their physico-chemical and biological properties in comparison with natural oligonucleotides, PNAs, MOs and HypNA-pPNA oligomers was initiated. Herein we report the results obtained in the investigation of properties of trans-l pHypNA oligomers in terms of biological stability, ability to penetrate into living cells and down-regulate gene expression in vitro and in vivo.
MATERIALS AND METHODS
Synthetic oligonucleotides and mimics
Synthesis of oligodeoxyribonucleotides was performed by the standard solid-phase phosphoramidite method using monomers purchased from Glen Research. Synthesis of oligoribonucleotides was carried out by the phosphoramidite method using 1-phenyl-1H-tetrazole-5-thiol as the activation agent as described (25). MOs were purchased from Gene Tools. Chain elongation in the automated solid phase synthesis of HypNA-pPNA and pHypNA oligomers was carried out using the properly protected dimer and monomer units, respectively, by the phosphotriester method using O-nucleophilic intramolecular catalysis according to the previously developed protocols (19,23). Fluorescein-labeled oligomers were synthesized on the 3′-fluorescein CPG-support (Glen Research). The removal of protective groups and isolation of oligomers was performed as described earlier (19,23). The identity of the oligomers obtained was confirmed by mass spectrometry (MALDI TOF).
Thermal denaturation experiments
Absorbance versus temperature curves of complexes formed by oligomers with the corresponding DNA, or RNA, templates were measured using an Ultrospec 2000 UV-VIS spectrophotometer (Pharmacia) equipped with a temperature control unit. Solutions of each oligomer (3–5 µM) were mixed in 100 mM NaCl/10 mM Tris–HCl (pH 7)/10 mM MgCl2 and heated at 95°C for 2 min. The mixture was allowed to cool slowly before data collection. The changes in absorbance were measured at 260 nm with a heating rate 0.5°C/min from 20 to 95°C. Melting temperatures (Tm) were taken to be the temperature of a half-dissociation and were obtained from a plot of the derivative of 1/T versus absorbance.
RNA production and in vitro translation
The plasmid pSPFLuc containing the sequence coding firefly luciferase was obtained from pSPluc+NF vector (Promega) by the insertion between its EcoRI and XbaI restriction sites of a synthetic poly-(A)30 region. The plasmid pRL-SV40 (Promega) coding Renilla luciferase and the plasmid pSPFLuc were linearized by the action of BamHI and XhoI enzymes, respectively, and purified on 1% agarose gel with the following electro-elution from the excised gel bands. Each purified plasmid (10 µg) was used as a template in 200 µl transcription reactions. Renilla and firefly luciferase mRNAs were transcribed using the RiboMax T7 and SP6 production systems (Promega), respectively, following the manufacturer's protocol. RNA transcripts were purified using mTRAP mRNA isolation system (Active Motif), which utilizes biotinylated homo-Thy HypNA-pPNA probe and Streptavidin-coated magnetic beads, as described (26).
In vitro translation assays were performed using nuclease-treated rabbit reticulocyte lysate (Promega) in 30 µl reaction mixtures containing 25 ng of each mRNA to prime translation reactions. Oligomers were added to mRNA in various concentrations prior to the addition of the lysate, without a pre-hybridization step. After 60 min incubation at 30°C in the presence, or absence, of the desired antisense oligomer, the translation level of the Renilla and firefly RNAs was evaluated by Dual-Luciferase Reporter Assay System (Promega) using BD Monolight 3096 microplate luminometer. The effect of antisense oligomers was evaluated by measuring the ratio between Renilla and firefly luciferases. Each data point was averaged over two replicates of three separate experiments.
Cellular uptake
In each experiment, two identical 24-well plates were prepared with the Phoenix ecotropic packaging cell line derived from 293T cells (7 × 104 cells per well) and cultured at 37°C for 16–20 h in DMEM (PanEco) supplemented with 10% of fetal bovine serum (FBS) purchased from Gibco-Life Technologies and penicillin-streptomycin (100 U/ml of each) without phenol red. Then, 3 h before transfection the culture medium was changed for DMEM/FBS without antibiotics. Fluorescently labeled mimic oligomers were prepared in 2 µM concentration in DMEM, subsequently diluted and added to the cells. For cationic lipids mediated delivery, complexes of oligomers with Lipofectamine-2000 (Invitrogen) were formed by the pre-incubation with lipofectamine (2 µl) in 250 µl of DMEM for 30 min. After 4–20 h incubation of cells with oligomers under the standard growth conditions, cells were washed with phosphate-buffered saline (PBS) with following incubation in DMEM/10% FBS for 2–6 h. In some experiments, oligomers were prepared in 100 µM chloroquine, or in 6 mM CaCl2, in DMEM, and added to the cells for incubation with the following washing and incubation as before. On this step, cells were analyzed by phase contrast and fluorescence microscopy, and images were taken with Olimpus CK40 inverted microscope by DP50 Olimpus CD camera. After that, the medium was removed; the monolayer washed 3 times with 1 ml PBS, and 200 µl PBS was added to each well, cells scraped and suspended. The suspension from each well was split in two 100 µl portions. One protion was used for direct fluorescence measurement on a Microplate fluorymeter FFM-01 (Cortec). Second portion was used for quantification of the cellular concentration of fluorescently labeled mimics as described (22).
Confocal microscopy
Phoenix Eco cells (10 × 103) were plated on a 4-well Lab-Tek chambered cover-glass (Nunc) in DMEM/10%FBS and cultured for 20 h. The medium was discarded and cells were incubated for 16 h with 250 µl of 0.5 µM solution oligomer in DMEM in the presence, or in the absence, of 100 µM chloroquine, or 6 mM CaCl2, for 20 h. After two washes, 200 µl of DMEM (without phenol red) was added into the wells for observation of living cells. The cells were subjected to microscopic studies without fixation using a Leica LCS TCS SP confocal system based on DMIRBE Leica inverted microscope with a HCX APO CS 63.0X1.32 objective. The Argon laser was used to excite fluorescein at 488 nm, and a HQ 515/30 filter was used for observation of the emission. The imaging conditions were kept constant for the observation of the different samples.
