Abstract
Heterotrimeric RecBCD enzyme unwinds and resects a DNA duplex containing blunt double-stranded ends and directs loading of the strand-exchange protein RecA onto the unwound 3′-ending strand, thereby initiating the majority of recombination in wild-type Escherichia coli. When the enzyme lacks its RecD subunit, the resulting RecBC enzyme, active in recD mutants, is recombination proficient although it has only helicase and RecA loading activity and is not a nuclease. However, E. coli encodes for several other exonucleases that digest double-stranded and single-stranded DNA and thus might act in consort with the RecBC enzyme to efficiently promote recombination reactions. To test this hypothesis, I inactivated multiple exonucleases (i.e., exonuclease I, exonuclease X, exonuclease VII, RecJ, and SbcCD) in recD derivatives of the wild-type and nuclease-deficient recB1067 strain and assessed the ability of the resultant mutants to maintain cell viability and to promote DNA repair and homologous recombination. A complex pattern of overlapping and sometimes competing activities of multiple exonucleases in recD mutants was thus revealed. These exonucleases were shown to be essential for cell viability, DNA repair (of UV- and γ-induced lesions), and homologous recombination (during Hfr conjugation and P1 transduction), which are dependent on the RecBC enzyme. A model for donor DNA processing in recD transconjugants and transductants was proposed.
AN exchange of genetic material between homologous DNA sequences is a fundamental process occurring in all living organisms, which enables both genomic stability and plasticity. The central recombination substrate is a single-stranded (ss) DNA region covered with a DNA strand-exchange protein of a RecA class of proteins (for reviews, see Kuzminov 1999; Chedin and Kowalczykowski 2002). It can originate from a gap in a DNA duplex or from processing of a double-stranded (ds) DNA end, which is more frequent and results in a 3′-ending single-stranded tail. Escherichia coli and some other bacteria use the RecBCD enzyme or its homologs to activate DNA duplexes for recombination (Chedin and Kowalczykowski 2002). This heterotrimeric helicase/nuclease processes blunt or nearly blunt dsDNA ends, occurring in a cell during normal growth (replication) or upon introduction of foreign DNA (by Hfr conjugation, infection by P1, T4, or λ-phage, etc.) or inflicted by exogenous agents that cause double-stranded breaks (DSBs) (UV, ionizing radiation, etc.) (reviewed in Arnold and Kowalczykowski 1999). It is not surprising, therefore, that E. coli devoid of RecBCD enzyme functions (recB and recC null mutants) is poorly viable (Capaldo et al. 1974), DNA repair deficient (Willets and Mount 1969), and unable to create new combinations of alleles (Emmerson 1968).
Biochemical studies showed that, upon binding to DNA duplex, the RecBCD enzyme unwinds DNA processively and concomitantly degrades both unwound strands, wherein a 3′-ending strand is degraded more vigorously than its complementary strand (Dixon and Kowalczykowski 1993). However, the activities of the RecBCD enzyme are changed upon its encounter and interaction with Chi (χ), an octanucleotide DNA sequence. While retaining its helicase activity, the Chi-modified enzyme exhibits attenuated nuclease activity of changed polarity (from dominant 3′–5′ to 5′–3′) and also starts facilitating RecA protein polymerization onto the unwound 3′-ending overhang (Anderson and Kowalczykowski 1997a,b). The resultant RecA nucleofilament then searches for a homologous DNA sequence and pairs with its complement, displacing the other strand and thus creating a structure called the D-loop. The D-loop may give rise to a Holliday junction and/or may initiate replisome assembly and subsequent recombination-dependent replication (reviewed in Kuzminov 1999).
Many genetic and biochemical studies indicate that interaction with a Chi site affects the RecD subunit (reviewed recently in Ðermić et al. 2006). Conversely, RecBC enzyme behavior resembles that of Chi-modified RecBCD enzyme. RecBC unwinds DNA, does not recognize the Chi site, and constitutively loads RecA protein onto the unwound 3′-ending strand (Churchill et al. 1999). There is, nevertheless, a marked difference between the two enzymes: namely, RecBC enzyme lacks any significant nuclease activity, whereas Chi-modified RecBCD is a 5′–3′ exonuclease (Anderson et al. 1997). Despite the lack of nuclease activity of RecBC enzyme, recD mutants are fully viable, DNA repair proficient, and even hyperrecombinogenic (Chaudhury and Smith 1984; Lovett et al. 1988).
Although RecBCD enzyme is the strongest exonuclease (ExoV) in E. coli, some other exonucleases are also active in that organism. SbcCD is also an exonuclease (of 3′–5′ polarity) that acts on dsDNA and an endonuclease that acts on ssDNA (Connelly et al. 1999). In addition, several other exonucleases in E. coli that act mainly on ssDNA (ssExos) exist. They include processive 5′–3′ exonuclease RecJ (Lovett and Kolodner 1989) and exonuclease I (ExoI) of 3′–5′ polarity (Lehman and Nussbaum 1964). Exonuclease X (ExoX) also digests in the 3′–5′ direction, but is distributive and attacks dsDNA too, albeit with lower affinity (Viswanathan and Lovett 1999). The processive exonuclease VII (ExoVII) resects both 3′- and 5′-ending single-strands (Chase and Richardson 1974).
Since the functions of some of these ssExos were shown to be moderately required for reactions dependent on the RecBCD enzyme (Miesel and Roth 1996; Razavy et al. 1996; Viswanathan and Lovett 1998, 1999), which itself is such a powerful nuclease, I wondered whether their functions might be even more required in RecBC-catalyzed reactions, meaning that cell viability, as well as DNA repair and homologous recombination proficiency of recD mutants, might also rely on exonuclease activity. Previous reports have shown that inactivation of RecJ exonuclease moderately affects viability and recombination in recD mutants, whereas UV survival is severely reduced in the double recD recJ mutant, relative to either single mutant (Lloyd et al. 1988; Lovett et al. 1988). However, it is known that ssExos play overlapping roles in the aforementioned recombination reactions, as well as in mismatch repair (Burdett et al. 2001; Viswanathan et al. 2001), mutational avoidance (Viswanathan and Lovett 1998), and stabilization of tandem repeats (Feschenko et al. 2003). Therefore, I inactivated single or multiple exonucleases in recD mutants to determine their roles in RecBC-dependent cell viability, DNA repair (in γ- and UV-irradiated bacteria), and homologous recombination (during Hfr conjugation and P1 transduction).
MATERIALS AND METHODS
Bacterial strains and growth conditions:
E. coli strains (Table 1) were grown in Luria–Bertani (LB) broth and on LB broth agar plates (Miller 1992). When required, media were supplemented with the appropriate antibiotics: 100 μg ml−1 ampicillin (Ap), 10 μg ml−1 tetracycline (Tc), 20 μg ml−1 kanamycin (Km), and 15 μg ml−1 chloramphenicol (Cm). New strains were constructed by P1 transduction as described earlier (Miller 1992). All experiments were done with exponentially growing cultures, grown at 37° until they reached an optical density at 600 nm (OD600) of 0.3.
TABLE 1.
