Abstract
The human population has increased greatly in size in the last 100,000 years, but the initial stimuli to growth, the times when expansion started, and their variation between different parts of the world are poorly understood. We have investigated male demography in East Asia, applying a Bayesian full-likelihood analysis to data from 988 men representing 27 populations from China, Mongolia, Korea, and Japan typed with 45 binary and 16 STR markers from the Y chromosome. According to our analysis, the northern populations examined all started to expand in number between 34 (18–68) and 22 (12–39) thousand years ago (KYA), before the last glacial maximum at 21–18 KYA, while the southern populations all started to expand between 18 (6–47) and 12 (1–45) KYA, but then grew faster. We suggest that the northern populations expanded earlier because they could exploit the abundant megafauna of the “Mammoth Steppe,” while the southern populations could increase in number only when a warmer and more stable climate led to more plentiful plant resources such as tubers.
HUMANS have expanded enormously in geographical range and numbers in the last 100,000 years, starting as a rare species confined to parts of Africa and ending with the current population of >6 billion distributed all over the world, but the details of these changes are poorly understood (Jobling et al. 2004). Historical records document a substantial demographic expansion within historical times, and also the complexity of the changes that have occurred, but are available only for the last few thousand years. Before this time, the archaeological record indicates that humans increased substantially in number when Neolithic transitions led to greater and more reliable food production after ∼10 thousand years ago (KYA), but provide only limited quantitative information. Genetic variation can also provide insights into past demography. Standard neutral models of evolution predict the extent and pattern of variation expected in a constant-sized population, but experimental data from the human population are often not consistent with such a model. For example, an overall excess of rare variants, reflected by negative values for Tajima's D (e.g., Akey et al. 2004), is commonly interpreted as a signal of demographic expansion, although the details of such an expansion remain unclear (Wall and Przeworski 2000; Ptak and Przeworski 2002). Genomewide analyses of short tandem repeats (STRs) have been interpreted as revealing an early expansion in Africa 49–640 KYA with no expansion outside Africa (Reich and Goldstein 1998), or alternatively a constant population size in Africa compared with expansions in Europeans and Africans (Kimmel et al. 1998). A larger-scale study using 377 loci in 52 populations suggested expansion in African farmers starting ∼35 KYA, in Eurasians ∼25 KYA, and in East Asians ∼18 KYA, but found no significant signal of growth in African hunter-gatherers or populations from Oceania and America (Zhivotovsky et al. 2003). The conflicting conclusions may reflect, in part, the complexity of the real events, so that it may not be useful to compare descriptions of demographic change summed over large geographical regions. Instead, studies at higher spatial resolution may be necessary to understand how the demography has changed in different ways at local levels.
Single loci can be influenced by stochastic variation and locus-specific selection, but nevertheless two of them, mitochondrial DNA (mtDNA) and the Y chromosome, are of particular interest because of the insights they can provide into female-specific and male-specific evolutionary patterns, respectively. Mismatch distributions of mtDNA sequences from populations around the world have suggested expansion, on average, ∼40 KYA (Sherry et al. 1994), while a phylogenetic star contraction method indicated expansion of the major Asian clades M and N ∼30 KYA (Forster et al. 2001). Studies of the Y chromosome have shown a strong signal of expansion beginning in the Paleolithic ∼18 KYA (7–41 KYA, Pritchard et al. 1999) or ∼22 KYA (8.5–50 KYA, Macpherson et al. 2004) worldwide, with limited variation between continents. In contrast, a detailed study of one country, Armenia, suggested a start of expansion in the Neolithic ∼4.8 KYA (2.0–11.1 KYA, Weale et al. 2001).
We want to understand the history of East Asia, including its male demographic history. Modern humans were present in Australia at ∼50 KYA and, despite a lack of direct archaeological evidence, may have reached the southern part of East Asia at about the same time (Jobling et al. 2004). Classical marker studies reveal a genetic distinction between northern and southern China, with a boundary corresponding approximately to the Yangtze River (Xiao et al. 2000). Some authors have suggested that modern East Asian populations are derived largely from a northward expansion of southern populations after the last glacial maximum (LGM) ∼18–21 KYA (Jin and Su 2000), while others have suggested a significant male contribution from Central Asia (Karafet et al. 2001). Despite these and other (Su et al. 1999; Deng et al. 2004) surveys of Y-chromosomal haplogroup distributions, we know little about the detailed demography of the region and how it compares between north and south. We now show that male demographic history differs substantially between the northern and southern parts of East Asia and link this to ecological differences between the regions in the Paleolithic period.
MATERIALS AND METHODS
Data set:
Nine hundred eighty-eight males belonging to 27 populations from China, Mongolia, Korea, and Japan were included in this analysis. The samples, and their typing with 16 Y-specific binary and 16 short tandem repeat (STR) markers, have been described previously (Zerjal et al. 2003; Xue et al. 2005). For this study, we typed hierarchically an additional 29 binary markers (M89, M8, M38, P33, M217, M93, M48, M61, M76, M147, M27, M214, M5, M128, M178, M119, M101, M50, M175, P31, M95, M88, M122, M121, M134, M164, M159, M113, and M7) using multiplexed primer-extension reactions (Paracchini et al. 2002) adapted for the ABI (Columbia, MD) Prism SNaPshot system (Hurles et al. 2005) according to the manufacturer's guidelines. As before, DYS19 was excluded from most analyses because it is duplicated in some individuals.