Inhibition of GFP expression in cells
The Phoenix Eco cells were maintained at <80% confluence in DMEM supplemented with 10% FBS and antibiotics. Cells were seeded 24 h before treatment at 40% confluence in 35 mm diameter culture dishes (5 × 105 cells per dish). Before the transfection, growth media was removed and replaced with 2 ml of fresh media without antibiotics. Cells in a dish were transfected by p2FP-RNAi dual-color vector (Evrogen) using 1 µg of the plasmid DNA and 2 µl of LFA per 2 ml of the media. Immediately after this, an aliquot of the preliminary prepared mixture of an antisense oligomer (10 µM) and LFA (20 µl) in 1 ml of DMEM, which was incubated for 20 min in the dark, was added to the cells to obtain the proper concentration of the oligomer. At different time points after transfection, growth media containing the transfection mixture was replaced with fresh media containing serum and antibiotics, and the cells were subjected to fluorescent imaging. For trypsination, cells were washed with PBS three times and trypsinized for 10 min at 37°C following with the addition of PBS to 2 ml. Fluorescent measurements of proteins production in the cells were performed in 100 µl aliquots of the cell suspension in the wells of the black polystyrene 96-well plate (Costar) using the fluorymeter with the corresponding filters for registration of TurboGFP fluorescence (λex 482nm; λem 538nm) or JRed fluorescence (λex 546nm; λem 607nm) and accumulative time 1000 ms. Relative fluorescence was calculated by dividing the mean fluorescence of the oligomer-treated sample by the mean fluorescence of a control sample without the treatment.
Embryos handling and microinjection of oligomers
All operations with Xenopus embryos were performed as described (27). From 2 to 6 nl of 0.25 mM oligomer solution in water was microinjected into the right dorsal blastomere of 2–4-cell stage embryos by Eppendorf FemtoJet microinjector. For cell lineage labeling, the samples were mixed with Fluoresceine-Lysinated-Dextran amine (FLD) (Molecular Probes).
RESULTS AND DISCUSSION
Synthesis and hybridization properties of pHypNA oligomers
With the aim in view to obtain molecules possessing increased binding affinity to nucleic acids, phosphono-PNA oligomers (pPNAs) with N-(2-hydroxyethyl)-phosphono glycine, or N-(2-aminoethyl)-phosphono glycine backbone were synthesized using the solid phase technique based on the phosphotriester oligonucleotide synthesis with the O-nucleophilic intra-molecular catalysis (15). Using the same approach and dimers as building blocks, chimeras composed of pPNA and PNA monomers (PNA-pPNAs) and hetero-oligomers with alternating pPNA and HypNA monomers (HypNA-pPNAs) were obtained (16,17). A HypNA monomer structure is based on trans-4-hydroxy-L-proline, and it represents conformationally constrained chiral PNA analogue, in which the β-C atom of a hydroxyethyl unit and the α-C atom of a glycine unit of the backbone are bridged by methylene group. The HypNA-pPNA mimic was the most perspective and demonstrated high water solubility in combination with strong binding to complementary DNA and RNA strands and high level of specificity. The melting temperatures of complexes formed by HypNA-pPNAs with nucleic acid targets are very close to those formed by classical PNAs (17,18). HypNA-pPNA compounds were shown to be effective antisense agents in experiments with cell cultures and living organisms (21–24). However the necessity to use preliminary prepared dimers as units for their chemical synthesis is inconvenient and restricts their wide application in molecular biology to some extent.
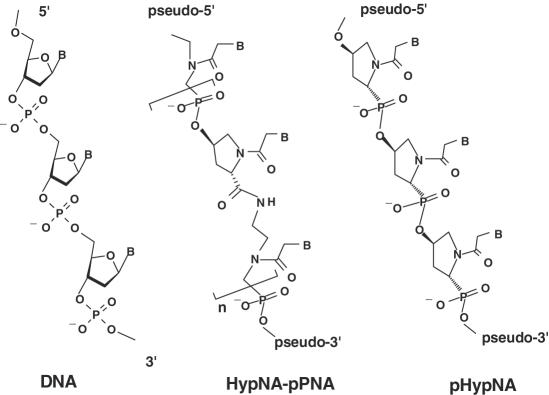
To continue our investigations, a novel type of DNA mimics, pHypNA oligomers, totally constructed from the chiral monomers derived from 4-hydroxyproline was recently designed (18,23,24). The pHypNA molecule represents conformationally constrained phosphono-PNA analogue (Figure 1). Earlier, the influence of a pHypNA monomer configuration on hybridization properties of the corresponding oligomers was investigated (23,24). Four various optically active Thy-pHypNA monomers were synthesized as well as oligomers totally constructed from monomers of the same chiral type. The results obtained in the study of the binding properties of these oligomers to complementary poly-dA and poly-rA targets have shown that oligomers totally constructed from pHypNA units of trans-l type exhibited the best hybridization characteristics, which were very close to those of PNAs and HypNA-pPNAs (23,24). Moreover, like other negatively charged PNA-analogues, pHypNAs demonstrated excellent solubility in water. Later, the corresponding trans-l adenine-, cytosine- and guanine-containing pHypNA monomers were synthesized and used for the solid-phase synthesis of trans-l pHypNA oligomers with mixed nucleobase sequences. As well as for other phosphonate-containing mimics, the synthesis of these oligomers was carried out by the phosphotriester method with O-nucleophilic intra-molecular catalysis. Procedures for the synthesis, deblocking and isolation of the mimics were essentially as described (19,20). From the ultraviolet-titration data and the mobility shift analysis in polyacrylamide gel electrophoresis, it was concluded that, similar to PNAs and other PNA-like mimics, pHypNA homo-pyrimidine sequences form triple helixes with complementary DNA (or RNA) targets, whereas oligomers with mixed nucleobase sequences form duplexes (23,24). It should be noted that the mobility of mimic oligomers and their complexes with the complementary oligonucleotides in a native polyacrylamide gel electrophoresis was different from the mobility of the iso-sequential natural oligonucleotides and their duplexes (Supplementary Figure 1).
Figure 1.
General chemical structures of HypNA-pPNA and pHypNA DNA mimics.
