Abstract
We investigate the origin and evolution of a mouse processed pseudogene, Makorin1-p1, whose transcripts stabilize functional Makorin1 mRNAs. It is shown that Makorin1-p1 originated almost immediately before the musculus and cervicolor species groups diverged from each other some 4 million years ago and that the Makorin1-p1 orthologs in various Mus species are transcribed. However, Mus caroli in the cervicolor species group expresses not only Makorin1-p1, but also another older Makorin1-derived processed pseudogene, demonstrating the rapid generation and turnover in subgenus Mus. Under this circumstance, transcribed processed pseudogenes (TPPs) of Makorin1 evolved in a strictly neutral fashion even with an enhanced substitution rate at CpG dinucleotide sites. Next, we extend our analyses to rats and other mammals. It is shown that although these species also possess their own Makorin1-derived TPPs, they occur rather infrequently in simian primates. Under this circumstance, it is hypothesized that already existing TPPs must be prevented from accumulating detrimental mutations by negative selection. This hypothesis is substantiated by the presence of two rather old TPPs, MKRNP1 and MKRN4, in humans and New World monkeys. The evolutionary rate and pattern of Makorin1-derived processed pseudogenes depend heavily on how frequently they are disseminated in the genome.
INTRONLESS processed pseudogenes result from the reverse transcription of mRNA followed by integration into the genome. This process is mediated through reverse transcriptase activity produced by retrotransposable elements such as LINE-1 in mammals (Skowronski and Singer 1986). More than 8000 processed pseudogenes are identified in the human genome (Zhang et al. 2003; Zhang and Gerstein 2004) and ∼5000 in the mouse genome (Zhang et al. 2004). The great majority of processed pseudogenes are derived from a limited number of genes that are highly transcribed in the germ cell line. In general, because of the lack of functional promoters, integrated processed pseudogenes are thought to be untranscribed and completely nonfunctional since their origins. Only occasionally may processed pseudogenes acquire promoter activity and be transcribed (Yano et al. 2004; Harrison et al. 2005). However, Cheng et al. (2005) reported unexpectedly high occurrences of transcribed processed pseudogenes (TPPs). In accord with this, quite a few examples of TPPs are known and some have gained new functions (neofunctionalization): the dihydrofolate reductase gene in humans (Chen et al. 1982; Vanin 1985), the chimerical gene jingwei in Drosophila (Long and Langley 1993; Balakirev and Ayala 2003; Long et al. 2003), the nitric oxide synthase (NOS) gene in the snail Lymnaea stagnalis (Korneev et al. 1999), chromodomain Y (CDY) in primates (Dorus et al. 2003), and most recently Makorin1-p1 in mice (Hirotsune et al. 2003).
The Makorin1 gene encodes an E3 ubiquitin ligase that can specifically digest telomerase (Kim et al. 2005) and is expressed ubiquitously in the mouse body, but most prominently in testis (Gray et al. 2000). The mutant that lacks sufficient amounts of Makorin1 mRNAs exhibits either lethality within 2 days of birth or polycystic kidneys and severe bone deformities if it survives. Surprisingly, such deficiencies also result from downregulated transcripts of a Makorin1-derived processed pseudogene, Makorin1-p1. Hirotsune et al. (2003) therefore hypothesized that Makorin1-p1 transcripts prevent the decay of functional Makorin1 mRNAs by titrating out destabilizing factors that bind to the 5′ part of Makorin1 and Makorin1-p1.
Following Hirotsune et al. (2003), Podlaha and Zhang (2004) defined an ∼700-bp functionally important region (B region) sandwiched by an ∼100-bp upstream A region and an ∼700-bp downstream C region. On the basis of these homologous sequences in various species in subgenus Mus, they came to the conclusion that the A + B region is more evolutionarily conserved than the C region. Unfortunately, they used two sets of PCR primers that were designed for amplifying the A + B and C regions, separately. While these primer sets should not have any serious problem in amplifying a single-copy gene, they are problematic in the presence of multiple Makorin1-p1-like sequences in the Mus genome. In the latter case, neither the orthologous relationships among the sequences nor the linkage relationships between the A + B and C region sequences become immediately discernible.