Data analyses:
Haplotypes for this haploid locus could be constructed simply from the combination of STRs and/or binary markers present in the same individual and their frequencies determined by counting. Analysis of molecular variance (AMOVA) was performed using Arlequin 2.0 (Schneider et al. 2000) and spatial AMOVA (SAMOVA) analysis using SAMOVA1.0 (Dupanloup et al. 2002). Spatial autocorrelation was carried out using autocorrelation index for DNA analysis (AIDA) (Bertorelle and Barbujani 1995). Inferences about Y-chromosomal lineage histories and demographies were made using the Bayesian analysis of trees with internal node generation program (BATWING) (Wilson et al. 2003). Populations (represented by 25–65 individuals) were analyzed individually using weakly informative prior distributions for N, the effective population size before expansion [gamma(1, 0.0001): mean = 10,000, SD = 10,000]; α, the rate of growth per generation [gamma(2, 400): mean = 0.005, SD = 0.0035]; and β, the time in coalescent units when exponential growth began [gamma(2, 1): mean = 2, SD = 1.41] (Wilson et al. 2003). A calibrated “evolutionary” mutation rate for Y-STRs (Zhivotovsky et al. 2004) was used as the basis for a per-locus mutation rate prior of gamma(1.47, 2130) (mean = 0.00069, SD = 0.00057) and was allowed to vary independently for each locus. This mutation rate was calibrated against two historical events (the divergence of the Maoris and Cook Islanders in the Pacific and the migration of the Bulgarian Roma from India to Europe), and thus our time estimates are also calibrated against these events and do not depend on assumptions about generation time. Binary markers (unique event polymorphisms, UEPs) were included under option 2, in which they condition only the tree structures possible. A total of 104 samples of the program's output representing 106 Markov chain Monte Carlo (MCMC) cycles were taken after discarding the first 3 × 103 samples as “burn-in,” and convergence was confirmed by examining longer runs of 108 MCMC cycles for four populations and finding the same posterior distributions. The influence of population sample size in the range 25–65 was investigated by randomly subsampling 25, 30, 40, or 50 individuals from the Outer Mongolian population with size 65 and found to be negligible. The 0.025, median, and 0.975 percentiles of the output samples were recorded. Regression analyses were carried out using SPSS 14; the stepwise criteria in multiple linear regression were the defaults, probability of F to enter ≤0.05 and probability of F to remove ≥0.10. A contour plot of expansion times was drawn using SigmaPlot version 9 with inverse square smoothing and a sampling proportion 0.5.
“Expansion” always refers to an increase in numbers rather than area and “expansion time” to the time when the increase started, unless otherwise stated.
RESULTS
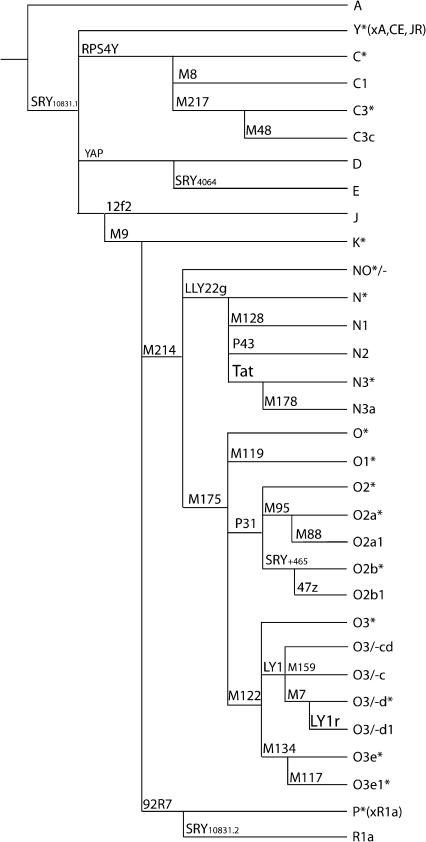
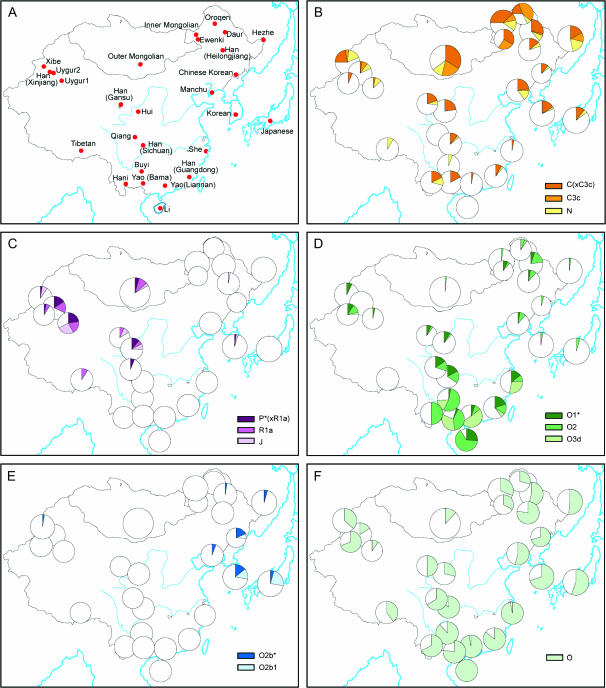
Approximately 1000 males from 27 East Asian populations were typed with 61 Y-chromosomal markers, and we first describe the basic properties of this data set. The 45 binary markers identified 31 haplogroups (including paragroups) in the sample, while the 15 STRs defined 730 different haplotypes (Figure 1, Table 1; see also supplemental Table 1 at http://www.genetics.org/supplemental/). Population diversities ranged from 0.60 to 0.94 for binary markers and from 0.84 to 1.00 for STRs (Table 2). There was considerable variation in the distribution of lineages between populations, but this did not correspond to the major ethnic distinction in the area, which is between the Han Chinese (>80% of the combined populations of China, Mongolia, Korea, and Japan) and the other populations. AMOVA analysis showed that only 1.8 and 0.5% of variation lay between Han and non-Han populations using binary and STR markers, respectively, and neither of these values was significantly greater than zero. There were, however, major geographical differences. Figure 2 shows that, despite the overall predominance of haplogroup O (56%), specific haplogroups were concentrated in each geographical region: C and N in the north; P and J in the west; O2b in the east; and O1*, O2*, and O3d in the south. We therefore wished to identify the most important elements of the geographical pattern in an objective way.
