Abstract
Anopheles gambiae is a major malaria vector in Africa and a popular model species for a variety of ecological, evolutionary, and genetic studies on vector control. Genetic manipulation of mosquito vectorial capacity is a promising new weapon for the control of malaria. However, the release of exotic transgenic mosquitoes will bring in novel alleles in addition to the parasite-inhibiting genes, which may have unknown effects on the local population. Therefore, it is necessary to develop methodologies that can be used to evaluate the spread rate of introduced genes in A. gambiae. In this study, the effects and dynamics of genetic introgression between two geographically distinct A. gambiae populations from western Kenya (Mbita) and eastern Tanzania (Ifakara) were investigated with amplified fragment length polymorphisms (AFLPs) and microsatellite markers. Microsatellites and polymorphic cDNA markers revealed a large genetic differentiation between the two populations (average FST = 0.093, P < 0.001). When the two strains were crossed in random mating between the two populations, significant differences in the rate of genetic introgression were found in the mixed populations. Allele frequencies of 18 AFLP markers (64.3%) for Mbita and of 26 markers (92.9%) for Ifakara varied significantly from F5 to F20. This study provides basic information on how a mosquito release program would alter the genetic makeup of natural populations, which is critical for pilot field testing and ecological risk evaluation of transgenic mosquitoes.
MALARIA, the most important parasitic disease in the world, results in 1–2 million deaths annually worldwide and in >90% of the deaths that occur in sub-Saharan Africa (World Health Organization 1998; Breman et al. 2004). The economic burden caused by malaria is enormous (Gallup and Sachs 2001). As the Plasmodium parasites become increasingly resistant to antimalarial drugs, and as insecticide-resistant anopheline mosquitoes become increasingly prevalent (White 2004), a strong need to develop new control measures exists. One promising and novel transmission-blocking approach is to use genetically modified mosquitoes that can effectively inhibit malaria parasite development (Collins et al. 2000; Alphey et al. 2002). Over the past several years, researchers have made remarkable progress in the development of mosquito germline transformation techniques (Catteruccia et al. 2000) and in the identification of parasite-inhibiting molecules (Beernsten et al. 2000; Blair et al. 2000; Jacobs-Lorena 2003; Moreira et al. 2004). However, little is known about the spread of transgenes in targeted natural mosquito populations (Braig and Yan 2001).
Ideally, the released transgenic mosquitoes should have a genetic background similar to that of the targeted populations in nature. Releasing individuals with a different genetic background would bring novel alleles, in addition to the parasite-inhibiting genes, to the target population. This may change the genetic makeup and fitness of the target population and thus may have unintended consequences on the transmission of malaria or other pathogens vectored by the anopheline mosquitoes. However, no information yet exists on how introducing mosquitoes with a different genetic background would change the genetic makeup of a local population. The aim of this study was to determine the dynamics of allele frequency and gene introgression when two genetically distinct mosquito populations were mixed.
Population hybridization and subsequent gene introgression have been investigated in plants (Arias and Rieseberg 1994; Mao et al. 1995; Rieseberg et al. 1995, 1999; Brar and Khush 1997). For example, high rates of population hybridization and gene introgression have been reported between the cultivated sunflower and its wild progenitor, Helianthus annuus (Arias and Rieseberg 1994; Rieseberg et al. 1995), but only 2.4% of the genome from the 10 rearranged linkages was transferred. Thus, chromosomal rearrangements appear to provide an effective mechanism for reducing or eliminating introgression in rearranged chromosomal segments. The rates of gene introgression vary among species of sunflowers. Rieseberg et al. (1999) reported a low rate of introgression in four sympatric H. petiolaris populations, with average marker frequencies per population ranging from 0.006 to 0.026.
In African malaria mosquitoes, gene introgression may be occurring between sympatric Anopheles gambiae and A. arabiensis species, but introgression is not uniform across the genome (Garcia et al. 1996). For example, X chromosome inversions do not introgress well due to selective elimination in hybrids and backcrosses. However, the consequence of hybridization on gene introgression within strains of the same species has not yet been studied.
This study investigated genetic introgression between two geographically distinct populations of A. gambiae mosquitoes. We used microsatellites and polymorphic cDNA markers to determine the degree of genetic differentiation and to test whether random or assortative mating occurred when populations are mixed. We then used amplified fragment length polymorphisms (AFLPs) to test dynamics of genetic introgression in different genome regions. This information aids understanding of the genetic structure of natural anopheline populations and helps develop strategies for field testing of transgenic mosquitoes in the future.
