Abstract
This report describes high-frequency germline gene targeting at two genomic loci in Drosophila melanogaster, y and ry. In the best case, nearly all induced parents produced mutant progeny; 25% of their offspring were new mutants and most of these were targeted gene replacements resulting from homologous recombination (HR) with a marked donor DNA. The procedure that generates these high frequencies relies on cleavage of the target by designed zinc-finger nucleases (ZFNs) and production of a linear donor in situ. Increased induction of ZFN expression led to higher frequencies of gene targeting, demonstrating the beneficial effect of activating the target. In the absence of a homologous donor DNA, ZFN cleavage led to the recovery of new mutants at three loci—y, ry and bw—through nonhomologous end joining (NHEJ) after cleavage. Because zinc fingers can be directed to a broad range of DNA sequences and targeting is very efficient, this approach promises to allow genetic manipulation of many different genes, even in cases where the mutant phenotype cannot be predicted.
TO discover the function of a gene, a geneticist typically isolates mutations in that gene and evaluates their effects. Large mutant collections exist for a number of model organisms, but some genes are not represented, and the nature of the alterations is not under the control of the experimenter. It is frequently desirable to be able to introduce targeted mutations to address specific questions about gene function. Particularly with the sequences of complete genomes of many organisms now available, gene identification is facilitated, but producing directed mutations is often still challenging. While genes or gene segments can be readily isolated and manipulated, methods to replace a chromosomal segment with the altered DNA by homologous recombination are frequently limited by low frequency of the desired event and competition with random integration (Vasquez et al. 2001).
In Drosophila melanogaster Rong and Golic (2000a) introduced a gene targeting procedure based on generation of a linear donor DNA in situ. Their approach was predicated on the observations in many situations that DNA ends are recombinagenic. In our view the efficiency of recombination between donor and target is still limited because the chromosomal target is largely inert. It is certainly true in model reactions that cleaving the target enhances the frequency of recombination with the donor (Rouet et al. 1994; Choulika et al. 1995; Smih et al. 1995; Cohen-Tannoudji et al. 1998; Donoho et al. 1998).
We have begun to employ hybrid proteins called zinc-finger nucleases (ZFNs) (Kim et al. 1996; Smith et al. 2000; Bibikova et al. 2001) as targetable cleavage reagents that have the potential to make double-strand breaks at arbitrarily chosen sites. These proteins consist of a DNA-binding domain composed of zinc fingers linked to a nonspecific DNA-cleavage domain derived from the restriction endonuclease, FokI. Each zinc finger contacts primarily 3 bp of DNA through interactions in the major groove, and fingers that recognize many of the 64 triplets have been isolated (Segal et al. 1999; Dreier et al. 2001, 2005; Liu et al. 2002; Segal 2002). Thus, in principle, zinc-finger domains can be designed to target a broad range of DNA sequences. In fact, designed zinc-finger combinations have been used successfully for directing synthetic transcription factors to specific targets (Jamieson et al. 2003; Blancafort et al. 2004), as well as for directing cleavage by ZFNs (Bibikova et al. 2002, 2003; Porteus and Baltimore 2003; Alwin et al. 2005; Porteus and Carroll 2005; Urnov et al. 2005).
Another feature of ZFNs is a requirement for dimerization of the cleavage domain (Smith et al. 2000; Bibikova et al. 2001). Because the dimer interface is quite weak, cleavage is best achieved by directing two proteins to neighboring sites. When both are bound, dimerization and cleavage ensue. This confers additional specificity on the cleavage reaction, since neither monomeric protein cuts DNA on its own, and the cleavage reagent is assembled at the target site.
In previous work we demonstrated that designed ZFNs can cleave the yellow (y) gene of Drosophila, leading to mutagenesis via nonhomologous end joining (NHEJ) (Bibikova et al. 2002), and that such cleavage enhances the frequency of homologous recombination (HR) with a donor DNA—i.e., gene targeting (Bibikova et al. 2003). (Throughout this article we refer to homologous gene replacement via HR as gene targeting and induced localized mutation via NHEJ as targeted mutagenesis.) In this report we describe our approach more fully, demonstrate its application to two additional genes, and report substantial improvements in targeting efficiency. The new targets, rosy (ry) and brown (bw), both affect eye color and were chosen, like y, for the ease of scoring their mutant phenotypes in these early tests of the generality of ZFN-induced targeting.
MATERIALS AND METHODS
Zinc-finger nucleases:
The design and construction of the ZFNs for the y gene, yA and yB, have been described previously (Bibikova et al. 2002). The coding sequence for each ZFN was linked to the hsp70 heat-shock promoter and introduced separately into the Drosophila genome on the P-element vector, pDM30 (FlyBase at http://flybase.bio.indiana.edu/). Multiple independent insertions were isolated for each nuclease and mapped to one of the three largest chromosomes. Syntenic pairs on chromosome 2 and on chromosome 3 were recovered by passage through the female germline and by screening by PCR using primers specific to the unique zinc-finger sequences of yA and yB. Several such pairs were isolated; the one reported in this study (AB10) is on chromosome 3. No attempt was made to determine the precise locations of the transgenes.
The ZFNs for the targets in bw and ry were constructed in the same fashion, but were cloned into Gateway vectors (Invitrogen, Carlsbad, CA). An entry vector, pEntrZFN, was constructed by cloning a ZFN into pENTR 2A. New fingers were introduced into pEntrZFN by cutting with NdeI and SpeI and ligating a PCR product encoding the new finger sequences flanked with these same restriction sites. ZFN sequences were transferred with Clonase (Invitrogen) into pCaSpeR-hs (http://thummel.genetics.utah.edu/), which had been modified with a Gateway RFB cassette to generate a pDEST vector.
Existing zinc-finger plasmids were used as templates for PCR with mutagenic primers that converted the codons for specificity-determining residues to those desired on the basis of tabulated data (Segal et al. 1999; Liu et al. 2002; Segal 2002) (see Table 1). Finger sequences for each triplet were chosen for their apparent specificity, as indicated in the published characterizations. The y and bw ZFNs are composed of fingers characterized in the Barbas lab (Segal et al. 1999; Segal 2002), while those for ry are mixtures of Barbas and Sangamo (Liu et al. 2002) fingers. No attempt was made to optimize the affinity or specificity of the zinc-finger sets. All of the zinc-finger frameworks were based on Zif268, except that of bwB, which is in the consensus CP-1 (Krizek et al. 1991; Segal 2002).
TABLE 1.
Zinc-finger sequences
| ZFN | Target (5′ → 3′) | Finger 1 | Finger 2 | Finger 3 |
|---|---|---|---|---|
| yA | GTG GAT GAG | RSDNLVR | TSGNLVR | RSDALVRa |
| yB | GCG GTA GGC | DPGHLVR | QSSSLVR | RSDERKRb |
| bwA | GAT GAT GGG | RSDKLVR | TSGNLVR | TSGNLVR |
| bwB | GAG GTG GGC | DPGHLVR | RSDELVR | RSDNLVR |
| ryA | GGC GTG GGA | QSGHLQRc | RSDALTRc | DPGHLVR |
| ryB | GTA GTA GCT | QSSDLTRc | QSSSLVR | QSSSLVR |
The specificity-determining residues [from position −1 to +6 in the standard notation (Segal 2002)] for each set of zinc fingers are given in the one-letter code. In reading the table, it is important to remember that the first finger binds the 3′-most DNA triplet. For example, the GAG triplet in the yA target is recognized by Finger 1 (RSDNLVR). Finger sequences without footnotes were derived and characterized in the Barbas lab (Segal et al. 1999).
This finger for GTG was obtained from the Barbas lab, but is not listed in their collection of derived fingers (Segal et al. 1999; Segal 2002).