Earlier, it was shown that the values of melting temperatures (Tm) of 16–18 bp pHypNA/DNA(RNA) duplexes are practically equivalent to those of duplexes formed by HypNA-pPNA oligomers (21,23,24). The examination of a salt concentration influence on melting temperatures of duplexes and triplexes formed by pHypNA mimics with the complementary DNA and RNA targets revealed that, similar to classic PNAs and HypNA-pPNAs, the stability of their complexes with nucleic acid targets are not dependent on ionic strength, and the decrease of a salt concentration has practically no effect on the stability of duplexes and triplexes formed by these mimics (23,24). The evaluation of the discrimination ability of pHypNA mimics in assays based on the hybridization technique revealed their high potential as bio-molecular probes (17,24). The hybridization of these oligomers with DNA and RNA targets occurred in a sequence specific manner, and the formation of complexes between mimic probes and non-complementary targets was not detected. The introduction of one mismatch in the center of a 16–18mer sequence gave a drop of 12–20°C in the melting temperature of a duplex depending on base mismatch position and the sequence (23,24). Some results on the investigation of hybridization ability and binding selectivity of the oligomers used in this study are shown in Table 1. As it can be seen from the table, the melting temperatures of duplexes formed by pHypNA oligomers with the RNA targets are higher than those formed by natural oligodeoxyribonucleotides. The introduction of two separately situated mismatches into the sequence of mimic oligomers leads to a significant destabilization of their complexes with the targets and gives a drop in Tm of 26–29°C for pHypNA/RNA duplexes and 37–39°C for pHypNA/DNA duplexes.
Table 1.
Sequences and hybridization properties of oligonucleotides and DNA mimics used in this study
| Oligomer sequence (target)a | Complementary oligonucleotidea | ODN Tm, °C (Δα;Δ)b | HypNA-pPNA Tm, °C(Δα;Δ)b | pHypNA Tm, °C (Δα;Δ)b |
|---|---|---|---|---|
| TCACTCAACACTCAC-Flu | d(GTGAGTGTTGAGTGA) | 57 | 58 | 55 |
| TGGCGTCGGTGACCAT (FLuc) | d(ATGGTCACCGACGCCA) | 64 | 65 | 67 |
| TGGCGTCGGTGACCAT (FLuc) | r(AUGGUCACCGACGCCA) | 56 | 73 | 72 |
| TGGaGTCGGcGACCAT (FLuc) | d(ATGGTCACCGACGCCA) | ∼25 (−36) | ∼26 (−39) | ∼28 (−39) |
| TGGaGTCGGcGACCAT (FLuc) | r(AUGGUCACCGACGCCA) | 32 (−24) | 48 (−25) | 45 (−27) |
| GCTCTCGTCGCTCTCCAT (GFP) | d(CATGGAGAGCGACGAGAGCGGC) | 69 | 65 | 63 |
| GCTCTCGTCGCTCTCCAT (GFP) | r(CAUGGAGAGCGACGAGAGC)dTT | 68 | 76 | 75 |
| GCTCTaGTCGCTaTCCAT (GFP) | d(CATGGAGAGCGACGAGAGCGGC) | 34 (−35) | 31 (−34) | ∼26 (−37) |
| GCTCTaGTCGCTaTCCAT (GFP) | r(CAUGGAGAGCGACGAGAGC)dTT | 49 (−19) | 51 (−25) | 49 (−26) |
| r(GCUCUCGUCGCUCUCCAUG)dTT (GFP) | r(CAUGGAGAGCGACGAGAGC)dTT | 86 | - | - |
| TGCGCTTTCTTTTGTCTG (Ras-dva) | r(CAGACAAAAGAAAGCGCA) | 60 | 64 | 67 |
| TGCtCTTTCTTcTGTCTG (Ras-dva) | r(CAGACAAAAGAAAGCGCA) | 35 (−25) | 43 (−21) | 38 (M29) |
aThe sequence is shown from 5′ to 3′end of an oligonucleotide, or from pseudo-5′ to pseudo-3′end of a mimic oligomer. Lowercase base represents a mismatch position.
bThe value in brackets shows the difference between the melting temperature of duplexes formed by the oligomers with matched and mismatched sequences with complementary targets.
Biological stability of pHypNAs
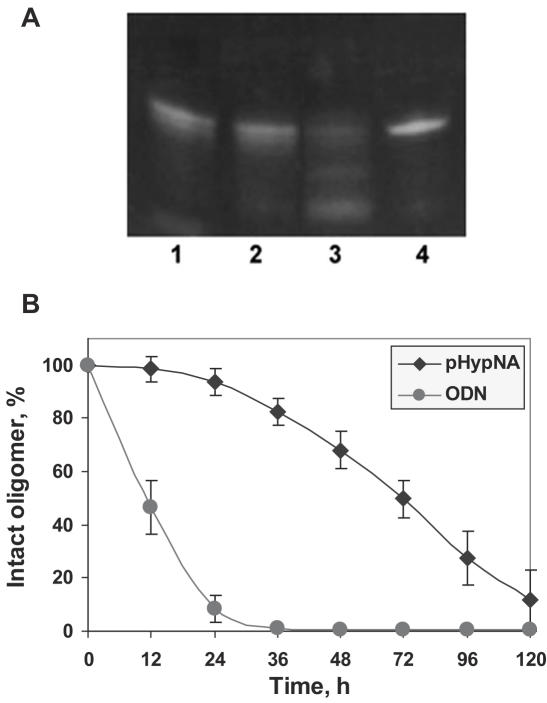
The ideal antisense molecule should not only effectively recognize a target, but also be stable in biological fluids. It was shown that classical PNAs are stable to the action of proteases, and they have a half-live in serum of ∼48 h (28). Earlier, we have shown that phosphonate-containing analogues of PNAs are highly stable to the action of nucleases (24). Although natural nucleobases are the components of pHypNA molecules, these compounds do not contain phosphodiester bonds found in the backbone of natural oligonucleotides. This is probably the reason why nucleases do not recognize this type of molecules as substrates (Supplementary, Figure 2). Based on the data that the major process of the oligonucleotide degradation occurs outside of the cell in the extra-cellular medium (29), we additionally examined thesusceptibility of pHypNAs to the degradation in DMEM containing 10% FBS. As PNA-related mimics do not interact with intercalating dyes, particularly with ethidium bromide (23), we used in this study the fluorescently labeled oligomers in conjunction with a mobility shift analysis in denaturing conditions (Figure 2). The results obtained have shown that >60% of the initial amount of the pHypNA oligomer was still stable after 48 h incubation, and the half-life of the pHypNAs can be estimated as ∼72 h. At the same time, >50% of the control natural 3′-fluorescein labeled oligodeoxyribonucleotide was cleaved after 12 h incubation at the same conditions, and it was practically fully degraded in 24 h. Similar stability of pHypNAs was observed in cytoplasmic and nuclear cell extracts (data not shown). Consistent with these observations, we concluded that the high stability of these mimics can allow them to preserve their action in cells for a prolonged period of time after a single administration.
