Abstract
Vertebrate and invertebrate dendrites are information-processing compartments that can be found on both central and peripheral neurons. Elucidating the molecular underpinnings of information processing in the nervous system ultimately requires an understanding of the genetic pathways that regulate dendrite formation and maintenance. Despite the importance of dendrite development, few forward genetic approaches have been used to analyze the latest stages of dendrite development, including the formation of F-actin-rich dendritic filopodia or dendritic spines. We developed a forward genetic screen utilizing transgenic Drosophila second instar larvae expressing an actin, green fluorescent protein (GFP) fusion protein (actin∷GFP) in subsets of sensory neurons. Utilizing this fluorescent transgenic reporter, we conducted a forward genetic screen of >4000 mutagenized chromosomes bearing lethal mutations that affected multiple aspects of larval dendrite development. We isolated 13 mutations on the X and second chromosomes composing 11 complementation groups affecting dendrite outgrowth/branching, dendritic filopodia formation, or actin∷GFP localization within dendrites in vivo. In a fortuitous observation, we observed that the structure of dendritic arborization (da) neuron dendritic filopodia changes in response to a changing environment.
NEURONS are morphologically complex cells with axons that can span several meters and dendrites that can branch extensively and contact thousands of other cells. Despite emanating from the same cell, axons and dendrites are dramatically different. While both axons and dendrites contain microtubules and F-actin, the organization of the cytoskeleton in these compartments is different. Axons contain microtubules oriented minus–plus from soma toward growth cone, while dendrites contain mixed polarity microtubules (Baas et al. 1988; Burton 1988). Parallel arrays of F-actin emerge from dendritic shaft microtubules (Rao and Craig 2000), in contrast to F-actin in axon growth cones that exists predominantly in crosslinked/meshwork arrays at the microtubule interface (for review, Dent and Gertler 2003). These divergent cytoskeletal architectures suggest that identifying genetic regulators of dendrite development requires not only relying on superimposing the axon compartment onto the dendrite but also studying the dendrites themselves.
During dendrite outgrowth, the actin and microtubule cytoskeletons must be continuously regulated. Dendrite branch points, which split a single dendrite in two, depend on regulated coordination of actin/microtubule interactions. Dendrite-branching patterns must be at least partially genetically determined as different neuronal subtypes possess characteristic but distinct branching patterns. Genetic determination of dendrite-branching patterns ranges from Drosophila (Bodmer and Jan 1987) to mammals (Cajal 1891), and therefore genetic approaches to analyzing dendrite development should yield significant insights.
During early dendrite development numerous apparently equivalent neurites are extended. Eventually, one process becomes the axon and grows more robustly than the others, while the other processes become dendrites. An early molecular event for dendrite specification requires the transport of minus-end-distal microtubules into dendrites that will distinguish them from plus-end-distal axonal microtubules (Baas et al. 1989). The CHO1/MKLP1 motor protein is required for this early event in dendritic maturation (Sharp et al. 1997; Yu et al. 1997). Antisense knockdown of CHO1/MKLP1 can prevent the formation of minus-end-distal microtubules or induce this loss of microtubule polarity in more mature dendrites (Yu et al. 2000). Experiments like these have relied mainly on reverse genetic approaches in vitro for identifying molecular regulators of dendrite outgrowth. Subsequently, attempts to design in vivo forward genetic assays have been developed for understanding dendrite outgrowth and branching particularly in Drosophila (Gao et al. 1999; for review, Gao and Bogert 2003). Genetic analysis of Drosophila dendrite development has temporally lagged behind axon developmental studies largely due to technical limitations. Drosophila embryos secrete a highly impermeable cuticle that presents technical obstacles for antibody-based dendrite visualization. Axons develop before dendrites and therefore this barrier did not impede antibody-based studies of early axon outgrowth, but novel approaches were required to examine dendrite development. Indeed, new approaches for in vivo analysis of dendrite development using green fluorescent protein (GFP) have been developed for genetic screens in Drosophila embryos (Gao et al. 1999). This past genetic screen (Gao et al. 1999) utilized soluble GFP and analyzed stage 16–17 embryos during the initial stages of dendrite outgrowth and branching.
We sought to develop genetic approaches for analysis of later stages of dendrite development. In mammalian development, following dendrite outgrowth and branching, numerous F-actin-rich structures, including dendritic spines and dendritic filopodia (Fifkova and Delay 1982), form and continuously emerge and retract on dendrites. Previously, we demonstrated that actin-rich dendritic filopodia can also be found on Drosophila dendrites during larval stages and that these structures also continuously emerge and retract on dendrites (Andersen et al. 2005). Forward genetic analysis of late stages of dendrite development have been limited because many assay systems used to analyze dendrite development examine neurons that lack actin-rich dendritic filopodia or dendritic spines on mature dendrites (Wu and Cline 1998). To identify late-stage genetic regulators of dendrite development, we utilized a previously generated (Verkhusha et al. 1999) and characterized (Andersen et al. 2005) transgenic reporter—actin, fused to GFP (actin∷GFP)—that we hypothesized would be readily amenable to forward genetic approaches in Drosophila. Forward genetic screens in Drosophila have been used successfully to identify molecular components required for processes as diverse as embryo segmentation (Wieschaus et al. 1984) and axon development (Seeger et al. 1993; Van Vactor et al. 1993).
We chose to assay later-stage dendrite development of Drosophila dendritic arborization (da) sensory neurons. These Drosophila neurons are large, few in number, and physically dispersed, allowing detailed visualization of intricate dendrite structure (Bodmer and Jan 1987). These neurons also spread underneath a transparent single-cell layer, allowing high-resolution imaging of dendritic structure of neurons expressing the actin∷GFP reporter in a rapid manner suitable for forward genetic approaches. Using this reagent and assay system, we conducted a visual anatomical screen for defects in larval dendrite development by assaying Drosophila second instar larvae bearing >4000 distinctly mutagenized chromosomes. In this initial genetic screen we assayed the X and second chromosomes for genes regulating larval dendrite development. From this novel screen we identified 13 recessive mutations composing 11 complementation groups affecting larval dendrite development. While multiple aspects of late stages of dendrite development could be altered, for simplicity these mutants are classified into one of three categories: branching/outgrowth, filopodia formation, or altered actin localization dendrite mutants. Surprisingly, while preparing for the screen, we observed that dendritic structure could change in response to changing culture conditions.