Figure 1.
Phylogeny of Y-chromosomal haplogroups detected in this study.
TABLE 1.
Haplogroup frequencies in East Asian populations
| Haplogroup | Daur | Ewenki | Hezhe | Hui | Manchu | Inner Mongolian | Oroqen | Uygur (Urumqi) | Uygur (Yili) | Xibe | Han (Harbin) | Han (Yili) | Korean (China) | Buyi | Hani | Li | Qiang | She | Tibetans | Yao (Bama) | Yao (Liannan) | Han (Chengdu) | Han (Lanzhou) | Han (Meixian) | Japanese | Korean (Korea) | Outer Mongolian | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y*(xA, CE, JR) | 2 | 2 | 2 | 1 | 8 | 1 | 1 | 2 | 1 | 3 | 2 | 25 | ||||||||||||||||
| C*(xC1, C3) | 0 | 1 | 3 | 4 | 1 | 1 | 6 | 16 | ||||||||||||||||||||
| C1 | 4 | 4 | ||||||||||||||||||||||||||
| C3*(xC3c) | 11 | 7 | 5 | 4 | 8 | 17 | 6 | 9 | 5 | 2 | 3 | 6 | 1 | 1 | 4 | 6 | 3 | 1 | 7 | 22 | 128 | |||||||
| C3c | 1 | 7 | 5 | 1 | 4 | 13 | 3 | 2 | 13 | 49 | ||||||||||||||||||
| D | 4 | 1 | 1 | 1 | 1 | 6 | 15 | 1 | 2 | 13 | 1 | 1 | 47 | |||||||||||||||
| E | 1 | 2 | 3 | |||||||||||||||||||||||||
| J | 3 | 8 | 3 | 1 | 3 | 1 | 2 | 21 | ||||||||||||||||||||
| K* | 1 | 1 | 2 | 1 | 5 | 2 | 3 | 1 | 1 | 1 | 1 | 19 | ||||||||||||||||
| NO*/− | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 9 | |||||||||||||||||||
| N* | 2 | 1 | 1 | 4 | 1 | 1 | 4 | 3 | 1 | 2 | 1 | 2 | 1 | 24 | ||||||||||||||
| N2 | 8 | 1 | 2 | 2 | 2 | 15 | ||||||||||||||||||||||
| N1 | 2 | 1 | 2 | 5 | ||||||||||||||||||||||||
| N3*(xN3a) | 1 | 1 | ||||||||||||||||||||||||||
| N3a | 3 | 6 | 2 | 2 | 1 | 4 | 18 | |||||||||||||||||||||
| O* | 1 | 1 | 2 | 1 | 5 | |||||||||||||||||||||||
| O1*(xO1a, O1b) | 2 | 2 | 3 | 1 | 1 | 3 | 1 | 3 | 1 | 9 | 5 | 5 | 1 | 2 | 5 | 2 | 7 | 53 | ||||||||||
| O2* | 1 | 1 | 1 | 3 | 1 | 2 | 3 | 3 | 1 | 1 | 1 | 13 | 1 | 5 | 1 | 1 | 39 | |||||||||||
| O2a* | 6 | 1 | 1 | 13 | 2 | 20 | 3 | 3 | 1 | 2 | 1 | 1 | 2 | 56 | ||||||||||||||
| O2a1 | 6 | 15 | 1 | 1 | 1 | 4 | 28 | |||||||||||||||||||||
| O2b* | 1 | 2 | 2 | 1 | 5 | 2 | 6 | 19 | ||||||||||||||||||||
| O2b1 | 2 | 11 | 6 | 19 | ||||||||||||||||||||||||
| O3*(xO3a–O3e) | 1 | 7 | 1 | 3 | 3 | 2 | 2 | 7 | 7 | 6 | 1 | 2 | 2 | 1 | 1 | 3 | 1 | 1 | 7 | 2 | 60 | |||||||
| O3/-cd* | 6 | 1 | 4 | 3 | 3 | 2 | 2 | 2 | 3 | 3 | 1 | 1 | 5 | 3 | 7 | 3 | 9 | 1 | 59 | |||||||||
| O3/-c | 1 | 1 | ||||||||||||||||||||||||||
| O3/-d | 1 | 1 | 6 | 10 | 12 | 18 | 2 | 50 | ||||||||||||||||||||
| O3/-d1 | 1 | 1 | 2 | 4 | ||||||||||||||||||||||||
| O3e*(xO3e1) | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 2 | 5 | 8 | 1 | 3 | 3 | 1 | 4 | 2 | 1 | 2 | 5 | 2 | 1 | 2 | 5 | 1 | 59 | |||
| O3e1*(xO3e1a) | 3 | 4 | 7 | 1 | 5 | 5 | 1 | 1 | 2 | 2 | 5 | 2 | 4 | 1 | 2 | 3 | 6 | 13 | 5 | 5 | 3 | 4 | 8 | 5 | 3 | 100 | ||
| P*(xR1a) | 4 | 6 | 6 | 1 | 1 | 2 | 1 | 3 | 24 | |||||||||||||||||||
| R1a | 2 | 7 | 6 | 2 | 3 | 2 | 6 | 28 | ||||||||||||||||||||
| Total | 39 | 26 | 45 | 35 | 35 | 45 | 31 | 31 | 39 | 41 | 35 | 32 | 25 | 35 | 34 | 34 | 33 | 34 | 35 | 35 | 35 | 34 | 30 | 35 | 47 | 43 | 65 | 988 |
TABLE 2.
Population statistics: observed variation and BATWING prior and posterior estimates
