Abstract
The idea that natural hybridization has served as an important force in evolutionary and adaptive diversification has gained considerable momentum in recent years. By combining genome analyses with a highly selective field experiment, we provide evidence for adaptive trait introgression between two naturally hybridizing Louisiana Iris species, flood-tolerant Iris fulva and dry-adapted I. brevicaulis. We planted reciprocal backcross (BC1) hybrids along with pure-species plants into natural settings that, due to a flooding event, favored I. fulva. As expected, I. fulva plants survived at much higher rates than I. brevicaulis plants. Backcross hybrids toward I. fulva (BCIF) also survived at significantly higher rates than the reciprocal backcross toward I. brevicaulis (BCIB). Survivorship of BCIB hybrids was strongly influenced by the presence of a number of introgressed I. fulva alleles located throughout the genome, while survivorship in the reciprocal BCIF hybrids was heavily influenced by two epistatically acting QTL of opposite effects. These results demonstrate the potential for adaptive trait introgression between these two species and may help to explain patterns of genetic variation observed in naturally occurring hybrid zones.
THE process of introgressive hybridization whereby hybrids act as intermediaries for gene flow via repeated backcrossing to pure-species plants was first described by Anderson and Hubricht (1938). Anderson (1949) later postulated that this introduction of novel genetic material could result in the transfer of adaptive traits. Since interspecific hybrids have been documented in a broad taxonomic array of plants and animals (Ellstrand et al. 1996; Dowling and Secor 1997), introgressive hybridization has the potential to promote adaptive evolution in a wide variety of organisms.
Although hybridization can theoretically result in the transfer of adaptations, several barriers to introgression can prevent this process from occurring. Species pairs have often evolved numerous prezygotic and postzygotic isolating mechanisms that greatly reduce the chance for hybrid formation (Schemske 2000; Ramsey et al. 2003; N. H. Martin and J. H. Willis, unpublished results). Clearly, however, if hybrids are to act as intermediaries for gene flow, prezygotic and postzygotic barriers must not be so strong that they completely exclude the opportunity for fertile hybrid formation. Those regions of the genome contributing to both prezygotic and postzygotic isolation will likely not cross species barriers since they will be selected against. Because there are multiple barriers to hybrid formation, and the genetic basis of each barrier is typically polygenic (reviewed by Coyne and Orr 2004), a large number of genes (and therefore a large number of entire chromosomal blocks due to linkage) are potentially resistant to introgression. Despite these strong impediments to hybridization and gene flow, a remarkable number of studies have documented the introgression of molecular markers across species boundaries (e.g., Arnold et al. 1987, 1991; Rieseberg et al. 1990; Martinsen et al. 2001; Abbott et al. 2003; Sweigart and Willis 2003; Payseur and Nachman 2005; Won and Hey 2005). Yet, it is still largely unknown whether the introgression of these molecular markers is due to their linkage with genes [or quantitative trait loci (QTL)] coding for selectively favored traits.
Numerous examples of adaptive trait introgression have been hypothesized in a variety of plants (Stutz and Thomas 1964; Arnold and Bennett 1993; Martinsen et al. 2001; Bradshaw and Schemske 2003), animals (Lewontin and Birch 1966; Parsons et al. 1993; Morrow et al. 2000), and microbes (Kroll et al. 1998; Machado and Ayala 2001; Smoot et al. 2002). However, in many of these examples it is difficult to distinguish between the effects of introgression and those of joint retention of ancestral character states (i.e., incomplete lineage sorting) and/or convergent evolution. One study that yielded strong evidence for adaptive introgression involved the analysis of the genetic architecture of morphological traits in hybridizing Helianthus annuus and H. debilis ssp. cucumerifolius (Kim and Rieseberg 1999). East Texas populations of H. annuus (i.e., H. annuus ssp. texanus) are hypothesized to be derived from introgression with H. debilis ssp. cucumerifolius (Heiser 1951). Kim and Rieseberg's (1999) study revealed 56 QTL that affected 15 divergent morphological traits between these two species. Of those QTL, only 11 appeared to be tightly linked to sterility/inviability factors. The remaining 45 revealed no linkage to reproductive barriers and the introgression of only a small number of these QTL could recover the phenotype of H. annuus ssp. texanus. Unfortunately, this study was not designed such that the fitness of alternate QTL states could be tested. However, given Rieseberg et al.'s (2003) demonstration of the potential for natural hybridization among Helianthus lineages to give rise to novel adaptations and thus new taxa, it will not be surprising if future analyses support the role of adaptive trait introgression in the derivation of H. annuus ssp. texanus.
For a definitive test of adaptive trait introgression, one needs to demonstrate that introgressed traits cause an increase in the fitness of individuals possessing those novel traits. Here, we test directly for adaptive trait introgression under natural conditions between the Louisiana Iris species, Iris fulva and I. brevicaulis. Of interesting historical note, we perform these tests for adaptive trait introgression in the same species complex cited by Edgar Anderson (1949) as the “typical example” in his classic monograph Introgressive Hybridization. This study was performed using reciprocal backcross (BC1) hybrids and pure-species plants that were exposed to highly selective field conditions (i.e., a flood). Extended natural submersion/flooding reduced greatly the survivorship of experimental plants. We performed genome scans using both single marker-trait associations and QTL analyses to answer the following questions: Did survivorship have a significant genetic component, and if so, what is the genetic architecture of survivorship? In particular, were a large number of regions scattered throughout the genome responsible for producing the observed patterns of survivorship, or are there only a few such regions? Finally, is survivorship enhanced by the introgression of heterospecific alleles, thus reflecting adaptive trait introgression?