Figure 2.
Analysis of the stability of the fluorescein-labeled pHypNA oligomer (Flu-1) by the electrophoresis in a 15% polyacrylamide gel in denaturing conditions. The oligomer (5 µM) was incubated at 37°C in 500 µl of DMEM/10% FBS. Aliquots (20 µl) were taken at each 24 h and frozen at −20°C until analysis by electrophoresis. (A) The image shows probes of the mimic oligomer taken after 48 h (1), 72 h (2), 120 h (3) and 0 h (4) incubation; the visualization was in UV-light at 365 nm. (B) The comparison of the stability of the fluorescein-labeled pHypNA oligomer and the control 3′-fluorescein-labeled oligodeoxyribonucleotide of the same sequence. Densitometric calculations of the amount of full-length oligomer were done using UVP Bio-Imaging system and LabWorks imaging software.
Silencing in a Cell-Free Translation System
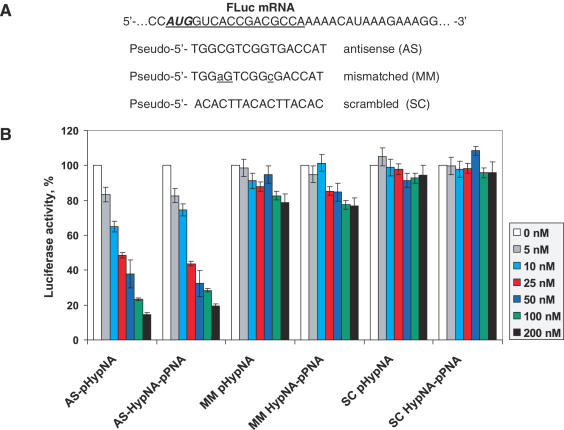
The initial evaluation of pHypNAs antisense properties against their targets has been performed in an in vitro translation assay with purified mRNA transcripts containing the coding sequences for firefly (FLuc) and Renilla (RLuc) luciferases. The pHypNA probe complementary to the start codon region of the firefly luciferase coding sequence and the corresponding HypNA-pPNA probe were synthesized, and the relative efficiency of these oligomers against the same transcript was tested. After the incubation in a cell-free translation system containing FLuc and RLuc mRNAs, the relative effect of antisense oligomers was evaluated by measuring the ratio between firefly and Renilla luciferase production in the presence and in the absence of oligomers. It was found that both types of oligomers exhibited strong antisense effect at 50–200 nM concentrations with up to 85% reduction of the FLuc production (Figure 3). The inhibition was dose-dependent and still observed at 5 nM of oligomers. Practically no reduction of RLuc activity was seen in any case (data not shown). The control oligomers with two mismatches and oligomers with scrambled sequences had no inhibitory effect under the same conditions.
Figure 3.
Dose-dependent inhibition of firefly luciferase translation in vitro by the mimic antisense oligomers targeted against the translational start site of FLuc mRNA. (A) Sequences of HypNA-pPNA and pHypNA oligomers designed to target the FLuc mRNA; (B) Analysis of firefly luciferase translation inhibition by the oligomers. Firefly luciferase activity was calculated relative to the activity in the absence of any oligomer and normalized respect to Renilla luciferase production.
Cellular uptake of pHypNA oligomers
In contrast to natural oligonucleotides and their phosphorothioate analogues and similar to PNAs, negatively charged PNA-like mimics do not activate RNase-H upon binding to RNA (22,24), and their ability to affect mRNAs is more likely to be directly associated with steric hindrances. Therefore significant amounts of such antisense oligomers must be delivered into the cells to block translation or redirect splicing, and an efficient delivery is very important to observe strong gene knockdown effect. Usually, antisense oligonucleotides and their analogues (e.g. phosphorothioates) are effectively delivered into eukaryotic laboratory cell lines via cationic liposome complexation (30). PNAs being uncharged molecules cannot directly give complexes with cationic liposomes, and their delivery via this route is possible using PNA–lipid conjugates, PNA–DNA hybrids or duplexes formed by PNAs with the complementary oligonucleotides (31). Also, a number of studies exploited PNA conjugates with cell-penetrating peptides (12,32) or other carriers such as 9-aminoacridine (31), but in some cases these systems deliver PNAs mainly to the lysosomal compartments of eukaryotic cells and not efficiently to the cytosolic and nuclear compartments, where they should act as antisense agents (12,33). Nevertheless, the results have been reported on the significant enhancement of classical PNAs antisense effect in the presence of calcium ions as well as lysosomotropic reagent chloroquine, presumably by facilitating endosomal release of antisense compounds (12,33,34).
In previous studies with HypNA-pPNA oligomers, it was shown that free HypNA-pPNAs are able to penetrate into cultured HS-68 and HeLa cells and distribute in the cytoplasm (22,23). The addition of cationic lipids, or a non-covalent peptide-based delivery system Pep-2, improves their delivery into living cells. Confocal microscopy evidence was obtained that in this case HypNA-pPNAs are localized mainly in the cytoplasm with some staining in nucleus (22).
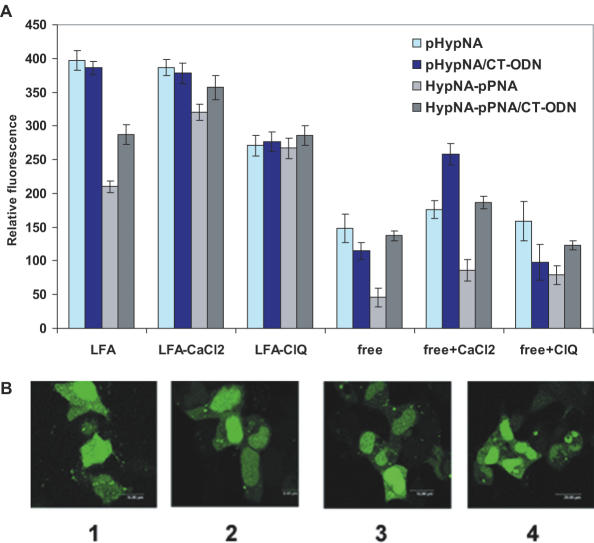
Delivery of a fluorescein-labeled pHypNA oligomer into eukaryotic cells was tested in several formats, particularly as a free oligomer, its duplex with the complementary oligodeoxyribonucleotide (CT-ODN) and those in combination with Lipofectamine-2000 (LFA). In control experiments, the Flu-labeled HypNA-pPNA oligomer was added to cells either free, or in the complex with CT-ODN. Transfection with the fluorescein-labeled pHypNA oligomer, or with its duplex with CT-ODN, in the presence of LFA resulted in a high level of fluorescence in treated cells after 4 h already. Maximum uptake was achieved after 20–24 h. However, the treatment with free pHypNA oligomer, or with that in combination with CT-ODN, resulted in much lower intracellular fluorescence (Figure 4A). Similar results were obtained with fluorescein-labeled HypNA-pPNA oligomer. Counter staining with propidium iodide indicated that the mimics did not show any cellular toxicity (data not shown).