The purpose of this article is threefold. First, we reexamine the origin and evolution of Makorin1-p1 in subgenus Mus. Since it turns out that Mus caroli occupies a critical phylogenetic position, special attention is paid to the isolation of the bona fide Makorin1-p1 ortholog and its testis transcription in this species. Second, since it appears that Makorin1-p1 is found only in subgenus Mus, we search for a Makorin1-p1-like processed pseudogene in rats and examine its cotranscription with Makorin1. Third, we hypothesize that under the circumstance that TPPs of Makorin1 appear one after another in the genome, they can evolve in a completely neutral fashion as Makorin1-p1, but under the opposite circumstance that TPPs rarely occur, certain existing TPPs must be prevented from accumulating detrimental mutations by negative selection. To test this hypothesis, we analyze Makorin1-derived processed pseudogenes in other mammals including primates and show some evidence for the conservation of the B relative to the C region.
MATERIALS AND METHODS
Genomic DNA sources and PCR amplification:
Mouse genomic DNAs used in this study are of M. m. musculus (BLG2/MsRbrc), M. m. castaneus (CAST/Ei), M. m. molossinus (MSM/MsRbrc), M. spretus (SPRET/Ei and SPR/RpRbrc), and M. caroli (Car/Rbrc). The RIKEN BioResource Center (BRC) (Tsukuba, Japan) provided the BLG2/MsRbrc (BRC no. 00653), SPR/RpRbrc (BRC No. 00208), and Car/Rbrc (BRC No. 00823) strains. The CAST/Ei and SPRET/Ei strains were purchased from the Jackson Laboratory (Bar Harbor, ME). Rat genomic DNAs used in this study are taken from Rattus norvegicus of strain Wister (Oriental Yeast Co.). Primate genomic DNAs are of four New World monkey species (Aotus trivirgatus, owl monkey; Ateles belzebuth, spider monkey; Cebus apella, tufted capuchin; and Saguinus oedipus, cotton-top tamarin) and of three prosimian species (Galagoides demidoff, Demidoff's galago; Galago moholi, South African galago; and Otolemur crassicaudatus, thick-tailed bush baby). Those DNAs were kindly provided by J. Klein (previously at Max-Planck Institute for Biology, Tübingen, Germany, and currently at Pennsylvania State University, University Park, PA). Makorin1 consists of eight exons spanning ∼23 kb in mice and 26 kb in humans, so we do not determine the genomic sequences of these primates.
Genomic DNAs are used as templates for PCR amplification of mouse Makorin1-p1 homologs (1472 bp) and primate Makorin1 processed pseudogenes (996 bp). The primers are designed in a region conserved between the processed pseudogenes and functional genes in mice and humans. Unlike Podlaha and Zhang (2004), these primers can amplify the B and C regions simultaneously. For mice, the upper primer sequence is 5′-CCCACAGTCGCTGCCCCGTC-3′ (5F1) and the lower is 5′-CAGCACCTGGAGTTTGAGAGG-3′ (3R1). For primates, the upper and lower primers are 5′-CATGACCTCTCTGACAGTCCG-3′ (5F2) and 5′-TCTGCTTGATGTTCCCAC-3′ (3R2), respectively. To confirm the Makorin1-p1 ortholog in M. caroli, we also use a different set of primers designed in Makorin1-p1 flanking regions (P3, 5′-CCACAAAGGGGCAGGCTGACGAAAC-3′ and P5, 5′-GGTTTCCATTGCTGTAAAGAGACACCACGACT-3′).
