Abstract
Chromatin insulators have been implicated in the regulation of higher-order chromatin structure and may function to compartmentalize the eukaryotic genome into independent domains of gene expression. To test this possibility, we used biochemical and computational approaches to identify gypsy-like genomic-binding sites for the Suppressor of Hairy-wing [Su(Hw)] protein, a component of the gypsy insulator. EMSA and FISH analyses suggest that these are genuine Su(Hw)-binding sites. In addition, functional tests indicate that genomic Su(Hw)-binding sites can inhibit enhancer–promoter interactions and thus function as bona fide insulators. The insulator strength is dependent on the genomic location of the transgene and the number of Su(Hw)-binding sites, with clusters of two to three sites showing a stronger effect than individual sites. These clusters of Su(Hw)-binding sites are located mostly in intergenic regions or in introns of large genes, an arrangement that fits well with their proposed role in the formation of chromatin domains. Taken together, these data suggest that genomic gypsy-like insulators may provide a means for the compartmentalization of the genome within the nucleus.
THE eukaryotic genome is organized into clusters of coexpressed genes that tend to be transcribed coordinately at specific times throughout development and/or the cell cycle, even though they may or may not be functionally related (Cohen et al. 2000; Caron et al. 2001; Spellman and Rubin 2002; Lercher et al. 2003; Li et al. 2005). The mechanisms responsible for partitioning the genome into domains containing groups of coexpressed genes are currently not well understood. Chromatin insulators may provide a strategy by which cells control the establishment and maintenance of independent transcriptional domains. Insulators are defined experimentally by two properties suggestive of their involvement in higher-order chromatin organization. First, insulators can block activation of transcription by an enhancer when located between the enhancer and the promoter, but they do not inactivate either element. Second, insulators can shield transgenes from position effects caused by surrounding chromatin. These two properties suggest that insulators may normally function to compartmentalize the genome into independent domains of gene expression (Capelson and Corces 2004; Felsenfeld et al. 2004).
Evidence suggesting that insulators play a role in the regulation of higher-order chromatin structure has been provided, in part, by the analysis of the gypsy insulator present in the Drosophila gypsy retrotransposon. This insulator is composed of a 340-bp sequence and several proteins, Suppressor of Hairy-wing [Su(Hw)], Modifier of (mgd4)2.2 [Mod(mdg4)2.2], and centrosomal protein 190 (CP190) (Parkhurst et al. 1988; Georgiev and Gerasimova 1989; Pai et al. 2004). A fourth protein component of the gypsy insulator dTopors may play a role in regulating its activity (Capelson and Corces 2005). The Su(Hw) protein binds to insulator DNA through a domain containing 12 zinc fingers (Dorsett 1990; Spana and Corces 1990). Results from biochemical experiments suggest that the binding site for the Su(Hw) protein consists of the sequence YRTTGCATACCY with a 6-bp invariant core motif TGCATA (Y = C or T, R = A or G) (Spana and Corces 1990). In total, there are 12 Su(Hw)-binding sites in the 340-bp gypsy insulator element, which are sufficient for the full insulator activity attributed to the gypsy retrotransposon (Geyer and Corces 1992; Scott et al. 1999). The Su(Hw) binding sites in gypsy are flanked by A/T tracts, and these sequences are also required for proper insulator function (Spana and Corces 1990). These A/T-rich sequences function as matrix attachment regions (MARs) (Nabirochkin et al. 1998) and may represent binding sites for DNA topoisomerase II or some other MARs-binding protein. Mod(mdg4) and CP190 have also been shown to be essential for gypsy insulator function, but they do not bind gypsy DNA directly and instead interact with the Su(Hw) protein and each other (Gerasimova et al. 1995; Pai et al. 2004).
Su(Hw) and its immediate interacting protein partner, Mod(mdg4)2.2, colocalize at >300 sites on polytene chromosomes of Drosophila (Gerasimova and Corces 1998; Gerasimova et al. 2000). These sites do not correspond to sites of insertion of the gypsy retrotransposon. Rather, they may represent genomic insulators, similar in sequence and function to the insulator present in gypsy, and may play a role in the organization and compartmentalization of the Drosophila genome. CP190, a third component of the insulator complex, associates with Su(Hw) and Mod(mdg4)2.2 at these sites, but it is also present at additional genomic locations perhaps through direct interaction with the DNA via its three zinc fingers (Pai et al. 2004). In diploid cells Su(Hw), Mod(mdg4)2.2, and CP190 insulator proteins colocalize at 20–25 large foci rather than in a diffuse pattern as would be expected from their widespread distribution in polytene chromosomes (Gerasimova and Corces 1998; Gerasimova et al. 2000; Pai et al. 2004). These foci, termed insulator bodies, are created by gypsy insulator proteins present at multiple insulator sites via association with each other and with the nuclear matrix (Gerasimova and Corces 1998; Gerasimova et al. 2000; Capelson and Corces 2004; Pai et al. 2004; K. Byrd and V. G. Corces, unpublished results). The aggregation of multiple insulator sites into large insulator bodies results in the formation of chromatin loops (K. Byrd and V. G. Corces, unpublished results). Endogenous gypsy-like insulators may thus play a key role in the formation of domains of higher-order chromatin structure and, as a consequence, they may be involved in the establishment of independent domains of gene expression.
To gain insight into the role of insulators in genome nuclear organization, we set out to identify and characterize gypsy-like Su(Hw)-binding sites present in the Drosophila genome. Characterization of these sequences allowed us to identify a gypsy-like consensus-binding site for Su(Hw). These sequences are mostly found as single copies but can also be found in clusters of two to six binding sites. Clusters of these binding sites are distributed throughout the genome almost exclusively in noncoding genomic regions or in large genes with very long introns. These genomic Su(Hw)-binding sites show insulator activity that is dependent on the number of binding sites present in each cluster, their genomic location, and possibly their proximity to other insulator sites. The results are consistent with a role for insulators in the organization of the genome within the eukaryotic nucleus.
MATERIALS AND METHODS
Drosophila stocks:
All stocks were maintained on standard yeast–agar medium at 18° or 25°. The following fly stocks were used for the experiments described in the results: Canton-S, w;Dr1/TM6 Δ2-3, y w; Sb1/TM6B, y w; Adv/CyO, Su(Hw)V/TM6B, Su(Hw)f/TM3,Sb1, Su(Hw)2/TM6B, and mod(mdg4)ul.
In vitro translation and immunoprecipitation of genomic Su(Hw)-binding sites:
The full-length Su(Hw) cDNA was cloned into the pCS2+MT vector. Protein was translated using a coupled rabbit reticulocyte transcription/translation system from Promega (Madison, WI) following instructions provided by the manufacturer.