This finger for GCG is a natural sequence from the transcription factor Zif268 (Pavletich and Pabo 1991).
These fingers are from the Sangamo collection (Liu et al. 2002).
A mutant version of yA was produced by substituting an alanine codon (GCC) for the aspartate codon (GAC) at position 175 in the nuclease active site. This was accomplished by PCR with primers carrying the desired sequence alteration. This sequence was substituted for that of yA in the pDM30 vector, and transformants were recovered after embryo injection. The final step in cloning of this variant was performed by Keyclone Technologies (Cincinnati).
Mutant donor DNAs:
y:
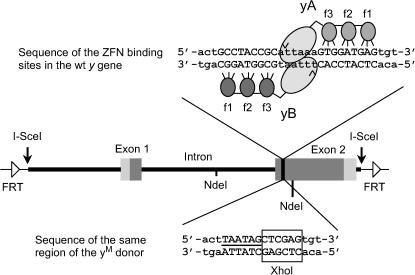
The plasmid pS/G (Geyer and Corces 1987) carries 8.0 kb of Drosophila chromosomal DNA, including the complete y+ gene. An internal 6.7-kb BglII fragment was excised and recloned into the BamHI site of pBluescript KS+. The designed mutation (see Figure 4) was created in a 2.1-kb NdeI fragment by PCR. Two overlapping primers carried the desired sequence changes, while two outside primers included NdeI sites in the y+ sequence flanking the ZFN target. Two separate reactions were performed, each with one central and one flanking primer. The products of these reactions were mixed and reamplified with the flanking primers. This product was phosphorylated with polynucleotide kinase and cloned into the EcoRV site of pBluescript KS+. The modified NdeI fragment was excised and exchanged for the corresponding segment of the cloned y+ BglII fragment (see Figure 4). The entire modified insert was then moved as a SalI + XbaI fragment to pBSN, a pBluescript derivative with two NotI sites flanking the insert. The modified gene, yM, was cloned as a NotI fragment into the P-element vector pw30, which carries a w+ marker and in which yM is flanked by recognition sites for FLP and I-SceI (Gong and Golic 2003). This was introduced into the genome by embryo injection. A single transformant in which the w+ transgene mapped to chromosome 3 was isolated; flies homozygous for this insertion are viable.
Figure 4.
Structure of the yM donor. The 6.7-kb segment of the y gene, with surrounding sequences, that was included in the donor is shown in the center of the diagram; introns and exons are labeled. Above this, the specific sequence targeted by ZFNs is shown, as in Figure 1. Below this is the sequence of this same region as modified in the yM donor. The two in-frame stop codons are underlined, and the new XhoI site is boxed. The locations of the two NdeI sites used in construction of the yM modification are also indicated.
ry:
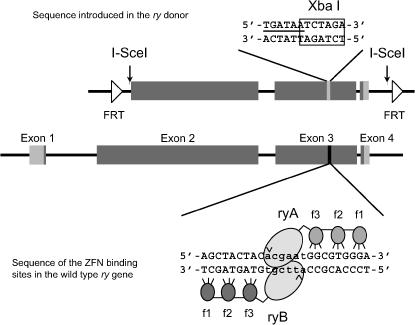
A similar protocol was used to generate the mutant ry donor. The starting sequence was the ry gene included in pDM30. The fragment incorporated into the donor vector stretches from the EcoRI site within exon 2 to the HindIII site beyond the end of exon 4; it has 4.16 kb of homology to the genomic locus (see Figure 1). In this case the diagnostic restriction site inserted was XbaI. The designed mutation was created in separate 5′ and 3′ segments with overlapping central PCR primers and outside primers carrying convenient restriction sites (sequences available upon request). The construct was cloned initially in pBluescript and then transferred as an XhoI–SacII fragment to pP{whiteOut2} (http://sekelsky.bio.unc.edu/research/vectors/vectors.html), where it was flanked by sites for FLP and I-SceI. Flies carrying the w1118 mutation were transformed with this construct by embryo injection. Eight independent transgenes that mapped to chromosomes 2 and 3 were used in subsequent experiments.
Figure 1.
The Drosophila ry gene showing the ZFN target and its modification in the ryM donor. The lower line shows the structure of the gene: exons are shown as boxes, with protein-coding sequences shaded and introns as lines. The sequence of the ZFN-targeted site is shown below with triplets bound by the zinc fingers in capital letters. Zinc fingers are illustrated as shaded ovals (f1, f2, f3), the FokI cleavage domains attached to each set of fingers as lightly shaded ovals, and the expected cleavage sites on each strand with carats. The 4.16-kb segment of the ry gene that was included in the donor is shown above. It is flanked by recognition sites for I-SceI and for FLP (FRTs). The specific modifications made in the donor are shown: the two in-frame stop codons are underlined, and the new XbaI site is boxed.
Recovery and analysis of germline mutants:
ZFNs alone:
Simple ZFN-induced mutations were recovered as described (Bibikova et al. 2002). For each target gene, flies carrying the individual ZFNs, under heat-shock promoter control, were crossed. Four days later, the parents were removed, and the vial carrying their larval and embryonic offspring was immersed in a water bath at the indicated temperature for 1 hr, allowed to cool, and then returned to an incubator at 25°. Emerging adults were screened for the phenotype indicating the presence of the ZFNs and then mated with partners that would reveal the corresponding germline mutations. In the case of y, each male was crossed with two or three attached-X [C(1)DX] females, and each female was crossed with two or three y (FM6) males. Males from the former crosses and both males and females from the latter were screened for yellow body color. These were further crossed with the same partners to propagate the new mutations. Yellow males were selected, and DNA was prepared from three flies for PCR, using PureGene (Gentra Systems, Minneapolis) as recommended by the manufacturer. Amplified fragments were sequenced. For bw, heat-shocked candidates were crossed with Df(2R)bw-HB132, FrdHB132/CyO, bw*. Flies carrying a new bw mutation were then backcrossed and collected over Df(2R)bw-HB132, FrdHB132 for PCR analysis. In the case of ry, the diagnostic partners carried two ry balancers: TM2 rySC and MKRS. Progeny from this cross were then crossed to flies homozygous for the ry506 deletion (mwh ry506 e) and collected for PCR analysis.
With donor:
Appropriate crosses were performed to bring together the ZFN genes, the donor DNA, and the genes for FLP and I-SceI. Flies with heat-inducible FLP + I-SceI transgenes on chromosome 2 were obtained from Yikang Rong and Kent Golic (Rong and Golic 2000a). For y, the yA and yB transgenes were on chromosome 3, and the cross was:
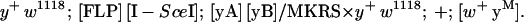 |
This configuration was preferable to the alternative in which one parent provided FLP, I-SceI, and yM, since carrying the donor sequence with FLP and I-SceI for multiple generations led to loss of yM, due to leaky expression of FLP and/or I-SceI (Golic and Golic 1996), even when the w+ marker was retained. (We have no direct evidence concerning leaky expression of ZFNs under heat-shock control, but we have not observed any deleterious effects of maintaining flies with these constructs at 18° or 25°, even when the same ZFNs were toxic upon heat shock.) The induction protocol and subsequent analysis were as described above. Emerging adults were screened for the appropriate phenotype [indicating the presence of the donor (w+) and the absence of the balancer (Sb+)]. After PCR, amplified fragments were tested for the presence of the diagnostic XhoI site, and some of the apparent NHEJ products (XhoI resistant) were sequenced.