I. fulva and I. brevicaulis are two species broadly sympatric throughout lowland swamps in the North American Mississippi River drainage system (Viosca 1935). Randolph et al. (1961) determined the haploid chromosome count for both species to be 21. These long-lived perennials reproduce both sexually and clonally, and both species have a mixed mating system with large, showy flowers that reveal differing pollination syndromes (Viosca 1935; Cruzan and Arnold 1994; Cruzan et al. 1994; Burke et al. 2000; Wesselingh and Arnold 2000). Both allopatric and sympatric populations can be encountered throughout the ranges of the species. In locally sympatric populations, a number of prezygotic reproductive isolating mechanisms restrict gene flow. First, divergent flowering phenologies limit the initial production of F1 hybrids, with I. fulva initiating flowering about a month earlier than I. brevicaulis in southern Louisiana populations (Cruzan and Arnold 1994). Second, divergent floral phenotypes affect pollinator behavior. I. fulva has a suite of floral traits commonly observed in other hummingbird-pollinated systems, including red floral parts and anthers that extend beyond the receptive stigma and is preferentially visited by this pollen vector (Emms and Arnold 2000; Wesselingh and Arnold 2000). I. brevicaulis, on the other hand, possesses floral characters presumably adapted for attracting bee pollinators, including blue petals and sepals with white and yellow nectar guides, recessed anthers, and sturdy flower components that serve as landing pads for bumblebees. Consistent with this floral display, I. brevicaulis is visited mainly by bumblebees (Wesselingh and Arnold 2000). Some amount of postzygotic isolation is also observed between these two species in the form of reduced hybrid viability and fertility (Cruzan and Arnold 1994; Burke et al. 1998; Bouck 2004). Finally, and of particular significance for this study, habitat isolation also exists. I. fulva occurs in flooded bayou margins and swamps, while I. brevicaulis is often found in drier, hardwood forests (Cruzan and Arnold 1993; Johnston et al. 2001). Despite the presence of multiple pre- and postzygotic barriers, these species form hybrid zones where they co-occur, and extensive introgression has been documented in natural populations (Cruzan and Arnold 1993; Johnston et al. 2001). It is, however, unclear whether this introgression is adaptive.
MATERIALS AND METHODS
Construction of mapping populations:
One wild-collected individual each from I. fulva (If174, collected from Terrebonne Parish, Louisiana) and I. brevicaulis (Ib72, collected from St. Martinville Parish, Louisiana) was used to make reciprocal, interspecific backcross (BC1) populations. To minimize the within-species genetic variation, clones of these parental individuals were used as both F1 parents and as recurrent backcross parents. F1 plants were produced using Ib72 as the maternal parent and If174 as the pollen parent. Two F1 plants, designated as F1(2) and F1(3), were used as pollen parents to produce the BC1 hybrids. F1(2) was used to pollinate several clones of If174, and F1(3) was used as the pollen donor for several clones of Ib72. Several hundred I. fulva backcross (BCIF) and I. brevicaulis backcross (BCIB) hybrid seeds were ultimately produced in 1999. Seeds were germinated and the resulting plants were maintained in greenhouses at the University of Georgia. In the fall of all subsequent years (through 2004), backcross, F1, I. fulva, and I. brevicaulis plants were repotted by removing a single rhizome. Plants that survived throughout these four years in the greenhouse were used in this experiment.
Construction of linkage maps:
Two independent linkage maps were constructed using the Iris retroelement (IRRE) transposon display marker system designed by Kentner et al. (2003) and the linkage mapping program Mapmaker 3.0 (Lander et al. 1987; Lincoln et al. 1992). Bouck et al. (2005) provide a detailed description of the map construction protocol. In short, BCIB genotypes were determined for 414 IRRE transposon display markers (N = 230 plants), and BCIF genotypes were determined for 309 IRRE markers (N = 120 plants). The two linkage maps are independent because transposon display markers possess dominant inheritance. The presence or absence of dominant I. fulva markers in BCIB individuals was ascertained, and these markers were used to create the I. fulva map. The presence or absence of dominant I. brevicaulis markers in BCIF individuals was ascertained, and these markers were utilized for creating the I. brevicaulis map. Linkage groups identified in both maps were labeled in order of the largest to the smallest lengths (in centimorgans, LG1–LG22). Shared designations between linkage groups (LGs) from the two maps do not imply homology. The I. fulva map (derived from BCIB hybrids) consists of 142 framework markers and 22 linkage groups, with an average marker spacing of 12 cM. The I. brevicaulis map (derived from BCIF hybrids) consists of 108 framework markers and 22 linkage groups, with an average marker spacing of 13 cM.
Assaying field survival:
The field experiment was performed in a hardwood forest and associated bayou habitats typical for I. brevicaulis and I. fulva, respectively. Two experimental plots were established bordering the Choupique Bayou located in the U. S. Army Corps of Engineers Atchafalaya Basin Floodway in Louisiana. The maximum elevation of the experimental populations was ∼4.9 m above mean sea level (msl), while the minimum elevation was ∼3.7 m above msl. On October 22–28, 2004, we separated three to four rhizomes from each BCIF and BCIB plant (207 and 239 individuals, respectively), as well as 30 rhizomes each from I. fulva and I. brevicaulis plants. We transplanted one to two of these rhizomes each into two separate rectangular plots located ∼400 m apart. Each plot spanned hardwood forest and bayou habitats. Within each plot we randomly assigned positions for the various genotypes, spacing them 0.5 m apart. At this site, small changes in elevation can have large effects on the amount of water availability for plants. Indeed, during this unusually wet season, all plants below 4.3 m msl were inundated continuously. Because of this, no plants below this level, regardless of genotype, survived. None of those individuals were included in this study. Plants above this elevation experienced short, periodic episodes during which they were exposed to the air, thus allowing establishment and growth. In total, 185 BCIB hybrid genotypes and 209 BCIF hybrid genotypes were planted above the 4.3 msl elevation. Since multiple replicates were used for each genotype, a total of 416 separate BCIB, 357 BCIF, 11 I. fulva, and 13 I. brevicaulis ramets were planted above the 4.3-m msl elevation and assayed for survival on April 19, 2005, when water levels had subsided enough so that plots were accessible. Chi-square 2 × 2 contingency tables were used to compare the survivorship of two sets of genotypic classes: I. fulva with I. brevicaulis and BCIB with BCIF. For QTL analyses (below) “survivorship” is defined as the proportion of all replicates above the 4.3-m msl elevation that survived the flooding.