MATERIALS AND METHODS
Fly stocks:
We use guidelines from FlyBase (http://www.flybase.org) for italicized gene names, as well as upper- or lowercase designations. Proteins are nonitalicized. Fly strains used were Gal4 109(2)80 (Gao et al. 1999), UAS-actin∷GFP (Verkhusha et al. 1999), and UAS-GMA (Dutta et al. 2002). Gal4 109(2)80 was recombined on the UAS-actin∷GFP (yw; Gal4-109(2)80, UAS-actin∷GFP) or the F-actin reporter UAS-GMA (yw; Gal4-109(2)80, UAS-GMA) chromosomes using standard meiotic recombination. The Gal4 109(2)80, UAS-actin∷GFP recombinant was chosen for most experiments for its prominent labeling of dendritic filopodia. Homozygous recombinant second instar larvae were used for all experiments unless otherwise indicated. All flies were maintained at 25° in yeast–cornmeal vials.
EMS mutagenesis and screening:
The Gal4 109(2)80 and UAS-actin∷GFP P elements were recombined onto the second chromosome and isogenized in the yw; BcGla/Cyo background. Mutations were induced by feeding males 15 mm EMS (Sigma, St. Louis) (Lewis 1968) for 20 hr following 5 hr of starvation at room temperature. For the X chromosome, mutagenized yw; Gal4 109(2)80, UAS-actin∷GFP males were mated en masse to FM7i [PActGFP]JMR3/Df(1)JA27; Gal4 109(2)80, UAS-actin∷GFP/Cyo females. Single progeny females (F1) of the genotype yw/FM7i; Gal4 109(2)80, UAS-actin∷GFP were each mated to four males of genotype FM7i; Gal4 109(2)80, UAS-actin∷GFP in a single cornmeal-yeasted vial. The absence of non-Bar male F2 progeny indicated a lethal X chromosome bearing line that could then be expanded for eventual screening. For the second chromosome, mutagenized yw; Gal4 109(2)80, UAS-actin∷GFP males were mated en masse to yw; BcGla/Cyo females. Single male F1 progeny were then backcrossed to six yw; BcGla/Cyo females in a single cornmeal-yeasted vial. F2 siblings of the genotype yw: Gal4 109(2)80, UAS-actin∷GFP/CyO were mated and the absence of non-Cyo F3 progeny indicated a line bearing a lethal second chromosome mutation.
To visually screen lines bearing lethal mutations on either chromosome, six females were mated to three males in a vial containing Drosophila Instant Media/“blue food” (Fisher, Pittsburgh) with a dab of yeast paste. This food produces larvae with greatly reduced gut auto-fluorescence as detected by either epiflourescent or confocal microscopy compared to standard cornmeal-reared larvae. Early second instar larvae were gently removed on day 5 or 6, rinsed in water, and immediately coverslipped in halocarbon oil (Sigma) and assayed for dendritic defects by epifluorescent microscopy. For each line, 25–30 larvae were screened before determining a phenotype, early lethality, or no phenotype.
Mapping:
As an initial mapping approach we took advantage of the duplication/deficiency kits (DK-1 and DK-2) in the Bloomington Stock Center. For mapping purposes, we reasoned that in most cases lethality and dendrite phenotypes would be caused by the same mutation. However, with mappable lethal mutations, we verified that the same deficiency uncovers both the lethality and the phenotype. For each lethal line, we crossed seven females to four males from the appropriate deficiency kit. Every cross for identifying a noncomplementing deficiency was done in duplicate and at least 50 progeny for each vial cross were scored. For the second chromosome, we identified 20 molecularly defined deficiencies of the Exelexis collection to increase coverage of the second chromosome.
Immunohistochemistry and microscopy:
Embryos were dechorionated with 50% bleach for 2 min and fixed with standard 4% paraformaldehyde/50% heptane followed by methanol extractions as described (Brenman et al. 2001). Larvae were dissected while submersed in room temperature 4% paraformaldehyde in phosphate buffered saline (PBS). The anterior and posterior ends of the larvae were dissected and discarded to allow penetration of fixative and antibodies to sensory neurons in abdominal segments. A single incision along the dorsal midline was made and gut tissue was removed to allow access to peripheral nervous system tissue. Dissected larvae were incubated in 4% paraformaldehyde/PBS for 1 hr at room temperature, washed with 0.1% Triton in PBS (PBST), and then incubated for 30 min in blocking solution containing 5% normal goat serum and PBST. Antibody incubations were done in 5% normal goat serum/PBST, and dilutions were mouse monoclonal anti-Futsch 1:200 (22C10, Iowa Hybridoma Bank, Iowa City, IA) and rabbit anti-GFP 1:1000 (Molecular Probes, Eugene, OR). The dissected larvae were incubated with primary antibodies overnight at 4° and washed three times with PBST. Secondary antibodies, cyanine 2 (Cy2)-conjugated goat anti-rabbit (Jackson ImmunoResearch, West Grove, PA) and cyanine 3 (Cy3)-conjugated goat anti-mouse (Jackson ImmunoResearch), were diluted 1:200 in 5% normal goat serum/PBST and incubated with samples for 4 hr at room temperature. Images were from abdominal hemi-segment A6, acquired under similar settings using a Zeiss LSM (Laser Scanning Microscope) 510 (Heidelberg, Germany) confocal microscope with a ×40 oil immersion lens. Briefly, system argon/helium–neon lasers were used to excite Cy2 and Cy3, respectively, and a 2-μm optical slice was taken. Images were sized and cropped with Adobe Photoshop (San Jose, CA), and placed into Adobe Illustrator (San Jose, CA) for labels and arrangement. For live animal imaging, homozygous recombinant X or second chromosome mutant second instar larvae were placed on an air-permeable membrane (cut-out 40-μm cell strainer mesh (Falcon, BD Biosciences, Franklin Lakes, NJ) on a glass slide directly over a 1-cm-diameter hole to allow air diffusion. Larvae were covered in Halocarbon oil 27 (Sigma, St. Louis) and gently coverslipped (22 × 50 mm; Fisher Scientific, Pittsburgh) to restrict movement, but not cause bursting of the body wall. Confocal images of dendrite morphology were obtained with a Zeiss LSM 510 confocal microscope by exciting the 488-nm argon line to excite GFP. Abdominal hemi-segment A6 was imaged for all experiments.
RESULTS
Dendrite development:
Previously, we conducted a forward genetic screen to identify mutations affecting early events in dendrite development, namely dendrite outgrowth and branching in Drosophila embryos (Gao et al. 1999). Subsequently, we documented later events in dendrite development, including the formation of F-actin-rich dendritic filopodia that could be observed with actin, GFP fusion protein (actin∷GFP) expressed in transgenic Drosophila (Andersen et al. 2005).