E. coli K-12 strains
| Strain | Relevant genotype | Source or derivation |
|---|---|---|
| AB1157 and derivatives | ||
| AB1157 | F−thr-1 ara-14 leuB6 Δ(gpt-proA)62 lacY1 tsx-33 supE44 galK2 λ− Rac−hisG4 rfbD1 mgl-5 rpsL31 kdgK51 xyl-5 mtl-1 argE3 thi-1 qsr− | Bachmann (1996) |
| DE100 | recD1903∷Tn10d(tet) | P1 DPB271 × AB1157 (Tcr) |
| DE1050 | recD1903∷Tn10d(tet) ΔxonA300∷cat | P1 STL2694 × DE100 (Cmr) |
| DE1051 | recD1903∷Tn10d(tet) ΔexoX1∷npt | P1 STL4525 × DE100 (Kmr) |
| DE1052 | recD1903∷Tn10d(tet) ΔxseA18∷amp | P1 STL4537 × DE100 (Apr) |
| DE1053 | recD1903∷Tn10d(tet) ΔsbcD300∷kan | P1 JJC260 × DE100 (Kmr) |
| DE1054 | recD1903∷Tn10d(tet) ΔxonA300∷cat ΔxseA18∷amp | P1 STL4537 × DE1050 (Apr) |
| DE1055 | recD1903∷Tn10d(tet) ΔxonA300∷cat ΔexoX1∷npt | P1 STL4525 × DE1050 (Kmr) |
| DE1056 | recD1903∷Tn10d(tet) ΔxonA300∷cat ΔsbcD300∷kan | P1 JJC260 × DE1050 (Kmr) |
| DE1057 | recD1903∷Tn10d(tet) ΔxseA18∷amp ΔexoX1∷npt | P1 STL4525 × DE1052 (Kmr) |
| DE1058 | recD1903∷Tn10d(tet) ΔxseA18∷amp ΔsbcD300∷kan | P1 JJC260 × DE1052 (Kmr) |
| DE1059 | recD1903∷Tn10d(tet) ΔxonA300∷cat ΔexoX1∷npt ΔxseA18∷amp | P1 STL4537 × DE1055 (Apr) |
| DE1060 | recD1903∷Tn10d(tet) ΔxonA300∷cat ΔxseA18∷amp ΔsbcD300∷kan | P1 JJC260 × DE1054 (Kmr) |
| DE1061 | recD1903∷Tn10d(tet) recJ2052∷Tn10kan | P1 STL113 × DE100 (Kmr) |
| DE1062 | recD1903∷Tn10d(tet) ΔxonA300∷cat recJ2052∷Tn10kan | P1 STL113 × DE1050 (Kmr) |
| DE1063 | recD1903∷Tn10d(tet) ΔxseA18∷amp recJ2052∷Tn10kan | P1 STL113 × DE1052 (Kmr) |
| RIK151 | recB1067 recD1903∷Tn10d(tet) | Jockovich and Myers (2001) |
| DE1081 | recB1067 recD1903∷Tn10d(tet) ΔxonA300∷cat | P1 STL2694 × RIK151 (Cmr) |
| DE1082 | recB1067 recD1903∷Tn10d(tet) ΔexoX1∷npt | P1 STL4525 × RIK151 (Kmr) |
| DE1083 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp | P1 STL4537 × RIK151 (Apr) |
| DE1084 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp ΔxonA300∷cat | P1 STL2694 × DE1083 (Cmr) |
| DE1085 | recB1067 recD1903∷Tn10d(tet) ΔexoX1∷npt ΔxonA300∷cat | P1 STL2694 × DE1082 (Cmr) |
| DE1086 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp ΔexoX1∷npt | P1 STL4525 × DE1083 (Kmr) |
| DE1087 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp ΔexoX1∷npt ΔxonA300∷cat | P1 STL2694 × DE1086 (Cmr) |
| DE1091 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp ΔxonA300∷cat ΔsbcD300∷kan | P1 JJC260 × DE1084 (Kmr) |
| DE1088 | recB1067 recD1903∷Tn10d(tet) recJ2052∷Tn10kan | P1 STL113 × RIK151 (Kmr) |
| DE1089 | recB1067 recD1903∷Tn10d(tet) ΔxonA300∷cat recJ2052∷Tn10kan | P1 STL113 × DE1081 (Kmr) |
| DE1090 | recB1067 recD1903∷Tn10d(tet) ΔxseA18∷amp recJ2052∷Tn10kan | P1 STL113 × DE1083 (Kmr) |
| DE101 | recB268∷Tn10 | Salaj-Šmic et al. (2000) |
| RIK123 | recB1067 | Jockovich and Myers (2001) |
| Other strains | ||
| Hfr3000 | Hayes PO1 proAB+ | Bachmann (1996) |
| IRB100 | As Hfr3000 but λ+ | Laboratory collection |
| BW6156 | Hfr PO3 proAB+ metB1 | M. Berlyn |
| IRB110 | As BW6156 but metB+ | P1 Hfr3000 × BW6156 (Met+) |
| DPB271 | MG1655 recD1903∷Tn10d(tet) | Laboratory collection |
| STL2694 | ΔxonA300∷cat | Viswanathan and Lovett (1998) |
| STL4525 | ΔexoX1∷npt | S. T. Lovett |
| STL4537 | ΔxseA18∷amp | S. T. Lovett |
| STL113 | recJ2052∷Tn10kan | Viswanathan and Lovett (1998) |
| JJC260 | ΔsbcD300∷kan | B. Michel |
Chromosomal recombination assays:
Inheritance of the chromosomal Pro+ marker was assessed by Hfr conjugational crosses as described in Ðermić et al. (2005). Log-phase cultures (OD600 of 0.3) of donor Hfr3000 or IRB110 and recipient cells (recD mutants and their exonuclease-deficient derivatives) were mixed at a ratio of 1:10 and allowed to mate for 30 min at 37°. Then proAB+ transconjugants were selected on minimal M9 agar plates (Miller 1992) containing glucose (0.4% v/v), vitamin B1 (1 μg ml−1), and all required amino acids except proline. The plates also contained streptomycin (100 μg ml−1) to counterselect against donors and were incubated at 37° for 24–48 hr. Recombination efficiency in each Hfr cross has been expressed in relation to the titer of donors. Recombination efficiencies in the crosses have been given relative to the recD control strains DE100 or RIK151. To monitor λ-prophage zygotic induction during Hfr crosses (as an internal control for the efficiency of DNA transfer), lysogenic IRB100 donor was mixed with recipients at a 1:10 ratio and incubated for 45 min at 37° (Ðermić and Trgovčević 1999). The mating mixture was serially diluted in 67 mm phosphate buffer (pH 7.0), and infective centers were determined by plating with Hfr3000 bacteria into LB broth top agar to LB broth agar plates and incubated at 37° for 24 hr. Since IRB100 does not transfer its DNA to Hfr3000 indicator bacteria due to surface exclusion, the titer of infective centers represents the number of infected transconjugants. This was confirmed in control crosses involving only the IRB100 donor and Hfr3000 indicator strain (without recipients), in which <0.1% infective centers per total donor titer was observed.
P1vir phage propagated on Hfr3000 were used to transfer the chromosomal Pro+ marker to recipients at a multiplicity of infection of 0.1, as described (Salaj-Šmic et al. 2000). A transducing mixture was incubated for 20 min at 37° and then centrifuged and resuspended in 67 mm phosphate buffer containing 5 mm sodium citrate. Pro+ transductants were selected on minimal M9 agar plates (Miller 1992) containing glucose (0.4%), vitamin B1 (1 μg ml−1), 5 mm sodium citrate, and all required amino acids except proline and counted after 24–48 hr of incubation at 37°. Recombination efficiency in each P1 cross has been expressed relative to the phage titer and then compared to the crosses with DE100 or RIK151 parental recD strains.
Irradiations:
Bacteria to be UV irradiated were serially diluted in 67 mm phosphate buffer (pH 7.0) and plated on LB broth agar plates. The plates were immediately irradiated with UV light (254 nm) from a low-pressure mercury germicidal lamp of UV crosslinker (Amersham Biosciences), at room temperature. The colonies of survivors were scored after 24–48 hr incubation at 37° in the dark.
Exponentially growing bacteria were irradiated with a 150 Gy dose of γ-rays from a 60Co source, with a dose rate of 10.1 Gy−1 at 0°. The γ-irradiated bacteria were serially diluted in 67 mm phosphate buffer and then plated on LB agar plates. After 24–48 hr incubation at 37°, the colonies of survivors were scored. The survival of irradiated bacteria has been expressed as a fraction of the unirradiated control and represented in relation to survival of recD parental strains DE100 or RIK151.
Cell viability and mass doubling time:
A fresh overnight culture was diluted 100-fold in fresh LB broth medium and incubated at 37° with aeration. Its OD600 (which is a measure of total cell concentration in a population) was determined at intervals with a Novaspec II colorimeter (Amersham Pharmacia Biotech). Bacterial cultures were incubated until they reached an OD600 of 0.3. Each bacterial culture was then serially diluted in 67 mm phosphate buffer and plated on LB broth agar plates to determine a viable cell count. Since in recD mutants, as in wild-type bacteria, each cell is able to develop a colony, parental recD mutant strains DE100 or RIK151 were references for assessing viabilities of their derivatives devoid of exonuclease(s), i.e., the fraction of their total cell titer that formed colonies.