Genomic DNA was isolated from the y2 w ct6 strain and sonicated to an average length of 500 bp. Double-stranded blunt-ended linkers were annealed using the oligonucleotides 5̃′-AGAGGACCTGCAGGTTCTTCC-3′ and 5′-GAAGAACCTGCAGGTCCTCT-3′. The linker-ligated DNA (1 pmol) was then used in the binding reaction containing the following components: 15 mm HEPES, pH 7.6, 100 mm KCl, 5 mm MgCl2, 25 μm ZnCl2, 100 μg/ml BSA, 1 mm DTT, 100 ng poly(dI-dC), 5 μl in vitro-translated c-myc epitope-tagged Su(Hw), and 5 μl of 9E10 α-myc antibody with 1 μl rabbit anti-mouse immunoglobulin G. Reactions were incubated on ice for 30 min. Previously swollen 50% vol/vol stock of protein A-sepharose (Pharmacia) was added to the binding mixture for immunoprecipitation. The solution was incubated at 4° for 1.5 hr with constant mixing. The beads were spun down and the pellet was washed with ice-cold washing buffer (15 mm HEPES, pH 7.6, 100 mm KCl, 5 mm MgCl2, 25 μm ZnCl2, 0.2% NP-40). Bound DNA was eluted from the beads by incubation at 45° with 200 μl of 0.5 m ammonium acetate, 5 mm EDTA, and 0.5% SDS. Eluted DNA was extracted once with phenol–chloroform and precipitated. DNA was then resuspended in 10 μl of TE. A 5-μl aliquot of recovered DNA was then amplified via PCR for 20 cycles using primer 5′-GAAGAACCTGCAGGTCCTCT-3′ and radiolabeled with [α-32P]dCTP. Part of the PCR amplification (5 μl) was used for the next round of binding. After the third and fifth cycles of enrichment, DNA was cloned into the pCR2.1 vector (Invitrogen, San Diego) and sequenced.
Electrophoretic mobility shift assays:
DNA was PCR amplified from vectors using appropriate primers. PCR products were end labeled using [γ-32P]dATP and T4 polynucleotide kinase. Purified probes were then used for electrophoretic mobility shift assays (EMSA). In vitro-translated c-myc-tagged Su(Hw) protein (5 μl) was added to 50,000 cpm of purified probe in a 20-μl final volume of binding reaction containing 15 mm HEPES, pH 7.6, 100 mm KCl, 5 mm MgCl2, 25 μm ZnCl2, 100 μg/ml BSA, 100 ng poly(dI-dC), and 1 mm DTT. As controls, 5 μl of rabbit reticulocyte lysate or luciferase protein was added to the probe instead of Su(Hw). To test the specificity of the interaction, 9E10 serum (5 μl) was added to visualize a supershift in the Su(Hw)–DNA complex. Binding reactions were incubated on ice for 20–30 min and then resolved on a 5% nondenaturing polyacrylamide gel. Gels were dried and subjected to autoradiography. For DNA competition analysis, a 113-bp DNA fragment of the gypsy retrotransposon insulator spanning nucleotides 671–775 and representing the first three Su(Hw)-binding sites was amplified using appropriate primers. The amplified product was labeled as described above. This probe was incubated with in vitro-translated Su(Hw) and binding was competed using 25-fold excess unlabeled gypsy DNA or DNA fragments containing genomic Su(Hw)-binding sites. The reaction products were resolved on a 5% polyacrylamide gel, which was then dried and subjected to autoradiography.
In situ hybridization and immunolocalization:
Probes for DNA in situ hybridization were labeled by a random priming reaction using dig-11-dUTP. Probes were purified by ethanol precipitation and resuspended in 4.0× SSC, 50% formamide, 1.0× Denhardt's, and 0.4 mg/ml of salmon sperm DNA. Third instar larvae were dissected in 1× PBS and 0.1% Triton-X. Salivary glands were isolated and transferred to 45% acetic acid for 1 min. The glands were then fixed for 4 min in a 1:2:3 mixture of lactic acid, water, and glacial acetic acid. The fixed glands were squashed, quickly frozen in liquid nitrogen, and the coverslip was removed. The slides were then placed immediately in chilled ethanol and gradually warmed to room temperature. To prepare the chromosomes for DNA fluorescence in situ hybridization (FISH), the slides were heat stabilized at 70° for 1 hr and subjected to an increasing concentration of ethanol series for dehydration. The chromosomes were then denatured in 0.07 m NaOH for exactly 3 min. Following denaturation, the chromosomes were subjected to another ethanol series and air dried. The DNA probe was denatured by boiling and rapidly chilling on ice. The probe was then applied to the slides and covered with a coverslip, and the edges were sealed with rubber cement. The slides were incubated for 12–15 hr at 37°. The rubber cement was then gently peeled off and slides were washed for 10 min each in 2×, 1×, and 0.5× SSC and rinsed three times in 1× PBS. Slides were blocked in 1× PBS containing 0.1% Tween-20 (PBST) and 3% BSA for 30 min at room temperature. Incubation with primary antibody was carried out overnight at 4°. Slides were washed in 1× PBST and incubated for 30 min at 37° with rhodamine-conjugated antidigoxygenin antibody (Boeringer Mannheim, Indianapolis) to detect FISH signals and goat anti-rat Alexa 477 (Molecular Probes, Eugene, OR). Slides were then washed with 1× PBST, rinsed in 1× PBS, incubated in DAPI, and rinsed with PBS. Slides were visualized under UV light with a Zeiss microscope using the Metamorph software package.
P-element-mediated germline transformation of Drosophila embryos:
DNA fragments containing genomic Su(Hw)-binding sites were subcloned from pCR2.1 clones containing the specific inserts. The y-454, X103, and 2L203 fragments were amplified from genomic DNA using PCR and primers previously described in Golovnin et al. (2003) or primers X103A-5′-GCGGCCGCCCCCTGATATTGGCC-3′, X103B-5′ GCGGCCGCGGGCTTAAGGTGCACCGAC-3′, 2L203A-5′-GCGGCCGCGTCGCCGCTCCCAGACG-3′, and 2L203B-5′-GCGGCCGCATTCGCATTCGAGTGGGGC-3′. Constructs were cloned into the NotI site of a modified yellow gene and introduced into the P-element transformation vector pCaSpeR-2 (pCaSpeR-2-yellow). To accomplish this, the SalI genomic fragment of the yellow gene was subcloned into the pCaSpeR-2 vector where the XbaI site was changed to an XhoI site to accommodate the insertion. The yellow gene was then modified so that the Eco47III site located 893 bp upstream of the yellow gene transcription start site was changed into a NotI site. All transgenes were introduced into w; Dr1/TM3, Sb Δ2–3 embryos as described in Rubin and Spradling (1983) and mapped to chromosomes following standard protocols. Transgenic animals were identified by the w+ phenotype due to the presence of the white gene in the transformation vector. Multiple independent insertions were obtained for each transgene construct.
RESULTS
Isolation of genomic Su(Hw)-binding sites:
A prominent feature of the gypsy retrotransposon is the presence of the repetitive sequence 5′-YRTTGCATACCY-3′ (Y = C or T; R = A or G), which is the binding site for the Su(Hw) protein; this sequence is repeated directly 12 times within the 350-bp gypsy insulator. The distance between two neighboring Su(Hw)-binding sites in gypsy varies from 14 to 23 bp and each element contains a 6-bp TGCATA invariant core. The spacer sequences are AT rich and have been shown to be important for Su(Hw) binding and full insulator activity (Spana et al. 1988; Spana and Corces 1990). With the sequencing of the Drosophila melanogaster genome (Adams et al. 2000), it is possible to predict putative Su(Hw)-binding sites and examine their genomic organization. Using an enhancer finding program, FlyEnhancer (http://flyenhancer.org) (Markstein and Levine 2002), we found a total of 131 sites in the genome that match the 12-bp consensus for Su(Hw) binding. A total of 129 of these sites are separated by sequences >10 kb long, and in only one case are two sites closely located, 1.3 kb apart (data not shown). Thus, most of these putative Su(Hw)-binding sites do not exist in clusters like those found in the gypsy insulator and may therefore fail to function as insulators, since at least 4 Su(Hw)-binding sites may be required for full insulator activity (Scott et al. 1999). It is possible that genomic Su(Hw)-binding sites are sufficiently different from those present in gypsy that they cannot be identified by sequence homology searches using the current Su(Hw)-binding consensus sequence.