Similar analyses were done for the ry target. In this case, the ryA and ryB transgenes were located on chromosome 3, and donor insertions on chromosomes 2 and 3 were used. Before experimental crosses were performed, it was necessary to mutate the ry gene marking the FLP insertion on chromosome 2 and to move all of the components from a w− background to a w+ background. Thus, male flies of the genotype [FLP, ry+] [I-SceI, v+]/sco; [ryA] [ryB]/+ were heat-shocked at 38° and crossed to CyO; TM2, rySC/MKRS females. Mutant flies of the genotype [FLP, ry−] [I-SceI, v+]/CyO; ry−/MKRS were recovered and used for further experiments. The experimental cross was then:
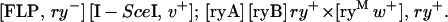 |
Heat shocks were performed at 37° or 38°, and subsequent analysis was performed as described above.
Three parameters are reported for the germline targeting experiments. First, we calculated the percentage of all females and males emerging from the heat shock that gave at least one new mutant offspring. Second, the average number of germline mutants per heat-shocked parent was determined, and an estimate of the percentage of all offspring that were mutant was made. Finally, the proportion of the new mutants that were products of HR and NHEJ was assessed. These figures appear most valuable to experimenters, who want to know, how many parents do I have to screen? And how many mutants of each type will I obtain?
Southern and Western blots:
Southern blot hybridizations were performed essentially as described by Gong and Golic (2003), using a DIG DNA labeling and detection kit supplied by Roche Diagnostics (Indianapolis). DNA was isolated from 10–15 adult male y mutant flies, digested with SalI or SalI + XhoI, subjected to electrophoresis in a 0.8% agarose gel, and transferred to a nylon membrane. This was probed with the 8-kb SalI fragment from pS/G, labeled with digoxygenin dNTPs, and developed as directed by the manufacturer. For the ry mutants, digestions were done with SalI and SalI + XbaI, and the probe was a 1.2-kb PCR fragment from pDM30, corresponding to sequences deleted by the ry506 mutation.
To monitor expression of the ZFNs, total protein was isolated in SDS sample buffer from 10 larvae at 3, 6, and 9 hr after heat shock. Samples were fractionated by electrophoresis in a 12% polyacrylamide gel, subjected to Western blotting with anti-FokI antiserum, obtained from S. Chandrasegaran, and developed with HRP-labeled goat anti-rabbit antiserum and a chemiluminescent substrate (Pierce Biotechnology, Rockford, IL).
RESULTS
Targeted NHEJ mutagenesis at bw and ry:
In previous experiments we produced new germline mutations in the Drosophila y gene by expression of a pair of ZFNs designed to target a sequence in that gene. To test whether the procedure could be extended to other genomic targets, we produced novel zinc-finger combinations for sites in bw (chromosome 2R 59E2-3) and ry (3R 87D9). These loci are autosomal and interstitial, whereas y is near a telomere on the X. The sequences of the sites chosen for targeting are shown in Table 1. In each case, the gene was searched for a sequence of the form 5′-(NNC)3N6(GNN)3-3′. This was based on the availability of zinc fingers that bind all of the GNN triplets with good affinity and specificity (Segal et al. 1999; Liu et al. 2002), the need to bind two three-finger proteins in opposite orientations to enforce dimerization of the cleavage domain (Smith et al. 2000), and the observation that a spacer of exactly 6 bp between binding sites is optimal (Bibikova et al. 2001).
The site identified in bw is in exon 7 and corresponds to codons 478–485 of the 675-amino-acid gene product. The site in ry lies in exon 3 and encompasses codons 1157–1164. This sequence is within the essential pterin–molybdenum cofactor-binding domain of the gene product (xanthine dehydrogenase), in which a number of null mutations have been characterized (Clark et al. 1986; Gray et al. 1991). Both bw and ry products function in the pathway leading to red eye pigment. We expected that cleavage-induced sequence alterations in either gene would be readily scored by the resulting mutant phenotype—i.e., brown eyes.
The ZFNs for each of these targets were introduced individually into the genome on P elements under the control of the hsp70 promoter. Insertions were mapped to specific chromosomes, and pairs were brought together in appropriate crosses (bwA × bwB; ryA × ryB). Offspring were heat-shocked 4 days after initiation of the cross. Eclosing adults were crossed to partners that would reveal new germline mutations in the gene of interest. Flies carrying the bwB transgene were unable to withstand heat-shock temperatures >35°. This is similar to the situation with the yA ZFN, which is discussed below. In the case of ry, neither ZFN was lethal at any heat-shock temperature, so inductions were performed at higher temperatures.
The data on ZFN-induced mutation frequencies at all three loci are presented in Table 2. As reported earlier for y, no germline mutants were recovered from females heat-shocked at 35°, while 5.7% of males yielded at least one new mutant, and mutants represented 0.4% of all offspring (Bibikova et al. 2002). Similarly at bw, no mutants emerged from female parents, and 2.4% of males gave mutants, representing ∼0.1% of all offspring.
TABLE 2.
ZFN-induced NHEJ mutation frequencies
| Female germline
|
Male germline
|
||||||
|---|---|---|---|---|---|---|---|
| Locus | hsT | Parents | Yielders | Mutants | Parents | Yielders | Mutants |
| y | 35° | 125 | 0 | 0 | 228 | 13 | 24 |
| bw | 35° | 228 | 0 | 0 | 245 | 6 | 6 |
| ry | 36.5° | 57 | 31 | 89 | 50 | 22 | 45 |
| 38° | 80 | 71 | 504 | 78 | 52 | 534 | |
For each experiment, the heat-shock temperature (hsT) is indicated. The number of female and male parents subjected to the heat shock is given, followed by the number that yielded at least one mutant offspring (Yielders), and the total number of mutants recovered. The data for y are from Bibikova et al. (2002).
At ry the yields of mutants were dramatically higher. At 36.5°, approximately half the heat-shocked parents, both females and males, produced at least one mutant offspring. The new mutants represent 3.5% (from females) and 0.9% (from males) of the total progeny. When the heat shock was performed at 38°, 89% of females and 67% of males yielded mutants, constituting 14.0 and 6.8% of total offspring, respectively. Since each offspring represents a single gamete, and therefore a single target locus, these frequencies also report the percentage of targets mutated in each case.
We conclude that zinc-finger combinations can be designed to target a variety of different sequences in disparate genes and chromosomal locations. Expression of the corresponding ZFN pairs leads to targeted cleavage and mutagenesis. In the best cases, the frequency of mutation is high enough to be practical for the recovery of new null alleles.
Gene targeting at ry:
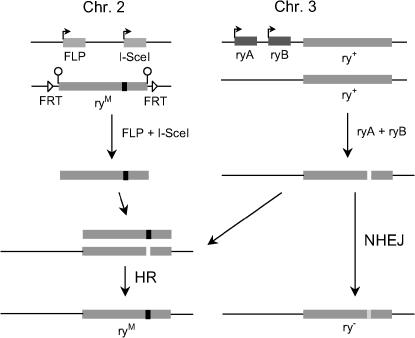
Because of the high frequency of cleavage and mutagenesis at ry, we were very eager to include a marked donor DNA and test the efficiency of gene targeting. The structure of the donor is shown in Figure 1 in relation to the ry gene. It has 4.16 kb of homology to the target, from a position within exon 2 to a point beyond the end of the gene. The ZFN target was replaced with two in-frame stop codons and an XbaI site as shown. These changes protect the donor from ZFN cleavage, ensure a null phenotype of the targeted product, and provide a convenient marker for molecular analysis. To present the donor in its most effective form (Bibikova et al. 2003), it was flanked with recognition sites for FLP and I-SceI. As demonstrated by Golic and co-workers, this allows for the generation of an extrachromosomal, linear molecule in situ (Rong and Golic 2000a; Gong and Golic 2003). The donor DNA was introduced into the genome on a P-element vector, and several different insertions were isolated.