Figure 4.
Cellular uptake and intracellular distribution of the fluorescein-labeled mimic oligomers (TCACTCAACACTCAC-Flu). (A) Fluorescent analysis of the delivery of oligomers (1 µM) into Phoenix Eco cells performed 20 h after the treatment under various delivery conditions: in the presence, or in the absence, of LFA, 6 mM CaCl2, and 100 µM chloroquine (ClQ). (B) Confocal microscopy images of the pHypNA oligomer uptake after 20 h incubated with unfixed Phoenix Eco cells in the presence of: LFA (1); LFA/6mM Ca2+ (2); CT-ODN/LFA (3) and LFA/100 µM ClQ (4).
To obtain a detailed determination of the intracellular localization of the pHypNAs, we examined the ability of a pHypNA oligomer to penetrate into eukaryotic cells by live-cell confocal microscopy. In these experiments, the Flu-pHypNA oligomer, or its duplex with the complementary oligodeoxyribonucleotide, were incubated with living cells in the presence of LFA. Confocal microscopy showed that in all cases pHypNA oligomer appeared to be evenly distributed in the cytoplasm with preferential staining of the nucleus, and the addition of lysosomotropic reagent chloroquine, or calcium ions, had no influence on the distribution. Figure 4B shows some examples of typical confocal microscopy images with a significant and uniform fluorescence in the cytosol and nucleus both in the presence and in the absence of chloroquine and calcium ions. Moreover, co-administration of chloroquine in 100 µM concentration had a negative influence on the cell viability, which was found to be 80–85% related to the viability of untreated cells and cells treated with CaCl2 (data not shown).
Gene silencing in cell culture
The experiments on down-regulation of protein production in living cultured cells by hydroxyproline-based mimic oligomers were initiated several years ago with HypNA-pPNA mimics. Antisense HypNA-pPNA 16–18mer oligomers demonstrated efficient and specific inhibition of green fluorescent protein (GFP) production in Escherichia coli cells and AChE-R production in rat brain cell culture in 0.5 µM concentration (35). The down-regulation of the levels of cyclin B1, a protein essential for cell cycle progression, was demonstrated for HypNA-pPNAs in 50–100 nM concentrations in HeLa cells, human fibroblasts HS 68 and 293 cells (22). Their antisense effect was analyzed in comparison with the effect of classical PNAs and phosphorothioate oligonucleotides and estimated as in 8.5-fold and 25-fold higher than in the case of PNA oligomer and phosphorothioate oligonucleotide, respectively (22). In recent reports, HypNA-pPNA oligomers were successfully used for the investigation of the mechanism of Mallory body formation in the liver cells of chronic liver diseases (36) as well as for down-regulation of various genes in zebrafish (21,37,38).
To compare the in vivo antisense effect of pHypNA oligomers with that of corresponding HypNA-pPNA oligomers and double-stranded siRNA (dsRNA), a dual-reporter system based on a novel mammalian expression vector p2FP-RNAi was used. This vector was recently designed for RNAi studies and encoded two fluorescent reporter proteins, TurboGFP and JRed-Neo. The genes for both proteins are under the control of an immediate early promoter of cytomegalovirus, and their codon usage is optimized for high expression in mammalian cells. TurboGFP is an improved variant of the green fluorescent protein CopGFP cloned from copepoda Pontellina plumata (39). It reveals bright green fluorescence, which can be clearly detected in cells within 4–8 h after transfection, and does not form aggregates in long-term cultures. Moreover, fast maturation in eukaryotic cells improves the ability to visualize gene expression and protein localization at early stages of biological processes. JRed is a novel red fluorescent protein obtained by mutagenesis of Anthomedusae jellyfish chromoprotein (39). After the transfection into eukaryotic cells, JRed function serves as a positive transfection marker, while TurboGFP stands as an indicator of gene silencing efficiency. In the presence of a functional antisense compound directed against TurboGFP, its expression is knocking down, whereas JRed expression remains unmodified. Thus, the increasing red/green fluorescence intensity ratio compared with control experiment indicates successful performance of antisense agent.