Time needed for a growing bacterial culture to double its OD600 represents its mass doubling time and was expressed in relation to mass doubling times of parental DE100 or RIK151 strains.
RESULTS
DNA repair in recD mutants devoid of activities of multiple exonucleases:
Two series of strains carrying the recD1903∷Tn10d(tet) mutation were examined for DNA repair: one derivative of standard rec+ strain AB1157 and the other of RIK123, a nuclease-deficient recB1067 derivative of AB1157. DNA repair capacity of these mutants was assessed by monitoring their (i) viability and mass doubling time (to indicate how efficiently they repair endogenous DNA damage formed during normal growth) and (ii) UV and γ-survival (to determine the efficiency of repairing lesions caused by exogenous genotoxic agents), as expressed in relation to the parental strains.
DNA repair in recB+ recD genetic background:
An isogenic set of recD mutant strains was constructed by replacing wild-type genes of multiple exonucleases (xonA codes for ExoI, xseA for a subunit of ExoVII, exoX for ExoX, sbcD for a subunit of SbcCD, and recJ for RecJ) with their null alleles by P1 transduction.
As shown in Table 2, when ssExos of 3′–5′ polarity of action (i.e., ExoI, ExoX, and ExoVII) were singly inactivated in recD genetic background, each of the four events monitored was left essentially unaffected. A combined inactivation of two exonucleases of 3′ polarity also did not produce strong effects, except in the RecD− ExoI− ExoVII− mutant, which had markedly reduced UV and γ-survival. About 70% fewer survivors were observed after 20 J m−2 of UV and 150 Gy of γ-rays, compared to the recD control.
TABLE 2.
Recombinational DNA repair, cell viability, and homologous recombination in recD mutants lacking activities of various exonucleases
| Relative cellular viability
|
Relative fractional survival
|
λ zygotic induction in Hfr cross
|
Relative recombination frequency
|
||||
|---|---|---|---|---|---|---|---|
| Strain | Description | UV (20 J m−2) | γ (150 Gy) | Pro+ transconjugants | Pro+ transductants | ||
| DE100 | RecD− | 1.0a | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DE1050 | RecD− ExoI− | 0.85 | 1.0 | 0.68 | 0.95 | 0.22 | 0.73 |
| DE1051 | RecD− ExoX− | 1.0 | 1.18 | 1.20 | 0.91 | 1.16 | 1.14 |
| DE1052 | RecD− ExoVII− | 0.98 | 0.95 | 0.93 | 0.93 | 1.07 | 1.28 |
| DE1057 | RecD− ExoX− ExoVII− | 0.96 | 0.82 | 0.62 | 0.79 | 0.97 | 1.18 |
| DE1055 | RecD− ExoI− ExoX− | 0.68 | 0.98 | 0.76 | 0.76 | 0.0089 | 0.31 |
| DE1056 | RecD− ExoI− SbcD− | 0.83 | 0.96 | 0.61 | ND | ND | ND |
| DE1054 | RecD− ExoI− ExoVII− | 0.81 | 0.33 | 0.29 | 1.0 | 0.78 | 1.07 |
| DE1059 | RecD− ExoI− ExoVII−ExoX− | 0.72 | 0.13 | 0.24 | 0.92 | 0.80 | 1.13 |
| DE1060 | RecD− ExoI− ExoVII− SbcD− | 0.56 | 0.09 | 0.12 | ND | ND | ND |
| DE1061 | RecD− RecJ− | 0.68 (0.86b; 1.0cd) | 0.00058 (0.0007c; ∼0.002b; ∼0.07d) | 0.18 (∼0.3d) | 0.91 (0.74c) | 0.135 (0.012c; 0.28d; 0.34b) | 0.75 (0.07c) |
| DE1062 | RecD− RecJ− ExoI− | 0.20 | 0.00008 | 0.00024 | 0.68 | 0.0063 | 0.13 |
| DE1063 | RecD− RecJ− ExoVII− | 0.009 | 0.00004 | <0.0001 | 0.62 | 0.00051 | <0.001 |
| DE101 | RecB− | 0.39 | 0.0013 | 0.0018 | 0.89 | 0.0051 | 0.0036 |
ND, not determined.
Cellular viability of 1.0 corresponds to 1.3 × 108 recD colony-forming units ml−1 at an OD600 of 0.3. UV and γ-survival of 1.0 correspond to 21 and 75 survivors/100 irradiated recD cells, respectively. Frequency 1.0 for λ zygotic induction corresponds to 41 infective centers/100 IRB100 donors. Recombination frequency of 1.0 corresponds to 22 Pro+ transconjugants/100 Hfr3000 donor cells and 6 × 10−5 Pro+ transductants per P1 plaque-forming unit. Recombination efficiency is corrected for the viability of the recipients. All values are averages of at least three independent experiments except for zygotic induction in Hfr crosses, which are averages of two determinations.
Reported by Lloyd et al. (1988).
Reported by Lovett et al. (1988).
Reported by Ivančić-Baće et al. (2005).
Inactivation of all three 3′–5′ ssExos further impaired all the events monitored, suggesting that these enzymes have overlapping roles in DNA repair in a recD genetic background. The quadruple RecD− ExoI− ExoVII− ExoX− mutant grew ∼15% more slowly and was ∼30% less viable than the control. The UV and γ-survival of the mutant were even more affected, being ∼8- and 4-fold lower than that of the control, respectively. A recD mutant deficient in activities of the ExoI, ExoVII, and SbcCD nucleases was about half as viable as the control and had ∼10-fold lower UV and γ-survival, suggesting that SbcCD nuclease is synergistic with ExoI (confirming an earlier report by Seigneur et al. 1999) and ExoVII. Unfortunately, I was unable to construct the quintuple RecD− ExoI− ExoVII− ExoX− SbcCD− mutant, lacking activities of the major 3′–5′ exonucleases, due to lack of suitable selective cotransducible markers. Together, these data show that redundant functions of ExoI, ExoVII, ExoX, and SbcCD exonucleases are required for an efficient DNA repair in the recD mutant background.
There are only two known ssExos of 5′–3′ polarity in E. coli, RecJ and ExoVII (which also possesses 3′–5′ activity). When recJ mutation was introduced into the recD strain, the resultant mutant became ∼30% less viable. A much stronger effect was observed on its UV survival, which was >1000-fold lower than that of the recD parental strain. This extreme UV sensitivity of the RecD− RecJ− mutant (even stronger than that of a recB mutant) is in agreement with some previous reports (Lloyd et al. 1988; Lovett et al. 1988), but is much stronger than the one reported recently (Ivančić-Baće et al. 2005). Its γ-survival was also reduced (∼5-fold), but not as much as UV survival.
Inactivation of ExoVII in the RecD− RecJ− mutant elicited striking effects. It took 4–5 days for newly constructed ExoVII− transconjugants to develop visible colonies on LB broth plates. The mass doubling time of the RecD− RecJ− ExoVII− mutant was ∼2.5-fold longer than that of the RecD− strain. The viability of that mutant was also remarkably low, <1% of its cells forming colonies. Both UV and γ-survival of the RecD− RecJ− ExoVII− mutant decreased at least 10,000-fold relative to the recD control, suggesting that recombinational repair of UV- and γ-induced lesions is abolished in the mutant. These results show that activities of RecJ and ExoVII are essential for DNA repair in the recD mutant background. This conclusion was corroborated by an observation that during subcultivation of the RecD− RecJ− ExoVII− mutant on LB broth plates, large-colony variants appeared. In addition to being UV resistant, these variants were also Tcs and unable to propagate T4 2 phage, suggesting that Tn10d(tet) was excised precisely from their recD gene, which regained its activity, rendering these bacteria RecD+ again and thus grossly independent of the functions of RecJ and ExoVII.
The above-described defects in DNA repair of the RecD− RecJ− mutant were amplified by inactivation of ExoI, too. Viability of the triple RecD− RecJ− ExoI− mutant was >3-fold lower than that of the double RecD− RecJ− mutant, while its UV and γ-survival decreased 10- and 100-fold, respectively, suggesting that ExoI and RecJ exonucleases have overlapping roles in the recD mutant, even though they have opposite polarities of action.