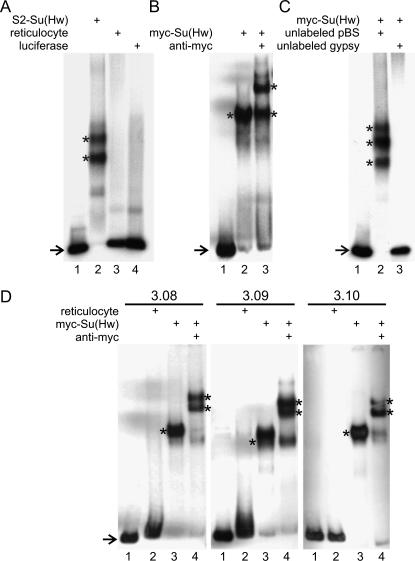
To investigate the possible role of Su(Hw) in chromatin organization, genomic Su(Hw)-binding sites were isolated following a modification of the procedure of Cuvier et al. (1998). A c-myc-tagged Su(Hw) protein was translated in vitro using a rabbit reticulocyte lysate. The c-myc-tagged protein appears to interact with gypsy insulator DNA with strength and specificity similar to that of native Su(Hw) protein purified from Drosophila S2 cells (Figure 1, A and B). The rabbit reticulocyte lysate alone or an in vitro-translated luciferase protein do not interact with gypsy DNA (Figure 1A). The c-myc–Su(Hw) complex can be supershifted with antibodies to the c-myc epitope, and a 100-fold excess of unlabeled gypsy DNA can completely compete off the binding of the radiolabeled probe (Figure 1, B and C). These results suggest that the myc-tagged Su(Hw) protein produced in vitro interacts specifically with gypsy DNA.
Figure 1.
Interaction of Su(Hw) with gypsy-like sequences. All lanes contain 32P-labeled gypsy DNA plus the proteins or extracts indicated in A–C. The location of the unbound gypsy DNA is indicated by an arrow and the location of the protein–DNA complexes is indicated by an asterisk. (A) Su(Hw) purified from Drosophila S2 cells binds specifically to the 351-bp gypsy insulator in EMSAs. The different lanes contain gypsy probe alone (lane 1), gypsy probe with the addition of S2-Su(Hw) (lane 2), gypsy probe with rabbit reticulocyte extract (lane 3), and gypsy probe with luciferase protein (lane 4). (B) Specificity of the myc–Su(Hw)–gypsy interaction. Gypsy probe (lane 1); gypsy DNA with in vitro-translated myc-tagged Su(Hw) (lane 2); and gypsy DNA with myc-tagged Su(Hw) and myc antibody, which causes a supershift in the mobility of the gypsy/myc–Su(Hw) complex (lane 3). (C) myc–Su(Hw) binds specifically to gypsy insulator DNA. Free gypsy probe (lane 1), gypsy probe plus a 100-fold excess unlabeled Bluescript (pBS) DNA (lane 2), and gypsy probe plus a 100-fold excess unlabeled gypsy DNA (lane 3). (D) myc–Su(Hw) can bind genomic DNA containing putative Su(Hw)-binding sites. DNA sequences 3.08, 3.09, and 3.10 were radiolabeled (lane 1 in each gel) and incubated with rabbit reticulocyte (lane 2), myc–Su(Hw) extract (lane 3), and myc–Su(Hw) with anti-myc antibody (lane 4).
To identify Su(Hw)-binding sites, genomic DNA was isolated from the fly strain y2 w ct6 carrying two gypsy-induced mutations in the yellow (y) and cut (ct) genes, respectively. The gypsy elements in this strain serve as internal positive controls for binding and immunoprecipitation experiments. The genomic DNA was sheared to ∼500-bp fragments and then ligated with linker DNA for linker-mediated PCR amplification. In vitro-translated c-myc-tagged Su(Hw) protein was bound to the sheared genomic DNA and then immunoprecipitated with monoclonal antibodies generated against the c-myc epitope. After several washes, immunoprecipitated DNA was recovered and amplified using primers designed in the linker region. Repeated cycles of protein binding, immunoprecipitation, and amplification allowed selection of specific Su(Hw) DNA-binding sequences from the pool of genomic DNA. The sheared DNA was radiolabeled during PCR amplification and quantified at each step to follow the enrichment of Su(Hw)-binding sites with each cycle.
Genomic DNA was cloned after the third and the fifth cycles of enrichment. Sixty-four independent clones (named 3.01–3.64) were isolated and sequenced from material obtained after the third cycle; 13 of these corresponded to gypsy retrotransposon insulator sequences, suggesting that this approach resulted in the isolation of true Su(Hw)-binding sites. Thirty independent clones (named 5.01–5.30) were sequenced from material isolated after five cycles of protein binding and immunoprecipitation, 19 of which corresponded to the gypsy retrotransposon. Among the non-gypsy clones, only 1 was identified twice and all the non-gypsy sequences from cycle 5 were different from those sequenced in cycle 3. The lack of overlap between the genomic DNA sequences in the clones isolated after three or five cycles is expected, since only a very small percentage of the total number of isolated clones was actually sequenced.
The Su(Hw) protein interacts specifically with genomic DNA fragments:
To test the specificity of the interaction between Su(Hw) and the DNA fragments isolated in the experiments just described, individual clones were tested for their ability to bind Su(Hw) protein. Probes made from DNA of individual clones were radiolabeled and subjected to EMSA with in vitro-translated c-myc Su(Hw) protein. Figure 1D shows a representative example with three different DNA fragments (3.08, 3.09, and 3.10). All three fragments can produce a gel shift when incubated with myc-Su(Hw) protein but not with the rabbit reticulocyte extract used to synthesize the protein. In addition, incubation of the DNA fragments with both c-myc-tagged Su(Hw) protein and a monoclonal c-myc antibody resulted in a supershift of the complex, suggesting that the observed shift is specifically caused by the interaction of myc–Su(Hw) with the labeled DNA. A similar EMSA analysis was carried out with 24 different clones isolated after the third cycle of enrichment. Twenty-three of these clones show a slower migrating band upon incubation with c-myc-tagged Su(Hw) and a supershift upon incubation with anti-myc antibody (data not shown), suggesting that the majority of the isolated fragments contain bona fide Su(Hw)-binding sites.