Each PCR reaction mixture was 25 μl in volume and contained 40–100 ng of genomic DNA, 1× Ex Taq buffer, 0.16 μm of dNTPs, 0.05 unit of TaKaRa (Berkeley, CA) Ex Taq, and 0.2 μm of each primer. The standard condition for PCR was as follows: denaturation at 94° for 4 min; 30 cycles of 94° for 30 sec, 58–64° for 1 min, and 72° for 1 min; and an additional extension at 72° for 4 min. The PCR products were purified using QIAquick PCR purification kits (QIAGEN, Valencia, CA), QIAquick gel extraction kits (QIAGEN), or ExoSAP-ITs (United States Biochemical, Cleveland). Purified products were either sequenced directly or cloned into pCR2.1 TOPO with TOPO TA cloning kits (Invitrogen, San Diego). Cloned products were purified with QIAprep Spin Miniprep (QIAGEN) and used as templates in subsequent sequencing reactions. Sequencing reactions were performed with BigDye Terminator v1.1 cycle sequencing kits (Applied Biosystems, Foster City, CA) and analyzed on an ABI PRISM 377 DNA sequencer (Applied Biosystems). To minimize sequencing errors, PCR primary products or cloned DNAs were sequenced two to six times in both directions. These sequences are deposited in the DNA Data Bank of Japan (DDBJ), and their accession numbers are AB219432–AB219448, AB239533, and AB239534.
RNA extraction and RT–PCR amplification:
Total RNAs were isolated from M. m. domesticus, M. spretus, and M. caroli testis by using NucleoSpin RNAII (Macherey-Nagel). In regard to rats and humans, total RNAs from testis were purchased from Stratagene (La Jolla, CA) and OriGene, respectively. These total RNAs were proven not to contain genomic DNAs and were used as templates for RT–PCR using TaKaRa RNA PCR kits (AMV) V. 3.0 (TaKaRa) according to the manufacturer's instruction. Three sets of upper and lower primers were used: 5F1 and 3R1, 5F2 and 3R2, and 5′-GGCGGCGGCTGGACGAACA-3′ (5F3) and 5′-TGGGCAGCATCCACTGGGTGTAGG-3′ (5R3). This third set of primers was originally designed for RT–PCR in Hirotsune et al. (2003) and used here as a control in our RT–PCR. Furthermore, three sets of upper and lower primers were used for the rat RT–PCR: 5F2 and 3R2, 5F3 and 3R3, and 5′-AAAGCTATCACCCATTGCTGC-3′ (5F4) and 5′-GAGTGCAAAGCTATCACCCAT-3′ (3R4). One set of upper and lower primers was used for the human RT–PCR: 5′-CGATAGTTGAAATGAATACAAGC-3′(5F5) and 5′-CTCCTCTCTCCTCCTCTTTACG-3′ (3R5).
RT–PCR products were cloned in the same way as mentioned above. Cloned DNAs were purified with the rolling circle amplification (RCA) method. Sequencing reactions were performed with the dye terminator method and analyzed on a MegaBACE1000 sequencer (Amersham Biosciences, Arlington Heights, IL). At least eight clones were sequenced in both directions. Since RT–PCR products of either functional Makorin1 or processed pseudogenes are nearly the same in size, we searched for such restriction recognition sites that can be distinguished in these different simultaneously amplified products. Each restriction enzyme reaction mixture was 50 μl in volume and contained 5 μg of PCR products, 1× universal buffer provided by manufacturers, and 6 units of each restriction enzyme. These mixtures were kept at 37° for 1 hr and the digested PCR products were separated through 1.5% agarose gel electrophoresis.
Sequence designations:
The Makorin1 sequences of M. m. domesticus (NM_018810), M. spretus (AY699805), and R. norvegicus (NW_047690) were retrieved from the NCBI database. For notational convenience, we subsequently designate the mouse Makorin1 and Makorin1-p1 as Mkrn1 and Mkrn1-p1, respectively. Furthermore, for a given species, we use a four-letter prefix to abbreviate the genus and (sub)species names. Thus, Makorin1 and Makorin1-p1 in M. m. domesticus are designated as Mudo Mkrn1 and Mudo Mkrn1-p1, respectively. The only exception is for M. m. castaneus for which we use Mucs to distinguish it from Muca that is kept for M. caroli. Processed pseudogenes of Mkrn1 other than Mkrn1-p1 are defined as Mkrn1-p2, -p3, and so on. Specifically, a newly found processed pseudogene in M. caroli is designated as Muca Mkrn1-p2.