MATERIALS AND METHODS
Mosquito rearing and maintenance:
Two A. gambiae strains (Mbita and Ifakara) were used in this study. The Mbita strain was from wild mosquitoes collected at Mbita Point (000 25′S, 340 13′E), western Kenya, and it has been maintained in the laboratory since 1999. The Ifakara strain was originally collected from Njage village, 70 km from Ifakara, southeast Tanzania, and it has been maintained under laboratory conditions since 1996. The two strains were maintained at Mbita Point Field Station, International Centre of Insect Physiology and Ecology, Kenya. The mosquito colonies were maintained at ambient tropical temperature (maximum average monthly temperature of 28.4° and minimum temperature of 15.0° during 1970–2000) and under artificial light conditions provided by fluorescent tubes. The detailed protocol for mosquito larvae and adult rearing is described in Menge et al. (2005).
Experimental population:
We mixed equal proportions of 1-day-old Ifakara and Mbita strains (100 Ifakara females, 100 Ifakara males, 100 Mbita females, and 100 Mbita males) in a 30 × 30 × 30-cm cage. The F1 larvae were reared to adulthood under similar conditions. Subsequent generations from F2 to F20 were obtained by mass mating, with a population size of ∼1000 individuals.
For genetic introgression analysis, we set up two populations that mixed the Mbita and Ifakara strains. Briefly, the F1 generation hybrids from the two strains were obtained by mixing a population (F0) of 100 1-day-old Ifakara females, 100 Ifakara males, 100 Mbita females, and 100 Mbita males together in a single cage. The F1 larvae were reared to adulthood under similar conditions. Subsequent generations, F2–F20, were obtained by allowing an equal number of males and females of the previous generation (100 each) to mate and start a subsequent generation. Two replicate experiments were set up following the same procedure.
Establishment of population for genetic mapping:
To determine the genome position of the molecular markers, we set up an F2 population using pairwise mating between an Mbita male and an Ifakara female and an F1 intercross. A total of 123 F2 individuals were used for linkage map construction.
Screening and genotyping of polymorphic AFLP markers:
We extracted genomic DNA from individuals in the mapping population (two parents and 123 individuals of the F2 generation) following the phenol/chloroform method (Severson 1997). All individuals were subjected to genotyping with AFLP markers using the methods described by Zhong et al. (2003). A total of 100 AFLP primer combinations were screened for polymorphism. The majority of the primer combinations (85 pairs) produced polymorphic fragments between the Mbita and Ifakara strains. We selected 18 pairs of AFLP primer combinations for segregation analysis of the F2 intercross populations on the basis of reproducibility and the extent of polymorphism (Table 1). Only polymorphic fragments that could be scored unambiguously were used for linkage map construction. With the 18 pairs of AFLP primer combinations, we genotyped the Ifakara and Mbita parental populations and the F1, F5, F10, F15, and F20 populations for a total of 482 individuals. Only clearly amplified fragments were scored (168 AFLP markers).
TABLE 1.
AFLP primer sequences and combinations used for Anopheles gambiae genome mapping and genetic introgression
| EcoRI primers (5′–3′) |
| L1: GACTGCGTACCAATTCACA |
| L2: GACTGCGTACCAATTCAAC |
| L3: GACTGCGTACCAATTCAAG |
| L4: GACTGCGTACCAATTCACC |
| L5: GACTGCGTACCAATTCACG |
| L7: GACTGCGTACCAATTCAGC |
| L8: GACTGCGTACCAATTCAGG |
| MseI primers(5′–3′) |
| B1: GATGAGTCCTGAGTAA CAC |
| B2: GATGAGTCCTGAGTAA CAT |
| A3: GATGAGTCCTGAGTAA CTC |
| A4: GATGAGTCCTGAGTAA CTA |
| A8: GATGAGTCCTGAGTAA CGA |
| A9: GATGAGTCCTGAGTAA CGT |
| A10: GATGAGTCCTGAGTAACGG |
| A18: GATGAGTCCTGAGTAA TAG |
| A19: GATGAGTCCTGAGTAATTC |
| A21: GATGAGTCCTGAGTAATTT |
| A27: GATGAGTCCTGAGTAATCC |
| Primer combinations used for genome mapping |
| L1/A3, L1A8, L1/A9, L1/A10, L1/A18, L1/A21 |
| L2/A18 |
| L3/A8, L3/A19 |
| L4/B1 |
| L5/B2 |
| L7/A3, L7/A4, L7/A18, L7/A27 |
| L8/A4, L8/A18, L8/A27 |
Screening and genotyping of polymorphic microsatellite markers:
To determine the degree of genetic differentiation between the Mbita and Ifakara populations, and to determine whether the two mosquito strains mate randomly when they are mixed in cages, we screened the two strains using 36 microsatellite markers (Zheng et al. 1996). Microsatellite analysis was conducted using the methods described in Nyanjom et al. (2003). Ten markers (AGXH503, AGXH131, and AGXH678 on chromosome X; AG2R7, AG2H187, AG2H786, AG2H603, and AG2H117 on chromosome II; and AG3H158 and AG3H29C on chromosome III) were polymorphic in the mating population and thus suitable for mapping. We also screened 13 cDNA markers (Della Torre et al. 1996) for polymorphism between the two strains, but only 1 marker (U50474) was found to be polymorphic. Using these 10 microsatellite loci and one cDNA locus as anchor markers, the AFLP linkage map developed in this study was integrated into the previously published genetic maps of A. gambiae (Della Torre et al. 1996; Zheng et al. 1996). To examine whether the mixed populations were mating randomly, we genotyped the F1 and F5 populations using five microsatellite markers for which the two strains exhibited the highest degree of genetic differentiation (AGXH131, AG2R7, AG2H117, AG2H187, and AG3H158).
Data analysis:
Chi-square analysis was conducted on all markers to test whether they were segregating according to the expected Mendelian ratio of 3:1 for dominant AFLP markers and 1:2:1 for codominant microsatellite and cDNA markers. Using MapMaker version 3.0 (Lincoln et al. 1992), a genetic linkage map based on AFLP, microsatellite, and cDNA markers was generated from the F2 mapping population derived from the cross between Mbita and Ifakara strains. A LOD score of 5.0 was used as a threshold value for defining a linkage group. The map distance was calculated using Kosambi's mapping function (Kosambi 1944).
To determine whether random mating occurred in the mixed cage populations, the Hardy–Weinberg equilibrium (HWE) was tested for each microsatellite locus using the probability test implemented in the GENEPOP software (Raymond and Rousset 1995). Significance of deviation from HWE was adjusted for multiple comparisons, using the sequential Bonferroni procedure (Rice 1989). We used Wright's FST (Weir and Cockerham 1984) to quantify genetic differentiation between the Mbita and Ifakara populations. This analysis of microsatellite genotype data was performed with FSTAT computer software (Goudet 1995).
Because AFLP loci segregate as dominant markers, they were scored as presence (1) or absence (0) in each population. The frequencies of the null recessive allele and of the dominant allele were estimated using the methods of Lynch and Milligan (1994). Analysis of molecular variance (AMOVA), which partitions the genotypic variance into components attributable to different hierarchical levels, was implemented in the software program ARLEQUIN (version 2.000; Schneider et al. 2000). Two population structure models were examined. In the first model, the two parental populations were defined as one group and AMOVA partitioned the total variance into components due to differences between parental populations and differences between individuals within parental populations. In the second model, the five progeny populations (F1, F5, F10, F15, and F20) were defined as a group to be compared with their parents, and thus AMOVA partitioned the total variance into components due to differences between parents and their progeny and differences between groups within populations. AMOVA was based on the pairwise squared Euclidian distances among the AFLP genotypes. The significance of the variance components at the different hierarchical levels was assessed with a permutation procedure (1000 permutations).
RESULTS
AFLP marker polymorphism in A. gambiae populations and linkage map:
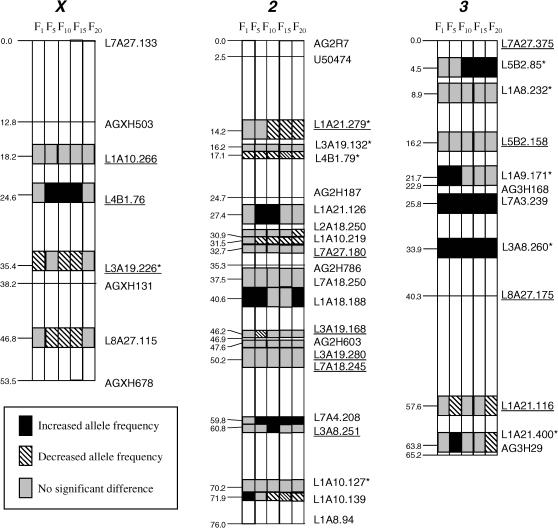
Eighteen AFLP primer combinations yielded 168 informative markers. Each primer combination generated an average of 9.3 informative markers. The L1A8 and L1A21 primer combinations produced the highest number of qualified polymorphic fragments (13 each), whereas the L8/A18 primer combination had the fewest polymorphic fragments (2). Among the 168 informative AFLP markers, 124 markers were assigned to three linkage groups (20 on chromosome X, 73 on chromosome II, and 31 on chromosome III). In addition, 10 microsatellite markers (3 on chromosome X, 5 on chromosome II, and 2 on chromosome III), and 1 cDNA marker (U50474) were mapped in relation to the AFLP markers (Figure 1). Linkage groups ranged in length from 53.5 to 76.0 cM. The total map consists of 194.7 cM, with a marker density of 1.44 markers/cM (194.7 cM for a total of 135 markers). However, only a partial linkage map consisting of 28 traceable AFLP markers, with notable change in allele frequency observed up to the F20 generation, is presented (Figure 1).