| Y-STR variation
|
BATWING output
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Location
|
Y-SNP diversity
|
Diversity
|
Nposterior
|
Expansion time (KYA)
|
αposterior
|
TMRCA (KYA)
|
|||||||||||||
| Population | Long °E | Lat °N | 15 STRs | 4 STRs | 3 STRs | ASD | 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | 2.5% | Median | 97.5% | |
| BATWING prior | 254 | 6940 | 36900 | 1.5 | 291.5 | 5138.3 | 0.0006 | 0.0042 | 0.0140 | ||||||||||
| Buyi | 105.7 | 26.0 | 0.810 | 0.988 | 0.958 | 0.866 | 64 | 203 | 585 | 1666 | 6.8 | 15.5 | 36.1 | 0.0011 | 0.0036 | 0.0090 | 18.7 | 34.6 | 70.4 |
| Hani | 101.9 | 23.5 | 0.767 | 0.995 | 0.865 | 0.825 | 75 | 168 | 453 | 1413 | 8.6 | 17.2 | 34.7 | 0.0018 | 0.0043 | 0.0098 | 18.1 | 31.6 | 61.8 |
| Li | 109.3 | 18.5 | 0.596 | 0.838 | 0.677 | 0.665 | 44 | 163 | 491 | 1462 | 4.6 | 12.4 | 35.5 | 0.0006 | 0.0036 | 0.0105 | 17.0 | 32.1 | 78.6 |
| She | 117.4 | 25.1 | 0.849 | 0.966 | 0.914 | 0.890 | 77 | 254 | 638 | 1773 | 6.0 | 17.6 | 46.6 | 0.0005 | 0.0024 | 0.0063 | 18.8 | 33.4 | 66.2 |
| Tibet | 91.1 | 29.6 | 0.682 | 0.990 | 0.956 | 0.936 | 145 | 298 | 868 | 2146 | 5.6 | 17.1 | 50.8 | 0.0007 | 0.0028 | 0.0103 | 24.7 | 44.4 | 85.2 |
| Yao (Bama) | 107.2 | 23.4 | 0.733 | 0.879 | 0.778 | 0.738 | 63 | 210 | 483 | 1072 | 1.1 | 11.9 | 44.6 | 0.0001 | 0.0011 | 0.0062 | 15.9 | 29.5 | 57.2 |
| Yao (Liannan) | 112.2 | 24.7 | 0.716 | 0.992 | 0.891 | 0.778 | 66 | 161 | 473 | 1571 | 5.7 | 15.6 | 36.4 | 0.0011 | 0.0039 | 0.0114 | 21.3 | 41.0 | 90.5 |
| Daur | 124.6 | 49.3 | 0.875 | 0.972 | 0.919 | 0.908 | 123 | 286 | 686 | 1791 | 13.4 | 26.4 | 59.4 | 0.0006 | 0.0019 | 0.0042 | 24.0 | 40.8 | 78.4 |
| Ewenki | 118.6 | 48.0 | 0.841 | 0.923 | 0.895 | 0.874 | 95 | 273 | 827 | 2127 | 4.2 | 22.7 | 57.7 | 0.0004 | 0.0017 | 0.0053 | 21.8 | 38.1 | 70.3 |
| Hezhe | 132.5 | 47.5 | 0.898 | 0.975 | 0.952 | 0.937 | 117 | 500 | 1181 | 2842 | 8.2 | 27.0 | 71.0 | 0.0003 | 0.0013 | 0.0034 | 25.4 | 43.7 | 80.2 |
| Hui | 105.5 | 36.6 | 0.941 | 0.985 | 0.958 | 0.881 | 115 | 197 | 452 | 1182 | 14.9 | 25.8 | 43.9 | 0.0017 | 0.0033 | 0.0063 | 20.8 | 34.5 | 58.1 |
| Inner Mongolian | 126.0 | 51.0 | 0.822 | 0.972 | 0.910 | 0.905 | 138 | 333 | 903 | 2029 | 17.7 | 34.3 | 68.3 | 0.0007 | 0.0017 | 0.0038 | 30.4 | 52.3 | 92.7 |
| Japanese | 134.0 | 34.2 | 0.842 | 0.997 | 0.938 | 0.909 | 117 | 224 | 632 | 1655 | 12.3 | 21.7 | 39.3 | 0.0019 | 0.0039 | 0.0074 | 24.7 | 40.2 | 69.4 |
| Korean (China) | 121.4 | 42.0 | 0.878 | 0.993 | 0.953 | 0.923 | 130 | 289 | 721 | 2657 | 8.8 | 26.5 | 56.8 | 0.0008 | 0.0024 | 0.0066 | 27.4 | 46.1 | 88.8 |
| Korean (Korea) | 127.0 | 37.5 | 0.898 | 0.991 | 0.956 | 0.932 | 123 | 240 | 602 | 1773 | 16.4 | 30.3 | 58.5 | 0.0010 | 0.0024 | 0.0048 | 28.5 | 48.3 | 86.6 |
| Manchu | 123.2 | 40.2 | 0.914 | 1.000 | 0.978 | 0.963 | 149 | 311 | 790 | 2361 | 11.8 | 22.5 | 45.1 | 0.0020 | 0.0046 | 0.0098 | 29.0 | 49.4 | 91.6 |
| Oroqen | 123.4 | 50.3 | 0.794 | 0.933 | 0.886 | 0.873 | 92 | 228 | 600 | 1600 | 8.9 | 22.7 | 53.4 | 0.0006 | 0.0022 | 0.0060 | 23.2 | 41.6 | 83.2 |