Identification of a new Su(Hw) consensus-binding site from genomic DNA:
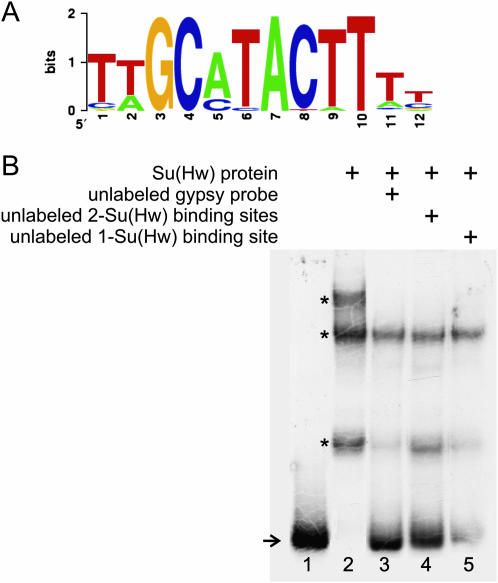
Using all of the isolated DNA fragments that interacted with Su(Hw), we carried out an in silico analysis of these sequences using the multiple motif discovery programs MEME (Multiple Em for Motif Elicitation) (Bailey and Elkan 1994) and BioProspector (Liu et al. 2001) to search for a consensus motif among all the fragments. The results of these two analyses are comparable but only those obtained with BioProspector are detailed. The BioProspector results were then used to generate a weighted consensus binding site with WebLogo, a sequence logo generator (Crooks et al. 2004) (Figure 2A). This consensus site was present in 41 of 55 non-gypsy isolated DNA sequences. This consensus motif is similar but not identical to the one present in the gypsy retrotransposon, which is itself variable. In the new consensus sequence derived from genomic Su(Hw)-binding sites, the central core contains two nucleotides different from those present in gypsy. In addition, the new consensus sequence contains heavily weighted thymine nucleotides in the 3′-end of this motif while the gypsy motif contains mostly cytosine nucleotides. Interestingly, and contrary to the large number of Su(Hw)-binding sites present in gypsy, isolated genomic DNA fragments contain only one or two consensus-binding sites for Su(Hw) and most are separated by greater distances than in gypsy.
Figure 2.
Sequence and binding strength of genomic Su(Hw)-binding sites. (A) The weighted consensus recognition sequence for gypsy-like genomic Su(Hw)-binding sites. (B) EMSA competition analysis. All lanes contain a truncated radiolabeled gypsy probe containing the first three Su(Hw)-binding sites found in gypsy. (Lane 1) Truncated-gypsy probe alone. (Lane 2) Truncated-gypsy probe with Su(Hw) protein. (Lane 3) Unlabeled truncated-gypsy DNA containing Su(Hw)-binding sites. (Lanes 4 and 5) Unlabeled DNA containing either one or two genomic binding sites for Su(Hw).
Using the new consensus sequence for genomic Su(Hw)-binding sites, YWGCMTACTTHY (Y = T or C, W = T or A, M = A or C, H = T, A or C), and the FlyEnhancer program, we carried out a search for similar sequences present in the D. melanogaster genome. We were able to identify >2500 Su(Hw)-binding sites conforming to the consensus sequence shown in Figure 2A. Many of these are present in the genome as individual sites whereas others are arranged in clusters that contain two to six binding sites. For example, there are 164 clusters with at least two motifs within a 2-kb span, with 18 of these clusters containing sites that are immediately adjacent or partially overlapping. When the permissible range is increased to 5 kb, the number of clusters containing two or more sites increases to 351. Of the 351 clusters, 48 contain three or more sites, with 9 clusters containing four sites, 4 clusters containing five sites, and 1 cluster containing six consensus-binding sites. All of these clusters are in closer proximity than all but one site found using the previously derived gypsy Su(Hw)-binding consensus.
Strength of the Su(Hw)–DNA interaction correlates with the number of binding sites:
The gypsy insulator found in the gypsy retrotransposon contains 12 binding sites for Su(Hw) interspersed with A/T-rich sequences. These A/T-rich sequences are important for high-affinity binding by Su(Hw) (Spana and Corces 1990) and have been shown to function as MARs (Nabirochkin et al. 1998). It has been shown previously that the strength of insulation by gypsy depends on the strengths of the enhancers and promoters tested as well as the number of binding sites for Su(Hw) (Scott et al. 1999). At least four binding sites were found to be necessary for an effect, but the binding sites used in this analysis did not include the adjacent MARs sequences. Since these sequences are essential for high-affinity binding of Su(Hw), it is possible that fewer than four binding sites are required when MARs are present in the DNA fragment. It is then possible that the genomic Su(Hw)-binding sites identified in the experiments described above can act as insulators in spite of containing only 2–3 or 1 binding site if MARs sequences are also present in the same DNA fragments. Alternatively, the genomic-binding sites may have a higher affinity for Su(Hw) than those present in gypsy, requiring fewer of these binding sites for full insulator activity.
To address this issue, we first tested whether the strength of the interaction between genomic DNA fragments and Su(Hw) is similar to that between Su(Hw) and gypsy DNA, and whether there is a correlation between the number of binding sites present in the genomic DNA fragment and the strength of their interaction with Su(Hw). To this end, we conducted competitive EMSAs using the newly isolated fragments and gypsy DNA. A radiolabeled truncated gypsy fragment carrying only the first three binding sites for Su(Hw) was incubated with c-myc-tagged Su(Hw) and the shifted band was competed with unlabeled genomic DNA. When radiolabeled gypsy DNA is competed with 25-fold excess of unlabeled cloned DNA containing either one or two genomic Su(Hw)-binding sites, these fragments are able to compete for binding of Su(Hw). One site is not as efficient as two sites, but two seem to work as efficiently as the gypsy DNA (Figure 2B). These results suggest that the affinity for Su(Hw) genomic-binding sites is comparable to that for the gypsy sequence.
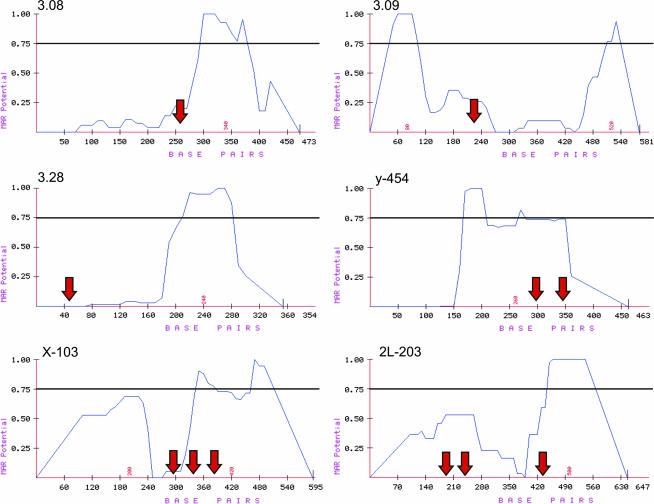
Since most DNA fragments capable of interacting with Su(Hw) have one to three binding sites for this protein and MAR sequences appear to be important for full activity of the insulator present in the gypsy retrotransposon, we tested the possibility that genomic DNA fragments containing binding sites for Su(Hw) also contain MARs. All the immunoprecipitated genomic fragments containing Su(Hw)-binding sites were analyzed for the presence of MARs using the MAR-Wiz program (Singh et al. 1997). These DNA fragments were found to contain sequences predicted to be MARs immediately adjacent to or encompassing the Su(Hw)-binding sites. Figure 3 shows examples for fragments 3.08, 3.09, 3.28, X-103, and 2L-203 (described in detail below and in supplemental data S1 at http://www.genetics.org/supplemental/), as well as a previously identified genomic gypsy-like insulator, y-454 (Golovnin et al. 2003; Parnell et al. 2003). The red arrow indicates the location of Su(Hw)-binding sites within the DNA sequence. Although the function of these predicted MARs sequences has not been tested, their consistent presence immediately adjacent to predicted Su(Hw)-binding sites suggests that they may play a role in the insulator function of these sites. The presence of MARs sequences may increase the activity of these predicted genomic insulators and may explain the relatively low frequency of clusters with more than three Su(Hw)-binding sites in the Drosophila genome.
Figure 3.