The overall scheme of the targeting experiment is shown in Figure 2. The ry+ target is on both chromosomes 3; one chromosome 3 also carries the ryA and ryB ZFN transgenes. The marked donor (ryM) is on one chromosome 2, and the genes for FLP and I-SceI are on the other. The latter genes and those for the ZFNs are under the control of the heat-inducible Drosophila hsp70 promoter. Expression of FLP and I-SceI excises the donor and makes it linear, in an ends-out configuration relative to the target (Figure 2). When the ry+ gene is cleaved by ryA + ryB, its integrity can be restored by NHEJ (which may be mutagenic), by HR with the donor, by HR with an intact gene on the sister chromatid or homologous chromosome, or by simple religation (the latter two of which would leave ry+). New germline mutations, whether created by NHEJ or HR, were recovered in appropriate crosses of the heat-shocked flies. After PCR amplification, products of HR were identified by the presence of the diagnostic XbaI site in the ryM sequence. Those lacking this site were presumed to arise by NHEJ, and sequence analysis of many such products confirmed this (see Table 3).
Figure 2.
Initial set-up and expected products of the gene-targeting experiments at ry. The target gene (ry+) is on chromosome 3, and one of these chromosomes carries transgenes for the ZFNs (ryA and ryB). (The locations of these transgenes are not known.) The transgenes for FLP and I-SceI are on one second chromosome; the ryM donor is on the other. Angled arrows indicate heat-inducible promoters. Upon heat shock, the ZFNs make a DSB in the ry+ gene, as illustrated (only one is shown for simplicity). Expression of FLP will excise the donor as an extrachromosomal circle, and coexpression of I-SceI will convert it to an ends-out linear molecule. The break at ry can be restored to wild type, or it can acquire a mutant sequence either by NHEJ or by HR with the donor.
TABLE 3.
Characterization of NHEJ mutations
| Locus | Total | Simple deletions | Simple insertions | Deletions + insertions |
|---|---|---|---|---|
| y | 161 | 83 | 18 | 60 |
| bw | 13 | 11 | 1 | 1 |
| ry | 74 | 40 | 29 | 5 |
Results of DNA sequencing of independent mutations at each locus.
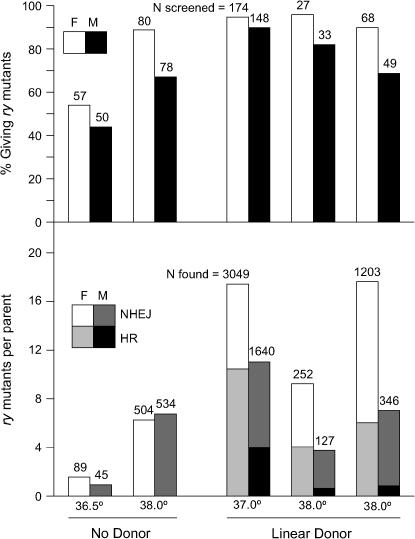
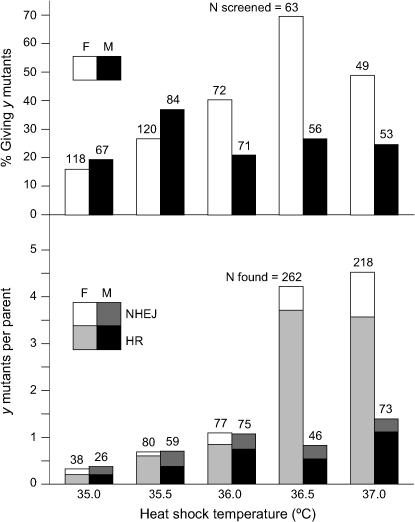
Figure 3 presents the results of three separate experiments and compares them to mutant yields in the absence of the donor. In the presence of a donor, nearly all of the heat-shocked flies, both males and females, gave at least one ry mutant offspring; some individuals gave >50. With a 37° heat shock, the average for all parents was 17.5 mutants from each female and 11.1 from each male. Molecular analysis was performed, using the XbaI site in the donor to distinguish HR from NHEJ products. Of 112 mutants from 39 female parents, 66 (59%) were products of HR, and 46 (41%) were from NHEJ. From 18 male parents, we identified 9 (30%) HR and 21 (70%) NHEJ products. These frequencies are included in Figure 3 (bottom).
Figure 3.
Gene targeting at ry. (Top) The percentage of heat-shocked flies that gave at least one ry mutant offspring is shown for both females (open bars) and males (solid bars). Above each bar in the histogram is the number of flies screened in each category. (Bottom) The number of ry mutants per parent, and the proportion that were the result of NHEJ and HR are given. Above each bar the total number of ry mutants recovered is shown for each case. Comparisons are made among independent experiments with ZFNs only (“No Donor,” data from Table 2) and those that included the ryM donor (“Linear Donor”) at several temperatures.
The yields and proportion of HR products were somewhat lower in two experiments with a 38° heat shock (Figure 3). In these experiments, siblings that lacked the ryA and ryB ZFNs, but carried the ryM donor, FLP, and I-SceI genes, were analyzed to determine what degree of stimulation was achieved above linear donor alone by target cleavage. Only 1 of 280 heat-shocked females, and none of 322 heat-shocked males yielded ry mutants. These figures are in the range of those reported by Golic and colleagues (Rong et al. 2002) at several genes. In comparison, target cleavage enhances the frequency of gene targeting substantially.
Some HR mutants were verified by Southern blot hybridization. There is a polymorphic XbaI site in the ry genes that we analyzed, which complicates the results. Nonetheless, 28 of 29 HR candidates yielded SalI and SalI + XbaI digests consistent with simple replacements (data not shown). The pattern from the final mutant was that expected for a duplication of the ry sequence, in which both copies carried the XbaI mutation from the donor. This may have been generated by recombination of the cleaved target with a circular or dimerized donor. Thus, it appears that the majority of HR events were simple replacements, as expected (Figure 2).
When multiple siblings were examined, it was quite common to find both HR and NHEJ mutants among the progeny of one parent. In some cases, more than one NHEJ sequence emerged from a single parent. This was true among the y mutants as well (below) and reflects the induction of independent mutagenic events.
Gene targeting at y:
Why are the yields of gene-targeting products so much higher at ry than those observed at y (Bibikova et al. 2003)? It seems likely that this is due to the level of expression of the ZFNs. Our initial studies at the y locus were done with heat shocks at 35° because of low viability at higher temperatures. In the course of those experiments, we noted that a pair of ZFNs inserted on chromosome 3 (AB10) survived higher heat-shock temperatures. Compared to siblings that did not inherit the ZFNs, AB10 flies showed no reduction in viability at 35.0° and 35.5° and yielded 35% as many offspring at 37.0°. As far as we are aware, the only differences between this and more sensitive strains are the integration sites of the ZFN transgenes; the sequences of the yA and yB genes should be identical. Experiments with a linear donor in AB10 flies were performed at increasing heat-shock temperatures.
The structure of the yM donor is shown in Figure 4; it is very similar in concept to that used for ry, but the diagnostic restriction site is for XhoI and the homology with the target is 6.7 kb. The yield of y mutants with the AB10 ZFN combination is shown in Figure 5. At 35.0° the numbers were quite close to those obtained previously (Bibikova et al. 2003). Between 15 and 20% of both males and females gave at least one y− offspring, and the average yield was between 0.3 and 0.4 mutants per heat-shocked parent. The percentage of total candidate progeny was estimated at 0.8% for females and 1.6% for males (the latter reflecting the fact that males have only one X chromosome). The difference between the genders was statistically significant (P = 0.0125).
Figure 5.
Gene targeting at y as a function of heat-shock temperature. Data are presented as in Figure 3, including the number of parents screened in each category (top) and the total number of new y mutants recovered (bottom).