| Outer Mongolian | 107.0 | 48.2 | 0.837 | 0.987 | 0.929 | 0.904 | 114 | 316 | 617 | 1409 | 13.9 | 26.3 | 54.6 | 0.0009 | 0.0022 | 0.0045 | 23.5 | 39.4 | 74.4 |
| Qiang | 103.5 | 31.4 | 0.919 | 0.998 | 0.975 | 0.930 | 115 | 252 | 681 | 1632 | 12.0 | 22.9 | 44.6 | 0.0015 | 0.0035 | 0.0073 | 26.3 | 44.9 | 79.7 |
| Uygur (Urumqi) | 87.6 | 43.8 | 0.899 | 1.000 | 0.985 | 0.976 | 174 | 344 | 1336 | 3673 | 12.7 | 30.6 | 64.1 | 0.0009 | 0.0025 | 0.0068 | 38.1 | 65.0 | 117.2 |
| Uygur (Yili) | 87.3 | 43.5 | 0.858 | 0.997 | 0.976 | 0.961 | 142 | 578 | 970 | 1911 | 18.0 | 28.6 | 48.1 | 0.0018 | 0.0034 | 0.0060 | 30.8 | 48.3 | 81.6 |
| Xibe | 81.7 | 44.0 | 0.922 | 0.995 | 0.970 | 0.955 | 134 | 282 | 735 | 2432 | 16.0 | 30.0 | 55.6 | 0.0012 | 0.0027 | 0.0051 | 28.4 | 49.5 | 92.2 |
Long, longitude; Lat, latitude.
Figure 2.
Geographical distributions of Y-chromosomal haplogroups. (A) Populations sampled. (B–F) Haplogroup frequencies: circle area is proportional to sample size and sector area to haplogroup frequency. (B–E) Haplogroups are sorted into those showing predominantly northern (B), western (C), southern (D), and eastern (E) distributions. (F) The overall frequency of the most common haplogroup, O.
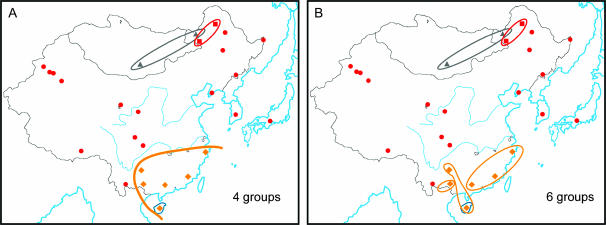
We based the subsequent analyses on the STR data unless otherwise indicated because of the problems in interpreting data from preascertained binary markers. SAMOVA analysis (Dupanloup et al. 2002) identifies, for a prespecified number of groups of populations, the geographical groups that are most differentiated from one another. Application of this method to the East Asian Y-STR data set using two or three groups distinguished small numbers of unusual populations, a finding that is readily understood from the high frequencies of the “star cluster” (Zerjal et al. 2003) and “Manchu cluster” (Xue et al. 2005) lineages in some northern populations, and reflects extreme expansions of individual patrilines within historical times. The use of four groups provided the most informative subdivision, with a cluster of six southern populations distinguished in addition to some of the northern ones (Figure 3A). This pattern corresponds well to the north–south distinction seen with classical markers and shows that, in this respect, the Y-chromosomal variation is typical of that on other chromosomes. The division of the sample into more groups led to further subdivisions in the south (e.g., Figure 3B). Spatial autocorrelation analysis (Bertorelle and Barbujani 1995), based on the binary marker variation, produced correlograms that indicated significant clinal patterns or long-distance differentiation (not shown). The north–south haplogroup structure is therefore a continuum rather than a sharp bipartite division. To understand it further, we have explored the characteristics of the populations in more detail, concentrating on the 22 non-Han populations because of the spread of the Han during historical times (Wen et al. 2004).