For mammals except primates, we use the same locus abbreviation as Mkrn in mice, but use Mkrnp for a pseudogene. To primates, we apply the standard nomenclature in humans (MKRN1, MKRNP1, etc). Supplement 1 at http://www.genetics.org/supplemental/ provides more information about the nomenclature, the accession numbers, the sequence lengths, and the p-distances (p is the number of nucleotide differences per site).
Data analyses:
Alignments were made by Clustal X (Thompson et al. 1997) and manually checked (supplement 2 at http://www.genetics.org/supplemental/). Any sites containing insertions or deletions (indels) were excluded in calculating the p-distances and constructing the neighbor-joining (NJ) tree of Saitou and Nei (1987). However, whenever necessary, the actual number of nucleotide substitutions was estimated from the p-distances (Jukes and Cantor 1969; Kimura 1980). Tree constructions and distance calculations were conducted in MEGA version 3.0 (Nei and Kumar 2000; Kumar et al. 2004). For the phylogenetic analysis, the maximum parsimony (MP) method by Fitch (1971) was also used.
RESULTS
Isolation of Mkrn1-derived processed pseudogenes in rodents:
Using the primer set of 5F1 and 3R1, we isolated several Mkrn1-p1-like sequences from three M. musculus subspecies, two strains of M. spretus, and one strain of M. caroli. Twenty-six informative sites in the alignment of these Mkrn1-p1-like sequences support the monophyletic relationships of the sequences sampled from the M. musculus subspecies. In the M. spretus strains, we found two sequences that differ from each other by 19 nucleotides (p = 1.4%) as well as the presence or absence of a 323-bp insertion. However, since this level of p-distances may be caused by polymorphism, these are regarded as alleles and designated as Musp Mkrn1-p1.a/b. Twenty-two informative sites support their orthologous relationships with a cluster of the Mkrn1-p1 sequences from the M. musculus subspecies.
In the M. caroli strain, we obtained three different sequences that are apparently homologous to Mudo Mkrn1-p1. However, all three are substantially different from the Mkrn1-p1 sequence that was reported by Podlaha and Zhang (2004). One sequence (1398 bp) shares an ATAC insertion with another processed pseudogene on chromosome 3 (Mudo Mkrn1-p2) in the M. m. domesticus database (NCBI, build 34.1). This sequence is therefore designated as Muca Mkrn1-p2. The remaining two sequences (1416 and 1419 bp) share a 15-bp insertion and a 24-bp deletion, but they differ from each other by several indels and at 11 nucleotide sites (p = 0.8%). We note that the M. caroli strain was not inbred and kept in the laboratory for only two generations. Taking this into account, we assume that these two sequences represent different alleles. The p-distances of these allele sequences from Mudo Mkrn1 are 3.6 and 3.7%, which are much smaller than p = 9.3% of Muca Mkrn1-p2. Importantly, there are eight informative sites that support the orthologous relationships of these alleles with Mudo Mkrn1-p1. We therefore tried to prove their orthology directly by examining the corresponding flanking regions of Mudo Mkrn1-p1. The PCR primer set (P3 and P5) is designed to amplify the ∼300-bp upstream and ∼200-bp downstream flanking regions. If the Mkrn1-p1 ortholog is present in M. caroli, the amplified sequence is expected to be ∼2.2 kb long and if not it becomes ∼500 bp long. The obtained PCR product is actually ∼2.2 kb long and contains a Mkrn1-p1-like sequence. This sequence is identical to one of the two alleles, Muca Mkrn1-p1.a, and both the 5′- and 3′-flanking regions show clear-cut evidence of orthology with Mudo Mkrn1-p1.