As the heat-shock temperature increased, the mutant yields improved quite dramatically, particularly in the female germline. At 36.5° 70% of females and 27% of males gave mutants (Figure 5, top), with an average of 4.2 mutants for each heat-shocked female parent and 0.8 for each male (Figure 5, bottom). We estimate that the proportion of all offspring that were mutant was ∼10% from females and 3% from males. At 37.0° we recovered 4.45 new y mutants/female parent and 1.4/male, corresponding to ∼11 and 5% of all candidate offspring. The distribution of mutants among parents was nonuniform, with some induced flies yielding none and others giving multiple mutants. The largest cluster size that we observed was 33 y offspring from a single AB10 female heat-shocked at 37°.
The trends with temperature, except that of the percentage of males giving y offspring, were highly significant (P < 0.0001). At the highest temperatures the superiority of females to males both in percentage giving new mutants (P < 0.0001 at 36.5° and P = 0.0013 at 37.0°) and in mutant offspring per parent (P < 0.0001 at both temperatures) was very significant.
Molecular analysis was performed on a number of the new y mutants, using the diagnostic XhoI site in the donor to distinguish HR from NHEJ products. Of 954 y mutants from all temperatures, 488 were subjected to molecular analysis. A total of 380 (78%) were HR products in which the yM sequence replaced y+ at its normal site on the X chromosome. The remaining 108 (22%) were apparent products of NHEJ (XhoI resistant); some of these were sequenced to confirm this identification (see below). The proportions of each type for all temperatures and both parent sexes are superimposed on the total mutant yields in Figure 5 (bottom). As was found earlier (Bibikova et al. 2003), the fraction of HR products was typically somewhat higher in the female than in the male germline. In the experiments of this study, 82% of mutants from females and 66% from males were products of HR.
The structure of the apparent gene replacements was confirmed in a number of cases by Southern blot analysis. Using a y gene probe, all 20 examples analyzed showed a single 10-kb SalI fragment that was cleaved to the expected 4- and 6-kb fragments by XhoI (data not shown). No evidence of multiple insertions nor remnants of the donor were seen.
Donor experiments were not conducted at the bw locus because of the high lethality of the bwB ZFN.
Analysis of NHEJ products:
We determined nucleotide sequences of many of the ZFN-induced mutations at y, bw, and ry, following amplification of the altered targets by PCR. Only rarely did amplification fail with primers located within a few hundred base pairs of the cleavage site. This indicates that large deletions are uncommon. The summary in Table 3 includes y− sequences reported earlier (Bibikova et al. 2002) and examples from experiments both with and without donor DNAs. All of the alterations are localized to the immediate region of the zinc-finger target, and they have characteristics seen previously for NHEJ in Drosophila and other organisms (Takasu-Ishikawa et al. 1992; Staveley et al. 1995; Beall and Rio 1996; Dray and Gloor 1997; Jeggo 1998; Gloor et al. 2000; van Gent et al. 2001).
Most of the deletions were quite small, with 70% being ≤5 bp. Insertions were also small, most being ≤6 bp. A fairly common insertion at both y and ry can be attributed to fill-in of the 4-bp 5′ overhang left by ZFN cleavage (Smith et al. 2000), followed by blunt end joining. Deletions with insertions seem to be more common at y than at the other loci, but the significance of this observation has not been tested. The vast majority of mutations altered the reading frame, and since the targets are all in coding sequence, this readily explains the mutant phenotypes. At both y and ry, a small number of deletions and insertions retained the original reading frame, but added or deleted one or a few amino acids. Since these were also selected for their mutant phenotype, this indicates that the ZFN targets code for structurally sensitive portions of these gene products.
Lethality of the yA nuclease:
In earlier experiments with y and all of those with bw, the temperature used for induction of the ZFNs was limited to 35° because flies carrying the yA or bwB transgene survived poorly after heat shocks at higher temperatures (Bibikova et al. 2002, 2003). The yB, bwA, ryA, and ryB ZFNs conferred no lethality at any temperature. It seemed likely that the lethality was due to excessive cleavage when particular ZFNs were overexpressed. To test this, we made a single amino acid substitution in the FokI cleavage domain of yA. Replacement of aspartate with alanine at position 450 of natural FokI abolished cleavage without affecting DNA binding (Waugh and Sauer 1993). We made the corresponding mutation (D175A) in the coding sequence for yA and introduced it into the genome behind a heat-shock promoter, exactly as was done for yA originally. Two independent transformants were isolated and tested for lethality after a 37° heat shock. Neither showed any reduction in viability relative to flies having no ZFN transgene. Western blot analysis confirmed that the mutant protein was expressed at a level comparable to that of the original yA nuclease (data not shown). This demonstrates that the lethality of yA was likely due to excessive cleavage and rules out the alternative hypothesis (Bibikova et al. 2002) that simple binding at one or more sites recognized by the monomeric protein interferes with an essential chromosomal function, since the nuclease mutant, as well as the active yA protein, should still bind. Because the cleavage domain must dimerize to cut DNA, we presume that, at high levels of expression, the yA zinc fingers bind to noncanonical sequences leading to cleavage at unanticipated sites.
DISCUSSION
ZFN-induced gene targeting:
In this study we tested our ability to design ZFNs that would target cleavage to three different chromosomal loci in Drosophila. In each case expression of the paired ZFNs at early stages of development led to the recovery of progeny carrying break-induced NHEJ mutations precisely at the desired target. When a linear, extrachromosomal donor DNA was included, ZFN-induced cleavage stimulated homologous recombination, leading to incorporation of the marked donor at the target locus.
High frequencies of ZFN-induced gene targeting were obtained in both the male and female germlines. At the ry locus, >90% of induced parents gave at least one mutant offspring, and >10 mutants were recovered on average from each parent. The yields were somewhat lower at y, but in the best case half or more of the induced parents yielded mutants, and several new mutants emerged from each parent. These frequencies are high enough that, if reproduced at other loci, it will be possible to screen for the desired targeted products even when the introduced alteration causes no obvious phenotype. These levels of gene replacement were obtained despite the facts that only a single copy of the donor was present in each diploid genome and that competing repair pathways were available.
Cleavage of the target clearly makes an important contribution to the efficiency of gene targeting. The comparison was made directly for the y locus in our earlier study (Bibikova et al. 2003), where, in experiments conducted with a 35° heat shock, ZFN expression in the presence of a linear donor elevated the yield of targeted HR products 15-fold in the female germline and 60-fold in the male germline. In this study, at ry the enhancement was even greater. Another relevant comparison is between our data and those obtained by Golic and co-workers, who achieved gene targeting by producing linear donor DNA in situ, but without target cleavage (Rong and Golic 2000b, 2001; Rong et al. 2002; Gong and Golic 2003). This comparison is appropriate, since we adopted Golic and co-workers' approach and materials for our experiments. They obtained their highest frequencies at y, with both ends-in and ends-out donors, and targeting was much more efficient in females than in males. As shown in Table 4, ZFN-induced HR at the y locus in females was >50-fold higher than that achieved with the linear donor alone. A comparable ZFN-induced frequency was attained in males, several hundred-fold higher than without target cleavage. The frequency of HR at ry, a locus not tested by Golic and co-workers, was even higher, with 25% of all offspring of females being new mutants and 15% the products of HR. Males yielded 14% new mutants and ∼4% HR.
TABLE 4.