MARs plots for DNA fragments containing genomic Su(Hw)-binding sites. The x-axis denotes position within the DNA sequences and the y-axis indicates the probability of MARs activity. Peaks >0.75 show areas of high MARs potential. Red arrows indicate the location of Su(Hw)-binding sites in DNA fragments containing one (3.08, 3.09, 3.28), two (y-454), or three (X-103, 2L-203) Su(Hw)-binding sites.
Putative Su(Hw)-binding sites colocalize with Su(Hw) protein on polytene chromosomes:
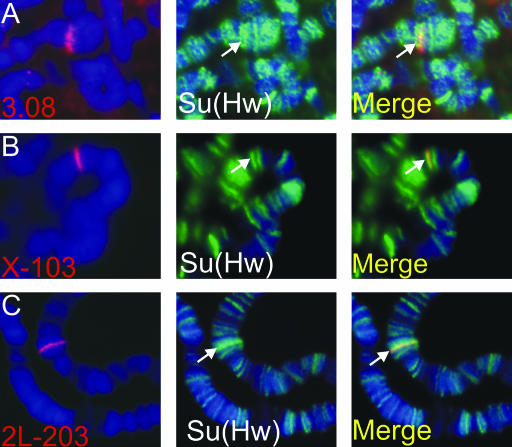
To determine if the immunoprecipitated and in silico-derived Su(Hw)-binding sites correspond to sites in the genome where the Su(Hw) protein is present, we performed FISH on polytene chromosomes. Genomic DNA fragments obtained by immunoprecipitation after incubation with Su(Hw) protein or derived from the in silico analysis were used for hybridization to polytene chromosomes followed by immunostaining with Su(Hw) antibody. A DNA fragment obtained from immunoprecipitation experiments (fragment 3.08) and two in silico-derived sequences (X-103 and 2L-203) were used in these experiments and the results are shown in Figure 4. In each case, there is complete overlap between the FISH and immunofluorescence signals, suggesting that, within the limits of resolution afforded by this technique, the DNA fragments tested belong to regions of the genome that interact with the Su(Hw) protein in vivo. There appears to be a correlation between the number of Su(Hw)-binding sites present in the DNA fragment and the intensity of the immunoflourescence signal associated with Su(Hw), with fragments containing only one predicted site localizing to regions of the genome with a weak Su(Hw) signal (Figure 4A). Fragments with three predicted binding sites appear to localize to regions containing some of the most intense immunofluorescence signals observed in the polytene chromosomes (Figure 4, B and C), suggesting that relatively few sites in the genome may contain clusters of more than three Su(Hw)-binding sites. These results support the conclusion that the binding sites for Su(Hw) identified experimentally or in silico are indeed occupied by Su(Hw) in vivo, giving credence to the computational approach for identifying insulator-binding sites.
Figure 4.
Predicted Su(Hw)-binding sites colocalize with Su(Hw) protein on polytene chromosomes. Fluorescently labeled genomic DNA fragments containing Su(Hw)-binding sites were used for DNA in situ hybridization on polytene chromosomes. Chromosomes were simultaneously labeled with antibodies against Su(Hw). The DNA is visualized with DAPI and shown in blue. The DNA FISH signals corresponding to probes 3.08 (A), X-103 (B), and 2L-203 (C) are shown in red. The location of Su(Hw) protein is shown in green. The overlapping signals for FISH and Su(Hw) protein appear yellow.
Genomic DNA fragments have insulator activity that is dependent on the presence of Su(Hw) and on the number of binding sites for this protein:
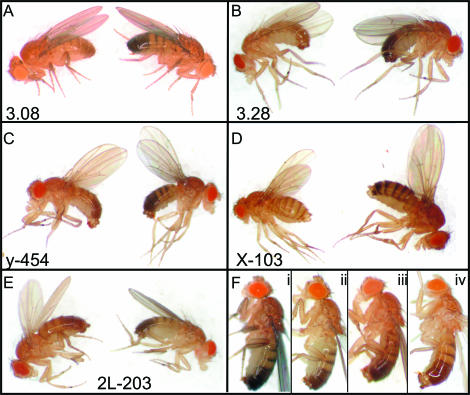
To test whether genomic DNA fragments containing Su(Hw)-binding sites are able to act as insulators in vivo, we employed an enhancer-blocking assay used extensively to study the gypsy insulator (Geyer et al. 1986). In this assay, DNA fragments to be tested for insulator activity are inserted into a plasmid carrying the yellow gene at a position between the wing and body enhancers and the yellow gene promoter. This system allows for quantitation of insulator strength by observing the coloration of the wings and body cuticle, while at the same time controlling for position effects by measuring the expression of the yellow gene in the bristles and other tissues not affected by the insulator. Using this assay, we tested newly identified DNA fragments 3.08 and 3.28, which contain single Su(Hw)-binding sites, and the in silico-derived sequences X-103 and 2L-203, which contain three binding sites. As a reference, we also tested a previously characterized genomic fragment, named y-454, which contains two Su(Hw)-binding sites and has been shown to display insulator activity (Golovnin et al. 2003; Parnell et al. 2003).
Plasmids containing each of the fragments described previously were inserted into the Drosophila genome using P-element-mediated transformation, and multiple independent transgenic lines were obtained for each construct. The phenotypes of transgenic lines are shown in Figure 5 and Table 1. The insertion of a full gypsy element, containing 12 binding sites for Su(Hw), in the yellow gene of the y2 allele results in a dramatic decrease in the pigmentation of the abdomen and wings (Geyer and Corces 1992). Flies carrying the yellow transgene with fragment 3.08, which contains a single Su(Hw)-binding site, show a phenotype similar to that of the gypsy-induced y2 allele in 1 of 7 (14%) transgenic lines (Figure 5A). Transgenic flies carrying the 3.28 fragment, which also contains a single Su(Hw)-binding site, show a y2-like phenotype in 1 of 6 (17%) of the lines examined (Figure 5B). These results suggest that DNA fragments containing 1 Su(Hw)-binding site display insulator function. Insulator activity depends on the genomic location of the transgene insertion but this dependence decreases as the number of Su(Hw)-binding sites present in the DNA fragment increases. When we tested fragments with additional Su(Hw)-binding sites in transgenic animals, the number of transgenic lines showing a strong insulator effect increased. For transgenic lines carrying the y-454 fragment, which contains 2 Su(Hw)-binding sites, 5 of 20 (25%) of the lines give a y2-like phenotype (Figure 5C). An additional 5 of 20 lines (25%) show a phenotype intermediate between that of y2 and wild type, ranging from weak to strong insulation (data not shown but similar to those displayed in Figure 5F and Table1). A similar result for this y-454 fragment was described by Golovnin et al. (2003) and Parnell et al. (2003). Transgenic lines carrying the X-103 and 2L-203 fragments, both of which contain 3 binding sites for Su(Hw), display a y2 phenotype in 3 of 10 (30%) and 5 of 15 (33%) lines, respectively (Figure 5, D and E), and an additional 5 of 10 (50%) and 8 of 15 (53%) show a range of intermediate phenotypes (Figure 5F shows a range for construct 2L-203). In total, DNA fragment y-454, containing two binding sites for Su(Hw) and a putative MARs site, gives 50% transgenic fly lines with a y2 or compromised yellow phenotype. Fragment X-103, containing 3 Su(Hw)-binding sites, gives 80% transgenic lines with a y2 or compromised yellow phenotypes whereas fragment 2L-203 gives an 87% frequency of y2 or compromised yellow phenotypes.