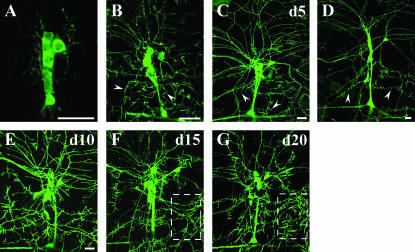
We used the Gal4/UAS system (Brand and Perrimon 1993), including Gal4 109(2)80 (Gao et al. 1999), to drive UAS-actin∷GFP expression to visualize dendritic filopodia on da neurons in the peripheral sensory nervous system (PNS) (Andersen et al. 2005). Gal4 109(2)80 is expressed by all multiple dendrite neurons within the PNS, including all da neurons, the single bipolar dendrite neuron, and the single tracheal-innervating dendrite neuron within the dorsal cluster of the PNS (Gao et al. 1999). We reasoned that the transgenic animals expressing actin∷GFP could be exploited to conduct a novel forward mutagenesis screen aimed at isolating genetic mutations that perturb later dendrite developmental events that occur predominantly during larval stages. We characterized this actin∷GFP reporter in da neuron dendrites during different developmental stages to gain insight for designing a forward screen. In embryos, only minimal actin∷GFP fluorescence can be visualized outside the cell body and even visualizing large dendritic branches is difficult (Figure 1A). During later larval development, however, increasing actin∷GFP fluorescence intensity can be detected in dendrites (Figure 1, B–D). Dendritic filopodia labeled with actin∷GFP first become visible during the first instar larval stage (Figure 1B). As the larva increases in size, dendrites must grow to cover an ever-increasing body-wall size. Previously, we observed that dendrites during embryonic development (stages 14–17) are highly dynamic, extending and retracting during outgrowth (Gao et al. 1999); however, we did not analyze larval dendrites. We investigated whether or not larval dendrites undergo pruning—a process whereby neurons extend many more dendrites than needed and subsequently retract a large subset of them. However, we did not observe extensive dendrite pruning; rather, a similar number of dendrites appear less dense during larval development as they must innervate a much larger growing body wall (note magnification, Figure 1, B–D). Between days 3 and 6 of larval development the number of branch points from the posterior projecting dendrite of neuron ddaA did not change significantly [11.0 branch points ± 2.03 (SD); 10 larvae counted for each day]. The relative stability of larval dendrites is in contrast to highly dynamic dendritic filopodia, which do extend and retract continuously during larval development and were characterized previously (Andersen et al. 2005). Dendritic filopodia greatly increase in number from first to second instar larval stages but increase only slightly from second to third instar larvae (Figure 1, C and D). The actin∷GFP reporter allows for epifluorescent visualization of dendritic filopodia and could therefore be utilized for forward or reverse genetic analysis of larval dendrite development in vivo.
Figure 1.
Development of Drosophila PNS da neurons visualized with actin∷GFP fusion protein. (A) Embryos have strong actin∷GFP fluorescence in cell bodies but very faint fluorescence in dendrites. (B) As first instar larvae develop, dendrite branches become clearly visible with actin∷GFP, and dendritic filopodia first become visible (arrowheads). (C) During the second instar larval stage, significantly more dendritic filopodia are apparent as small spike-like structures become visible with actin∷GFP (arrowheads). (D) During the third instar larval stage similar numbers of dendritic filopodia are visible (arrowheads) compared to second instar larvae; however, dendrites must elongate to cover a much larger body wall and therefore appear less dense. Note the decreasing magnification (A–D). (E–G) Larvae that develop under older, previously churned culture conditions have increasingly thickened and longer filopodia are visualized with actin∷GFP. Day 15 (F, d15) and day 20 (G, d20) larvae demonstrate thicker and longer filopodia (outlined box) compared to day 10 (E, d10) or day 5 (C, d5) larvae. Bars, 20 μm. (All genotypes are yw; 109 (2) 80, UAS-actin∷GFP with A6 hemi-segment dorsal cluster sensory neurons used for imaging; anterior is toward the left and dorsal is toward the top.)
Dendritic structure and the environment:
Before conducting the genetic screen, we wanted to evaluate and control for any variable that could potentially skew the results, including cohort competition and larval density. Initially, we examined larvae at different ages in culture vials to determine whether cohort competition would affect the percentage of mutant-to-nonmutant genotypes as the culture became older. Instead, we made a fortuitous but striking observation: the structure of da neuron dendritic filopodia changes according to the age of the culture. Larvae developing in older, previously churned cultures demonstrated dendritic filopodia that became increasingly thickened, longer, and more intensely labeled with actin∷GFP (Figure 1, E–G). This phenomenon was not due to the age of the parents as transferring old adults to fresh cultures resulted in dendritic filopodia structure similar to that of the progeny of younger parents. In addition, a similar phenomenon was observed in larvae reared in multiple food media, including cornmeal, yeasted-cornmeal, instant media (blue food), and yeasted–instant media. Larvae with the older culture dendritic filopodia phenotype did not recover when transplanted to fresh medium during the 2–3 days before initiation of puparium formation from the second instar larval stage (data not shown). The dendrites themselves, however, did not show any obvious changes under changing environmental conditions. The mechanisms underlying the changes of dendritic filopodia in response to the environment are unknown; however, for this study we determined that we would not screen any mutants from cultures older than 6 days.
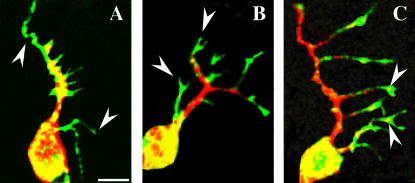
Dendrites are essentially packets of microtubules in the dendrite shaft (Baas et al. 1988) and packets of F-actin in dendritic filopodia or spines (Fifkova and Delay 1982). After characterizing the actin-based compartments in da neuron dendrites, we wanted to characterize the microtubule-based events that occur in da neuron dendrites during development. We utilized an F-actin fluorescent reporter consisting of a fragment of Moesin fused to GFP (GMA) (Dutta et al. 2002). Visualization of the microtubule network was done by immunostaining against Futsch, a MAP-1B-like protein that labels neuronal microtubules (Hummel et al. 2000) (Figure 2). Simultaneous visualization of GMA revealed prominent F-actin-rich growth-cone-like structures at the tips of da neuronal dendrites in embryos (Figure 2B). As embryonic dendrite outgrowth proceeds, “floating microtubules” can be observed in these F-actin-rich growth-cone-like structures (Figure 2C). Interestingly, “floating microtubules” were first identified in growing axon collateral branches and believed to be transported microtubule polymer (Gallo and Letourneau 1999). We reasoned that this genetic screen could also potentially identify mutations affecting the coupling of actin/microtubule interactions in dendrites arising either early in embryos or late in larvae during dendrite development.