QTL analysis of survivorship:
Separate genome scans for QTL affecting survivorship were performed on each BC1 map using composite interval mapping (CIM; Zeng 1993, 1994) followed by refinement with the multiple interval mapping method (MIM; Kao et al. 1999; Zeng et al. 1999). Both analyses were implemented in the program Windows QTL Cartographer version 2.5 (WinQTLCart 2.5; Basten et al. 2005). CIM tests were performed at 1-cM steps, with a 10-cM window size to exclude closely linked cofactors. Three cofactors for each mapping population tested were chosen by the CIM program through forward and backward regression. Genomewide threshold values were determined at the 5% level by running 1000 permutations for each map (Churchill and Doerge 1994; Doerge and Churchill 1996). MIM tends to have more power and precision for detecting QTL (Kao et al. 1999), so QTL detected in CIM analysis were then used as an initial model for MIM. If no QTL were found with CIM searches, we used forward and backward regression selection on all markers with partial r2 probability set at 0.01 as the initial model for MIM. MIM was used to refine QTL positions, search for additional QTL, search for epistatic effects among detected QTL, and finally estimate individual QTL effects and the proportion of phenotypic variance explained by the QTL. Tests in MIM were performed at 1-cM steps, with Bayesian information criterion (BIC) model selection and the penalty function set to c(n) = ln(n). The 1-LOD support limits for each QTL were calculated around the most likely QTL position.
Single-marker analysis:
In addition to multiple interval mapping, we performed single-point regression analyses on all mapped markers at P < 0.05 significance levels. These analyses were also performed in WinQTLCart 2.5 (Basten et al. 2005). These tests allowed us to determine the direction of the effects of each marker (positive indicates increase in survivorship; negative indicates decrease in survivorship) and whether marker–trait association was significant.
For each of the backcross hybrid populations, we counted the number of positive and the number of negative marker–trait associations (regardless of whether the associations were significant, and ignoring linkage). We then utilized these observed data in χ2 tests, with the expected numbers of positive and negative associations being equal, to determine whether introgressed markers generally promote or reduce survivorship in each backcross. We also asked whether linkage groups tended to promote or reduce survivorship. We assigned linkage groups as being either positively or negatively associated with survivorship (on the basis of whether a majority of a linkage group's markers revealed either positive or negative associations) and then performed χ2 tests with the expected number of positive and negative associations being equal. Both of these approaches have weaknesses. Analyzing all markers as if they were independent (the first χ2 test described) ignores the fact that multiple markers are likely linked to the same phenotypic QTL, while analyzing linkage groups independently (the second χ2 test described) ignores the fact that multiple QTL may be present on the same linkage group. However, if both tests reveal the same patterns, we conclude that there is likely an underlying and related biological cause.
RESULTS
Survivorship:
The time period during which this study was conducted was unusually wet, and extremely high water levels in the Atchafalaya River basin resulted in repeated and prolonged submersion of Iris ramets for over 3 months. This had a severe impact on survivorship, such that, of 1040 experimental ramets planted below 4.3 m msl, none survived in either plot. None of those plants are included in this analysis. Ramets planted in both plots above this elevation are combined for the survivorship, QTL, and single-marker analyses. Even ramets planted above 4.3 m msl and included in this study survived at very low rates such that, of 798 ramets, only 59 (7.39%) survived.
We found significant differences among the four genotypic classes (i.e., I. fulva, I. brevicaulis, BCIB, and BCIF) in frequency of survivorship. Only 11 I. fulva plants and 13 I. brevicaulis plants were assayed for survival in this experiment. Despite this low sample size, it was sufficient to reveal significant differences in survivorship between the two species. Chi-square 2 × 2 contingency tests revealed that I. fulva survived at a significantly higher frequency (3 of 11 individuals survived) than I. brevicaulis (0 of 13 individuals; χ2 = 4.05, 1 d.f., P = 0.044; Table 1). Furthermore, I. fulva survivorship was significantly higher than either BCIB (χ2 = 8.86, 1 d.f., P = 0.003; Table 1) or BCIF hybrids (χ2 = 3.95, 1 d.f., P = 0.047; Table 1). Consistent with the parental responses, backcross hybrids toward I. fulva survived at significantly higher rates than reciprocal hybrids toward I. brevicaulis (χ2 = 4.05, 1 d.f., P = 0.048; Table 1).
TABLE 1.
I. fulva, I. brevicaulis, BCIB, and BCIF (BC1 hybrids toward either species) that lived or died in two experimental plots
| Cross | Alive | Dead | Fraction survival |
|---|---|---|---|
| BCIB | 23 | 393 | 0.055288 |
| BCIF | 33 | 325 | 0.092179 |
| I. fulva | 3 | 8 | 0.272727 |
| I. brevicaulis | 0 | 13 | 0 |
QTL identification:
In the BCIF mapping population, CIM identified two putative QTL that exceeded the permutation-based detection threshold of likelihood ratio = 9.6. They were located at 45.5 cM on linkage group 6 (LG6, QTL 1) and at 18.4 cM on linkage group 13 (LG13, QTL 2, Figure 1). These QTL were used as the initial model for MIM. MIM did not detect any additional significant QTL. However, MIM did reveal significant negative epistasis between these two additive QTL using a very stringent BIC model selection criterion (Table 2).