Figure 3.
SAMOVA analysis illustrating the geographical divisions identified when four (A) or six (B) groups are specified.
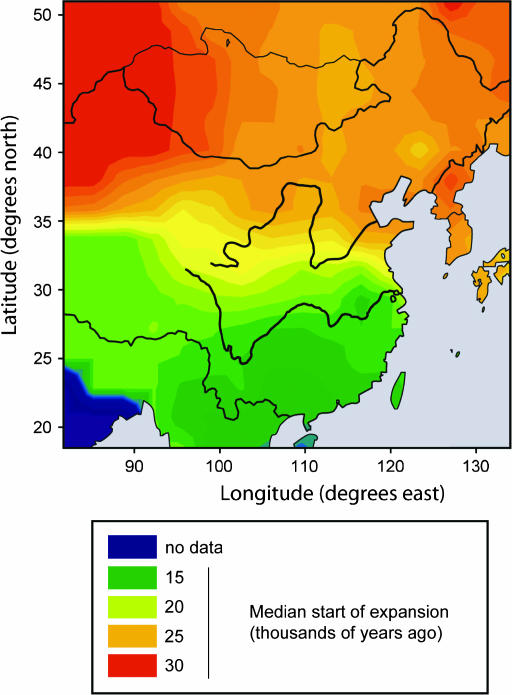
A simple property of a population is the variation it contains, and this can be expressed in a number of ways. A widely used measure, diversity, is so high when 15 STRs are used that the differences between populations are small (Table 2) and difficult to interpret. Reducing the number of STRs to an arbitrary four or three (Table 2, supplemental Figure 1 at http://www.genetics.org/supplemental/) produces a wider range of diversity values, and these are notably higher in the north than in the south. An alternative measure of variation within a population, average squared distance (ASD), shows a similar pattern. BATWING analysis allows demographic parameters of the populations to be explored. Using a model where the population size remains constant for a period and then begins to expand exponentially, we estimated, for each population, posterior values of (1) the effective population size during the constant period, Nposterior; (2) the time at which growth began; (3) the rate of growth per generation, α; and (4) the time to the most recent common ancestor (TMRCA) of the population (Table 2). We again noted substantial variation with latitude. Median Nposterior was higher in the north, the expansion began earlier, the rate of growth was slower, and the TMRCA was longer. Although all of these variables correlated significantly with latitude when examined individually in regression analyses (Table 3), the highest was with expansion time (adjusted R2 = 0.68), compared with 0.40 for the next highest, ASD. Unsurprisingly, a stepwise multiple regression analysis identified expansion time as the best predictor of north–south distance, and only α increased this significantly to reach an adjusted R2-value of 0.75. Thus earlier expansion time in the north and, to a lesser extent, more rapid expansion in the south, account best for the observed north–south differences. We display the expansion times as a contour plot in Figure 4, where the consistent difference between north and south is apparent. Figure 4 suggests, however, that the highest correlation of population expansion may not be with distance due north–south, but with distance along an axis tilted slightly northwest–southeast, and further examination showed that a tilt of ∼10° in fact gave the highest R2-value (0.71 compared with 0.69).
TABLE 3.
Regression analysis
| Independent variable | R2 | Adjusted R2 | P-value |
|---|---|---|---|
| Simple regression (dependent variable: latitude) | |||
| Three STRs | 0.41 | 0.38 | 0.001 |
| ASD | 0.43 | 0.40 | 0.001 |
| Nposterior | 0.30 | 0.27 | 0.008 |
| Expansion time | 0.69 | 0.68 | 0.000 |
| α | 0.24 | 0.20 | 0.022 |
| TMRCA | 0.33 | 0.29 | 0.005 |
| Stepwise multiple regression | |||
| Model 1. Expansion time | 0.69 | 0.68 | 0.000 |
| Model 2. Expansion time + α | 0.87 | 0.75 | 0.000 |
Figure 4.
Contour plot showing the distribution of expansion times. Demographic expansion began earlier in the north than in the south.