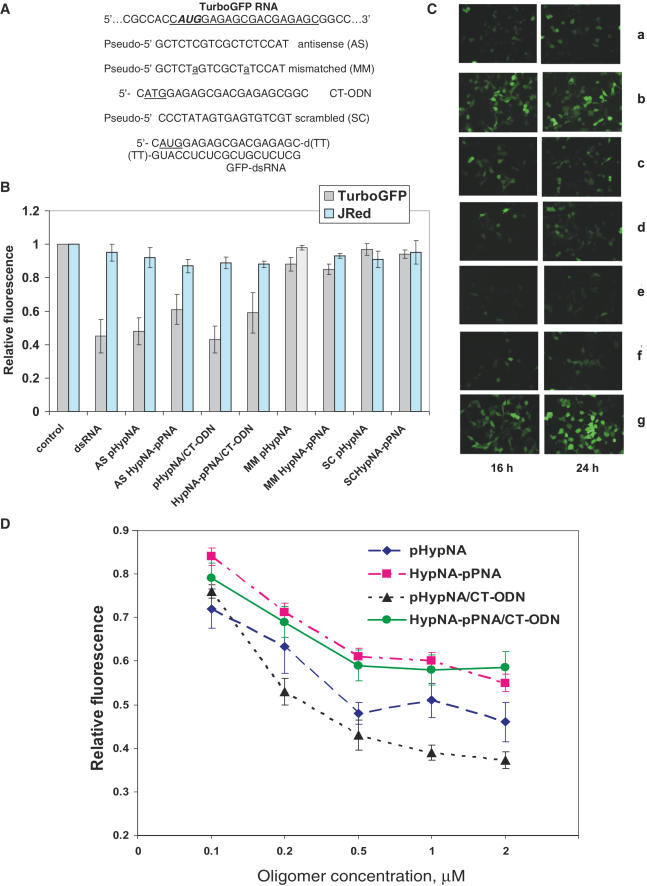
To test a potential of pHypNAs as TurboGFP gene knockdown agents, the antisense pHypNA and HypNA-pPNA oligomers directed against the start codon region of TurboGFP mRNA as well as control mismatched and scrambled oligomers were used (Figure 5). Phoenix Eco cells were transfected with p2FP-RNAi vector mixed with the oligomers in the presence of LFA. Treatment with antisense oligomers produced a dose-dependent decrease of green fluorescence in the cells compared with the untreated control, whilst practically no reduction in JRed activity was seen in any case. Mismatched and scrambled oligomers were inactive in the assays. A significant level of reduction of TurboGFP was seen at 0.5–1 µM concentration of antisense oligomers. The comparison of mimic effectiveness revealed that pHypNA oligomer was even more effective than the corresponding HypNA-pPNA oligomer in the gene knockdown tests (Figure 5). Both antisense oligomers were still able to suppress the target gene expression at a concentration of 100 nM that is in accordance with the data published earlier for HypNA-pPNAs (22). As a variant, antisense oligomers were delivered into the cells as duplexes with the complementary oligodeoxyribonucleotide. In the presence of LFA, the use of duplexes had some positive effect on the antisense activity of oligomers, probably owing to faster delivery into the cells in comparison with single-stranded mimics. The addition of 6 mM Ca2+ into a culture medium had a positive influence on the antisense activity of oligomers, and in this case the sufficient level of TurboGFP reduction can be also achieved without the addition of LFA to the antisense oligomer before the cell transfection (data not shown). The comparison of antisense effect of the pHypNA oligomer with that of the synthetic dsRNA directed against TurboGFP revealed that 0.5 µM mimic oligomer was practically as active as 0.2 µM dsRNA (Figure 5). Cell viability was >90% for all experiments with mimics (data not shown). It should be noted that dsRNA used in this study was iso-sequential to antisense pHypNA and HypNA-pPNA oligomers. It was directed against the start codon region of TurboGFP, and its target site on mRNA was not optimized.
Figure 5.
Analysis of the reduction of TurboGFP levels by antisense mimic oligomers in Phoenix Eco cells transfected with p2FP-RNAi vector. Co-transfection of oligomers was performed in the presence of LFA. The cells transformed with the vector alone were analyzed as controls. (A) Sequences of oligomers designed to target the translational start site of the TurboGFP mRNA. (B) Analysis of green and red fluorescence intensity of the cells treated with 0.5 µM oligomers, or with 0.2 µM dsRNA, after 24 h. The average data of three separate experiments are shown. (C) Fluorescent microscopy images of the cells transfected with p2FP-RNAi and treated with 0.5 µM mimic oligomer samples for 16 and 24 h. Panels show examples of cells treated with 0.2 µM dsRNA (a); mismatched pHypNA oligomer (b); duplex of antisense pHypNA oligomer with CT-ODN (c); duplex of antisense HypNA-pPNA oligomer with CT-ODN (d); antisense pHypNA oligomer (e) and antisense HypNA-pPNA oligomer (f). Panel (g) shows cells untreated with oligomers. (D) Effect of the antisense oligomer concentration on the TurboGFP production in cells. Fluorescence was measured 24 h after the transfection. Average data points from three independent experiments are shown.
Furthermore, the time course for the suppression of TurboGFP expression by the oligo-mimics was investigated. An effective silencing of the target gene for up to 3 days was observed for both types of antisense mimics (data not shown). Maximal inhibition level of this protein was found to be at 24–48 h post-transfection for pHypNA and HypNA-pPNA antisense mimics, whereas for dsRNA it was estimated as 24 h.
Gene knockdown in a living model organism
The next step of the pHypNAs evaluation as antisense agents is the examination of their potency for down regulation of genes in living organisms. In previous years, MOs were successfully utilized in the living models, particularly Danio rerio (zebrafish) and Xenopus laevis, for genome-wide, sequence-based, reverse genetic screens during embryonic development. Negatively charged HypNA-pPNA oligomers were evaluated as an alternative to MOs for inhibition of gene expression in zebrafish embryos, and it was shown that they are effective and specific in vivo translational inhibitors in over a range of developmental stages (37,38). It was observed that HypNA-pPNA 18mer displayed comparable potency to 25mer MOs as gene knockdown agents and demonstrated higher sequence specificity and greater mismatch stringency than MOs. Another key advantage of HypNA-pPNAs for gene knockdown studies is the lack of nonspecific effects, and doses of HypNA-pPNA, which are high enough to produce strong loss-of-function phenotypes, produced few or no non-specific effects.
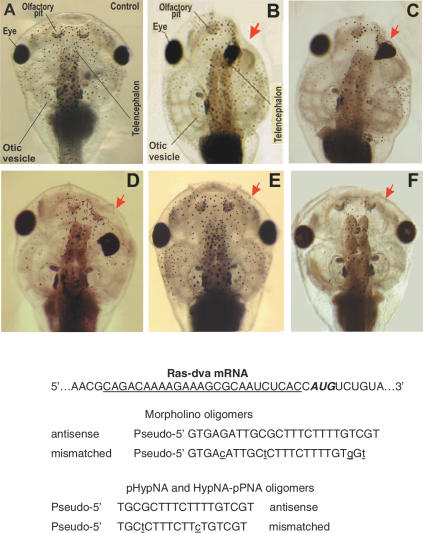
The gene knockdown efficiency of pHypNAs in comparison with MOs and HypNA-pPNAs was estimated in Xenopus embryos. Recently, the gene for a novel Ras-like GTPase Ras-dva was identified, and it was reported that Ras-dva and its homologues in other species constitute a novel family of Ras-like small GTPases (27). This protein is expressed during X.laevis neurulation in a very restricted area surrounding the anterior margin of the neural plate. The sequences of antisense oligomers were designed against the mRNA 5′-untranslated region, which is conservative in both pseudo-alleles of the X.laevis Ras-dva. The down-regulation of its functioning by microinjecting MO antisense 25mer into early embryos resulted in severe head development abnormalities on the injected side of 40–62% of the developing tadpoles. These include a reduction of the forebrain, olfactory pits, otic vesicles, branchial arches and malformations of the head cartilages. Similar abnormalities were observed for 60–85% tadpoles after microinjecting antisense pHypNA, or HypNA-pPNA, oligomers (Figure 6). The specificity of the effects obtained was confirmed by the fact that neither head nor trunk abnormalities were observed when the control oligomers with mismatched and scrambled sequences were injected. These results show that the negatively charged pHypNA mimics can act as specific antisense inhibitors of gene function in living model organisms. The experiments on the examination of antisense properties of pHypNA oligomers in zebrafish embryos are currently in progress.