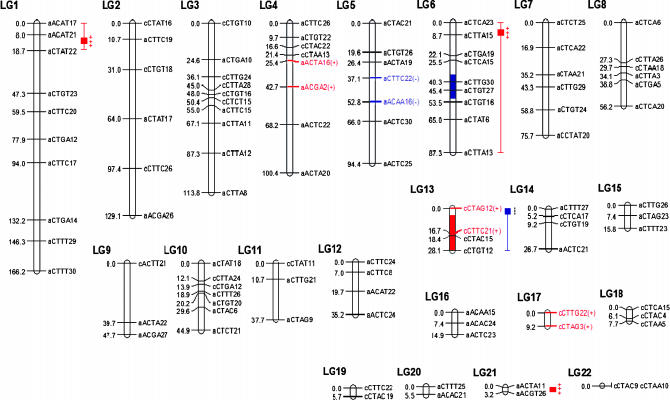
Figure 1.
Linkage map of dominant I. brevicaulis IRRE retrotransposon display markers segregating in the F1 hybrid used to produce BCIF hybrids. Markers whose text is colored reveal significant transmission-ratio distortion (Bouck et al. 2005). Significant QTL for survival in greenhouse conditions are denoted (with 2-LOD confidence intervals) to the right of the marker names (Martin et al. 2005). QTL for survival in natural conditions (this study) are denoted by colored bar segments (2-LOD confidence intervals). “Red” represents regions where introgressed (hybrid/heterozygous) regions are favored, while “blue” represents regions where recurrent (parental/homozygous) regions are favored.
TABLE 2.
QTL for survivorship in a flooded field experiment: estimates of locations, effects, and interactions in a backcross population (BCIF) of I. fulva × I. brevicaulis
| QTL no. | Linkage group (location) | Nearest marker | P-valuea | Effectb | PVEc |
|---|---|---|---|---|---|
| QTL 1 (additive effect) | LG6 (45.5 cM) | aCTGT27 | 0.009 | −0.2522 | 0.1381 |
| QTL 2 (additive effect) | LG13 (18.4 cM) | cCTAC15 | 0.041 | 0.2577 | 0.1085 |
| QTL1 × QTL2 (epistatic effect) | — | — | — | −0.5731 | 0.2124 |
Results were obtained by MIM. QTL previously identified by CIM were used as the initial model for MIM analysis.
Significance level of nearest marker.
Effect of the QTL on survivorship.
The proportion of the phenotypic variance explained by the QTL.
The main effects of each of the two QTL were relatively large, but in opposite directions with additive effects of QTL1 and QTL2 being −25.22% and +25.77%, respectively (Table 2). In other words, QTL1 was associated with decreased survivorship when introgressed I. brevicaulis alleles were present, while QTL2 promoted increased survivorship with introgressed I. brevicaulis alleles (Figure 1, Table 2). The significant negative epistatic interaction detected between the two QTL (measured to be −57.31%, Table 2) indicates that having both alleles decreases survivorship beyond that explained by a simple additive model. No dominance effects could be tested for due to the crossing design. All combined, the additive and epistatic effects accounted for 45.89% of the total phenotypic variance explained: the additive effects for QTL1 and QTL2 accounting for 13.81% and 10.85%, respectively, and the epistatic interaction accounting for 21.24% (Table 2). No significant QTL were detected using the BCIB mapping population.
Interpretation of QTL mapping studies such as this one are complicated by the fact that if sample size is low, the power to detect QTL of small effect is diminished (Beavis 1994, 1998; Xu 2003). A sample size of 185 independently derived BCIB hybrid genotypes and 209 BCIF hybrid genotypes was utilized in the present mapping study. Of those genotypes, only a small proportion survived. Given this reduced sample size, it is likely that additional QTL not detected in this study due to a lack of power affect survivorship. The reduced sample size also probably explains the inability to detect significant QTL in the BCIB mapping population. However, even under designs where power is low, detected QTL most likely reflect loci that affect the phenotypic trait under investigation (Lynch and Walsh 1998). Thus, the two QTL identified in the BCIF mapping population should be considered a highly conservative, minimum estimate of the total number affecting survivorship. In cases of low power, the effect sizes of detected QTL can be significantly overestimated (Beavis 1994). This can result in false conclusions that phenotypic variation is controlled by a few QTL of major effect (Beavis 1994). However, as with the number of QTL that affect survivorship in this study, we consider the phenotypic variance explained by each of the QTL, as well as that calculated for their epistatic interaction, to reflect a biological reality and a hypothesis for future analyses.
Single-marker analysis:
In BCIF hybrids, the two QTL were located very close to significant single markers whose effects were in the same direction. QTL1 spanned marker aCTGT27 on LG6, while QTL2 spanned marker cCTAC15 on LG13 (Table 2, Figure 1). There were also significant single-marker associations located on LG1 (effect, +), LG3 (+), LG10 (no effect, −), and LG18 (−), but no significant QTL were detected on these linkage groups by MIM. Since mapping studies often have reduced power to detect significant QTL, when in fact QTL exist, we view these significant single-marker associations as candidate regions wherein QTL may exist, but were undetectable at the stringent LOD thresholds in MIM.
We used χ2 tests to examine whether introgression of I. brevicaulis markers into a mostly I. fulva background tended to promote or reduce survivorship (see materials and methods) in the BCIF population. Introgressed I. brevicaulis alleles promoted survivorship in BCIF individuals, but at marginal significance (χ2 = 3.77, 1 d.f., P = 0.052; Table 3). In addition, tests for I. brevicaulis linkage groups promoting or reducing survivorship found no significant trend (χ2 = 1.19, 1 d.f., P = 0.275; Table 3).
TABLE 3.