Comparison of gene-targeting frequencies
| Mutants as % of progeny
|
||||||
|---|---|---|---|---|---|---|
| Female germline
|
Male germline
|
|||||
| Target | Reference | hsT | HR | NHEJ | HR | NHEJ |
| Intact targets | ||||||
| y | Rong and Golic (2000a) | 38° | 0.2 | — | 0.01 | — |
| pug | Rong and Golic (2001) | 38° | 0.02 | — | 0.006 | — |
| y | Gong and Golic (2003) | 38° | 0.26, 0.12 | — | — | — |
| Cleaved targets | ||||||
| y | Bibikovaet al. (2003) | 35° | 0.39 | 0.15 | 1.4 | 0.8 |
| y | This study | 37° | 7.0 | 1.9 | 3.6 | 1.0 |
| ry | This study | 37° | 15.0 | 10.0 | 4.2 | 9.8 |
| 38° | 9.0 | 14.0 | 1.3 | 7.6 | ||
ZFN lethality:
In the cases of the y and bw targets, one of the two designed ZFNs was lethal when induced at high heat-shock temperatures. This lethality was shown to be due to excessive cleavage, presumably at noncanonical sites and presumably reflecting imperfect recognition specificity by the zinc fingers in question. Nonetheless, we were able to moderate the heat shock and find conditions that balanced lethality with effective target cleavage.
For y, we found a combination of yA and yB transgenes that allowed induction at higher temperatures. Since it is unlikely that the yA coding sequence is altered, we suspect that the improved viability is due to a chromosomal position effect on its expression. An inherently lower level of yA expression would be protective, but might still allow the smaller amount produced to be captured effectively at the canonical target in conjunction with excess (nonlethal) yB. Alternatively, the tissue specificity of yA expression may be altered such that effective levels are produced in the germline, but tolerable levels in the tissue(s) are responsible for its lethal effects.
Avoiding toxic side effects will be an important part of developing the ZFNs for broad use in gene targeting. One prospect is picking another target in or near the gene of interest. Another is redesigning the zinc fingers for greater specificity. An example of this approach is the use of additional fingers; Urnov et al. (2005) found that the use of well-designed four-finger ZFNs allowed targeting of the human IL-2Rγ gene without detectable cell loss. One could also conceivably modify the cleavage domain to reduce cutting by individual ZFNs. Finally, it might be possible to enhance the cell's or organism's ability to withstand nontargeted cleavage by enhancing repair capabilities or reducing the tendency to undergo apoptosis.
Effects of temperature and sex:
We were able to increase the targeting frequencies over those that we reported previously (Bibikova et al. 2003) by raising the temperature of the heat shock used to induce target cleavage and donor excision. Because all four transgenes—the two ZFNs, FLP, and I-SceI—were under heat-shock control, the levels of all four enzymes presumably increased at higher temperatures. For this reason it is difficult to say whether the beneficial effect is attributable to a single factor—target cleavage, donor excision, donor linearization—or a combination thereof. The fact that the frequency of NHEJ also rises with temperature indicates that an increase in ZFN production is at least an important contributing factor.
The effect of temperature was particularly dramatic in the female germline. At y the targeting frequency was higher in males at 35° (Bibikova et al. 2003), but females were considerably more productive at higher temperatures (Figure 5). In the case of ry, only high temperatures were used, and females gave higher yields. What is responsible for this sex difference? It is possible that the hsp70 promoter has a steeper temperature dependence in the female germline. It is also possible that male and female germline cells respond differently to increased levels of DSBs. The effect is not specific to the X chromosome, since autosomal loci behaved similarly to y: bw at low heat-shock temperature and ry at high temperature. The ultimate higher mutation frequency in females than in males and the higher proportion of HR events may reflect a greater propensity for DSB repair by homologous recombination in the female germline. The sex-specific differences may also reflect aspects of germline differentiation in males and females at the time of ZFN induction (Lin 1997). Whatever the explanation, the very high efficiencies observed at y in females and in both genders at ry demonstrate that the presence of a homologous chromosome does not interfere with interaction of the cleaved target with the linear donor, which was present in only a single copy.
In experiments at ry, we observed somewhat higher overall mutation frequency and a higher proportion of HR at 37° than at 38°. We can envision several possible explanations: (1) it is possible that the HR machinery is somewhat less active after the higher temperature induction; (2) the rate of NHEJ or of accurate ligation may be higher, so more double-strand breaks (DSBs) are captured by these competing repair processes; (3) the lifetime of the donor may be shorter, perhaps due to an increased rate of degradation. It is important to keep in mind, however, that the duration of the heat shock is only 1 hr, so presumably most of the repair proceeds at 25° regardless of the induction temperature. Thus, it is likely that the differences reflect the physiological response to stress, rather than temperature sensitivity of a specific repair process.
It seems clear that the targeting events that we observed occurred in premeiotic germline cells. Individual heat-shocked flies often gave multiple offspring with the same mutation, indicating clonal derivation from a single event. This was true for both NHEJ and HR products. Although there is essentially no crossing over in Drosophila male meiosis (Ashburner et al. 2005), premeiotic cells in the male germline are capable of supporting homologous recombination between donor and target sequences. The relationship between the mechanism of female meiotic recombination and the events seen here is not clear.
Mechanism of gene targeting:
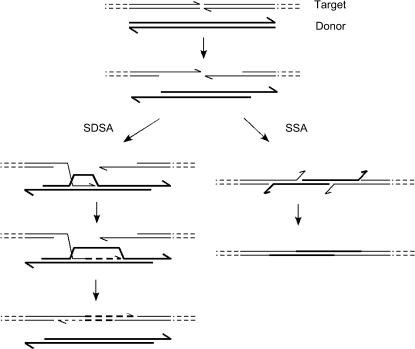
What is the molecular mechanism by which the HR events proceed? Several models can account for the observed products, and we illustrate two plausible ones in Figure 6. In essentially all cell types, molecular ends are resected by 5′-to-3′ exonuclease action, and we show this as the first step. Examination of repair products following DSB generation by P-element excision led Engels and colleagues to prefer a model called synthesis-dependent strand annealing (SDSA) (Nassif et al. 1994). In this process (Formosa and Alberts 1986; Pâques and Haber 1999) one of the single-stranded 3′ tails invades homologous sequences, where synthesis is primed from the free 3′-end. The extended end then withdraws and anneals to complementary sequences exposed by resection of the other, noninvading end, and the junctions are completed by DNA polymerase, nuclease, and ligase. Any homologous sequence can serve as template in this scheme: a sister chromatid, the homologous chromosome, or an ectopic donor. This mechanism is adequate to explain the gene-targeting products that we see following cleavage by the ZFNs.
Figure 6.
Mechanisms of homologous recombination that could explain gene targeting after ZFN cleavage, illustrated for the case of a linear donor. The target sequence is shown as thin lines, the donor as thick lines, with each line representing one DNA strand. After cleavage by the ZFNs, the target and donor ends are resected by a 5′-to-3′ exonuclease activity. In the SDSA mechanism (left), one of the resulting 3′ single-stranded tails invades homologous sequence in the donor and begins to copy (dashed line). After some synthesis, this end withdraws and pairs with the single-stranded tail from the other end at the original break, and any remaining gaps are filled by further DNA synthesis. In the SSA mechanism (right), resected ends from the target and linear donor anneal to each other by simple base pairing. Excess DNA is removed by nuclease action, and the junctions are completed by a combination of DNA synthesis and ligation. Because SDSA uses internal sequences of the donor as a template, it would work equally well with circular or integrated donor configurations, whereas SSA requires molecular ends on the donor as well as on the target.