I also constructed the quadruple RecD− RecJ− ExoI− ExoVII− mutant, which was so crippled that after several days of incubation at 37° it developed barely visible colonies. When restreaked on fresh plate, no progeny colonies appeared, suggesting that this mutant is not viable.
DNA repair in recB1067 recD genetic background:
To ascertain that the effects reported here are not influenced by putative nuclease activity of the RecBC enzyme, I inactivated exonucleases in a recD derivative of a nuclease-deficient recB1067 mutant. The recB1067 mutation, as well as the recB1080 mutation, causes a single amino acid change in the unique nuclease center of the RecBCD enzyme that renders the mutant enzyme devoid of nuclease and RecA-loading activities, while its helicase activity remains intact (Yu et al. 1998b; Anderson et al. 1999; Wang et al. 2000). Both RecB1067C and RecB1080C enzymes, active in the recD derivatives of these mutants, are nuclease free but helicase and RecA loading proficient (Amundsen et al. 2000; Ivančić-Baće et al. 2005). Since both biochemical and genetic studies showed that recB1067 and recB1080 mutations exert indistinguishable phenotypes (Wang et al. 2000; Jockovich and Myers 2001), the former was used for experiments.
As shown in Table 3 and supplemental Table S6 at http://www.genetics.org/supplemental/, inactivation of 3′–5′ ssExos produced rather moderate effects on the events tested (cell viability, UV and γ-survival) unless all three of them were inactivated, or in the RecB* RecD− ExoI− ExoVII− SbcCD− mutant, whose γ-survival was 0.12 and whose UV survival was 0.029 of the recB1067 recD control.
TABLE 3.
Recombinational DNA repair, cell viability, and homologous recombination in recB1067 recD mutants lacking activities of major single-strand DNA exonucleases
| Relative cellular viability
|
Relative fractional survival
|
λ zygotic induction in Hfr cross
|
Relative recombination frequency
|
||||
|---|---|---|---|---|---|---|---|
| Strain | Description | UV (20 J m−2) | γ (150 Gy) | Pro+ transconjugants | Pro+ transductants | ||
| DE1080 | RecB*a RecD− | 1.0b | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| DE1081 | RecB* RecD− ExoI− | 0.72 | 0.67 | 0.44 | 0.87 | 0.12 | 0.55 |
| DE1088c | RecB* RecD− RecJ− | 0.53 | 0.000008 | 0.011 | 0.74 | 0.015 | 0.50 |
| DE1089 | RecB* RecD− RecJ− ExoI− | 0.02 | ND | 0.00005 | 0.63 | 0.0011 | 0.008 |
| DE1090 | RecB* RecD− RecJ− ExoVII− | 0.012 | ND | 0.00006 | 0.59 | 0.0007 | <0.0005 |
ND, not determined.
RecB* designates a nuclease-deficient recB1067 mutation.
Cellular viability of 1.0 corresponds to 1.3 × 108 recB1067 recD colony-forming units ml−1 at an OD600 of 0.3. UV and γ-survival of 1.0 correspond to 9.5 and 63 survivors/100 irradiated recB1067 recD cells, respectively. Frequency 1.0 for λ zygotic induction corresponds to 41 infective centers/100 IRB100 donors. Recombination frequency of 1.0 corresponds to 10.5 Pro+ transconjugants/100 Hfr3000 donor cells and 9.5 × 10−5 Pro+ transductants per P1 plaque-forming unit. Recombination efficiency is corrected for the viability of the recipients. All values are averages of at least three independent experiments except for zygotic induction in Hfr crosses, which are averages of two determinations.
Ivančić-Baće et al. (2005) reported a cellular viability of 0.75, a UV fractional survival of 0.0003, a γ fractional survival of ∼0.001, and a relative conjugal recombination of 0.013.
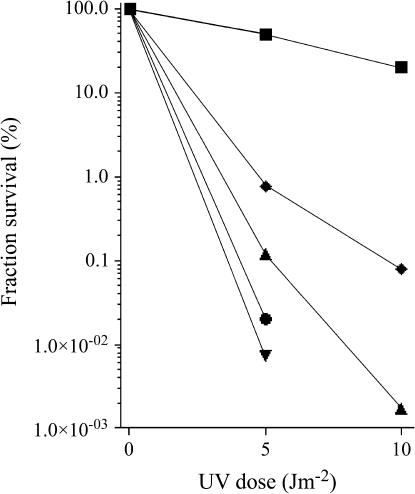
A single mutation in RecJ exonuclease conferred lower viability and extremely decreased UV (∼100,000-fold at 20 J m−2) and γ-survival (∼100-fold) to the resulting recB1067 recD recJ mutant (Table 3). The effects of RecJ mutation on DNA repair in a recB1067 recD genetic background were thus stronger than those on DNA repair in a recB+ recD background, in accord with earlier studies (Jockovich and Myers 2001; Ivančić-Baće et al. 2005). Inactivation of ExoI nuclease in the recB1067 recD recJ mutant also caused a more extreme phenotype compared to the RecD− RecJ− ExoI− mutant. The RecB* RecD− RecJ− ExoI− mutant had only ∼2% viable cells, grew ∼2.6-fold slower than its parental strain, and was extremely sensitive to γ-rays (Table 3). Its UV survival was so poor that it had to be irradiated with <20 J m−2. At 5 J m−2, ∼5000-fold fewer RecB* RecD− RecJ− ExoI− mutant cells survived compared to recB1067 recD bacteria (Figure 1). Severely damaged DNA repair observed in this mutant is similar to the phenotype of the RecB* RecD− RecJ− ExoVII− mutant, which also needed to be irradiated with lower doses of UV light to score any survivor colonies (Figure 1). Its cells had ∼2.7-fold longer mass doubling time and heavily impaired viability and survived 150 Gy of γ-rays much worse (>10,000-fold) than the recB1067 recD control strain (Table 3).
Figure 1.
UV survival of the recB1067 recD mutant strain RIK151 (▪) and its ssExo-deficient derivatives DE1088 (RecB* RecD− RecJ−) (▴), DE1089 (RecB* RecD− RecJ− ExoI−) (•), and DE1090 (RecB* RecD− RecJ− ExoVII−) (▾); DE101 (♦) is a recB-deficient mutant. Fraction survival is given as a fraction of unirradiated culture. Each value is a mean of two independent experiments.
Together, these data show that DNA repair in both recD and recB1067 recD genetic backgrounds requires overlapping activities of ExoI, ExoVII, ExoX, SbcCD, and RecJ exonucleases. However, in the recB1067 recD mutant, lack of function of the two major ssExos, ExoI and RecJ exonuclease, was less efficiently replaced by the other exonucleases.
Homologous recombination in recD mutants devoid of activities of multiple exonucleases:
The same sets of mutant strains were used for monitoring the inheritance of the chromosomal marker proAB+ in Hfr conjugational and P1 transductional crosses.
Homologous recombination in recB+ recD genetic background:
As shown in Table 2, the recombination efficiency in both assays was moderately reduced by ExoI inactivation, suggesting that a functional ExoI is required for an efficient conjugational and transductional recombination. The triple RecD− ExoI− ExoX− mutant exhibited even more reduced conjugational (∼110-fold reduced, approximating that of the recB mutant) and transductional recombination (∼3-fold less efficient), suggesting that in the RecD− ExoI− mutant the majority of residual conjugational recombination activity and a fraction of remnant transductional recombination is maintained by ExoX. However, inactivation of ExoVII improved recombination in both ExoI− and ExoI− ExoX− mutants, rendering the triple RecD− ExoI− ExoVII− and the quadruple RecD− ExoI− ExoX− ExoVII− mutants as proficient in P1 transduction as the recD control, while in conjugational crosses about four-fifths of the recombination level of the recD mutant was observed. These results suggest that for RecBC-catalyzed chromosomal recombination ExoI has a dominant role among 3′–5′ ssExos. ExoX only partially complemented ExoI deficiency, and ExoVII could not substitute for ExoI and ExoX functions; in fact it seems to interfere with recombination in the recD mutant background, which is in accord with an earlier report wherein ExoVII was found to reduce recombination efficiency in the ExoI− derivative of wild-type strain (Viswanathan and Lovett 1998).