Markers that reveal positive associations (introgressed alleles favored) or negative associations (introgressed alleles disfavored) with survivorship on each of the 22 linkage groups
| BCIB
|
BCIF
|
|||
|---|---|---|---|---|
| Linkage group | I. fulva alleles favored | I. fulva alleles disfavored | I. brevicaulis alleles favored | I. brevicaulis alleles disfavored |
| 1 | 2 | 11 | 10 (3) | 0 |
| 2 | 11 (1) | 1 | 0 | 6 |
| 3 | 14 (2) | 0 | 10 (5) | 0 |
| 4 | 5 | 2 | 7 | 1 |
| 5 | 4 | 1 | 2 | 5 |
| 6 | 7 | 3 | 0 | 9 (4) |
| 7 | 6 | 5 | 6 | 0 |
| 8 | 2 (1) | 3 | 2 | 4 |
| 9 | 4 | 1 | 3 | 0 |
| 10 | 4 | 1 | 0 | 7 (1) |
| 11 | 7 | 1 | 2 | 1 |
| 12 | 3 | 3 | 4 | 0 |
| 13 | 6 (2) | 0 | 4 (2) | 0 |
| 14 | 3 | 2 | 3 | 1 |
| 15 | 4 | 0 | 3 | 0 |
| 16 | 1 | 4 | 0 | 3 |
| 17 | 4 | 0 | 1 | 1 |
| 18 | 3 | 1 | 0 | 3 (1) |
| 19 | 4 | 0 | 2 | 0 |
| 20 | 3 | 0 | 2 | 0 |
| 21 | 4 | 0 | 2 | 0 |
| 22 | 0 | 2 | 0 | 2 |
| Total markers | 101 | 41 | 63 | 43 |
| Linkage groups | 17 | 4 | 13 | 8 |
Linkage groups having the same number do not imply homology. In BCIB hybrids, introgressed I. fulva alleles promoted survival significantly more often than did recurrent I. brevicaulis alleles (χ2 = 25.35, d.f. = 1, P < 0.0000005, linkage groups χ2 = 8.05, d.f. = 1, P < 0.005). In BCIF hybrids, no significant difference existed between recurrent I. fulva alleles or introgressed I. brevicaulis alleles in their ability to promote survival (χ2 = 3.77, P = 0.052, linkage groups χ2 = 1.19, d.f. = 1, P = 0.275). The number of markers revealing significant associations (P < 0.05) using single-marker linear regressions are in parentheses.
While MIM identified no QTL that significantly affected survivorship in the BCIB population, significant single-marker associations were found on LG2, LG3, LG8, and LG13. All of these significant associations were in the same direction, such that introgressed I. fulva alleles promoted survivorship (Table 3), indicating candidate regions for QTL affecting survivorship. Chi-square tests of the direction of the effect of markers (i.e., positive indicating increased survivorship and negative indicating decreased survivorship) also suggested that introgressed I. fulva alleles significantly promoted survivorship (χ2 = 25.35, 1 d.f., P < 0.0000005). Furthermore, linkage groups with predominantly I. fulva markers promoted survivorship significantly more than did linkage groups with predominantly recurrent I. brevicaulis markers (χ2 = 8.05, 1 d.f., P < 0.005; Table 3).
DISCUSSION
A large body of evidence has now been amassed that supports the notion that introgressive hybridization is “one of the commonest” (Anderson and Hubricht 1938) results of natural hybrid formation. Whether introgressive hybridization results in (or from) the transfer of adaptive traits across species boundaries is less clear. The development of molecular techniques has made it relatively easy to demonstrate the introgression of molecular markers across hybrid zones (Rieseberg et al. 1990; Arnold et al. 1991; Martinsen et al. 2001; Sweigart and Willis 2003), but this does not necessarily imply that their introgression was adaptive, since mutations that cause no fitness differences are presumably much more prevalent than those that cause an increase in fitness in a novel background (Rieseberg and Wendel 1993). Other studies have demonstrated the introgression of phenotypic traits across hybrid zones and species barriers (plants: Stutz and Thomas 1964; Arnold and Bennett 1993; Martinsen et al. 2001; Abbott et al. 2003; Bradshaw and Schemske 2003; animals: Lewontin and Birch 1966; Parsons et al. 1993; Morrow et al. 2000; microbes: Kroll et al. 1998; Machado and Ayala 2001; Smoot et al. 2002). However, it is often unclear what the adaptive value of those traits are.
The permeability of species genomes to introgression will depend on the degree to which hybrids are reproductively isolated from their parents. Often, a number of prezygotic and postzygotic isolating mechanisms interact to greatly reduce hybridization and subsequent gene flow (Schemske 2000; Ramsey et al. 2003; Coyne and Orr 2004; N. H. Martin and J. H. Willis, unpublished results). Furthermore, the genetic architecture of each barrier will affect the permeability of genomes. Genes and QTL that act to reduce hybridization, either via prezygotic or postzygotic means, have a greatly reduced chance of crossing species boundaries since they will be selected against. On the other hand, if novel, heterospecific genes and QTL serve to increase the fitness of individuals carrying those new alleles, then they will have a much greater chance of introgressing into a novel genetic background. Here, we demonstrate that regions of the Louisiana Iris genome are likely to introgress across species boundaries and that introgression results from fitness advantages (i.e., increased survivorship in highly selective, flooded conditions) transferred concurrently with those chromosomal regions.
Several independent lines of evidence indicate that I. fulva plants have adaptations that allow them to better tolerate flooded environments relative to I. brevicaulis. First, in this study, I. fulva individuals survived extended flooding while no I. brevicaulis plants survived (Table 1); although the sample size is relatively small, this result is significant. Furthermore, our findings are consistent with numerous prior data that indicate I. fulva's tolerance and I. brevicaulis's lack of tolerance to flooding (Viosca 1935; Cruzan and Arnold 1993; Johnston et al. 2001). Second, BCIF hybrids, in which 75% of the genome, on average, is composed of I. fulva alleles, survived at significantly higher rates than did BCIB hybrids. This also corresponds well with previous experiments showing that I. fulva-like hybrids occupy wetter environments than do I. brevicaulis-like hybrids (Viosca 1935; Cruzan and Arnold 1993; Johnston et al. 2001). Third, although I. fulva alleles make up, on average, only 25% of the genome of BCIB hybrids, this appears to be sufficient to allow for their increased survival relative to pure I. brevicaulis plants (although this is not significant due to the extremely low sample size of pure I. brevicaulis ramets planted into the field). The fourth line of evidence (and certainly the most compelling) suggesting that I. fulva alleles confer adaptations in a largely I. brevicaulis genetic background comes from the single-marker tests performed in this study. Throughout the BCIB hybrid genome, I. fulva alleles tended to promote survivorship over I. brevicaulis alleles. Of 22 linkage groups, 17 revealed a majority of markers that were associated with increased survivorship when I. fulva alleles were present. However, only six loci on 4 linkage groups revealed significant associations. All of these data lead us to the conclusion that not only is I. fulva adapted to wet environments, but also the traits responsible for those adaptations can be transferred via introgressive hybridization from I. fulva into I. brevicaulis. Our findings of relatively few QTL that are significantly associated with flood tolerance are consistent with studies involving rice cultivars. In Oryza, the ability to survive in completely submerged conditions for extended periods of time is a trait that is controlled by one or a few loci of major effect and by some minor-effect modifier loci (Mohanty and Khush 1985; Sinha and Saran 1988; Haque et al. 1989; Sripongpangkul et al. 2000; Toojinda et al. 2003).