We found that the configuration of the donor strongly affects its utilization for HR (Bibikova et al. 2003). An integrated donor was quite inefficient, an excised circular donor was better, and an extrachromosomal linear donor was best. This may reflect how readily ends created at the target can find the donor and use it as a template for SDSA. In the case of the linear donor, an alternative mechanism may also come into play. When all the participants have ends, recombination can proceed by single-strand annealing (SSA) (Lin et al. 1984; Carroll 1996) (Figure 6). One SSA event at the left end of the donor and another at its right end would be sufficient to incorporate it into the target. This process could certainly occur with the linear donor, but may also participate when I-SceI is not present if the circular donor is occasionally broken by some other means. Because neither strand invasion nor DNA synthesis is required for SSA, this mechanism has enzymatic requirements different from SDSA (Pâques and Haber 1999). It should be possible to distinguish them in the gene-targeting protocol by use of well-chosen DNA repair mutants.
Even as the frequency of HR products increased, there was always a measurable level of NHEJ mutagenesis. In many cases, NHEJ products represented a minority of the total, so there would be no difficulty in screening for a desired HR event. Since some genetic requirements for NHEJ have been defined—e.g., involvement of DNA ligase 4, Xrcc4, and the Ku70/Ku80 heterodimer (van Gent et al. 2001)—it may be possible to reduce this background even further by mutation or by transient inhibition of the unique factors. There is evidence, however, that at least two NHEJ pathways are present in some cell types, only one of which depends on known components. In some assays, the overall frequency of NHEJ was not much reduced by elimination of DNA ligase 4 or a Ku subunit, although the nature of the products was affected (Liang et al. 1996; Kabotyanski et al. 1998; Verkaik et al. 2002; Ma et al. 2003; Smith et al. 2003; Yu and Gabriel 2003; Guirouilh-Barbat et al. 2004).
ZFN-induced targeting in Drosophila:
Application of the ZFN approach to other genes in Drosophila should be quite straightforward. The chosen target sequence can be searched for sites that can be attacked with combinations of existing zinc fingers. Because the fingers that bind GNN triplets have proved most effective and the optimum distance between binding sites is 6 bp (Bibikova et al. 2001), we typically search for a target of the form (NNC)3N6(GNN)3. The required fingers can be located in the literature (Segal et al. 1999; Liu et al. 2002), and the corresponding coding sequences created by synthesis (Segal 2002) or by modification of existing cloned sequences. Fingers directed to ANN and CNN triplets have also been described (Dreier et al. 2001, 2005), but they appear to have less specificity in simple binding analyses, and few of them have been tested in vivo. Linkage of finger sets to the FokI cleavage domain and transfer to a transformation vector are facilitated by the Gateway constructs that we have produced (materials and methods). Simple heat-induced expression of a pair of ZFNs is sufficient to create targeted mutations, while inclusion of an intentionally modified homologous donor DNA allows introduction of altered sequences of any desired form.
Although it appears likely that a broad range of targets can be attacked successfully with this approach, a number of issues remain to be explored regarding the potential of ZFN-induced gene targeting in Drosophila.
How much homology is required between donor and target for efficient gene replacement? Conceivably, donors even smaller than the 6.7- and 4.2-kb examples described here could be used.
How far from the cleavage site can a change in the donor be located and still be incorporated efficiently at the target? When a break in the target was induced by P-element excision, Gloor et al. (1991) found that polymorphisms lying some distance from the break were captured at lower, but still useful, frequencies. When both donor and target have ends, the answer will depend on the mechanism for recombination, since at least one example of SSA showed essentially equal incorporation over substantial distances (Carroll et al. 1994; Lehman et al. 1994).
Can nontranscribed sequences be targeted as effectively as those in known genes? Potential effects of chromatin structure on zinc-finger binding (Liu et al. 2001) have not yet been explored.
Can the procedure be simplified by injecting the necessary components into Drosophila embryos, much as P-element injections are performed? This requires both finding means to deliver the ZFNs and the donor DNA and the cooperation of embryonic pole cells in supporting homologous recombination.
Applications of ZFNs to other systems:
Because DSBs stimulate HR and NHEJ in essentially all cell types, the ZFN approach to enhanced gene targeting should be broadly useful (Porteus and Carroll 2005). Initial applications to plants (Lloyd et al. 2005; Wright et al. 2005) and mammalian cells (Porteus and Baltimore 2003) have been reported. Particularly intriguing is the report demonstrating efficient manipulation of the human IL-2Rγ gene in cultured cells (Urnov et al. 2005). These investigators made several modifications of the basic technique and achieved very high frequencies in targeting this gene of therapeutic importance. They used a pair of four-finger ZFNs based two-finger units (Moore et al. 2001) and an expanded library of recognition capabilities, and the binding sites were separated by only 5 bp. While issues of efficient delivery, broad applicability, specificity, and toxicity remain to be explored more thoroughly, these results suggest that ZFNs will find wide utility in stimulating gene targeting.
Acknowledgments
We are grateful to S. Chandrasegaran for providing the anti-FokI antiserum, to Wei Gong and Kent Golic for the gift of the pw30 vector and advice on Southern blots, to Jeff Sekelsky for providing pP{whiteOut2}, to Ana Bozas and Josh Checketts for help with DNA isolation and analysis, to Jason Morton for advice, to John Staton for help with some steps of cloning, and to Scott Pendley for initiating the construction of the mutant yA gene. Gary Drews, Kent Golic, and David Segal made helpful comments on various versions of the manuscript. This work was supported by National Institutes of Health award R01 GM58504 (to D.C.) and in part by the University of Utah Cancer Center support grant.
References
- Alwin, S., M. B. Gere, E. Gulh, K. Effertz, C. F. Barbas, III et al., 2005. Custom zinc-finger nucleases for use in human cells. Mol. Ther. 12: 610–617. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., K. G. Golic and R. S. Hawley, 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Beall, E. L., and D. C. Rio, 1996. Drosophila IRBP/Ku70 corresponds to the mutagen-sensitive mus309 gene and is involved in P-element excision in vivo. Genes Dev. 10: 921–933. [DOI] [PubMed] [Google Scholar]
- Bibikova, M., D. Carroll, D. J. Segal, J. K. Trautman, J. Smith et al., 2001. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 21: 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, M., M. Golic, K. G. Golic and D. Carroll, 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova, M., K. Beumer, J. K. Trautman and D. Carroll, 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed]
- Blancafort, P., D. J. Segal and C. F. Barbas, III, 2004. Designing transcription factor architectures for drug discovery. Mol. Pharmacol. 66: 1361–1371. [DOI] [PubMed] [Google Scholar]
- Carroll, D., 1996. Homologous genetic recombination in Xenopus: mechanism and implications for gene manipulation. Prog. Nucleic Acid Res. Mol. Biol. 54: 101–125. [DOI] [PubMed] [Google Scholar]
- Carroll, D., C. W. Lehman, S. Jeong-Yu, P. Dohrmann, R. J. Dawson et al., 1994. Distribution of exchanges upon homologous recombination of exogenous DNA in Xenopus laevis oocytes. Genetics 138: 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulika, A., A. Perrin, B. Dujon and J.-F. Nicolas, 1995. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. H., A. J. Hilliker and A. Chovnick, 1986. Genetic analysis of the right (3′) end of the rosy locus in Drosophila melanogaster. Genet. Res. 47: 109–116. [DOI] [PubMed] [Google Scholar]
- Cohen-Tannoudji, M., S. Robine, A. Choulika, D. Pinto, F. El Marjou et al., 1998. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol. Cell. Biol. 18: 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoho, G., M. Jasin and P. Berg, 1998. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cell. Mol. Cell. Biol. 18: 4070–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray, T., and G. B. Gloor, 1997. Homology requirements for targeting heterologous sequences during P-induced gap repair in Drosophila melanogaster. Genetics 147: 689–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreier, B., R. R. Beerli, D. J. Segal, J. D. Flippin and C. F. Barbas, III, 2001. Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J. Biol. Chem. 276: 29466–29478. [DOI] [PubMed] [Google Scholar]