Since the present results revealed complex interactions among 3′–5′ ssExos, I surmised that the negative effect of ExoVII on recombination might be caused by its interference with some additional exonuclease(s), whose functions are required when ExoI is inactive. To test this speculation, I inactivated SbcCD nuclease, which has already been shown to participate in DNA repair in recD mutants, together with several other 3′–5′ exonucleases (see Table 2). This protein has a 3′–5′ exonuclease activity and also an endonuclease activity (Connelly et al. 1999). The inheritance of the chromosomal proAB+ marker was examined in Hfr crosses with another donor (IRB110) to prevent transfer of wild-type sbcCD alleles to recipients. As shown in Table 4, I obtained essentially the same results as those presented in Table 2 concerning the effect of ExoI and ExoVII on recombination efficiency in the recD mutant. About a fourfold reduction of recombination in the RecD− ExoI− mutant was again shown to depend on functional ExoVII. Inactivation of SbcCD nuclease did not markedly influence recombination in RecD−, RecD− ExoVII−, and RecD− ExoI− mutants. However, when SbcCD was inactive in the RecD− ExoI− ExoVII− mutant, its restored recombination became over twofold less efficient, suggesting that functional ExoVII prevents SbcCD nuclease from compensating for the missing activity of ExoI. Unfortunately, due to a lack of suitable cotransducible selective markers, I was unable to construct the RecD− ExoI− ExoX− ExoVII− SbcCD− mutant and check its recombination proficiency. Nevertheless, the results presented show that overlapping and sometimes competing activities of several 3′–5′ exonucleases are required for homologous recombination in recD background.
TABLE 4.
Interaction between ExoVII and SbcCD exonucleases affects conjugational recombination in recD mutants
| Recipient strain | Description | Relative cellular viability | Relative recombination frequency |
|---|---|---|---|
| DE100 | RecD− | 1.0a | 1.0b |
| DE1050 | RecD− ExoI− | 0.88 | 0.24 |
| DE1053 | RecD− SbcCD− | 1.05 | 0.81 |
| DE1052 | RecD− ExoVII− | 0.91 | 1.09 |
| DE1058 | RecD− ExoVII− SbcCD− | 0.83 | 0.88 |
| DE1056 | RecD− ExoI− SbcCD− | 0.81 | 0.23 |
| DE1054 | RecD− ExoI− ExoVII− | 0.76 | 0.93 |
| DE1060 | RecD− ExoI− ExoVII− SbcCD− | 0.55 | 0.41 |
Cellular viability of 1.0 corresponds to 1.2 × 108 colony-forming units ml−1 at an OD600 of 0.3.
Recombination frequency of 1.0 corresponds to 48 Pro+ transconjugants/100 IRB110 donor cells. Recombination efficiency is corrected for the viability of the recipients. All values are averages of two independent determinations.
To determine a role for 5′–3′ ssExos in chromosomal recombination in recD background, RecJ exonuclease was inactivated singly and in combination with ExoVII. As shown in Table 2, the RecD− RecJ− mutant recombined proAB+ marker ∼7-fold less efficiently in conjugational crosses and ∼1.5-fold in transduction. This conjugational recombination efficiency falls into a range observed earlier for that mutant, while the transductional recombination is somewhat higher (Lloyd et al. 1988; Lovett et al. 1988; Ivančić-Baće et al. 2005). The triple RecD− RecJ− ExoVII− mutant was completely devoid of recombination activity; both conjugational and transductional recombinations were reduced >1000-fold, resembling the recA mutant. In addition to, as shown above, synergy between RecJ and ExoVII, I also noted synergy between RecJ and ExoI. The triple RecD− RecJ− ExoI− mutant thus also showed reduction in both conjugational (>100-fold) and transductional (∼8-fold) recombination.
Homologous recombination in a recB1067 recD genetic background:
The inheritance of the chromosomal proAB+ marker was also monitored in conjugational and transductional crosses involving recB1067 recD-derived recipients deficient in activities of several ssExos, as shown in Table 3 and supplemental Table S6 at http://www.genetics.org/supplemental/. As in recD mutant, inactivation of ExoI in the recB1067 recD background reduced recombination moderately in both assays. This reduction of recombination efficiency was again shown to depend on ExoVII, because the RecB* RecD− ExoI− ExoVII− mutant recombined more efficiently; its conjugational recombination increased ∼4-fold and its transductional recombination ∼1.5-fold. The recombination deficiency of the RecB* RecD− ExoI− mutant was further increased by inactivation of ExoX. Conjugational recombination of the resulting RecB* RecD− ExoI− ExoX− mutant was reduced ∼100-fold and its transductional recombination ∼5-fold, compared to the recB1067 recD strain. This reduction was again greatly alleviated by inactivation of ExoVII.
Relative to the recB1067 recD control strain, RecJ inactivation caused an ∼75-fold reduction in conjugational recombination (comparable to an earlier report of Ivančić-Baće et al. 2005) and an ∼2-fold reduction in transductional recombination. The remaining recombination was again shown to rely on functions of ExoVII, since the RecB* RecD− RecJ− ExoVII− mutant was completely devoid of both conjugational and transductional recombination (reduced >1000-fold, as in recA mutants). The recombination in the recB1067 recD recJ mutant also depended on ExoI; its ExoI− derivative displayed further reduction in both conjugational (∼10-fold) and transductional (∼50-fold) recombination compared to the recB1067 recD recJ mutant.
In summary, homologous recombination in both recD and recB1067 recD genetic backgrounds was shown to depend on the overlapping and sometimes competing activities of ExoI, ExoVII, ExoX, SbcCD, and RecJ exonucleases.
DISCUSSION
This study has shown that cell viability, DNA repair, and homologous recombination in E. coli recD mutants are totally dependent on synergistic activities of multiple exonucleases. The RecBC enzyme is generally considered nuclease free. Indeed, several studies failed to detect any significant exonuclease activity of RecBC (Palas and Kushner 1990; Masterson et al. 1992; Korangy and Julin 1993; Anderson et al. 1997; Churchill et al. 1999). However, in some assays the RecBC enzyme exhibited either weak endonuclease activity or exonuclease activity on single-stranded DNA (Palas and Kushner 1990; Yu et al. 1998a). A genetic analysis was therefore performed in the recD derivative of the nuclease-deficient recB1067 mutant (Wang et al. 2000; Jockovich and Myers 2001), as well as in the recB+ recD background to assure that the putative nuclease activity of the enzyme does not interfere with the effects seen upon inactivation of single or multiple exonucleases in these mutants. The analysis included determination of cell viability, sensitivity to DNA-damaging agents (such as UV- and γ-irradiation), and homologous recombination (after Hfr conjugation and P1 transduction) in the resulting mutants.
Remarkably, the (RecB*) RecD− RecJ− ExoVII− mutants, defective in both 5′–3′ ssExos, had extremely low viability and prolonged mass doubling time. They were also completely deficient in homologous recombination and DNA repair, resembling recA mutants. These processes in recD mutants thus absolutely depend on degradation of 5′-ending ss tail. For that, RecJ has a dominant role, which can be partially substituted for by ExoVII. The extent of RecJ function substitution varied depending on the event assayed, indicating specialization of the function of ssExos [e.g., the roles of RecJ exonuclease in processing blocked replication forks (Courcelle and Hanawalt 1999) and in the RecF recombinational pathway). Moreover, redundant roles for RecJ and ExoI ssExos were also revealed, although they exhibit opposite polarities of action. ExoVII inactivation abolished a remaining minor activity in the RecD− RecJ− ExoI− mutant, rendering the resulting RecD− RecJ− ExoI− ExoVII− mutant nonviable. These results thus indicate that efficient RecBC-dependent reactions involve resection of both unwound strands by synergistic actions of ssExos, apparently to prevent their reannealing. This is surprising since only the requirement for degradation of a 5′-ending strand (by a RecJ exonuclease alone) has been predicted in recD mutants. Although earlier studies reported that ssDNA tails unwound by the RecBC(D) enzyme are degraded by ExoI and RecJ exonucleases (Rinken et al. 1992; Korangy and Julin 1994; Razavy et al. 1996), this study shows for the first time that the synergistic roles of these (and some other) exonucleases are indispensable for RecBC-dependent reactions.