In this study, the physiological mechanisms by which flood-tolerant Iris hybrids survive are unknown. Braendle and Crawford (1999) suggested that the primary cause of mortality among perennial plants (such as Iris) due to flooding is anoxia. The ability to survive long periods of oxygen deprivation requires sufficient energy reserves, prevention of cytoplasmic acidosis, and anaerobic utilization of stored starches (Braendle and Crawford 1999). Furthermore, while plants are in anaerobic conditions, cell injury can occur, and sufficient cellular repair mechanisms are presumably needed to prevent further damage when the plants reenter an aerobic environment. We were able to visually examine the Iris plants that succumbed to long periods of submergence. Apparently, the plants were unable to produce new growth in submerged conditions, as no (or few) new leaves were present. Furthermore, when the ramets were once again exposed to aerobic conditions, they quickly decomposed. We plan to use future analyses to define some of the physiological mechanisms that provide differential survivorship among the parental and hybrid genotypes.
Intriguingly, we also found evidence that introgression of I. brevicaulis alleles into I. fulva can increase the latter's fitness in flooded, i.e., non-I. brevicaulis-like, habitats. Thus, our QTL analysis provides strong evidence that I. brevicaulis alleles from at least one genomic region (QTL2) increase survivorship—relative to the I. fulva alleles—in a flooded environment. Specifically, substituting an I. fulva allele with an I. brevicaulis allele increases survivorship by >25%. The effect of QTL2 is surprising since no pure I. brevicaulis replicates survived the extensive flooding. Yet it is apparent that this species does contain regions that promote survivorship in flooded conditions, at least when placed on an I. fulva genetic background.
The ability to survive flooded conditions is but one component of fitness. To understand better patterns of introgression in Louisiana Iris hybrid zones, it is necessary to examine the genetic architecture of a number of fitness components across a variety of environments. In a previous study, we examined survivorship under greenhouse conditions for these same mapping populations (Martin et al. 2005). Interestingly, we found opposite patterns of survivorship, with BCIB hybrids surviving at much higher rates than BCIF hybrids in the greenhouse. The divergent patterns of survivorship seen in the two experiments are consistent with the hypothesis that the relatively dry environment in the greenhouse selected for alleles from the flood-intolerant I. brevicaulis and against alleles from the flood-tolerant I. fulva. The opposite result under the flooded field conditions would thus reflect the alternate selective regime. In the greenhouse, survivorship of BCIF hybrids was affected by at least four QTL (three revealing increased survivorship with introgressed I. brevicaulis alleles, and one increasing survivorship with recurrent I. fulva alleles). We have indicated these regions on Figure 1. [We provide only the linkage map derived from BCIF hybrids, since we were unable to detect significant QTL for the BCIB map either in this study or in the previous greenhouse study (Martin et al. 2005).] Two of the “greenhouse” QTL have confidence intervals overlapping the QTL for survivorship in this field study (Figure 1). It is unknown whether these QTL are due to different linked genes or whether they are the same genes that act oppositely in different environments. Combining these two studies with an analysis of segregation distortion (Bouck et al. 2005; regions of segregation distortion are also noted in Figure 1), it is clear that the Louisiana Iris genome contains numerous regions that are permeable to introgression, and in fact introgression may be selectively favored.
Are the regions that appear to promote increased fitness with the introgression of heterospecific alleles likely to introgress due to positive selection in naturally occurring hybrid zones? We suggest that this is extremely likely. First, we have shown that several heterospecific alleles promote survivorship (under greenhouse and natural field conditions) in both I. fulva and I. brevicaulis genetic backgrounds. Second, introgression between these two species in nature is well documented (Arnold 1993; Cruzan and Arnold 1993). While some proportion of this introgression almost certainly involves neutral loci, any selectively advantageous alleles are likely to introgress across the species' boundaries (Barton and Hewitt 1985). In the case of I. brevicaulis and I. fulva hybrid zones, we predict that the QTL found to promote tolerance to flooded environments are candidates for introgression, since they will presumably be positively selected under flooded conditions. This prediction is testable by assaying markers associated with the QTL in natural hybrid populations. In addition, field experiments are underway to determine whether other fitness-related characters are candidates for introgression. For example, traits such as seed set are certainly adaptive, and we hope to QTL map these fitness components under natural conditions. This should help determine whether introgression of these traits, and thereby the production of introgressed individuals with increased fitness, is likely. Finally, studies confirming the introgression in natural hybrid zones—of those same selectively advantageous regions identified by the present (and future) analyses—would provide further support for Anderson's (1949) use of the Louisiana Irises as his “typical example” of the evolutionary effects from introgressive hybridization, including the intertaxonomic transfer of adaptive traits.