Finally, to examine whether or not a Mkrn1-p1-like pseudogene exists in the rat genome, we surveyed the genome database using Rano Mkrn1 as a query. Although neither a Mkrn1-p1 nor a Mkrn1-p2 ortholog was found, there is one Mkrn1-derived processed pseudogene on chromosome 9 (Rano Mkrnp1). However, since the sequence is only partial in the database (336 bp corresponding to the C region), we determined the almost entire nucleotide sequence (998 bp). The p-distances of Rano Mkrnp1 from Rano Mkrn1 are merely 3.2%.
Sequence evolution of Mkrn1-derived processed pseudogenes:
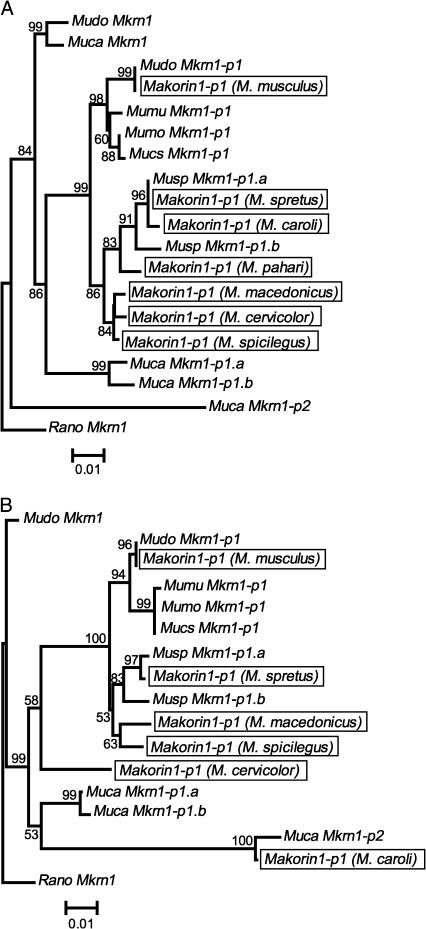
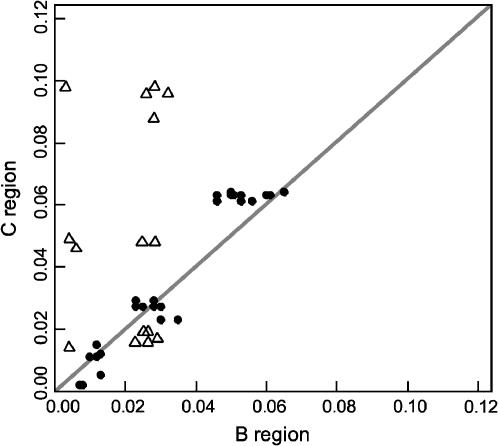
We made a phylogenetic analysis of our B and C region sequences of Makorin1-derived processed pseudogenes together with those reported by Podlaha and Zhang (2004) (Figure 1). In the B region, their M. caroli sequence clusters with the M. spretus sequences, whereas, in the C region, it clusters with Muca Mkrn1-p2. Thus, it is unlikely that the M. caroli sequence in Podlaha and Zhang (2004) is an ortholog of Makorin1-p1 and that the B and C region sequences are physically linked. Moreover, in the B region, the M. cervicolor and M. pahari sequences are embedded within the cluster of the sequences from the Palearctic musculus species group (M. m. musculus, domesticus, castaneus, and M. spretus). Using all the Mkrn1-p1 sequences, we further tested if functional constraints against the B region exist. Figure 2 shows the scatter diagram of the p-distances between the B and C regions in the pairwise comparisons of all available Mkrn1-p1 sequences. It is evident that the p-distances in the B region are almost the same as those in the C region in our data set, strongly arguing against the conservation of the B region relative to the C region (cf. Podlaha and Zhang 2004). Because of these abnormalities, we exclude the sequences reported by Podlaha and Zhang (2004) and use our own in the following analyses.