Figure 2.
Dendritic arborization neuron dendrite outgrowth proceeds with sequential regulation of F-actin and microtubule domains. (A) An F-actin in vivo reporter, GMA (green), strongly labels tips of growing dendrites (arrowheads), while microtubules (red) visualized with anti-Futsch (MAP-1B-like) immunostaining (22C10) are initially restricted to the cell body and nascent dendrites. (B) Later, growth-cone-like GMA-labeled processes (arrowheads, green) are prominently discernible at tips of growing dendrites. (C) During later stages as dendrites branch, pockets of Futsch immunoreactivity can be seen immersed within the F-actin-rich dendrite growth cone. Images are from stage 15 embryos. Bar, 5 μm.
Screening:
After gaining information about variables affecting dendrite development, we designed a mutagenesis scheme utilizing ethyl methanesulfonate (EMS) to induce chromosomal DNA lesions (details in materials and methods). EMS induces DNA transition mutations and has been used extensively for forward mutagenesis screens in Drosophila (Nusslein-Volhard and Wieschaus 1980; Seeger et al. 1993; Van Vactor et al. 1993). Using information from characterization of the actin∷GFP reporter (Figure 1), we evaluated the larval stage for screening that would optimize the number of mutants. If we were able to obtain significant numbers of mutants with a simpler straightforward traditional loss-of-function screen, we did not conduct a more labor-intensive genetic mosaic screen, especially since there have been no prior larval dendrite screens. Very few dendritic filopodia can be found on first instar larvae da neuron dendrites; therefore we analyzed the earliest possible developmental stage that could be screened with abundant filopodia, the second instar larval stage. In addition, we previously observed that activated CaMKII altered the turnover and dynamics of dendritic filopodia formation and maintenance (Andersen et al. 2005). This suggested that screening first instar larvae might miss mutations that produce phenotypes that become more prevalent over time. We focused our visual screen primarily on abdominal hemi-segment 6 da neurons of the dorsal cluster of the PNS. This segment and cluster are simultaneously easy to visualize and are enriched for class III da neurons (Grueber et al. 2003), which contain the highest densities of dendritic filopodia (Andersen et al. 2005). However, all phenotypes that we detected during the screen were also found in all abdominal hemi-segment clusters on all filopodia-bearing neurons.
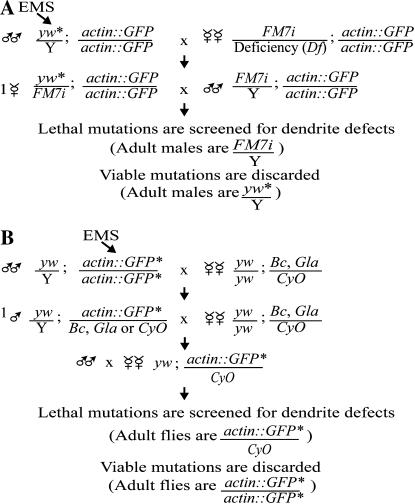
The screen was conducted as a standard recessive F2 screen for the second chromosome and a standard hemizygous recessive screen for the X chromosome (Figure 3). From the outset, we determined that we would keep only mutant lines from the screen that demonstrated phenotypes with at least 50% penetrance and could be confirmed by at least one other observer in a subsequent generation. As a first-pass screen for mutations affecting larval dendrite development we screened ∼1000 individually mutagenized chromsomes bearing lethal mutations on the X and ∼3000 lines on the second chromosome. All the mutants that we isolated had a penetrance of at least 80% among mutant progeny of any given genotype (see Table 1). This is similar to the penetrance that we obtained from a past embryonic dendrite screen (Gao et al. 1999). For mutants with multiple alleles, expressivity between alleles was nearly identical (e.g., bleb and projectile). Furthermore, each phenotype that we observed was similarly expressive from larva to larva and maintained the same phenotype after multiple generations. Approximately 15 lines initially isolated as mutants by a single observer were discarded after additional analysis by other investigators, as the penetrance was <50% or the original phenotype could not be verified in subsequent generations.
Figure 3.
Forward genetic mutagenesis scheme using EMS to induce DNA mutations. (A) A standard hemizygous recessive screen for the X chromosome. FM7i is a “green balancer” expressing GFP in a pattern distinct from the actin∷GFP chromosome, allowing genotyping of progeny by epifluorescent visualization. (B) A standard recessive F2 screen for the second chromosome. Progeny can be genotyped by the presence of one or two insertions of the actin∷GFP transgene detected by epifluorescent visualization. Heterozygous or homozygous mutant second chromosomes correspond to one or two actin∷GFP insertions, respectively.
TABLE 1.
Classification of mutations on the X and second chromosomes affecting larval dendrites
| Mutants | Major dendritic phenotypes | Class | Alleles | % penetrance (n ≥ 20) | Cytological interval |
|---|---|---|---|---|---|
| aspiny | Reduced filopodia formation | 2 | 1 | 90 | 17A1–18A2 Dp(1;Y)W39a |
| bleb | actin∷GFP accumulation in dendrites (reduced dendrite outgrowth) | 3 | 2 | 100/100 | 1F2–2B1 Dp(1;Y)y[2]67g19.1a |
| bristly | Densely clustered filopodia | 2 | 1 | 80 | N.U.b |
| camouflaged | Reduced actin∷GFP localization in filopodia | 3 | 1 | 100 | N.U.b |
| lackluster | Reduced actin∷GFP in dendrites and filopodia | 3 | 1 | 100 | 26D3–26F7 |
| lengthy | Increased filopodia length (abnormal dendrite outgrowth) | 2 | 1 | 100 | 46D7–47F15 |
| mace | Densely clustered filopodia | 2 | 1 | 100 | 38B3–40A3 |
| polka dot | actin∷GFP accumulation in dendrites | 3 | 1 | 100 | 36C2–37B10 |
| projectile | Long filopodia (abnormal branching) | 2 | 2 | 90/90 | N.U.b |
| punctate | Abnormal actin∷GFP accumulation in dendrite patches | 3 | 1 | 100 | 21E3–22B7 |
| scanty | Reduced dendrite outgrowth and branching | 1 | 1 | 100 | 34B12–35C1 |
Cytological interval indicates the extent of the chromosomal deficiency that uncovers the mutation. Secondary mutant phenotypes are indicated by parentheses.