Acknowledgments
We thank S. Wessler, R. Mauricio, and E. Kentner for important contributions to the linkage mapping projects that form the basis for this analysis. We thank S. Cornman, J. Ross-Ibarra, E. Kuntz, and S. Small for helpful comments on previous versions of the manuscript. R. H. Martin and Y. Z. Martin provided invaluable assistance in the field. J. Ross-Ibarra, J. H. Willis, R. Doerge, and Z. B. Zeng provided invaluable discussions concerning QTL analyses. We thank B. Osburghouse and the Army Corps of Engineers Atchafalaya Basin Floodway System for field sites and field assistance. The present study was supported by two National Science Foundation grants to M.L.A. (DEB-0074159 and DEB-0345123) and by a gift from M. Caillet.
References
- Abbott, R. J., J. K. James, R. I. Milne and A. C. M. Gillies, 2003. Plant introductions, hybridization and gene flow. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, E., 1949. Introgressive Hybridization. John Wiley & Sons, New York.
- Anderson, E., and L. Hubricht, 1938. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 25: 396–402. [Google Scholar]
- Arnold, M. L., 1993. Iris nelsonii: origin and genetic composition of a homoploid hybrid species. Am. J. Bot. 80: 577–583. [DOI] [PubMed] [Google Scholar]
- Arnold, M. L., and B. D. Bennett, 1993. Natural hybridization in Louisiana irises: genetic variation and ecological determinants, pp. 115–139 in Hybrid Zones and the Evolutionary Process, edited by R. G. Harrison. Oxford University Press, Oxford.
- Arnold, M. L., D. D. Shaw and N. Contreras, 1987. Ribosomal RNA-encoding DNA introgression across a narrow hybrid zone between two subspecies of grasshopper. Proc. Natl. Acad. Sci. USA 84: 3946–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, M. L., C. M. Buckner and J. J. Robinson, 1991. Pollen-mediated introgression and hybrid speciation in Louisiana irises. Proc. Natl. Acad. Sci. USA 88: 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, N. H., and G. M. Hewitt, 1985. Analysis of hybrid zones. Annu. Rev. Ecol. Syst. 16: 113–148. [Google Scholar]
- Basten, C. J., B. S. Weir and Z-B. Zeng, 2005. QTL Cartographer, Version 2.5. Department of Statistics, North Carolina State University, Raleigh, NC.
- Beavis, W. D., 1994. The power and deceit of QTL experiments: lessons from comparative QTL studies, pp. 250–266 in Proceedings of the 49th Annual Corn and Sorghum Industry Research Conference. American Seed Trade Association, Washington, DC.
- Beavis, W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, New York.
- Bouck, A. C., 2004. The Genetic Architecture of Reproductive Isolation in Louisiana Irises. University of Georgia, Athens, GA.
- Bouck, A. C., R. Peeler, M. L. Arnold and S. R. Wessler, 2005. Genetic mapping of species boundaries in Louisiana Irises using IRRE retrotransposon display markers. Genetics 171: 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw, H. D., and D. W. Schemske, 2003. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature 426: 176–178. [DOI] [PubMed] [Google Scholar]
- Braendle, R., and R. M. M. Crawford, 1999. Plants as amphibians. Perspect. Plant Ecol. Evol. Syst. 2: 56–78. [Google Scholar]
- Burke, J. M., S. E. Carney and M. L. Arnold, 1998. Hybrid fitness in the Louisiana Irises: analysis of parental and F1 performance. Evolution 52: 37–43. [DOI] [PubMed] [Google Scholar]
- Burke, J. M., M. R. Bulger, R. A. Wesselingh and M. L. Arnold, 2000. Frequency and spatial patterning of clonal reproduction in Louisiana Iris hybrid populations. Evolution 54: 137–144. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Cruzan, M. B., and M. L. Arnold, 1993. Ecological and genetic associations in an Iris hybrid zone. Evolution 47: 1432–1445. [DOI] [PubMed] [Google Scholar]
- Cruzan, M. B., and M. L. Arnold, 1994. Assortative mating and natural selection in an Iris hybrid zone. Evolution 48: 1946–1958. [DOI] [PubMed] [Google Scholar]
- Cruzan, M. B., J. L. Hamrick, M. L. Arnold and B. D. Bennett 1994. Mating system variation in hybridizing Irises: effects of phenology and floral densities on family outcrossing rates. Heredity 72: 95–105. [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, T. E., and C. L. Secor, 1997. The role of hybridization and introgression in the diversification of animals. Annu. Rev. Ecol. Syst. 28: 593–619. [Google Scholar]
- Ellstrand, N. C., R. Whitkus and L. H. Rieseberg, 1996. Distribution of spontaneous plant hybrids. Proc. Natl. Acad. Sci. USA 93: 5090–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms, S. K., and M. L. Arnold, 2000. Site-to-site differences in pollinator visitation patterns in a Louisiana iris hybrid zone. Oikos 91: 568–578. [Google Scholar]
- Haque, Q. A., D. Hille Ris Lambers, N. M. Tepora and Q. D. dela Cruz, 1989. Inheritance of submerance tolerance in rice. Euphytica 41: 247–251. [Google Scholar]
- Heiser, C. B., Jr., 1951. Hybridization in the annual sunflowers: Helianthus annuus × H. debilis var. cucumerifolius. Evolution 5: 42–51. [Google Scholar]
- Johnston, J. A., R. A. Wesselingh, A. C. Bouck, L. A. Donovan and M. L. Arnold, 2001. Intimately linked or hardly speaking? The relationship between genotype and environmental gradients in a Louisiana Iris hybrid population. Mol. Ecol. 10: 673–681. [DOI] [PubMed] [Google Scholar]
- Kao, C. H., Z-B. Zeng and R. D. Teasdale, 1999. Multiple interval mapping for quantitative trait loci. Genetics 152: 1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentner, E. K., M. L. Arnold and S. R. Wessler, 2003. Characterization of high-copy-number retrotransposons from the large genomes of the Louisiana Iris species and their use as molecular markers. Genetics 164: 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. C., and L. H. Rieseberg, 1999. Genetic architecture of species differences in annual sunflowers: implications for adaptive trait introgression. Genetics 153: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, J. S., K. E. Wilks, J. L. Farrant and P. R. Langford, 1998. Natural genetic exchange between Haemophilus and Neisseria: intergeneric transfer of chromosomal genes between major human pathogens. Proc. Natl. Acad. Sci. USA 95: 12381–12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Lewontin, R. C., and L. C. Birch, 1966. Hybridization as a source of variation for adaptation to new environments. Evolution 20: 315–336. [DOI] [PubMed] [Google Scholar]
- Lincoln, S., M. Daly and E. S. Lander, 1992. Constructing Genetic Maps With MAPMAKER/EXP 3.0. Whitehead Institute Technical Report, Whitehead Institute, Cambridge, MA.