- Dreier, B., R. P. Fuller, D. J. Segal, C. Lund, P. Blancafort et al., 2005. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in construction of artificial transcription factors. J. Biol. Chem. 280: 35588–35597. [DOI] [PubMed] [Google Scholar]
- Formosa, T., and B. M. Alberts, 1986. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47: 793–806. [DOI] [PubMed] [Google Scholar]
- Geyer, P. K., and V. G. Corces, 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1: 996–1004. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., N. A. Nassif, D. M. Johnson-Schlitz, C. R. Preston and W. R. Engels, 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253: 1110–1117. [DOI] [PubMed] [Google Scholar]
- Gloor, G. B., J. Moretti, J. Mouyal and K. J. Keeler, 2000. Distinct P-element excision products in somatic and germline cells of Drosophila melanogaster. Genetics 155: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golic, M. M., and K. G. Golic, 1996. A quantitative measure of the mitotic pairing of alleles in Drosophila melanogaster and the influence of structural heterozygosity. Genetics 143: 385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, W. J., and K. G. Golic, 2003. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. USA 100: 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M., A. Charpentier, K. Walsh, P. Wu and W. Bender, 1991. Mapping point mutations in the Drosophila rosy locus using denaturing gradient gel blots. Genetics 127: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat, J., S. Huck, P. Bertrand, L. Pirzio, C. Desmaze et al., 2004. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell 14: 611–623. [DOI] [PubMed] [Google Scholar]
- Jamieson, A. C., J. C. Miller and C. O. Pabo, 2003. Drug discovery with engineered zinc finger proteins. Nat. Rev. Drug Discov. 2: 361–368. [DOI] [PubMed] [Google Scholar]
- Jeggo, P. A., 1998. DNA breakage and repair. Adv. Genet. 38: 185–218. [DOI] [PubMed] [Google Scholar]
- Kabotyanski, E. B., L. Gomelsky, J.-O. Han, T. D. Stamato and D. B. Roth, 1998. Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res. 26: 5333–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.-G., J. Cha and S. Chandrasegaran, 1996. Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc. Natl. Acad. Sci. USA 93: 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B. A., B. T. Amann, V. J. Kilfoil, D. L. Merkle and J. M. Berg, 1991. A consensus zinc finger peptide: design, high-affinity metal binding, a pH-dependent structure, and a His to Cys sequence variant. J. Am. Chem. Soc. 113: 4518–4523. [Google Scholar]
- Lehman, C. W., S. Jeong-Yu, J. K. Trautman and D. Carroll, 1994. Repair of heteroduplex DNA in Xenopus laevis oocytes. Genetics 138: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, F., P. J. Romanienko, D. T. Weaver, P. A. Jeggo and M. Jasin, 1996. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl. Acad. Sci. USA 93: 8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F.-L., K. Sperle and N. Sternberg, 1984. Model for homologous recombination during transfer of DNA into mouse L cells: role for the ends in the recombination process. Mol. Cell. Biol. 4: 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H., 1997. The Tao of stem cells in the germline. Annu. Rev. Genet. 31: 455–491. [DOI] [PubMed] [Google Scholar]
- Liu, P.-Q., E. J. Rebar, L. Zhang, Q. Liu, A. C. Jamieson et al., 2001. Regulation of an endogenous locus using a panel of designed zinc finger proteins targeted to accessible chromatin regions. Activation of vascular endothelial growth factor A. J. Biol. Chem. 276: 11323–11334. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Z. Q. Xia, X. Zhong and C. C. Case, 2002. Validated zinc finger protein designs for all 16 GNN DNA triplet targets. J. Biol. Chem. 277: 3850–3856. [DOI] [PubMed] [Google Scholar]
- Lloyd, A., C. L. Plaisier, D. Carroll and G. N. Drews, 2005. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.-L., E. M. Kim, J. E. Haber and S. E. Lee, 2003. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 23: 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M., A. Klug and Y. Choo, 2001. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc. Natl. Acad. Sci. USA 98: 1437–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif, N. A., J. Penney, S. Pal, W. R. Engels and G. B. Gloor, 1994. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14: 1613–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques, F., and J. E. Haber, 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich, N. P., and C. O. Pabo, 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å resolution. Science 252: 809–817. [DOI] [PubMed] [Google Scholar]
- Porteus, M. H., and D. Baltimore, 2003. Chimeric nucleases stimulate gene targeting in human cells. Science 300: 763. [DOI] [PubMed]
- Porteus, M. H., and D. Carroll, 2005. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 23: 967–973. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. a Gene targeting by homologous recombination in Drosophila. Science 288: 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. b Site-specific recombination for the genetic manipulation of transgenic insects, pp. 53–75 in Insect Transgenesis: Methods and Applications, edited by A. M. Handler and A. A. James. CRC Press, Boca Raton, FL.
- Rong, Y. S., and K. G. Golic, 2001. A targeted gene knockout in Drosophila. Genetics 157: 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16: 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet, P., F. Smih and M. Jasin, 1994. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 14: 8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, D. J., 2002. The use of zinc finger peptides to study the role of specific factor binding sites in the chromatin environment. Methods 26: 76–83. [DOI] [PubMed] [Google Scholar]
- Segal, D. J., B. Dreier, R. R. Beerli and C. F. Barbas, III, 1999. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl. Acad. Sci. USA 96: 2758–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smih, F., P. Rouet, P. J. Romanienko and M. Jasin, 1995. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 23: 5012–5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J., M. Bibikova, F. G. Whitby, A. R. Reddy, S. Chandrasegaran et al., 2000. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 28: 3361–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J., E. Riballo, B. Kysela, C. Baldeyron, K. Manolis et al., 2003. Impact of DNA ligase IV on the fidelity of end joining in human cells. Nucleic Acids Res. 31: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staveley, B. E., T. R. Heslip, R. B. Hodgetts and J. B. Bell, 1995. Protected P-element termini suggest a role for inverted-repeat-binding protein in transposase-induced gap repair in Drosophila melanogaster. Genetics 139: 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu-Ishikawa, E., M. Yoshihara and Y. Hotta, 1992. Extra sequences found at P element excision sites in Drosophila melanogaster. Mol. Gen. Genet. 232: 17–23. [DOI] [PubMed] [Google Scholar]
- Urnov, F. D., J. C. Miller, Y.-L. Lee, C. M. Beausejour, J. M. Rock et al., 2005. Highly efficient endogenous gene correction using designed zinc-finger nucleases. Nature 435: 646–651. [DOI] [PubMed] [Google Scholar]
- van Gent, D. C., J. H. J. Hoeijmakers and R. Kanaar, 2001. Chromosome stability and the double-strand break connection. Nat. Rev. Genet. 2: 196–206. [DOI] [PubMed] [Google Scholar]
- Vasquez, K. M., K. Marburger, Z. Intody and J. H. Wilson, 2001. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. USA 98: 8403–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaik, N. S., E. E. Esveldt-van Lange, D. van Heemst, H. T. Bruggenwirth, J. H. J. Hoeijmakers et al., 2002. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 32: 701–709. [DOI] [PubMed] [Google Scholar]
- Waugh, D. S., and R. T. Sauer, 1993. Single amino acid substitutions uncouple the DNA binding and strand scission activities of FokI endonuclease. Proc. Natl. Acad. Sci. USA 90: 9596–9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, D. A., J. A. Townsend, R. J. Winfrey, Jr., P. A. Irwin, J. Rajagopal et al., 2005. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 44: 693–705. [DOI] [PubMed] [Google Scholar]
- Yu, X., and A. Gabriel, 2003. Ku-dependent and Ku-independent end-joining pathways lead to chromosomal rearrangements during double-strand break repair in Saccharomyces cerevisiae. Genetics 163: 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]