Duplication that rescues the mutation.
Mutation not uncovered by the deficiency kit.
It is not until one actually starts to obtain mutants that one can determine the frequency of obtaining mutants and the range of phenotypes. In a past screen focused on da neuron dendrites in embryos, approximately one mutant was obtained for every 126 lines screened (Gao et al. 1999). In this screen the rate of obtaining mutants was approximately one for every 300 lines screened. This likely reflects the later developmental stage of assay and inability to score strict embryonic lethal mutations. Mutations producing earlier dendrite phenotypes that did not survive until the second instar larval stage would not have been detected in this screen. Approximately one-third of all lethal mutations in Drosophila are embryonic lethals (Nusslein-Volhard et al. 1984) and this could explain our lower frequency of isolating mutants during second instar larval stages.
Phenotypes:
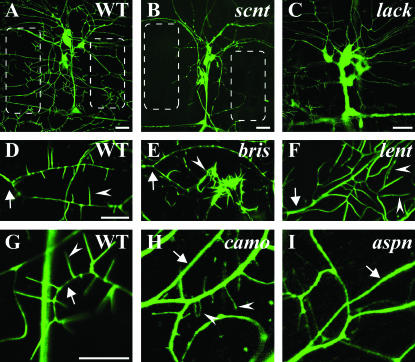
During the visual screening process we realized that mutations affecting larval dendrite development could be roughly classified into one of three groupings (Table 1). The initial stage of dendrite outgrowth and branching is followed temporally by dendritic filopodia formation and enriched actin∷GFP localization in dendrites and dendritic filopodia. Mutants could generally be described as defective in one of these three developmental events. The mutant class representing defects in dendrite outgrowth and branching (class 1) was the least represented. Mutants representing later stages of dendrite development, including abnormal formation of dendritic filopodia (class 2) and altered localization of actin∷GFP within dendrites (class 3), were the most prevalent classes. This is not surprising as the screen was designed to isolate mutations affecting these later events in dendrite development. In fact, for most of actin∷GFP localization and dendrite formation mutants, embryonic defects in dendrites could not be detected (data not shown). Representative mutant phenotypes are shown (Figure 4) and illustrate specificity of some of these phenotypes to one or two aspects of larval dendrite development. While the scanty (scnt) mutant displays dramatic decreases in dendrite outgrowth, the camouflaged (camo) mutant has normal outgrowth and branching but demonstrates very faint actin∷GFP localization in filopodia (Figure 4). Outcrossing the mutation onto a new reporter background failed to increase actin∷GFP accumulation in filopodia (data not shown), suggesting that the mutation is not in the reporter itself. The bristly (bris) mutant demonstrates normal dendrite outgrowth, branching, and actin localization, but dramatic increases in filopodia numbers (Figure 4D). In contrast, aspiny has normal dendritic branching and outgrowth, but decreased filopodia numbers (Figure 4I). While the general classification is somewhat crude and does not preclude a single gene mutation from producing multiple dendrite phenotypes and having multiple genetic functions, it ascribes most mutant phenotypes well (Table 1).
Figure 4.
Representative second instar larval dendrite phenotypes of the three mutant classes: class 1, dendrite outgrowth/branching; class 2, dendritic filopodia formation; class 3, actin∷GFP localization. (A, D, and G) Representative images of actin∷GFP localization, dendritic branching/outgrowth, and filopodia formation in wild-type larvae. (B) The scnt mutant demonstrates a class 1 phenotype with a major reduction in dendrite outgrowth and branching. Note the regions devoid of dendrites in scanty (B, outlined boxes) compared to wild type (A, outlined boxes). (C) The lackluster (lack) mutant (class 3) displays relatively faint localization of actin∷GFP in dendrites and filopodia compared to cell bodies (visible with increased image exposure; compare ratios to A). (E) The bris mutant represents a class 2 phenotype displaying greatly increased filopodia formation at the tip of neuron ddaA. However, dendritic branching is unaffected and the same dendrite branch can be observed in both bristly (E, arrow) and wild type (D, arrow). (F) The lengthy (lent) mutant (class 2) displays longer filopodia (arrowheads) at the tip of neuron ddaA. (H) The camo mutant represents a class 3 mutant containing filopodia only faintly labeled by actin∷GFP in a ddaA neuron (visible with increased image exposure). Note the ratios of fluorescent intensity in wild-type dendrites (G, arrow) to filopodia (G, arrowhead) compared to the fluorescent intensity of camouflaged dendrites (H, arrow) to filopodia (H, arrowheads), indicating decreased actin∷GFP localization in camouflaged mutants. (I) The aspiny (aspn) mutant (class 2) displays greatly reduced filopodia numbers, particularly evident on a dendrite (arrow) devoid of filopodia. Bars, 20 μm (A–C) and 10 μm (D and G).
We also investigated whether genetic interactions could be detected between any of the mutants. All trans-heterozygous interlocus alleles between mutants resulted in second instar larvae with dendrites and dendritic filopodia of a wild-type appearance (data not shown). Double homozygous (hemizygous) mutants with a combination of scanty and any other mutant revealed the scanty mutation to be dominant: the progeny appeared scanty. This may be attributable to the scanty phenotype arising earliest among the mutants during dendrite development. For other double-mutant combinations—except for aspiny and camo—the mutant phenotypes had characteristics of both loci. For mutants that affected filopodia formation only (class 2), camo appeared dominant. Interestingly, on previously churned/old food cultures all mutants continued to display the initially identified phenotype (Figures 4–6) in an additive fashion with the old culture phenotype except for camo and scanty, which both appeared resistant to older culture phenotypes.
Figure 6.