Figure 5.
Genomic Su(Hw)-binding sites have insulator activity that is dependent on Su(Hw) protein. DNA fragments containing one (3.08 and 3.28), two (y-454), or three (X-103 and 2L-203) binding sites for Su(Hw) were tested for their ability to inhibit promoter–enhancer interactions in the yellow gene. A representative example is given for each series of the transgenic insulating lines. Male flies are shown except in D where female flies are displayed. In all cases, the insulating activity is visible in both sexes. In each panel, the fly on the left contains the transgene in the background of a wild-type su(Hw) gene, whereas the fly on the right is mutant for su(Hw); only the phenotypes for su(Hw)2/su(Hw)V mutants are shown but other allele combinations, su(Hw)2/su(Hw)f and su(Hw)f/su(Hw)V, gave similar results. (A) Effect of a single Su(Hw)-binding site present in fragment 3.08 on the yellow enhancer-blocking assay (left). The wings and body cuticle appear yellow. When placed in a su(Hw) mutant background, the coloration of the wing and body reverts back to a wild-type phenotype (right). (B) A similar analysis carried out with fragment 3.28 (one binding site) also shows insulating activity (left) that is dependent on Su(Hw) (right). (C–E) Transgenic animals carrying DNA fragments y-454 (two binding sites), X-103 (three binding sites), and 2L-203 (three binding sites) in the absence (leftmost in each panel) or presence of mutations in su(Hw) (rightmost in each panel). (F) Adult flies from different transgenic lines carrying DNA fragment 2L-203 showing a series of insulator-induced phenotypes ranging from weak (i) to strong (iv).
TABLE 1.
Phenotypes of transgenic lines
| No. of transgenic lines demonstrating:
|
|||||||
|---|---|---|---|---|---|---|---|
| DNA construct | No. of Su(Hw)-binding sites | i. Weak insulation | ii. Moderate insulation | iii. Good insulation | iv. Strong insulation | Total no. of transgenic lines | Total no. showing insulation (%) |
| 3.08 | 1 | 6 (86) | 1 (14) | 7 | 14 | ||
| 3.28 | 1 | 5 (83) | 1 (17) | 6 | 17 | ||
| y-454 | 2 | 10 (50) | 2 (10) | 3 (15) | 5 (25) | 20 | 50 |
| X-103 | 3 | 2 (20) | 2 (20) | 3 (30) | 3 (30) | 10 | 80 |
| 2L-203 | 3 | 2 (13) | 4 (27) | 4 (27) | 5 (33) | 15 | 87 |
i, ii, iii, and iv correspond to flies in Figure 5F. Numbers in parentheses are percentages.
To confirm that the yellow phenotypes detected in the transgenic lines are caused by the presence of Su(Hw)-binding sites, we tested whether the insulator effect is dependent on the Su(Hw) protein by crossing all the fly lines that give a y2 phenotype with strains carrying mutations in the su(Hw) gene. The yellow phenotypes for all transgenic lines were tested in the mutant background of three different allelic combinations of su(Hw) mutations: su(Hw)f/su(Hw)2, su(Hw)f/su(Hw)V, and su(Hw)V/su(Hw)2. In all cases, the y2 phenotype of transgenic lines reverted back to wild type in all three trans-heterozygous combinations of su(Hw) mutations (Figure 5, A–E). In addition, these same transgenic lines show a reversion of the mutant yellow phenotype in the background of the mod(mdg4)ul mutation (data not shown). These data strongly suggest that the genomic Su(Hw)-binding sites do function as an insulator and that this function is dependent on Su(Hw) protein.
Genomic distribution of Su(Hw)-binding sites:
DNA fragments containing genomic Su(Hw)-binding sites can act as insulators, suggesting that they may play a role in the establishment of independent domains of gene expression and higher-order chromatin organization. A prediction from this conclusion is that the DNA fragments containing Su(Hw)-binding sites identified either experimentally or in silico would be located preferentially in genomic regions devoid of genes and that they would separate areas of gene-rich sequences. To test this hypothesis, we mapped the Su(Hw)-binding sites identified above to the Drosophila genome using FlyEnhancer and FLYBLAST. Since clusters of two or more Su(Hw)-binding sites are likely to have stronger insulator activity and therefore to play a more important role in genome organization, we concentrated our analysis on clusters rather than on single sites.
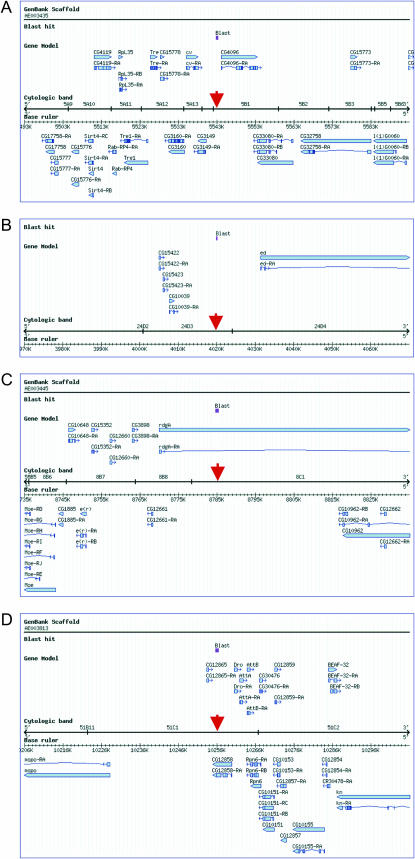
A typical example of the results of this analysis is shown in Figure 6, and a complete description of the genomic location of each of these sites is presented in the supplemental data in supplemental Figures S1 and S2 at http://www.genetics.org/supplemental/. Approximately 55% of the clusters of Su(Hw)-binding sites identified are located in intergenic regions. In some cases, the intergenic region is short (Figure 6A), but more often the clusters of Su(Hw)-binding sites are located in regions 10–20 kb long that are completely devoid of genes (Figure 6B). In each of these examples, the location of Su(Hw)-binding sites is indicated by a red arrow. In this type of arrangement, the insulator may separate two domains of gene-rich sequences, each containing several closely arranged genes, possibly preventing interactions between regulatory elements (e.g., enhancers, repressors, silencers) located on either side of the insulator. An additional 41% of clusters of Su(Hw)-binding sites are found separating gene-rich regions, but located within long genes with at least one very large intron (Figure 6C). For example, the Su(Hw)-binding site cluster shown in Figure 6C is located in a 70-kb intron of the rdgA gene, which itself is >90 kb in length. This is also the case for other genes containing Su(Hw)-binding sites in intron regions (see supplemental data in Figure S2 at http://www.genetics.org/supplemental/). The average length of X-linked genes containing intronic Su(Hw)-binding sites is 62 kb vs. the average Drosophila gene, which ranges in size from 3 to 5 kb. Although these clusters of Su(Hw)-binding sites are found within transcribed regions of genes, they may not affect expression of the gene itself, since the presence of the gypsy insulator in an intron of the yellow gene does not interfere with its transcription (Geyer and Corces 1992). Therefore it is likely that these intronic clusters of Su(Hw)-binding sites play a role similar to those found in regions devoid of genes, blocking enhancer–promoter communications between genes on either side of the insulator without interfering with the transcription of the gene that they occupy. The remaining 4% of the clusters of Su(Hw)-binding sites identified are found very close to or within exon-coding regions of genes (Figure 6D). Overall, the preferential localization of clusters of Su(Hw)-binding sites in regions largely devoid of protein-coding sequences or between gene-rich regions is consistent with a role for these insulators in organizing the genome into independent domains of gene expression.