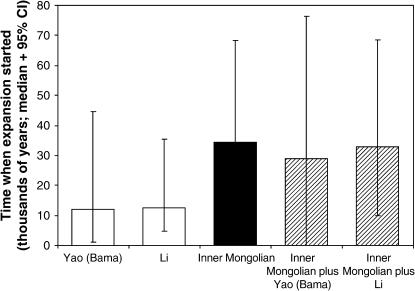
The demographic model used is simple: it assumes that each population is independent and that a constant phase is followed by exponential growth. The other demographic models available in BATWING, constant population size or continuous expansion, are not informative about the expansion time. To explore one consequence of departure from the model used, we investigated artificial population mixtures constructed from combinations of the populations showing the earliest expansion (Inner Mongolians) and those showing the most recent [Yao (Bama) or Li]. The artificial population mixtures showed an early expansion time equivalent to that of the Inner Mongolians (Figure 5), demonstrating that the signature of early expansion is not obliterated by admixture.
Figure 5.
Effect of artificial mixing of population data on estimated expansion time. Median values are plotted, together with their 95% confidence intervals.
DISCUSSION
We consider how our findings on East Asian male variation compare with previous studies and the implications of our work for the understanding of the demographic history of the region.
The distribution of Y-chromosomal haplogroups in East Asia has been extensively documented (e.g., Jin and Su 2000; Karafet et al. 2001; Deng et al. 2004), but these observations have raised questions about the relationship of northern and southern populations that remain unanswered. Su et al. (1999) typed 19 binary markers, 12 of which were chosen because they were already known to be variable in East Asia, and found higher diversity in the south than in the north and that the northern lineages were a subset of the southern ones, leading them to suggest that the northern populations were derived from the south by northward migrations. In contrast, Karafet et al. (2001) used a larger set of 52 binary markers ascertained mainly because of their variation in worldwide populations and discovered higher diversity (mean pairwise differences) in the north and that the northern lineages were not a subset of the southern ones. They concluded that a contribution to the northern populations from Central Asia was likely. The use of preascertained binary markers introduces a bias into estimates of diversity, but STRs are essentially free of this bias because they are variable in all populations. In our samples, STR diversity and ASD measurements were higher in the north than in the south (Table 2), a finding that is not easily reconciled with a largely or exclusively southern origin for the northern populations. It has been suggested that some populations, such as Hui, Uygurs, and Mongolians, have recent admixture with Central Asia and so reliance on them may give a false impression (Shi et al. 2005), but our findings are common to most populations from the north (Table 2).
Our most striking observation was the demographic contrast between north and south, which was explained largely by the variation in the start of population expansion (Tables 2 and 3; Figure 4). Despite the simplified demographic model and wide confidence intervals in the BATWING estimates (Table 2), the median values exhibit a simple and striking pattern: all of the northern estimates lie between 22 and 34 KYA, while all of the southern estimates are between 12 and 18 KYA. These suggest that the northern populations started to expand before the LGM (∼18–21 calendar KYA), while the southern populations started to expand after it. These time estimates are calibrated against historical events (Zhivotovsky et al. 2004) and so do not depend on the assumption of a particular male generation time, but nevertheless are uncertain, and so any interpretation based on them must be regarded with caution. Importantly, however, they are affected little by extensive admixture (Figure 5) and in such a case reflect the earlier expansion time. While extreme northern latitudes were inhospitable to early humans, Siberia has an extensive Upper Paleolithic archaeological record (Kuzmin and Orlova 1998) and a highly productive environment stretched across Asia. This showed an abundance of large animals and has been called the “Mammoth Steppe” (Guthrie 1990). Expansion times calculated in the same way for the Central Asian populations described by Zerjal et al. (2002), excluding those showing recent severe bottlenecks, lay between 24 (13–45) and 36 (16–74) KYA, like those of the northern populations from East Asia. We therefore propose that this cold but rich environment allowed the demographic expansion of populations who learned to exploit the profuse animal resources, and these people contributed in sufficient numbers to the ancestry of the northern populations we have tested to leave a signature in their paternal lineages. In contrast, this environment did not extend to the southern region, and the populations based there expanded only after the end of the LGM as the climate became warmer and more stable. The large-scale use of underground tubers is thought to have begun in the south as early as 15 KYA (Tong 2004), and it is notable that population expansion was subsequently more rapid there. The survival of this distinct demographic signature provides further evidence for the genetic differentiation between north and south and lack of extensive gene flow, leading to a genetic boundary seen initially in classical marker studies (Xiao et al. 2000).
Our conclusions, of course, refer only to the time when expansion began and do not conflict with the notion that population numbers increased much further during Neolithic and historical times. They do, however, illustrate the value of demographic studies at high spatial resolution: a similar analysis of a combined East Asian sample would lead to the conclusion that population growth began at ∼30 KYA [in remarkable agreement with the mtDNA estimate (Forster et al. 2001)] and would miss an important distinction. Further detailed genetic studies of demography in other parts of the world are now needed.
Acknowledgments
We thank all sample donors for their contributions to this work and all those who helped with sample collection and Andrew Flint, Tim Cutts, and Mike Shield for setting up and maintaining BATWING. This work was supported by a Joint Project from the National Natural Science Foundation in China and The Royal Society in the United Kingdom, by Key Teacher support from the Education Office of Heilongjiang Province, and by The Wellcome Trust.