Figure 1.
AFLP allele frequency change patterns in the mixed population of the Ifakara and Mbita strains of Anopheles gambiae. Solid markers represent significantly higher allele frequency than that of the Mbita parent strain, and hatched markers are those with significantly lower frequencies at P < 0.001 after a sequential Bonferroni correction. Shaded markers show no significant change in allele frequencies from that of the Mbita parent strain. An asterisk (*) indicates markers with alleles specific to one parent strain. Microsatellite markers (names beginning with AG) are used as anchors for AFLP marker orientation. AFLP markers are designated by the EcoRI primer name, the MseI primer name, and the molecular size of the fragment. For example, marker L7A27.133 represents the 133-bp fragment amplified by EcoRI primer L7 and MseI primer A27, whereas L1A10.266 represents the 266-bp fragment amplified by EcoRI primer L1 and MseI primer A10.
Genetic and HWE analyses:
Mbita and Ifakara populations were significantly different, as revealed by the 14 codominant microsatellite markers (FST = 0.093, P < 0.001). Ten markers (AGX1D1, AGX H131, AG2R7, AG2H117, AG2H187, AG2H79, AG2H46, AG3H158, AG3H29C, and AG3H33C) exhibited a large genetic differentiation, with the FST-value ranging from 0.061 to 0.266 (Table 2). Five of these were thus used to test whether the mixed populations exhibited any assortative mating. A random-mating test was based on genotype data of 50 individuals from each population (a total of 300 individuals). Among the microsatellite markers used, the AGXH131 locus was in good agreement with HWE in all the tested populations (parents, F1, and F5), whereas the AG2H187 locus exhibited significant departures from the HWE in all populations (Table 3). Because not all markers consistently showed significant deviation from HWE in F1 and F5 populations (Table 3), there was no evidence to suggest that assortative mating had occurred in the mixed populations.
TABLE 2.
Number of microsatellite alleles (n) and FST values indicate genetic differentiation between the Ifakara and Mbita strains of A. gambiae
| Chromosome location
|
n
|
||||
|---|---|---|---|---|---|
| Marker | Ifakara | Mbita | FST | P | |
| AGX1D1 | X: 1D | 4 | 5 | 0.081 | <0.001 |
| AGXH503 | X: (4) | 3 | 3 | 0.026 | 0.094 |
| AGXH131 | X: 3 | 3 | 3 | 0.266 | <0.001 |
| AG2R7 | 2L | 2 | 3 | 0.146 | <0.001 |
| AG2H117 | 2L | 3 | 3 | 0.061 | <0.001 |
| AG2H187 | 2R | 2 | 2 | 0.129 | <0.001 |
| AG2H79 | 2R | 7 | 5 | 0.122 | <0.001 |
| AG2H46 | 2L: 7A | 7 | 10 | 0.106 | <0.001 |
| AG2H603 | 2L: 25 | 3 | 3 | 0.000 | 0.896 |
| AG2H786 | 2R: 12 | 2 | 3 | 0.018 | 0.075 |
| AG3H158 | 3R | 4 | 4 | 0.148 | <0.001 |
| AG3H29C | 3R: 29C | 4 | 3 | 0.228 | <0.001 |
| AG3H33C | 3R: 33C | 2 | 9 | 0.045 | <0.001 |
TABLE 3.