Figure 6.
Organization of Su(Hw)-binding sites in the Drosophila genome. Representative examples of the location of Su(Hw)-binding sites with respect to the location of neighboring genes. The red arrow indicates the location of the Su(Hw)-binding sequences. (A) An example of an intergenic Su(Hw)-binding site cluster separating gene-rich regions. (B) An example of an intergenic Su(Hw)-binding site cluster separating genes >20 kb apart. (C) An example of an intragenic insulator sequence located within the intron of the large rdgA gene. (D) An example of a genomic insulator located, at least partially, within the coding region of an annotated gene. A, B, C, and D each display a 100-kb genomic region.
DISCUSSION
To gain insights into the possible role of chromatin insulators in genome compartmentalization, we carried out a search for insulator sites associated with the Su(Hw) protein, which is the DNA-binding component of the insulator present in the gypsy retrotransposon. This protein is widely present in Drosophila polytene chromosomes at sites predicted to be functional gypsy-like insulators, endogenous to the fly genome and distinct from that present in the gypsy retrotransposon. Using several cycles of immunoprecipitation and PCR, we were able to isolate a collection of DNA fragments containing genomic, gypsy-like Su(Hw)-binding sites. In silico analyses were then used to identify a consensus Su(Hw)-binding sequence that was used to search the entire Drosophila genome for similar sites. Three sites thus identified were shown to interact in vitro with Su(Hw) and to localize to regions of polytene chromosomes where this protein is present. In addition, these sites have insulator activity dependent on the number of predicted Su(Hw)-binding sites and the genomic location of transgene insertion. Finally, mapping the genomic Su(Hw)-binding sites demonstrate that they are found in intergenic and nonprotein coding sequences separating gene-rich domains. These results suggest that gypsy-like sequences with insulator activity are widely distributed throughout the Drosophila genome, raising the question of whether the gypsy retrovirus acquired insulator sequences from the host genome or vice versa. Although the results presented here do not distinguish between these two alternatives, it is possible that, as is the case in vertebrate retroviruses, gypsy obtained these sequences from the Drosophila genome. This may have offered gypsy an evolutionary advantage, as the genome of the virus then may have been protected from the repressive effect of adjacent sequences when inserted into the Drosophila genome.
Studies in a variety of organisms have established that, throughout development or during the cell cycle, coexpressed genes tend to be localized in physically adjacent positions along the chromosome, although a mechanism for establishing these domains remains elusive (Cohen et al. 2000; Caron et al. 2001; Lercher et al. 2002; Spellman and Rubin 2002; Li et al. 2005). Information suggesting that insulator elements may be involved in the establishment of independent chromatin domains has been obtained in several different systems. For example, FISH analysis using nuclear halo preparations resulted in the visualization of DNA located between two genomic gypsy insulator sites as a loop, with the insulator sites at the ends of the two stems attached to the nuclear matrix (K. Byrd and V. G. Corces, unpublished results). Placement of a new insulator in the middle of the loop resulted in the formation of two smaller loops, supporting the involvement of gypsy insulator components in the formation of chromatin loops that may correspond to gene expression domains. Similarly, the specialized chromatin structure (scs) and scs's insulators of Drosophila have been visualized forming loops using the chromatin conformation capture (3C) technique (Blanton et al. 2003). In a third example involving the vertebrate CTCF insulator, this protein was shown to copurify with the nuclear matrix and to associate with the nucleolus via interactions with nucleophosmin, suggesting a potential for the formation of chromatin loops similar to those seen with gypsy (Yusufzai and Felsenfeld 2004; Yusufzai et al. 2004). Therefore, chromatin loop formation may be a common mechanism used by different insulators to regulate enhancer–promoter interactions and functionally compartmentalize the genome. However, a correlation between the distribution of genes in the genome and the arrangement of chromatin insulators at the genome level has not been previously established. Data presented here on the genomic organization of gypsy-like insulators may support a role for these sequences in the establishment of chromatin domains that could explain the existence of clusters of coexpressed genes.
The total number of predicted Su(Hw)-binding sites in the D. melanogaster genome is >2500. Most of these are present in single copy. However, 351 clusters with 2 or more sites are found throughout the fly genome. This number roughly corresponds to the number of bands of the Su(Hw) protein observed by immunofluorescence analysis of polytene chromosomes (Gerasimova et al. 1995). The number of Su(Hw) sites per cluster is lower than the 12 sites present in gypsy and lower than the 4 sites shown to be required for insulator function in an enhancer-blocking assay (Scott et al. 1999), raising the question of whether the clusters of Su(Hw)-binding sites found in the Drosophila genome are functional insulators. Interestingly, genomic Su(Hw)-binding sites are closely associated with MARs, as is also the case for the sites present in the gypsy retrotransposon insulator. The presence of MARs may strengthen the insulator activity of genomic Su(Hw)-binding sites and lower the requirement for the number of sites needed for insulator function. In support of this conclusion, single insulator sites as well as clusters of 2 or 3 sites show insulator activity in an enhancer-blocking assay. The strength of the enhancer-blocking effect of these genomic insulators is similar to that observed with the gypsy insulator containing 12 binding sites for Su(Hw), but the insulator activity is dependent of the genomic context of the transgene insertion site. For example, DNA fragments containing single Su(Hw) sites show full insulator activity in 14–17% of transgenic lines whereas fragments with two Su(Hw)-binding sites show full or partial insulator activity in 50% and fragments with three sites show partial-to-full insulator activity in 80% of the lines. These data indicate that insulator activity is dependent on the number of Su(Hw)-binding sites present as well as the genomic location of the transgene. The fact that, for a given number of Su(Hw)-binding sites, some transgenic lines display insulator activity whereas other do not suggests that the genomic location of the insertion site is important. This effect is unlikely to be caused by specific sequences or chromatin structure at the insertion site affecting the expression of the yellow gene, since we did not observe a correlation between genomic location and the expression of the adjacent white gene present in the transformation vector (data not shown). Instead, the dependence of insulator function on the genomic location may correlate with the distance between the insertion site and other gypsy-like genomic insulators. This conclusion is supported by the finding that the variability in insulator activity as a function of the location of the insertion site declines as the number of Su(Hw)-binding sites present in the test fragment increases.
The observed correlation among insulator activity, genomic location, and number of Su(Hw)-binding sites agrees well with proposed mechanisms to explain insulator function and further supports the role of insulators in establishing chromatin domains. It has been proposed that insulators function by creating chromatin loops via interactions between individual insulator sites that coalesce at specific nuclear locations, forming large aggregates of insulator sites named insulator bodies (Gerasimova and Corces 1998). In the case of gypsy, the interaction between individual insulators may take place through the BTB-containing proteins Mod(mdg4)2.2 and CP190, which in turn attach to the insulator via interactions with Su(Hw) (Pai et al. 2004). If this is the case, one would predict that the ability of a particular insulator to interact with neighboring ones would depend on its relative distance to its neighbors and on the number of Su(Hw) molecules present. As the number of insulator-binding sites increases, so does the potential to find and interact with another insulator via the bridges created by Mod(mdg4)2.2 and CP190. The frequency and strength of these interactions are also a function of the distance between individual insulator sites in the genome, explaining why some transgenic lines display insulator effects while others fail to do so. The proportion of transgenic lines showing full insulator activity is only 14–17% in cases in which only one Su(Hw)-binding site is present in the DNA fragment tested. This observation does not imply that single Su(Hw)-binding sites in the genome do not function as insulators. It is possible that these sites are sufficiently close to other Su(Hw)-binding sites that a single one is sufficient for insulator function. Alternatively, these sites may be located in the genome between genes regulated by weak enhancers, such that a single site may not be enough to disrupt enhancer–promoter interactions in the yellow gene used in our analysis but may suffice in the context of the regulatory sequences present in its normal genomic environment.