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Machado, C. A., and F. J. Ayala, 2001. Nucleotide sequences provide evidence of genetic exchange among distantly related lineages of Trypanosoma cruzi. Proc. Natl. Acad. Sci. USA 98: 7396–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, N. H., A. C. Bouck and M. L. Arnold, 2005. Loci affecting long-term hybrid survivability in Louisiana Irises: implications for reproductive isolation and introgression. Evolution 59: 2116–2124. [PubMed] [Google Scholar]
- Martinsen, G. D., T. G. Whitham, R. J. Turek and P. Keim, 2001. Hybrid populations selectively filter gene introgression between species. Evolution 55: 1325–1335. [DOI] [PubMed] [Google Scholar]
- Mohanty, H. K., and G. S. Khush, 1985. Diallel analysis of submergence tolerance in rice, Oryza sativa L. Theor. Appl. Genet. 70: 467–473. [DOI] [PubMed] [Google Scholar]
- Morrow, J., L. Scott, B. Congdon, D. Yeates, M. Frommer et al., 2000. Close genetic similarity between two sympatric species of Tephritid fruit fly reproductively isolated by mating time. Evolution 54: 899–910. [DOI] [PubMed] [Google Scholar]
- Parsons, T. J., S. L. Olson and M. J. Braun, 1993. Unidirectional spread of secondary sexual plumage traits across an avian hybrid zone. Science 260: 1643–1646. [DOI] [PubMed] [Google Scholar]
- Payseur, B. A., and M. W. Nachman, 2005. The genomics of speciation: investigating the molecular correlates of X chromosome introgression across the hybrid zone between Mus domesticus and Mus musculus. Biol. J. Linn. Soc. 84: 523–534. [Google Scholar]
- Ramsey, J., H. D. Bradshaw and D. W. Schemske, 2003. Components of reproductive isolation between the monkeyflowers Mimulus lewisii and M. cardinalis (Phrymaceae). Evolution 57: 1520–1534. [DOI] [PubMed] [Google Scholar]
- Randolph, L. F., J. Mitra and I. S. Nelson, 1961. Cytotaxonomic studies of Louisiana Irises. Bot. Gaz. 123: 125–133. [Google Scholar]
- Rieseberg, L. H., and J. Wendel, 1993. Introgression and its consequences in plants, pp. 70–109 in Hybrid Zones and the Evolutionary Process, edited by R. Harrison. Oxford University Press, New York.
- Rieseberg, L. H., S. Beckstrom-Sternberg and K. Doan, 1990. Helianthus annuus ssp. texanus has chloroplast DNA and nuclear ribosomal RNA genes of Helianthus debilis ssp. cucumerifolius. Proc. Natl. Acad. Sci. USA 87: 593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., O. Raymond, D. M. Rosenthal, Z. Lai, K. Livingstone et al., 2003. Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301: 1211–1216. [DOI] [PubMed] [Google Scholar]
- Schemske, D. W., 2000. Understanding the origin of species. Evolution 54: 1069–1073. [Google Scholar]
- Sinha, M. M., and S. Saran, 1988. Inheritance of submergence tolerance in lowland rice. Oryza 25: 351–354. [Google Scholar]
- Smoot, L. M., D. D. Franke, G. McGillivary and L. A. Actis, 2002. Genomic analysis of the F3031 Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius by PCR-based subtractive hybridization. Infect. Immun. 70: 2694–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripongpangkul, K., G. B. T. Posa, D. W. Senadhira, D. Brar, N. Huang et al., 2000. Genes/QTLs affecting flood tolerance in rice. Theor. Appl. Genet. 101: 1074–1081. [Google Scholar]
- Stutz, H. C., and L. K. Thomas, 1964. Hybridization and introgression in Cowania and Purshia. Evolution 18: 183–195. [Google Scholar]
- Sweigart, A. L., and J. H. Willis, 2003. Patterns of nucleotide diversity in two species of Mimulus are affected by mating system and asymmetric introgression. Evolution 57: 2490–2506. [DOI] [PubMed] [Google Scholar]
- Toojinda, T., M. Siangliw, S. Tragoonrung and A. Vanavichit, 2003. Molecular genetics of submergence tolerance in rice: QTL analysis of key traits. Ann. Bot. 91: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viosca, P. J., 1935. The Irises of southeastern Louisiana: a taxonomic and ecological interpretation. Bull. Am. Iris Soc. 57: 3–56. [Google Scholar]
- Wesselingh, R. A., and M. L. Arnold, 2000. Pollinator behaviour and the evolution of Louisiana Iris hybrid zones. J. Evol. Biol. 13: 171–180. [Google Scholar]
- Won, Y. J., and J. Hey, 2005. Divergence population genetics of chimpanzees. Mol. Biol. and Evol. 22: 297–307. [DOI] [PubMed] [Google Scholar]
- Xu, S., 2003. Theoretical basis of the Beavis effect. Genetics 165: 2259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1993. Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc. Natl. Acad. Sci. USA 90: 10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, Z-B., C-H. Kao and C. J. Basten, 1999. Estimating the genetic architecture of quantitative traits. Genet. Res. 4: 279–289. [DOI] [PubMed] [Google Scholar]