Inbreeding coefficients (FIS) in parental populations and in F1 and F5 mixed populations
| F1
|
F5
|
|||||
|---|---|---|---|---|---|---|
| Microsatellite marker | Mbita | Ifakara | Population 1 | Population 2 | Population 1 | Population 2 |
| AGXH131 | 0.22 | 0.08 | 0.22 | 0.04 | 0.17 | 0.22 |
| AG2R7 | 0.07 | — | 0.14** | −0.03 | −0.01 | 0.12* |
| AG2H117 | 0.05 | −0.14 | 0.00 | −0.04 | −0.32*** | 0.23* |
| AG2H187 | 0.78*** | 0.80*** | 0.85*** | 0.93*** | 0.95*** | 0.74*** |
| AG3H158 | 0.00 | −0.07 | 0.32*** | 0.21 | 0.53*** | 0.09*** |
P < 0.05, **P < 0.01, and ***P < 0.001 for the Hardy–Weinberg equilibrium test.
Genetic differentiation between the parental population and the F1–F20 generations:
AMOVA results indicated a strong differentiation between the Ifakara and Mbita populations. Partitioning the variation within and between populations using AMOVA showed that 10.0% of the genetic variability existed between the two populations (FST = 0.0994, P < 0.001; Table 4). A comparison of parents and progeny populations showed that 4.3% of the variation existed between the two groups (P = 0.47). Approximately 7.8% of the variation was partitioned among the progenies (FST = 0.0125; P < 0.001). These results suggest that parental strains affect introgression rates of AFLP alleles.
TABLE 4.
Partitioning of molecular variance within and among parent populations (Ifakara and Mbita) and mixed populations (F1–F20 generations) using the analysis of molecular variance (AMOVA) method
| Source of variation | d.f. | SS | MS | VC | % | P-value |
|---|---|---|---|---|---|---|
| Between Ifakara and Mbita | 1 | 117.29 | 117.29 | 2.39 | 10.00 | <0.001 |
| Among progeny populations | 4 | 713.28 | 178.32 | 1.95 | 7.80 | <0.001 |
| Among parents and progeny populations | 1 | 265.15 | 265.15 | 1.11 | 4.29 | 0.047 |
| Within populations | 475 | 10,831.04 | 22.80 | 22.80 | 88.01 | <0.001 |
SS, sums of squares; MS, mean squares; VC, variance component; %, apportionment of genetic variability; P-value, level of significance based on 1000 permutations.
Pairwise genetic differentiation (FST) between the parents and progeny generations was highly significant (Table 5), with a maximum FST of 0.1889 (between Ifakara and the F15 generation) and a minimum FST of 0.0574 (between the F15 and F20 generations). It should be noted that the pairwise genetic difference between Ifakara and the progeny generations (mean FST = 0.1584) is bigger than the pairwise genetic difference between Mbita and the progeny generations (mean FST = 0.0892). The F15 generation showed the highest FST values (0.1889 for Ifakara and 0.1229 for Mbita). These results indicate that the Mbita strain contributed more alleles to the progeny of the mixing populations.
TABLE 5.
Pairwise genetic differentiation index (FST) among parental and progeny populations based on AFLP data
| Ifakara | Mbita | F1 | F5 | F10 | F15 | F20 | |
|---|---|---|---|---|---|---|---|
| Mbita | 0.0994 | — | |||||
| F1 | 0.1110 | 0.0371 | — | ||||
| F5 | 0.1671 | 0.0904 | 0.0724 | — | |||
| F10 | 0.1752 | 0.0955 | 0.1010 | 0.0613 | — | ||
| F15 | 0.1889 | 0.1229 | 0.1127 | 0.0793 | 0.0430 | — | |
| F20 | 0.1496 | 0.1001 | 0.0971 | 0.0788 | 0.0706 | 0.0574 | — |
All FST-values are statistically significant at P < 0.001.
Allele frequency changes in the progeny of the mixing populations:
Significant differences in allele frequency were found between the two parental populations and five progeny populations (FST = 0.0924, P < 0.001). Starting from the F1 up to the F20 generations, 18 of the 28 markers were found to have significantly changed allele frequency (Figure 1). Frequencies of alleles specific to the Mbita strain were significantly changed from F5 to F20 in 18 markers (64.3%), including 9 with increased allele frequency and 7 with decreased allele frequency. The allele frequency of 1 marker (L1A21.400) first increased from F1 to F5 and then decreased from F10 to F20. Another marker (L1A10.139) had increased allele frequency at F1 and then decreased from F5 to F20 (Figure 1). However, frequencies of alleles specific to the Ifakara strain changed significantly from F5 to F20 in 26 markers (92.9%); most of them showed decreased frequency. These results reveal that Mbita alleles showed higher genetic introgression and spread rate in the progeny of this mixing population in A. gambiae.