Exonucleases of 3′–5′ polarity (e.g., ExoI, ExoVII, ExoX, and SbcCD) also had overlapping roles for DNA repair, cell viability, and, mostly, for homologous recombination in recD mutants. Nevertheless, ExoVII prevented SbcCD [and possibly some other exonuclease(s)] to substitute for the missing ExoI activity in both recombination assays, indicating that exonucleases are not always synergistic in E. coli; their functions are rather competing in some processes. This finding is in accord with the previously reported hyperrecombination phenotype of ExoVII-deficient wild-type derivatives (Chase and Richardson 1977; Viswanathan and Lovett 1998). ExoVII thus exhibits either recombinogenic or antirecombinogenic functions, depending on a genetic context. In recD mutants lacking a 5′–3′ exonuclease activity of RecJ, ExoVII is required for recombination and repair, whereas in mutants devoid of activities of 3′–5′ ssExos (ExoI and ExoX), ExoVII interferes with recombination. Such behavior may mean that a 5′–3′ exonuclease activity of the enzyme is more potent than its 3′–5′ activity. Indeed, an earlier report showed that during methyl-directed mismatch repair, a 5′–3′ nuclease activity of ExoVII is much more effective than its 3′–5′ nuclease activity (Viswanathan et al. 2001). Additionally/alternatively, recombination reactions might specifically require stronger 3′–5′ exonuclease activity compared to the 5′–3′ one. Interestingly, ExoI and SbcCD exonucleases also have ambivalent roles: they are required for recombination in recD mutants, as shown here, whereas in a recBC background, their function is antirecombinogenic (reviewed in Kuzminov 1999).
When comparing exonuclease requirements in recD and recB1067 recD mutant backgrounds, nearly identical trends emerge, the only difference being more pronounced dependence on the two prominent ssExos (ExoI and RecJ) in the latter mutant. These data and the sickly phenotype of RecJ− ExoVII− derivatives in both backgrounds (suggesting their complete dependence on 5′–3′ exonuclease activity of ssExos) thus argue against an assumption that the RecBC enzyme retains some exonucleolytic activity. Rather, I favor an alternative explanation, namely that the RecB1067C enzyme is more processive than RecBC, meaning that longer single-stranded tails produced by the former enzyme require more potent ssExos (i.e., ExoI and RecJ) for efficient processing. This correlation extends to reactions catalyzed by even more processive nuclease-deficient RecB1067CD and RecB1080CD enzymes, which are more dependent on RecJ (Jockovich and Myers 2001) and ExoI (D. Ðermić, unpublished data) than those performed by the RecB1067C or RecBC enzyme.
In contrast to the above-described essential roles of multiple exonucleases in the recD genetic backgrounds, their functions are much less required in the wild-type background. A triple, wild-type-derived ExoI− ExoVII− ExoX− mutant does not show increased UV sensitivity (Viswanathan and Lovett 1999). A double mutant, lacking activities of the major ssExos RecJ− and ExoI−, survives UV irradiation almost as efficiently as its parental wild-type strain (Viswanathan and Lovett 1999) and shows moderate (∼10-fold) reduction in conjugational recombination (Viswanathan and Lovett 1998). Inactivation of 5′–3′ ssExos (RecJ and ExoVII) confers only moderate effect on UV survival (∼10-fold decrease at 20 J m−2) and recombination proficiency in the wild-type background (Viswanathan and Lovett 1998), whereas its RecD− counterpart is completely devoid of homologous recombination and DNA repair. Furthermore, while the RecD− RecJ− ExoI− ExoVII− mutant is nonviable, the RecJ− ExoI− ExoVII− derivative of the wild-type strain has normal viability (Harris et al. 1998), revealing a crucial role of exonuclease activity for viability of E. coli, in accord with a previous report (Miranda and Kuzminov 2003). The RecJ− ExoI− ExoVII− mutant is also only moderately UV sensitive (Viswanathan and Lovett 1998). These data thus show that exonuclease activity is essential for RecBC(D)-dependent recombination reactions. As RecBC enzyme lacks exonuclease activity (ExoV−), other exonucleases are called into action to replace it, which makes RecBC-dependent recombination reactions mechanistically very different and yet conceptually analogous to those catalyzed by RecBCD. Much stronger effects of the inactivation of various exonucleases in recD mutants compared to those in the wild-type background, indicate that the main role of exonucleases in homologous recombination in E. coli is in the presynaptic phase (which is catalyzed by either the RecBC or the RecBCD enzyme), whereas their role in the postsynaptic phase of recombination (which is independent of the two enzymes and thus is unlikely to be different in the two backgrounds) seems less important. Conversely, as shown in Table 5, recombination in recD mutants (a RecY pathway; Mahajan 1988) shares the requirement for (5′–3′) exonuclease activity with the other pathways for homologous recombination initiation in E. coli. A minor role of ssExos in the postsynaptic phase may reflect a general feature of recombination, namely that displaced strands in the recombination intermediate are stabilized by some alternative means other than degradation.
TABLE 5.
Comparison of recombination pathways in E. coli
| Recombination pathway
|
||||
|---|---|---|---|---|
| RecY | RecBCD | RecF | RecE | |
| Operative in genetic background | recD | Wild type | recBC sbcBC | recBC sbcA |
| Helicase | RecBC | RecBCD | RecQ (UvrD, HelD) | — |
| Exonuclease(s) | RecJ, ExoVII, ExoI, ExoX, SbcCD | RecBCD | RecJ | ExoVIII, RecJ |
| Facilitation of strand-exchange protein loading by: | RecBC | RecBCD | RecF, O, R | RecTa |
| Recombinational exchanges focused at: | DNA ends | Chi sites | DNA ends | DNA ends |
RecT is itself a strand-exchange protein.
In fact, 5′–3′ exonuclease activity is ubiquitous in the initiating phase of recombination reactions catalyzed by a broad range of organisms, including viruses (λ's Redα is a 5′–3′ dsDNA exonuclease; Muniyappa and Radding 1986), bacteria (RecJ and ExoVII large subunit orthologs are found in almost all bacterial genomes sequenced to date, as cited in Viswanathan et al. 2001), and eukaryotes (DSB repair in Saccharomyces cerevisiae includes resection of a 5′-ending strand; Sun et al. 1991). The 5′-ending strand resection in yeast is catalyzed by a Mre11/Rad50/Xrs2 complex (an SbcCD homolog) and by ExoI and Rad27 exonucleases (Moreau et al. 2001). Their redundancy is analogous to that of E. coli RecJ and ExoVII nucleases, which emphasizes the necessity of examining multiple mutants for phenotypic effects.
In addition to different requirements for various exonucleases in recD and wild-type backgrounds, differences among DNA metabolic processes assayed were also noted. This is not surprising since each of these processes involves different DNA substrates, which may be processed by different mechanisms. For instance, nuclease inactivation in recD recipients affected P1 transductional recombination much more mildly than conjugational recombination. During Hfr-mediated conjugation, a linear single strand is transferred to a recipient cell, where its complement is synthesized in a discontinuous way. As a result, a long double-stranded linear fragment of donor chromosomal DNA, bordered by a region of ∼50 kbp of F plasmid DNA at the leading end and a 3′ ss tail of variable length at the distal end, arises in the recipient (for a review, see Zechner et al. 2000). During P1 infection, a double-stranded linear fragment of ∼100 kbp of donor chromosomal DNA is sporadically introduced into a recipient cell (for a review, see Masters 1996). The linear dsDNA fragment that is introduced into a recipient cell during conjugation or transduction is a suitable substrate for RecBC(D) binding because it has blunt or nearly blunt ends. The majority of recombination exchanges (at least two, or a higher even number) between this fragment and the circular recipient chromosome are thus catalyzed by the RecBC(D) enzyme.