Figure 6.
Images of Xenopus tadpole heads as seen from the dorsal side and the sequences of MO, pHypNA and HypNA-pPNA oligomers used in Ras-dva mRNA translation inhibitory assays. The injection of antisense oligomers results in severe abnormalities of the head structure. The side of injection is shown by red arrow. (A) A control tadpole head; (B, C and D) heads of tadpoles developed from the embryos microinjected with the antisense MO, pHypNA and HypNA-pPNA oligomers, respectively; (E and F) heads of tadpoles developed from the embryos microinjected with the mismatched pHypNA and HypNA-pPNA oligomers.
CONCLUSIONS
The major aim of this study was to investigate the properties and evaluate the potency of a novel type of antisense compounds, trans-l pHypNAs, representing negatively charged conformationally constrained phosphono-PNA analogues. These DNA mimics combine high hybridization and discrimination characteristics with good water solubility and biological stability as well as the ability to penetrate cell membranes. The efficacy of pHypNAs as antisense agents was demonstrated in a cell-free system, living cells and a living model organism. These compounds demonstrated high antisense activity in a cell-free system, and the degree of inhibition of gene expression by pHypNAs in cultured cells was higher than that with HypNA-pPNAs. The results obtained in gene knockdown experiments in Xenopus embryos characterize pHypNAs as useful tools and a valuable alternative to MOs and HypNA-pPNAs for reverse genetic studies. Also, it is important to note that pHypNA oligomers can be easily synthesized from 4 monomers, and therefore they are more accessible compounds than HypNA-pPNAs, the synthesis of which needs the availability of 16 dimers. Attributes described above are important with respect to a potential therapeutic utilization of the pHypNA compounds as they promote gene silencing in living cells for a prolonged period of time. Based on the antisense potency of pHypNAs and their superiority comparing the earlier proposed by us HypNA-pPNAs, we can conclude that pHypNA mimics are attractive compounds for broad applications in life science research and drug discovery.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr E. Cytrin (Institute on Biology of Organisms Development) for expert performance of confocal microscopy. The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ito M., Kawano K., Miyagishi M., Taira K. Genome-wide application of RNAi to the discovery of potential drug targets. FEBS Lett. 2005;579:5988–5995. doi: 10.1016/j.febslet.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Uprichard S.L. The therapeutic potential of RNA interference. FEBS Lett. 2005;579:5996–6007. doi: 10.1016/j.febslet.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorsett Y., Tuschl T. siRNA: applications in functional genomics and potential as therapeutics. Nature Rev. Drug Discov. 2005;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 4.Achenbach T.V., Brunner B., Heerrneier K. Oligonucleotide-based knockdown technologies: antisense versus RNA interference. Chem. Bio. Chem. 2003;4:928–935. doi: 10.1002/cbic.200300708. [DOI] [PubMed] [Google Scholar]
- 5.Kurreck J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 2003;270:1628–1644. doi: 10.1046/j.1432-1033.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 6.Koshkin A.A., Singh S., Nieisen P.K., Rajwanshi V., Kumar R., Meldgaard M., Olsen C., Wengel J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 7.Jepsen J.S., Wengel J. LNA-Antisense rivals siRNA for gene silencing. Curr. Opin. Drug Discov. Devel. 2004;7:188–194. [PubMed] [Google Scholar]
- 8.Summerton J., Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 9.Egholm M., Buchardt O., Christensen L., Behrens C., Freier S., Driver D., Berg R., Kim S., Norden B., Nielsen P. PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 10.Bergman F., Bannwarth W., Tam S. Solid phase synthesis of directly linked PNA-DNA-hybrids. Tetrahedron Lett. 1995;36:6823–6826. [Google Scholar]
- 11.Koppelhus U., Awasthi S., Zachar V., Holst H., Ebbeson P., Nielsen P. Cell-dependent differential cellular uptake of PNA, peptides and PNA–peptide conjugates. Antisense Nucleic Acid Drug Dev. 2002;12:51–63. doi: 10.1089/108729002760070795. [DOI] [PubMed] [Google Scholar]
- 12.Turner J., Ivanova G., Verbeure B., Williams D., Arzumanov A., Abes S., Lebleu B., Gait M. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein D., Foster E., Huang S., Weller D., Summerton J. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 1997;7:151–157. doi: 10.1089/oli.1.1997.7.151. [DOI] [PubMed] [Google Scholar]
- 14.Summerton J., Stein D., Huang S.B., Matthews P., Weller D.D., Partridge M. Morpholino and phosphorothioate antisense oligomers compared in cell-free and in cell systems. Antisense Nucleic Acid Drug Dev. 1997;7:63–70. doi: 10.1089/oli.1.1997.7.63. [DOI] [PubMed] [Google Scholar]
- 15.Van der Laan A., Stromberg R., van Boom J., Kuyl-Yeheskiely E., Efimov V., Chakhmakhcheva O. An approach towards the synthesis of oligomers containing a N-2-hydroxyethyl-aminomethyl phosphonate backbone: a novel PNA analogue. Tetrahedron Lett. 1996;37:7857–7860. [Google Scholar]