DISCUSSION
This study describes methods that can be used to evaluate the spread rate of introduced genes in a target population. We used 28 AFLP markers that exhibited a large difference in allele frequencies between two geographically distinct strains of A. gambiae to investigate genetic introgression dynamics when the mosquito populations were mixed. We found that alleles specific to the Mbita strain had a higher genetic introgression and spread rate than those specific to the Ifakara strain. This study provides basic information on how release of mosquitoes with a genetic background different from that of a targeted population may alter the genetic makeup of the targeted population. This is critical for ecological risk evaluation of future release of transgenic mosquitoes.
Microsatellite markers were used as anchor markers to produce an AFLP linkage map and to determine whether any assortative mating occurred in our experimental populations. Due to high frequencies of allele sharing between the Kenyan and Tanzanian A. gambiae populations, we could not use the microsatellite markers to track allele frequency changes in the mixed populations. Therefore, we screened AFLP markers to identify strain-specific molecular markers or traceable markers that exhibited highly contrasted frequencies between the two strains. AFLP analysis is a DNA fingerprinting technique that gives several hundred fragments in a single PCR reaction, and it is possible that some strain-specific fragments may be identified (Rieseberg et al. 1999). AFLP markers have been widely used in genetic diversity and genetic mapping studies and are useful for studying genetic introgression in wild populations (Maughan et al.1996; Zhu et al. 1998; Yan et al. 1999; Zhong et al. 2004). AFLP's dominant nature is a disadvantage because each marker is represented by only two alleles, one of them being unamplifiable (null), thereby decreasing their accuracy in allele frequency estimation in comparison with codominant markers (Lynch and Milligan 1994). However, considerable progress has been made in the development of algorithms that provide less biased estimators, especially at low null-allele frequencies (Lynch and Milligan 1994; Zhivotovsky 1999). In this study, the AFLP procedures proved to be a useful tool for assessing genetic variability and genetic introgression in A. gambiae because band profiles with the primer combinations were reproducible and followed Mendelian inheritance patterns. We identified a substantial number of polymorphic, strain-specific, traceable markers that are useful for studying gene introgression in mixed mosquito populations. Our results indicated that allele frequency change varied among genome regions, as shown by significant increases in allele frequencies from generation F5 to F20 in some of the AFLP loci and by demonstrable decreases in other loci.
Unlike the western Africa A. gambiae populations that often exhibit a higher level of chromosomal inversion polymorphisms, the eastern Africa population has lower chromosomal inversion variability (Coluzzi et al. 2002). In our study, the two strains (Ifakara and Mbita) were derived from different divisions of the gene pool from two areas in eastern Africa; they were separated by a high genetic differentiation (FST > 0.1). Knowledge of the karyotype of these two populations would have been helpful, especially for interpreting the genetic diversity results from microsatellite or AFLP markers. However, this information is not available for the particular strains that we used.
Our results are consistent with other findings (Della Torre et al. 1997) that gene introgression due to exotic mosquito introduction occurs at different rates across the mosquito genome. The alleles of the Mbita strain showed a higher introgression rate than those of the Ifakara strain. This may be because the study was conducted at Mbita Point in Kenya where the Mbita mosquito strain originated. It may be better adapted to the local climatic or rearing conditions and thus exhibited higher fitness than the Ifakara strain from Tanzania. The rates of gene spread in the subsequent mixed populations depended on genome location, fitness of the hybrid generations and the parental populations, the selection pressure on different loci, and other factors (Besansky et al. 1994; Della Torre et al. 1997; Black and Lanzaro 2001; Mezzera and Largiadèr 2001). Using the same two strains of A. gambiae, we found that population hybridization led to increased fecundity, increased adult longevity, larger body size, and increased blood-feeding ability up to the F10 generation (Menge et al. 2005). Increased hybrid fitness would favor intermediate allele frequencies in mixed populations.
In conclusion, we have used AFLP methods to evaluate the spread rate of introduced genes in A. gambiae populations. We found that 28 mapped AFLP markers showed a significant difference in allele frequencies between the Mbita and Ifakara populations and that allele frequencies of 18 markers changed significantly from generation F5 to F20 in the mixed populations. The methods developed in this study may be adapted for pilot semifield studies on the gene introgression effects of transgenic mosquito release programs. An urgent need exists for methods and schemes that will be useful for evaluating the ecological and public health impacts of transgenic mosquito release programs. Field trials should be conducted only if the likelihood of achieving public health benefits is high while undesirable ecological or environmental impacts are very low (Enserink 2002). Studies in fully contained laboratory conditions and semifield systems are important first steps (Scott et al. 2002). Further studies are needed to understand the factors influencing the spread rate of introduced genes in seminatural and natural conditions.