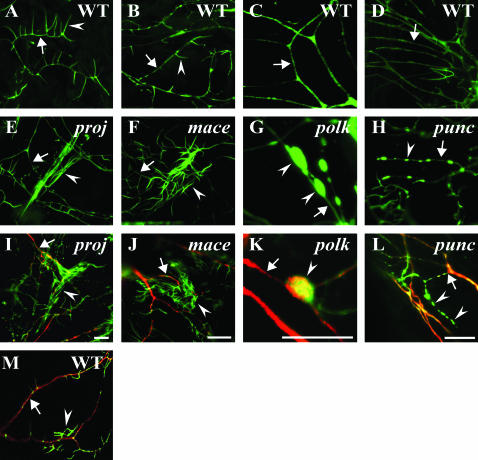
Mutations affecting late-stage dendrite development on the second chromosome. (A–D) Representative dendritic segments imaged from wild-type second instar larvae. In most images, arrows denote dendrite shafts and arrowheads indicate filopodia. (E–H) Corresponding representative dendritic segments imaged from equivalent neurons in mutant second instar larvae. (I–L) Antibody immunostaining of second instar larvae colabeled for actin∷GFP (green, anti-GFP antibody) and the microtubule-binding protein Futsch (red, 22C10 antibody). (E and I) The projectile (proj) mutant displays abnormally long bundled filopodia (arrowhead) that run parallel to each other at the end of the dendrite (arrow). (F and J) The mace mutant exhibits an unusually dense increase in filopodia number (arrowhead) at the end of ddaA neuron dendrites (arrow). (G and K) The polka dot (polk) mutant displays large accumulations of actin∷GFP in dendrites (G, arrowheads) and a diminishment in the microtubule-associated protein, Futsch, near these varicosities (K, arrow). The dendrite swellings contain both actin∷GFP and Futsch (arrowhead, K). (H and L) The punctate (punc) mutant contains dendrites with actin∷GFP puncta and long actin∷GFP-containing processes (arrowheads, H and L), but these processes do not contain Futsch. These processes, however, clearly emerge from a Futsch-immunopositive dendrite (L, arrow). (M) Wild-type dendrites (arrow, red) and filopodia (arrowhead, green) of neuron ddaA immunostained against Futsch (red, 22C10) and actin∷GFP from a region equivalent to proj (I). Bars, 10 μm.
Figure 5.
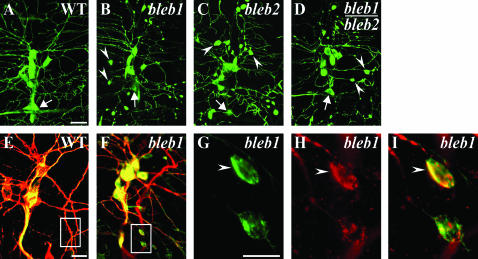
The bleb mutation leads to increased accumulation of actin∷GFP restricted to dendritic compartments. (A) Representative image of wild-type dorsal cluster neurons and dendrites with the bipolar dendrite neuron indicated (arrows, A–D) as a reference landmark. Representative phenotypes of different alleles of bleb as homozygotes (B and C) or trans-heterozygotes (D) demonstrate nearly identical phenotypes with large accumulations of actin∷GFP in dendrite varicosities (B–D, arrowheads). (E and F) Immunostaining of wild type (E) or bleb1 (F) with 22C10 (red, anti-Futsch, MAP-1B-like) and anti-GFP (green, actin∷GFP). Wild-type larvae never demonstrate enlarged swellings in dendrites (E, boxed region) in contrast to the unusual colocalization of microtubule and actin markers in dendritic swellings of bleb mutants (F, boxed region), which are more clearly visible upon higher magnification (G–I, arrowheads). Normally, the actin and microtubule markers are almost entirely distinct in dendrites. Bars, 20 μm (A and E) and 10 μm (G).
Given the intricate relationship between actin and microtubules in developing embryonic dendrites (Figure 2), we chose to colabel mutant larval dendrites against Futsch (MAP1B-like ortholog) (Hummel et al. 2000) and actin∷GFP. While only an initial characterization, these results provide glimpses of perturbed actin–microtubule interactions in larval dendrites. Normally in da larval dendrites, compartments containing F-actin and microtubules almost completely exclude each other (Andersen et al. 2005). In both the bleb and polka dot mutants, an unusual colocalization of actin∷GFP and Futsch is observed in varicosities with surrounding dendrites decreased for Futsch immunostaining (Figures 5 and 6). In contrast, in mutants including projectile and mace, the Futsch-immunostained microtubules in dendrites appear normal with defects detectable only with actin∷GFP at the tips of dendrites. The punctate mutation leads to unusual actin∷GFP patches in dendrites and an occasional neuritic outgrowth containing actin∷GFP but lacking Futsch immunoreactivity. This initial analysis demonstrates distinct aspects of altered cytoskeletal regulation differentially altered by different mutations.
DISCUSSION
Forward genetic approaches to address fundamental questions in biology have been especially invaluable for analysis of Drosophila melanogaster development. The most prominent forward genetic screen in Drosophila was perhaps the most elegant in its simplicity, essentially asking the question, How many genes does it take to make a proper epidermis (Nusslein-Volhard and Wieschaus 1980; Nusslein-Volhard et al. 1984; Wieschaus et al. 1984)? Many well-studied and clinically important molecular regulators and pathways were initially identified in this screen. Since that time, many Drosophila researchers have attempted to devise philosophically similar screens to answer their own particular questions of interest. We are interested in identifying molecular regulators of larval dendrite development and piloted a smaller-scale screen to ask whether we could identify some of the genes necessary to form and maintain larval dendrites.
As the first forward genetic screen using actin∷GFP in transgenic animals to analyze larval dendrite development, our pilot screen examined mutations on two chromosomes. We screened ∼4000 lines on two of the four major Drosophila chromosomes. We estimated that we would need to screen 20,000–30,000 individually mutagenized lines spread across these four chromosomes to obtain >90% of the EMS mutable loci that could produce a phenotype. On the basis of a Poisson distribution with ∼76% of the lines that we generated carrying at least one lethal mutation, we estimate that we screened >5000 lethal mutations compared to a total gene estimate of ∼13,000 in Drosophila (Rubin et al. 2000).
Future experiments:
Isolating additional mutations that produce larval dendrite phenotypes can be addressed by one of many possible solutions. The degree of saturation can be easily addressed in the future by simply screening more lines as alleles obtained increases linearly with lines screened, while loci saturate (Nusslein-Volhard et al. 1984). Alternatively, circumventing early lethality could also be accomplished by generating genetic mosaics particularly using heat-shock FLP/FRT/MARCM (mosaic analysis with a repressible cell marker)-based assay systems (Xu and Rubin 1993; Lee and Luo 1999). Genetic mosaics have been generated to analyze candidate genes using soluble GFP for da neurons in embryos and larvae (Grueber et al. 2002; Sweeney et al. 2002). As a first attempt at a novel screen, we were interested in obtaining only a few genes that could provide entry points to elucidate important genetic pathways. We screened lines bearing only lethal chromosomal mutations, but we did not know that only lethal mutations would produce detectable larval dendrite phenotypes. However, part of our rationale in screening chromosomes bearing lethal mutations was the relative ease of identifying molecular lesions in genes produced by EMS-generated lethal mutations compared to viable mutations. In the future, we believe molecular identification of the mutations already isolated in this screen will provide significant insight into larval dendrite development.