Insulators have been shown to form chromatin loops in the nucleus and, as a consequence, it has been hypothesized that their role is to partition the genome into functional units or domains such that the expression of genes located in one domain is independent of regulatory sequences present in a different domain (K. Byrd and V. G. Corces, unpublished results). Insulators could then play a role in controlling transcription at a global level, establishing certain organization of the chromatin fiber that would be required for subsequent regulation by standard transcription-factor-mediated mechanisms. The outcome of such an organization is that genes within one domain would be coexpressed, a prediction that has been confirmed by genomewide transcription profiling studies (Cohen et al. 2000; Caron et al. 2001; Lercher et al. 2002; Spellman and Rubin 2002; Li et al. 2005). Such a role for insulators would require a very specific arrangement of these sequences within the eukaryotic genome. Data presented here show that this is indeed the case. The precise cytological location of Su(Hw) sites using FISH analysis has been studied in three cases and found to overlap with the location of Su(Hw) bands detected by immunofluorescence. Assuming a similar overlap for other gypsy-like insulators, the cytological distribution of these sequences in polytene chromosomes should be similar to that of Su(Hw), with an exclusive euchromatic arrangement at the boundaries between bands and interbands. At the DNA sequence level, clusters of Su(Hw)-binding sites with insulator activity are located preferentially in intergenic and nonprotein-coding regions. Of these clusters, 55% are solely intergenic whereas 41% are located in introns of very large genes. The intragenic location of these intronic insulators does not preclude their ability to exert their function without inappropriately affecting expression of the gene in which they reside, as insulators have been shown to be permissive for transcription when inserted into an intron as long as their presence does not preclude enhancer–promoter interactions. This arrangement of Su(Hw)-binding sites within the Drosophila genome and their ability to operate as insulators in an enhancer-blocking assay suggest that these sequences are bona fide endogenous gypsy-like insulators and may have a role in organizing the genome into functional transcription domains.
Acknowledgments
We thank M. Rohrbaugh, T. Brandt, C. Karam, and E. Lei for critical comments on this manuscript. This work was supported by National Institutes of Health (NIH) National Research Service Award GM75604 to E.R. and U. S. Public Health Service Award GM35463 from the NIH to V.G.C.
References
- Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne et al., 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- Bailey, T. L., and C. Elkan, 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, pp. 28–36 in Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed]
- Blanton, J., M. Gaszner and P. Schedl, 2003. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 17: 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2004. Boundary elements and nuclear organization. Biol. Cell 96: 617–629. [DOI] [PubMed] [Google Scholar]
- Capelson, M., and V. G. Corces, 2005. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol. Cell 20: 105–116. [DOI] [PubMed] [Google Scholar]
- Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins et al., 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Cohen, B. A., R. D. Mitra, J. D. Hughes and G. M. Church, 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26: 183–186. [DOI] [PubMed] [Google Scholar]
- Crooks, G. E., G. Hon, J. M. Chandonia and S. E. Brenner, 2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier, O., C. M. Hart and U. K. Laemmli, 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18: 7478–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, D., 1990. Potentiation of a polyadenylylation site by a downstream protein-DNA interaction. Proc. Natl. Acad. Sci. USA 87: 4373–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenfeld, G., B. Burgess-Beusse, C. Farrell, M. Gaszner, R. Ghirlando et al., 2004. Chromatin boundaries and chromatin domains. Cold Spring Harbor Symp. Quant. Biol. 69: 245–250. [DOI] [PubMed] [Google Scholar]
- Georgiev, P. G., and T. I. Gerasimova, 1989. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol. Gen. Genet. 220: 121–126. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., and V. G. Corces, 1998. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell 92: 511–521. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., D. A. Gdula, D. V. Gerasimov, O. Simonova and V. G. Corces, 1995. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell 82: 587–597. [DOI] [PubMed] [Google Scholar]
- Gerasimova, T. I., K. Byrd and V. G. Corces, 2000. A chromatin insulator determines the nuclear localization of DNA. Mol. Cell 6: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., C. Spana and V. G. Corces, 1986. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 5: 2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovnin, A., I. Birukova, O. Romanova, M. Silicheva, A. Parshikov et al., 2003. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development 130: 3249–3258. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., A. O. Urrutia and L. D. Hurst, 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31: 180–183. [DOI] [PubMed] [Google Scholar]
- Lercher, M. J., T. Blumenthal and L. D. Hurst, 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13: 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., B. Lee and L. Zhang, 2005. Genome-scale analysis of positional clustering of mouse testis-specific genes. BMC Genomics 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., D. L. Brutlag and J. S. Liu, 2001. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput., 127–138. [PubMed]
- Markstein, M., and M. Levine, 2002. Decoding cis-regulatory DNAs in the Drosophila genome. Curr. Opin. Genet. Dev. 12: 601–606. [DOI] [PubMed] [Google Scholar]
- Nabirochkin, S., M. Ossokina and T. Heidmann, 1998. A nuclear matrix/scaffold attachment region co-localizes with the gypsy retrotransposon insulator sequence. J. Biol. Chem. 273: 2473–2479. [DOI] [PubMed] [Google Scholar]
- Pai, C. Y., E. P. Lei, D. Ghosh and V. G. Corces, 2004. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 16: 737–748. [DOI] [PubMed] [Google Scholar]
- Parkhurst, S. M., D. A. Harrison, M. P. Remington, C. Spana, R. L. Kelley et al., 1988. The Drosophila su(Hw) gene, which controls the phenotypic effect of the gypsy transposable element, encodes a putative DNA-binding protein. Genes Dev. 2: 1205–1215. [DOI] [PubMed] [Google Scholar]
- Parnell, T. J., M. M. Viering, A. Skjesol, C. Helou, E. J. Kuhn et al., 2003. An endogenous suppressor of hairy-wing insulator separates regulatory domains in Drosophila. Proc. Natl. Acad. Sci. USA 100: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. M., and A. C. Spradling, 1983. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 11: 6341–6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K. C., A. D. Taubman and P. K. Geyer, 1999. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics 153: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, G. B., J. A. Kramer and S. A. Krawetz, 1997. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 25: 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana, C., and V. G. Corces, 1990. DNA bending is a determinant of binding specificity for a Drosophila zinc finger protein. Genes Dev. 4: 1505–1515. [DOI] [PubMed] [Google Scholar]
- Spana, C., D. A. Harrison and V. G. Corces, 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2: 1414–1423. [DOI] [PubMed] [Google Scholar]
- Spellman, P. T., and G. M. Rubin, 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai, T. M., and G. Felsenfeld, 2004. The 5′-HS4 chicken {beta}-globin insulator is a CTCF-dependent nuclear matrix-associated element. Proc. Natl. Acad. Sci. USA 101: 8620–8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai, T. M., H. Tagami, Y. Nakatani and G. Felsenfeld, 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13: 291–298. [DOI] [PubMed] [Google Scholar]