References
- Akey, J. M., M. A. Eberle, M. J. Rieder, C. S. Carlson, M. D. Shriver et al., 2004. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2: e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle, G., and G. Barbujani, 1995. Analysis of DNA diversity by spatial autocorrelation. Genetics 140: 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., B. Shi, X. He, Z. Zhang, J. Xu et al., 2004. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J. Hum. Genet. 49: 339–348. [DOI] [PubMed] [Google Scholar]
- Dupanloup, I., S. Schneider and L. Excoffier, 2002. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11: 2571–2581. [DOI] [PubMed] [Google Scholar]
- Forster, P., A. Torroni, C. Renfrew and A. Röhl, 2001. Phylogenetic star contraction applied to Asian and Papuan mtDNA evolution. Mol. Biol. Evol. 18: 1864–1881. [DOI] [PubMed] [Google Scholar]
- Guthrie, R. D., 1990. Frozen Fauna of the Mammoth Steppe. University of Chicago Press, Chicago.
- Hurles, M. E., B. C. Sykes, M. A. Jobling and P. Forster, 2005. The dual origin of the Malagasy in Island Southeast Asia and East Africa: evidence from maternal and paternal lineages. Am. J. Hum. Genet. 76: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L., and B. Su, 2000. Natives or immigrants: modern human origin in East Asia. Nat. Rev. Genet. 1: 126–133. [DOI] [PubMed] [Google Scholar]
- Jobling, M. A., M. E. Hurles and C. Tyler-Smith, 2004. Human Evolutionary Genetics. Garland Science, New York/Abingdon, UK.
- Karafet, T., L. Xu, R. Du, W. Wang, S. Feng et al., 2001. Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am. J. Hum. Genet. 69: 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, M., R. Chakraborty, J. P. King, M. Bamshad, W. S. Watkins et al., 1998. Signatures of population expansion in microsatellite repeat data. Genetics 148: 1921–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin, V. Y., and L. A. Orlova, 1998. Radiocarbon chronology of the Siberian Paleolithic. J. World Prehist. 12: 1–53. [Google Scholar]
- Macpherson, J. M., S. Ramachandran, L. Diamond and M. W. Feldman, 2004. Demographic estimates from Y chromosome microsatellite polymorphisms: analysis of a worldwide sample. Hum. Genomics 1: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini, S., B. Arredi, R. Chalk and C. Tyler-Smith, 2002. Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res. 30: e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K., M. T. Seielstad, A. Perez-Lezaun and M. W. Feldman, 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Ptak, S. E., and M. Przeworski, 2002. Evidence for population growth in humans is confounded by fine-scale population structure. Trends Genet. 18: 559–563. [DOI] [PubMed] [Google Scholar]
- Reich, D. E., and D. B. Goldstein, 1998. Genetic evidence for a Paleolithic human population expansion in Africa. Proc. Natl. Acad. Sci. USA 95: 8119–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin: A Software for Population Genetics Data Analysis. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva.
- Sherry, S. T., A. R. Rogers, H. Harpending, H. Soodyall, T. Jenkins et al., 1994. Mismatch distributions of mtDNA reveal recent human population expansions. Hum. Biol. 66: 761–775. [PubMed] [Google Scholar]
- Shi, H., Y. L. Dong, B. Wen, C. J. Xiao, P. A. Underhill et al., 2005. Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3–M122. Am. J. Hum. Genet. 77: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, B., J. Xiao, P. Underhill, R. Deka, W. Zhang et al., 1999. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last Ice Age. Am. J. Hum. Genet. 65: 1718–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, E., 2004. Southern Civilization (in Chinese). Chongqing Press, Chongqing, China.
- Wall, J. D., and M. Przeworski, 2000. When did the human population size start increasing? Genetics 155: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weale, M. E., L. Yepiskoposyan, R. F. Jager, N. Hovhannisyan, A. Khudoyan et al., 2001. Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum. Genet. 109: 659–674. [DOI] [PubMed] [Google Scholar]
- Wen, B., H. Li, D. Lu, X. Song, F. Zhang et al., 2004. Genetic evidence supports demic diffusion of Han culture. Nature 431: 302–305. [DOI] [PubMed] [Google Scholar]
- Wilson, I. J., M. E. Weale and D. J. Balding, 2003. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. Ser. A 166: 155–188. [Google Scholar]
- Xiao, C. J., L. L. Cavalli-Sforza, E. Minch and R. F. Du, 2000. Geographic distribution maps of human genes in China (in Chinese). Yi Chuan Xue Bao 27: 1–6. [PubMed] [Google Scholar]
- Xue, Y., T. Zerjal, W. Bao, S. Zhu, S.-K. Lim et al., 2005. Recent spread of a Y-chromosomal lineage in Northern China and Mongolia. Am. J. Hum. Genet. 77: 1112–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal, T., R. S. Wells, N. Yuldasheva, R. Ruzibakiev and C. Tyler-Smith, 2002. A genetic landscape reshaped by recent events: Y-chromosomal insights into central Asia. Am. J. Hum. Genet. 71: 466–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal, T., Y. Xue, G. Bertorelle, R. S. Wells, W. Bao et al., 2003. The genetic legacy of the Mongols. Am. J. Hum. Genet. 72: 717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky, L. A., N. A. Rosenberg and M. W. Feldman, 2003. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am. J. Hum. Genet. 72: 1171–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky, L. A., P. A. Underhill, C. Cinnioglu, M. Kayser, B. Morar et al., 2004. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]