Filopodia as biological mediators:
While this screen focused only on dendritic filopodia on neurons, filopodia are found on many diverse cell types from neurons to fibroblasts and may mediate diverse functions from cell migration to tissue organogenesis. In general, filopodia function to increase the surface area of a cell for milieu sampling and/or to provide a specialized signaling compartment and point of contact. In Drosophila, filopodia have been demonstrated to mediate Notch-mediated lateral inhibition during nervous system specification (De Joussineau et al. 2003) and extend from postsynaptic muscle (myopodia) to contact and guide axon growth cones (Ritzenthaler et al. 2000). While some (class III) da neurons contain abundant filopodia (Grueber et al. 2003), the electrophysiological function of these da neurons is not well understood. However, several groups have demonstrated a potential function of these neurons during thermosensation and some forms of mechanosensation (Liu et al. 2003; Tracey et al. 2003). Functional analysis of these neurons will likely be an active research area in the future and some of these neurons will likely have currently unknown additional functions. One could speculate that regulation of dendritic filopodia density or length could modulate signaling consequences by altering surface area exposed to the environment. Intriguingly, observations that neural structure can change in response to a changing environment (Figure 1) suggest that the same stimulus could evoke a different response over time.
Dendrite cytoskeleton:
Surprisingly, little is known about molecular regulation of the neuronal dendrite cytoskeleton, despite the striking segregation of microtubules in dendrite shafts from F-actin in dendritic spines or dendritic filopodia. Many studies have examined high-resolution subcellular localization of cytoskeletal regulators in fibroblasts (Hall 1998), cell lines, or axonal compartments, which can contain large growth cones particularly in vitro. It is not clear which molecules identified as cytoskeletal mediators in fibroblasts will be applicable to dendrites, although undoubtedly many molecular mediators function in both contexts. However, even within a neuron the axonal and dendritic compartments display unique biochemical and functional properties, suggesting that some dendritic regulators need to be identified in forward genetic screens of dendrite development such as the screen described in this study. We believe that our screen demonstrates one successful approach for identifying dendrite regulators in vivo, without sorting through the vast portfolio of known cytoskeletal regulators or inferring function from a different cellular context.
Classes of phenotypes:
During the screen we were successful in identifying mutations that could largely be classified into one of three phenotypic categories. Of course this does not exclude the probability that multiple cell types during multiple stages of development could require a particular gene function. Mutations affecting dendrite outgrowth and branching were the least-represented mutant class in the screen. This is not surprising, as genes required for early events in dendrite development would likely have earlier unmasked gene functions in other cell types as well. At this point we do not know whether this particular screen may be enriched for isolating mutations preferentially affecting maternally contributed gene products. Eventually, molecular identification of some of these genes should answer that question.
Two classes of mutations may more likely implicate genes for larval dendrite development that may have been missed by earlier screens using soluble GFP (Gao et al. 1999). These mutations predominantly affect actin∷GFP localization or dendritic filopodia morphology/distribution in da neurons. Embryos do not have dendritic filopodia; therefore mutations that predominantly affect filopodia without altering dendrite outgrowth or branching would be difficult to isolate from embryonic dendrite screens. Some of these two classes of mutations also appear the most specific. For instance, mace and projectile have largely normal dendrite branching but have excessive filopodia formation at the tips of dendrites. While these mutations dramatically affect actin-based structure, there are no obvious defects with microtubule-based dendrite shafts. In contrast, other mutations such as bleb and polka dot have perturbations in both microtubule-based and actin∷GFP-based processes. In particular, bleb mutants display a highly unusual colocalization of actin∷GFP and Futsch immunostaining in dendritic varicosities. In the future, more detailed analysis of selected mutants will be done with additional antibody immunostainings and time-lapse analysis with additional in vivo reporters.
Real-time imaging:
Understanding how perturbed cytoskeletal regulation may lead to the observed phenotypes can be addressed in the future with time-lapse analysis. The mutants identified in this screen can be analyzed with multiple fluorescently tagged reporters in live animals to reveal when phenotypes arise and how the cytoskeleton is affected. For example, GMA and cherry∷tubulin (Shaner et al. 2004) could be introduced into mutant backgrounds to observe the F-actin and microtubule compartments simultaneously to determine how different genetic lesions alter the cytoskeleton. Further, photo-bleaching and fluorescence recovery after photo-bleaching experiments could exploit these fluorescently tagged markers to determine if different genetic mutations affect cytoskeletal turnover. Similar approaches have demonstrated that activated CaMKII leads to increased actin turnover in da neuron dendrites (Andersen et al. 2005). Drosophila provides the powerful combination of genetics with high-resolution visualization techniques in live animals to begin to make insights into the complex cell biological regulation of dendritic structure. Developing a convenient assay system to discover and identify genes with new molecular/genetic functions is the essence of exploiting Drosophila genetics to better understand our own biology. Even if the exact anatomical function is distinct from Drosophila to humans, most molecular-signaling pathways, including those associated with human disease (Bernards and Hariharan 2001) and those that impinge on the cytoskeleton (Rubin et al. 2000), are conserved. A forward genetic screen is the first step toward providing the identities of these molecular players in potentially conserved pathways.
Acknowledgments
We thank D. Kiehart for the gift of the GMA fly line, H. Oda for actin∷GFP lines, the Bloomington Stock Center, and the Iowa Hybridoma Bank for monoclonal antibodies. This work was supported by awards to J.E.B. from the Searle Scholars Program and Whitehall Foundation for Neurobiology. A multiphoton/confocal microscope imaging facility funded by P30NS45892 was used extensively.