- 16.Efimov V., Choob M., Buryakova A., Kalinkina A., Chakhmakhcheva O. Synthesis and evaluation of some properties of chimeric oligomers containing PNA and phosphono-PNA residues. Nucleic Acids Res. 1998;26:566–575. doi: 10.1093/nar/26.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Efimov V., Buryakova A., Chakhmakhcheva O. Synthesis of polyacrylamides N-substituted with PNA-like oligonucleotide mimics for molecular diagnostic applications. Nucleic Acids Res. 1999;27:4416–4426. doi: 10.1093/nar/27.22.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Efimov A., Buryakova A., Choob M., Chakhmakhcheva O. Phosphonate analogues of peptide nucleic acids and related compounds: synthesis and hybridization properties. Nucleosides Nucleotides. 1999;18:1393–1396. [Google Scholar]
- 19.Efimov V., Chakhmakhcheva O. Synthesis of DNA mimics representing HypNA-pPNA hetero-oligomers. Meth. Mol. Biol. 2005;288:147–163. doi: 10.1385/1-59259-823-4:147. [DOI] [PubMed] [Google Scholar]
- 20.Efimov V., Fernandez J., Archdeacon D., Archdeacon J., Chakhmakhcheva O., Buryakova A., Choob M., Hondorp K. 2005. Patent US-6,962,906 B2. [Google Scholar]
- 21.Wickstrom E., Choob M., Urtishak K., Tian X., Sternheim N., Talbot S., Archdeacon J., Efimov V., Farber S. Sequence specificity of alternating hydroxyprolyl/phosphono peptide nucleic acids against zebrafish embryo mRNAs. J. Drug Targeting. 2004;12:363–372. doi: 10.1080/10611860412331285242. [DOI] [PubMed] [Google Scholar]
- 22.Morris M., Chaloin L., Choob M., Archdeacon J., Heitz F., Divita G. Combination of a new generation of PNAs with a peptide-based carrier enables efficient targeting of cell cycle progression. Gene Therapy. 2004;11:757–764. doi: 10.1038/sj.gt.3302235. [DOI] [PubMed] [Google Scholar]
- 23.Efimov V., Chakhmakhcheva O. Phosphono-PNAs: synthesis, properties and applications. Coll. Symp. Series. 2002;5:136–144. [Google Scholar]
- 24.Efimov V., Klykov V., Chakhmakhcheva O. Phosphono peptide nucleic acids with a constrained hydroxyproline-based backbone. Nucleosides, Nucleotides Nucleic Acids. 2003;22:593–599. doi: 10.1081/NCN-120021962. [DOI] [PubMed] [Google Scholar]
- 25.Efimov V., Kalinkina A., Chakhmakhcheva O. New activators for the phosphoramidite oligonucleotide synthesis. Bioorgan. Khim.(Rus) 1996;22:149–152. [PubMed] [Google Scholar]
- 26.Phelan D., Hondorp K., Choob M., Efimov V., Fernandez J. Messenger RNA isolation using novel PNA analogues. Nucleosides, Nucleotides Nucleic Acids. 2001;20:1107–1111. doi: 10.1081/NCN-100002499. [DOI] [PubMed] [Google Scholar]
- 27.Tereshina M., Zaraisky A., Novoselov V. Ras-dva, a member of novel family of small GTPases, is required for the anterior ectoderm patterning in the Xenopus laevis embryo. Development. 2006;133:485–494. doi: 10.1242/dev.02207. [DOI] [PubMed] [Google Scholar]
- 28.Uhlmann E., Peyman A., Breipohl G., Will D. PNA: Synthetic polyamide nucleic acids with unusual binding properties. Angew. Chem. Int. Ed. 1998;37:2796–2823. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2796::AID-ANIE2796>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Akhtar S., Kole R., Juliano R. Stability of antisense DNA oligonucleotide analogs in cellular extracts and sera. Life Sci. 1991;49:1793–1801. doi: 10.1016/0024-3205(91)90480-y. [DOI] [PubMed] [Google Scholar]
- 30.Bennett C., Chiang M.-Y., Chan H., Shoemaker J., Mirabelli C. Cationic lipids enhance cellular uptake and activity of phosphorothioate antisense oligonucleotides. Mol. Pharmacol. 1992;41:1023–1033. [PubMed] [Google Scholar]
- 31.Shiraishi T., Nielsen P. Down-regulation of MDM2 and activation of p53 in human cancer cells by antisense 9-aminoacridine-PNA (peptide nucleic acid) conjugates. Nucleic Acids Res. 2004;32:4893–4902. doi: 10.1093/nar/gkh820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gait M. Peptide-mediated cellular delivery of antisense oligonucleotides and their analogues. Cell. Mol. Life Sci. 2003;60:1–10. doi: 10.1007/s00018-003-3044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shiraishi T., Pankratova S., Neilsen P. Calcium ions effectively enhance the effect of antisense peptide nucleic acids conjugated to cationic Tat and oligoarginine peptides. Chem. Biol. 2005;12:923–929. doi: 10.1016/j.chembiol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Corey D. Calcium liberates PNAs from endosomes. Chem. Biol. 2005;12:864–865. doi: 10.1016/j.chembiol.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Efimov V., Chakhmakhcheva O., Wickstrom E. Synthesis and application of negatively charged PNA analogues. Nucleosides, Nucleotides Nucleic Acids. 2005;24:1853–1874. doi: 10.1080/15257770500268830. [DOI] [PubMed] [Google Scholar]
- 36.Nan L., Wu Y., Bardag-Gorce F., Li J., French B., Wilson L., Nguyen S., French S. RNA interference of VCP/p97 increases Mallory body formation. Exp. Mol. Pathol. 2005;78:1–9. doi: 10.1016/j.yexmp.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 37.Krawitz P., Haffner C., Fluhrer R., Steiner H., Schmid B., Haass C. Differential localization and identification of a critical aspartate suggest non-redundant proteolytic functions of the presenilin homologues SPPL2b and SPPL3. J. Biol. Chem. 2005;280:39515–39523. doi: 10.1074/jbc.M501645200. [DOI] [PubMed] [Google Scholar]
- 38.Wickstrom E., Urtishak K., Choob M., Tian X., Sternheim N., Cross L., Rubinstein A., Farber S. Downregulation of gene expression with negatively charged peptide nucleic acids (PNAs) in zebrafish embryos. Methods Cell Biol. 2004;77:137–158. doi: 10.1016/s0091-679x(04)77008-9. [DOI] [PubMed] [Google Scholar]
- 39.Shagin D., Barsova E., Yanushevich Y., Fradkov A., Lukyanov K., Labas Y., Ugalde J., Meyer A., Nunes J., Widder E., et al. GFP-like proteins as ubiquitous Metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.