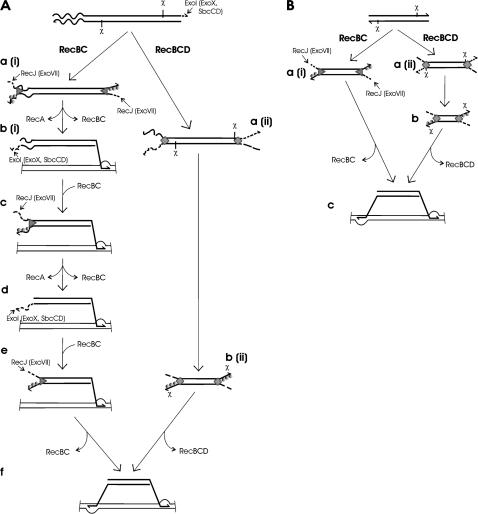
A model for donor DNA processing in recD and wild-type transconjugants and transductants has been presented. As shown in Figure 2A, a(i) and a(ii), RecBC and RecBCD enzymes load onto conjugationally transferred DNA and begin its unwinding from both ends. RecBCD-catalyzed unwinding is accompanied by degradation of unwound DNA. Since a region of F plasmid DNA bordering the leading end does not contain a Chi site (Smith 1991; Manwaring et al. 1999), it is completely degraded [Figure 2A, a(ii)]. Strong degradation occurs until the two RecBCD enzymes interact with Chi sites, which causes their modification, their nuclease activity being downregulated and switched from 3′–5′ to 5′–3′ polarity. Also, the Chi-modified RecBCDs facilitate RecA protein loading onto unwound 3′-ending overhangs, thereby producing nucleoprotein filaments, the central recombination intermediates [Figure 2A, b(ii)]. These RecA-coated filaments then invade homologous regions on a recipient chromosome and pair with their complements, thus creating a pair of D-loops (Figure 2A, f). For clarity, subsequent steps of recombination intermediate processing are not shown in Figure 2.
Figure 2.
Model for transferred donor DNA (depicted by thick lines) processing in recD and wild-type Hfr transconjugants (A) and P1 transductants (B). Wavy and thin lines represent F plasmid DNA and recipient chromosome, respectively. RecA, RecBC, and RecBCD proteins are shown as a circle, triangle, and rectangle, respectively. Details of the reactions at each stage are described in the text.
On the other hand, RecBC is a less processive helicase, which constitutively loads RecA protein onto the unwound 3′-tail. As RecBC is nuclease free, a 5′-ending strand unwound by it will be degraded by RecJ exonuclease (or ExoVII when RecJ is inactive) [Figure 2A, a(i)]. While at the distal end of transferred DNA these reactions produce a nucleoprotein filament that searches for its homologous DNA and then creates a synapsis [Figure 2A, b(i)], processing of the proximal end is more complicated. Initial reactions, RecBC binding to and unwinding of a DNA duplex, RecA polymerization on an unwound 3′-ending strand, as well as resection of its complementary strand by RecJ (or ExoVII), proceed as those at the distal end [Figure 2A, a(i)]. However, since F plasmid DNA is heterologous to the recipient DNA, a nucleoprotein filament created by RecBC enzyme at the proximal end is unable to give rise to synapsis. This RecBC-catalyzed nucleoprotein filament is thus of no use for recombination; and even more, it is stable because RecA protein protects it from degradation by ExoI (Churchill et al. 1999). Therefore, RecBC processing of nonhomologous DNA results in nonproductive and stable RecA presynaptic filament that inhibits further processing of that DNA end by preventing binding of another RecBC molecule onto it. If that were all, then conjugational recombination would be practically impossible to carry out by RecBC enzyme. However, recD mutants are recombination proficient, meaning that any recombination block is overcome in their cells. This is likely due to the dynamic nature of RecA nucleofilament, from which RecA protein dissociates in a process dependent on ATP hydrolysis (Menetski et al. 1990; Rehrauer and Kowalczykowski 1993). So, once RecA is dissociated from the nucleofilament, this 3′-ending tail becomes susceptible to degradation by ExoI (or, in its absence, by ExoX and SbcCD), leading to its resection until a flush dsDNA end is recreated [Figure 2A, b(i)]. This end is an entry site for the RecBC enzyme, which then starts a new cycle of DNA processing (Figure 2A, c). If its in vitro ability to unwind ∼3 kbp of DNA duplex (an estimate from the data in Korangy and Julin 1993) reflects its processivity in a cell, at least 15 successive rounds of DNA processing might be required for RecBC to pass through F DNA and reach donor chromosomal DNA. Then a new—this time productive—round of DNA processing would result in synapsis, like that at the distal end (Figure 2A, e and f). For clarity, just two cycles of F plasmid DNA processing are shown in Figure 2A. These sequential rounds of DNA processing certainly take some time and thus this model predicts that recombining Hfr DNA fragment would be more stable in recD mutants than in wild-type transconjugants. Lloyd and Buckman (1995) indeed noted an increased half-life of transferred Hfr DNA in recD transconjugants.
In contrast, DNA processing reactions during P1 transduction in recD mutants should be simpler due to the absence of heterologous DNA sequences at the ends of incoming DNA, as shown in Figure 2B. Both ends of a fragment are bound and DNA duplex is unwound by two RecBC enzymes, which constitutively facilitate RecA protein polymerization onto the unwound 3′-ssDNA tail, while its complementary, 5′-ending strand is digested by RecJ nuclease (or by ExoVII in RecJ− mutant) [Figure 2B, a(i)].
The RecBCD enzyme, on the other hand, does not require activity of any other nuclease [Figure 2B, a(ii)]. Upon interaction with a Chi site, the enzyme degrades the 5′-ending strand and directs RecA loading on the 3′ strand unwound by it (Figure 2B, b). The RecA-coated 3′-ending overhangs then pair with homologous regions of the recipient chromosome, which should proceed alike in both wild-type and recD transductants (Figure 2B, c).
The model described above readily explains stronger requirements for multiple exonucleases during conjugational crosses in recD mutants compared to transduction, arguing further for their role in the presynaptic phase of recombination.
In summary, this study shows that exonuclease activity is essential for RecBC(D)-dependent recombination reactions, meaning that strong ExoV activity of RecBCD is not merely an obstacle, but rather an essential part (along with helicase and RecA-loading activity) of the recombinogenic function of the enzyme. The missing ExoV activity in recD mutants is efficiently replaced by functions of several exonucleases, indicating their redundancy, which confers robustness to recombination in E. coli. A more detailed characterization of the roles of exonucleases in ExoV− backgrounds other than recD (e.g., recBC sbcBC, recBC sbcA, recB1080, and recB1067) is underway.
Acknowledgments
I am grateful to Susan T. Lovett, Benedicte Michel, Mary Berlin, and Richard S. Myers for providing bacterial strains. I am indebted to Mirjana Petranović for her encouragement and interest in this work. I also thank W. L. Ragland for correction of the English text. This work was supported by the Croatian Ministry of Science and Technology, grant 0098071.
References
- Amundsen, S. K., A. F. Taylor and G. R. Smith, 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. USA 97: 7399–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D. G., and S. C. Kowalczykowski, 1997. a The recombination hot spot, Chi, is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11: 571–581. [DOI] [PubMed] [Google Scholar]
- Anderson, D. G., and S. C. Kowalczykowski, 1997. b The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell 90: 77–86. [DOI] [PubMed] [Google Scholar]
- Anderson, D. G., J. J. Churchill and S. C. Kowalczykowski, 1997. Chi-activated RecBCD enzyme possesses 5′-3′ nucleolytic activity, but RecBC enzyme does not: evidence suggesting that the alteration induced by Chi is not simply the ejection of the RecD subunit. Genes Cells 2: 117–128. [DOI] [PubMed] [Google Scholar]
- Anderson, D. G., J. J. Churchill and S. C. Kowalczykowski, 1999. A single mutation, RecBD1080A, eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J. Biol. Chem. 274: 27139–27144. [DOI] [PubMed] [Google Scholar]
- Arnold, D. A., and S. C. Kowalczykowski, 1999. RecBCD helicase/nuclease, in Encyclopedia of Life Science. Nature Publishing Group, London (http://www.els.net).
- Bachmann, B. J., 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, pp. 2460–2488 in Escherichia coli and Salmonella: Cellular and Molecular Biology, edited by F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low et al. American Society for Microbiology, Washington, DC.