References
- Andersen, R., Y. Li, M. Resseguie and J. E. Brenman, 2005. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J. Neurosci. 25: 8878–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas, P. W., J. S. Deitch, M. M. Black and G. A. Banker, 1988. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc. Natl. Acad. Sci. USA 85: 8335–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas, P. W., M. M. Black and G. A. Banker, 1989. Changes in microtubule polarity orientation during the development of hippocampal neurons in culture. J. Cell Biol. 109: 3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards, A., and I. K. Hariharan, 2001. Of flies and men: studying human disease in Drosophila. Curr. Opin. Genet. Dev. 11: 274–278. [DOI] [PubMed] [Google Scholar]
- Bodmer, R., and Y. N. Jan, 1987. Morphological differentiation of the embryonic peripheral neurons in Drosophila. Roux's Arch. Dev. Biol. 196: 69–77. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., and N. Perrimon, 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Brenman, J. E., F. B. Gao, L. Y. Jan and Y. N. Jan, 2001. Sequoia, a tramtrack-related zinc finger protein, functions as a pan-neural regulator for dendrite and axon morphogenesis in Drosophila. Dev. Cell 1: 667–677. [DOI] [PubMed] [Google Scholar]
- Burton, P. R., 1988. Dendrites of mitral cell neurons contain microtubules of opposite polarity. Brain Res. 473: 107–115. [DOI] [PubMed] [Google Scholar]
- Cajal, S. R., 1891. Sur la structure de lecorce cerebrale de quelques mammiferes. La Cellule 7: 124–177. [Google Scholar]
- De Joussineau, C., J. Soule, M. Martin, C. Anguille, P. Montcourrier et al., 2003. Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature 426: 555–559. [DOI] [PubMed] [Google Scholar]
- Dent, E. W., and F. B. Gertler, 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40: 209–227. [DOI] [PubMed] [Google Scholar]
- Dutta, D., J. W. Bloor, M. Ruiz-Gomez, K. VijayRaghavan and D. P. Kiehart, 2002. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis 34: 146–151. [DOI] [PubMed] [Google Scholar]
- Fifkova, E., and R. J. Delay, 1982. Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J. Cell Biol. 95: 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo, G., and P. C. Letourneau, 1999. Different contributions of microtubule dynamics and transport to the growth of axons and collateral sprouts. J. Neurosci. 19: 3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. B., and B. A. Bogert, 2003. Genetic control of dendritic morphogenesis in Drosophila. Trends Neurosci. 26: 262–268. [DOI] [PubMed] [Google Scholar]
- Gao, F. B., J. E. Brenman, L. Y. Jan and Y. N. Jan, 1999. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 13: 2549–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber, W. B., L. Y. Jan and Y. N. Jan, 2002. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129: 2867–2878. [DOI] [PubMed] [Google Scholar]
- Grueber, W. B., L. Y. Jan and Y. N. Jan, 2003. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112: 805–818. [DOI] [PubMed] [Google Scholar]
- Hall, A., 1998. Rho GTPases and the actin cytoskeleton. Science 279: 509–514. [DOI] [PubMed] [Google Scholar]
- Hummel, T., K. Krukkert, J. Roos, G. Davis and C. Klambt, 2000. Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26: 357–370. [DOI] [PubMed] [Google Scholar]
- Lee, T., and L. Luo, 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lewis, E. B., 1968. Method of feeding ethyl methane sulfonate to Drosophila males. Dros. Inf. Serv. 43: 193. [Google Scholar]
- Liu, L., O. Yermolaieva, W. A. Johnson, F. M. Abboud and M. J. Welsh, 2003. Identification and function of thermosensory neurons in Drosophila larvae. Nat. Neurosci. 6: 267–273. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard, C., and E. Wieschaus, 1980. Mutations affecting segment number and polarity in Drosophila. Nature 287: 795–801. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard, C., E. Wieschaus and K. Kluding, 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux's Arch. Dev. Biol. 193: 267–282. [DOI] [PubMed] [Google Scholar]
- Rao, A., and A. M. Craig, 2000. Signaling between the actin cytoskeleton and the postsynaptic density of dendritic spines. Hippocampus 10: 527–541. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, S., E. Suzuki and A. Chiba, 2000. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 3: 1012–1017. [DOI] [PubMed] [Google Scholar]
- Rubin, G. M., M. D. Yandell, J. R. Wortman, G. L. Gabor Miklos, C. R. Nelson et al., 2000. Comparative genomics of the eukaryotes. Science 287: 2204–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger, M., G. Tear, D. Ferres-Marco and C. S. Goodman, 1993. Mutations affecting growth cone guidance in Drosophila: genes necessary for guidance toward or away from the midline. Neuron 10: 409–426. [DOI] [PubMed] [Google Scholar]
- Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer et al., 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22: 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sharp, D. J., W. Yu, L. Ferhat, R. Kuriyama, D. C. Rueger et al., 1997. Identification of a microtubule-associated motor protein essential for dendritic differentiation. J. Cell Biol. 138: 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, N. T., W. Li and F. B. Gao, 2002. Genetic manipulation of single neurons in vivo reveals specific roles of flamingo in neuronal morphogenesis. Dev. Biol. 247: 76–88. [DOI] [PubMed] [Google Scholar]
- Tracey, W. D., R. I. Wilson, G. Laurent and S. Benzer, 2003. painless, a Drosophila gene essential for nociception. Cell 113: 261–273. [DOI] [PubMed] [Google Scholar]
- Van Vactor, D., H. Sink, D. Fambrough, R. Tsoo and C. S. Goodman, 1993. Genes that control neuromuscular specificity in Drosophila. Cell 73: 1137–1153. [DOI] [PubMed] [Google Scholar]
- Verkhusha, V. V., S. Tsukita and H. Oda, 1999. Actin dynamics in lamellipodia of migrating border cells in the Drosophila ovary revealed by a GFP-actin fusion protein. FEBS Lett. 445: 395–401. [DOI] [PubMed] [Google Scholar]
- Wieschaus, E., C. Nusslein-Volhard and G. Jurgens, 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. Roux's Arch. Dev. Biol. 193: 296–307. [DOI] [PubMed] [Google Scholar]
- Wu, G. Y., and H. T. Cline, 1998. Stabilization of dendritic arbor structure in vivo by CaMKII. Science 279: 222–226. [DOI] [PubMed] [Google Scholar]
- Xu, T., and G. M. Rubin, 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117: 1223–1237. [DOI] [PubMed] [Google Scholar]
- Yu, W., D. J. Sharp, R. Kuriyama, P. Mallik and P. W. Baas, 1997. Inhibition of a mitotic motor compromises the formation of dendrite-like processes from neuroblastoma cells. J. Cell Biol. 136: 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., C. Cook, C. Sauter, R. Kuriyama, P. L. Kaplan et al., 2000. Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J. Neurosci. 20: 5782–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]