Acknowledgments
Two anonymous reviewers provided constructive criticisms and suggestions. This work was supported by National Institutes of Health grant D43 TW01505 and by a United Nations Development Programme/World Bank/World Health Organization Special Programme for Research and Training in Tropical Diseases grant A10429.
References
- Alphey, L., C. B. Beard, P. F. Billingsley, M. Coetzee, A. Crisanti et al., 2002. Malaria control with genetically manipulated insect vectors. Science 298: 119–121. [DOI] [PubMed] [Google Scholar]
- Arias, D. M. L., and H. Rieseberg, 1994. Gene flow between cultivated and wild sunflowers. Theor. Appl. Genet. 89: 655–660. [DOI] [PubMed] [Google Scholar]
- Beernsten, B. T., A. A. James and B. M. Christensen, 2000. Genetics of mosquito vector competence. Microbiol. Mol. Biol. Rev. 64: 115–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky, N. J., J. R. Powell, A. Caccone, D. M. Hamm, J. A. Scott et al., 1994. Molecular phylogeny of the Anopheles gambiae complex suggests genetic introgression between principal malaria vectors. Proc. Natl. Acad. Sci. USA 91: 6885–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, W. C. T., and G. C. Lanzaro, 2001. Distribution of genetic variation among chromosomal forms of Anopheles gambiae s.s: Introgressive hybridization, adaptive inversions, or recent reproductive isolation? Insect Mol. Biol. 10: 3–7. [DOI] [PubMed] [Google Scholar]
- Blair, C. D., Z. N. Adelman and K. E. Olsen, 2000. Molecular strategies for interrupting arthropod-borne virus transmission by mosquitos. Clin. Microbiol. Rev. 13: 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig, H. R., and G. Yan, 2001. The spread of genetic constructs in natural insect populations, pp. 251–314 in Genetically Engineered Organisms: Assessing Environmental and Human Health Effects, edited by D. K. Letourneau and B. E. Burrows. CRC Press, Washington, DC.
- Brar, D. S., and G. S. Khush, 1997. Alien introgression in rice. Plant Mol. Biol. 35: 35–47. [PubMed] [Google Scholar]
- Breman, J. G., M. S. Alilio and A. Mills, 2004. Conquering the intolerable burden of malaria: what's new, what's needed: a summary. Am. J. Trop. Med. Hyg. 71: 1–15. [PubMed] [Google Scholar]
- Catteruccia, F., T. Nolan, T. G. Loukeris, C. Blass, C. Savakis et al., 2000. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature 405: 959–962. [DOI] [PubMed] [Google Scholar]
- Collins, F. H., L. Kamau, H. A. Ranson and J. M. Vulule, 2000. Molecular entomology and prospects for malaria control. B. WHO 78: 1412–1423. [PMC free article] [PubMed] [Google Scholar]
- Coluzzi, M., A. Sabatini, A. Della Torre, M. A. Di Deco and V. Petrarca, 2002. A polytene chromosome analysis of the Anopheles gambiae species complex. Science 298: 1415–1418. [DOI] [PubMed] [Google Scholar]
- Della Torre, A., G. Favia, G. Mariotti, M. Coluzzi and K. D. Mathiopoulos, 1996. Physical map of the malaria vector Anopheles gambiae. Genetics 143: 1307–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della Torre, A., L. Merzagora, J. R. Powell and M. Coluzzi, 1997. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics 146: 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink, M., 2002. Malaria: ecologists see flaws in transgenic mosquito. Science 297: 30–31. [DOI] [PubMed] [Google Scholar]
- Gallup, J. L., and J. D. Sachs, 2001. The economic burden of malaria. Am. J. Trop. Med. Hyg. 64: 85–96. [DOI] [PubMed] [Google Scholar]
- Garcia, B. A., A. Caccone, K. D. Mathiopoulos and J. R. Powell, 1996. Inversion monophyly in African anopheline malaria vectors. Genetics 143: 1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet, J., 1995. FSTAT version 1.2: a computer program to calculate F-statistics. J. Hered. 86: 485–486. [Google Scholar]
- Jacobs-Lorena, M., 2003. Interrupting malaria transmission by genetic manipulation of anopheline mosquitoes. J. Vector Borne Dis. 40: 73–77. [PubMed] [Google Scholar]
- Kosambi, D. D., 1944. The estimation of map distance from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lincoln, S., M. Daly and E. Lander, 1992. Constructing Genetic Maps With MAPMAKER/EXP 3.0. Whitehead Technical Report, Ed. 2. Whitehead Institute, Cambridge, MA.