- Burdett, V., C. Baitinger, M. Viswanathan, S. T. Lovett and P. Modrich, 2001. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. USA 98: 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldo, F., G. Ramsey and S.D. Barbour, 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 118: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase, J. W., and C. C. Richardson, 1974. Exonuclease VII of Escherichia coli: mechanism of action. J. Biol. Chem. 249: 4553–4561. [PubMed] [Google Scholar]
- Chase, J. W., and C. C. Richardson, 1977. Escherichia coli mutants deficient in exonuclease VII. J. Bacteriol. 129: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A. M., and G. R. Smith, 1984. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc. Natl. Acad. Sci. USA 81: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin, F., and S. C. Kowalczykowski, 2002. A novel family of regulated helicases/nucleases from Gram-positive bacteria: insights into the initiation of DNA recombination. Mol. Microbiol. 43: 823–834. [DOI] [PubMed] [Google Scholar]
- Churchill, J. J., D. G. Anderson and S. C. Kowalczykowski, 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of χ, resulting in constitutive recombination activation. Genes Dev. 13: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, J. C., E. S. de Leau and D. R. Leach, 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27: 1039–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle, J., and P. C. Hanawalt, 1999. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 262: 543–551. [DOI] [PubMed] [Google Scholar]
- Ðermić, D., and Ž. Trgovčević, 1999. Specific effects of a recB mutation on the HfrH strain of Escherichia coli. J. Bacteriol. 181: 1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ðermić, D., E. Halupecki, D. Zahradka and M. Petranović, 2005. RecBCD enzyme overproduction impairs DNA repair and homologous recombination in Escherichia coli. Res. Microbiol. 156: 304–311. [DOI] [PubMed] [Google Scholar]
- Ðermić, D., E. Ðermić, D. Zahradka, M. Petranović and N. Lerš, 2006. Gamma-irradiated RecD overproducers become permanent recB−/C− phenocopies for extrachromosomal DNA processing due to prolonged titration of RecBCD enzyme on damaged Escherichia coli chromosome. Biochimie 88 (in press). [DOI] [PubMed]
- Dixon, D. A., and S. C. Kowalczykowski, 1993. The recombinational hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 73: 87–96. [DOI] [PubMed] [Google Scholar]
- Emmerson, P. T., 1968. Recombination deficient mutants of Escherichia coli K12 that map between thyA and argA. Genetics 60: 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschenko, V. V., L. A. Rajman and S. T. Lovett, 2003. Stabilization of perfect and imperfect tandem repeats by single-strand DNA exonucleases. Proc. Natl. Acad. Sci. USA 100: 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, R. S., K. J. Ross, M. J. Lombardo and S. M. Rosenberg, 1998. Mismatch repair in Escherichia coli cells lacking single-strand exonucleases ExoI, ExoVII, and RecJ. J. Bacteriol. 180: 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivančić-Baće, I., E. Salaj-Šmic and K. Brčić-Kostić, 2005. Effects of recJ, recQ, and recFOR mutations on recombination in nuclease-deficient recB recD double mutants of Escherichia coli. J. Bacteriol. 187: 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockovich, M. E., and R. S. Myers, 2001. Nuclease activity is essential for RecBCD recombination in Escherichia coli. Mol. Microbiol. 41: 949–962. [DOI] [PubMed] [Google Scholar]
- Korangy, F., and D. A. Julin, 1993. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 32: 4873–4880. [DOI] [PubMed] [Google Scholar]
- Korangy, F., and D. A. Julin, 1994. Efficiency of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 33: 9552–9560. [DOI] [PubMed] [Google Scholar]
- Kuzminov, A., 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63: 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, I. R., and A. L. Nussbaum, 1964. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase). J. Biol. Chem. 239: 2628–2636. [PubMed] [Google Scholar]
- Lloyd, R. G., and C. Buckman, 1995. Conjugational recombination in Escherichia coli: genetic analysis of recombinant formation in Hfr × F− crosses. Genetics 139: 1123–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. G., M. C. Porton and C. Buckman, 1988. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol. Gen. Genet. 212: 317–324. [DOI] [PubMed] [Google Scholar]
- Lovett, S. T., and R. D. Kolodner, 1989. Identification and purification of a single-strand-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 86: 2627–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, S. T., C. Luisi-DeLuca and R. D. Kolodner, 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 120: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan, S. K., 1988. Pathways of homologous recombination in Escherichia coli, pp. 87–140 in Genetic Recombination, edited by R. Kucherlapati and G. R. Smith. American Society of Microbiology, Washington, DC.
- Manwaring, N. P., R. A. Skurray and N. Firth, 1999. Nucleotide sequence of the F plasmid leading region. Plasmid 41: 219–225. [DOI] [PubMed] [Google Scholar]
- Masters, M., 1996. Generalized transduction, pp. 2421–2441 in Escherichia coli and Salmonella: Cellular and Molecular Biology, edited by F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low et al. American Society of Microbiology, Washington, DC.
- Masterson, C, P. E. Boehmer, F. McDonald, S. Chaudhuri, I. D. Hickson et al., 1992. Reconstitution of the activities of the RecBCD holoenzyme of Escherichia coli from the purified subunits. J. Biol. Chem. 267: 13564–13572. [PubMed] [Google Scholar]
- Menetski, J. P., D. G. Bear and S. C. Kowalczykowski, 1990. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. USA 87: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel, L., and J. R. Roth, 1996. Evidence that SbcB and RecF pathway functions contribute to RecBCD-dependent transductional recombination. J. Bacteriol. 178: 3146–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. H., 1992. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Miranda, A., and A. Kuzminov, 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163: 1255–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, S., E. A. Morgan and L. S. Symington, 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa, K., and C. M. Radding, 1986. The homologous recombination system of phage λ. Pairing activities of β protein. J. Biol. Chem. 261: 7472–7478. [PubMed] [Google Scholar]
- Palas, K. M., and S. R. Kushner, 1990. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J. Biol. Chem. 265: 3447–3454. [PubMed] [Google Scholar]
- Razavy, H., S. K. Szigety and S. M. Rosenberg, 1996. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics 142: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehrauer, W. M., and S. C. Kowalczykowski, 1993. Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268: 1292–1297. [PubMed] [Google Scholar]
- Rinken, R., B. Thoms and W. Wackernagel, 1992. Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J. Bacteriol. 174: 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salaj-Šmic, E., D. Ðermić, K. Brčić-Kostić, G. Čogelja-Čajo and Ž. Trgovčević, 2000. In vivo studies of the Escherichia coli RecB polypeptide lacking its nuclease center. Res. Microbiol. 151: 769–776. [DOI] [PubMed] [Google Scholar]
- Seigneur, M., S. D. Ehrlich and B. Michel, 1999. recD sbcB sbcD mutants are deficient in recombinational repair of UV lesions by RecBC. J. Bacteriol. 181: 6220–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., 1991. Conjugational recombination in E. coli: myths and mechanisms. Cell 64: 19–27. [DOI] [PubMed] [Google Scholar]
- Sun, H., D. Treco and J. W. Szostak, 1991. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell 64: 1155–1161. [DOI] [PubMed] [Google Scholar]
- Viswanathan, M., and S. T. Lovett, 1998. Single-strand DNA exonucleases in Escherichia coli: roles in repair and mutation avoidance. Genetics 149: 7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, M., and S. T. Lovett, 1999. Exonuclease X of Escherichia coli. A novel 3′-5′ DNase and Dnaq superfamily member involved in DNA repair. J. Biol. Chem. 274: 30094–30100. [DOI] [PubMed] [Google Scholar]
- Viswanathan, M., V. Burdett, C. Baitinger, P. Modrich and S. T. Lovett, 2001. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem. 276: 31053–31058. [DOI] [PubMed] [Google Scholar]
- Wang, J., R. Chen and D. A. Julin, 2000. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J. Biol. Chem. 275: 507–513. [DOI] [PubMed] [Google Scholar]
- Willets, N. S., and D. W. Mount, 1969. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J. Bacteriol. 100: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., J. Souaya and D. A. Julin, 1998. a The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not for the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 95: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., J. Souaya and D. A. Julin, 1998. b Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283: 797–808. [DOI] [PubMed] [Google Scholar]
- Zechner, E. L., F. de la Cruz, R. Eisenbrandt, A. M. Grahn, G. Koraimann et al., 2000. Conjugative-DNA transfer processes, pp. 87–174 in The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread, edited by C. M. Thomas. Harwood Academic Publishers, Amsterdam.