- Lynch, M., and B. G. Milligan, 1994. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 3: 91–99. [DOI] [PubMed] [Google Scholar]
- Mao, L., Q. Zhou, X. P. Wang, H. Hu and L. H. Zhu, 1995. RFLP analysis of the progeny from Oryza alta Swallen x Oryza sativa L. Genome 38: 913–918. [DOI] [PubMed] [Google Scholar]
- Maughan, P., M. Saghai-Maroof, G. Buss and G. Huestis, 1996. Amplified fragment length polymorphism (AFLP) in soybean: species diversity, inheritance, and near-isogenic line analysis. Theor. Appl. Genet. 93: 392–401. [DOI] [PubMed] [Google Scholar]
- Menge, D. M., T. Guda, D. Zhong, A. Pai, G. Yan et al., 2005. Fitness consequences of Anopheles gambiae population hybridization. Malaria J. 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzera, M., and C. R. Largiadèr, 2001. Comparative analysis of introgression at three marker classes: a case study in a stocked population of brown trout. J. Fish Biol. 59(Suppl. A): 289–305. [Google Scholar]
- Moreira, L. A., J. Wang, F. H. Collins and M. Jacobs-Lorena, 2004. Fitness of anopheline mosquitoes expressing transgenes that inhibit Plasmodium development. Genetics 166: 1337–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyanjom, S. R. G., H. Chen, T. Gebre-Michael, E. Bekele, J. Shililu et al., 2003. Population genetic structure of Anopheles arabiensis mosquitoes in Ethiopia and Eritrea. J. Hered. 94: 457–463. [DOI] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. GENEPOP (version 1.2): a population genetics software for exact test and ecumenicism. J. Hered. 86: 248–249. [Google Scholar]
- Rice, W. R., 1989. Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Rieseberg, L. H., C. R. Linder and G. J. Seiler, 1995. Chromosomal and genic barriers to introgression in Helianthus. Genetics 141: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., J. Whitton and K. Gardner, 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152: 713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., D. Roessli and L. Excoffier, 2000. Arlequin: A Software for Population Genetics Data Analysis, Ver. 2.000. Genetics and Biometry Lab, Department of Anthropology, University of Geneva, Geneva.
- Scott, T. W., W. Takken, B. G. J. Knols and C. Boete, 2002. The ecology of genetically modified mosquitoes. Science 298: 117–119. [DOI] [PubMed] [Google Scholar]
- Severson, D. W., 1997. RFLP analysis of insect genomes, pp. 309–320 in The Molecular Biology of Insect Disease Vectors, edited by J. M. Crampton, C. B. Beard and C. Louis. Chapman & Hall, London.
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- White, N. J., 2004. Antimalarial drug resistance. J. Clin. Invest. 113: 1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 1998. Fact Sheet 94. World Health Organization, Geneva.
- Yan, G., J. Romero-Severson, M. Walton, D. D. Chadee and D. W. Severson, 1999. Population genetics of the yellow fever mosquito in Trinidad: comparisons of amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) markers. Mol. Ecol. 8: 951–963. [DOI] [PubMed] [Google Scholar]
- Zheng, L., M. Q. Benedict, A. J. Cornel, F. H. Collins and F. C. Kafatos, 1996. An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae. Genetics 143: 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky, L. A., 1999. Estimating population structure in diploids with multilocus dominant DNA markers. Mol. Ecol. 8: 907–913. [DOI] [PubMed] [Google Scholar]
- Zhong, D., A. Pai and G. Yan, 2003. Quantitative trait loci for susceptibility to tapeworm infection in the red flour beetle. Genetics 165: 1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, D., A. Pai and G. Yan, 2004. AFLP-based genetic linkage map for the red flour beetle (Tribolium castaneum). J. Hered. 95: 53–61. [DOI] [PubMed] [Google Scholar]
- Zhu, J., M. D. Gale, S. Quarrie, M. T. Jackson and G. J. Bryan, 1998. AFLP markers for the study of rice biodiversity. Theor. Appl. Genet. 96: 602–611. [Google Scholar]