Figure 1.
The NJ trees for the B region (A) and the C region (B) of Makorin1 and its processed pseudogenes in mice. The sequences in Podlaha and Zhang (2004) are boxed. Both p-distance-based trees are rooted by Rano Mkrn1. The B and C regions compared are 609 and 581 bp long, respectively. Regarding Muca Mkrn1, the B region is taken from the NCBI database (AH014504), whereas the C region sequence determined here is 342 bp long and excluded in B. The number near each node represents the bootstrap value in 10,000 resamplings, but <50% bootstrap values are suppressed. The bar at the bottom shows the number of nucleotide differences per site. The abbreviations of the genus and species names are: Mudo (Mus m. domesticus), Mumu (Mus m. musculus), Mucs (Mus m. castaneus), Mumo (Mus m. molossinus), Musp (Mus spretus), Muca (Mus caroli), and Rano (Rattus norvegicus).
Figure 2.
The scatter diagram of the p-distances between the B and C regions in the pairwise comparison of the Mkrn1-p1 orthologs. Solid circles show the comparisons based on our sequences, whereas open triangles show those based on Podlaha and Zhang (2004). The conserved pattern of the B region in the latter comparisons is largely attributed to the dubious sequences of M. caroli and M. cervicolor.
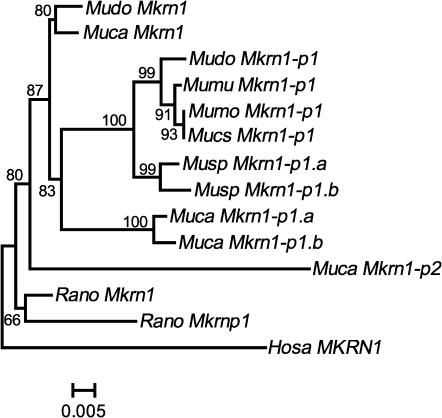
Since the B and C regions in our sequences are simultaneously amplified and evolved with similar rates (Figure 2), we use them collectively, but do not use the A region that is specific to and is not amplified for most Mkrn1-p1's. Figure 3 shows that the topological relationships among the Mkrn1-p1 orthologs in the NJ tree are consistent with those obtained from other genes (Suzuki et al. 2004). Importantly, it indicates that Mkrn1-p1 emerged before the Southeast Asian cervicolor species group (M. cervicolor and M. caroli) diverged from the Palearctic musculus species group and that the emergence of Mkrn1-p2 predated that of Mkrn1-p1. Moreover, it is clear that the rat processed pseudogene (Rano Mkrnp1) is clustered with Rano Mkrn1 and therefore of distinct origin from either Mkrn1-p1 or Mkrn1-p2 in mice.
Figure 3.
The p-distance-based NJ trees for Mkrn1 and its processed pseudogenes in mice and rats. The tree is rooted by Hosa MKRN1. The MP tree is topologically the same as the NJ tree. The number of sites in this comparison is 931 bp. The number close to each node represents the bootstrap value in 10,000 resamplings. The scale bar is given at the bottom. The abbreviations of the genus and species names are the same as those in Figure 1.
Dating the origin of Mkrn1-p1:
To date the origin of Mkrn1-p1, we noted the accelerated CpG substitution rate (Li et al. 1981; Gojobori et al. 1982) and first considered the CpG and non-CpG sites, separately. There are 16 CpG dinucleotide sites in the B region and 20 in the C region of both Mudo Mkrn1 and Rano Mkrn1. When these sequences are compared with Mudo and Musp Mkrn1-p1, it is possible to infer along which branches in the tree of (((Mudo Mkrn1-p1 and Musp Mkrn1-p1), Mudo Mkrn1), Rano Mkrn1), the C to T, the G to A, or all other kinds of substitutions occurred at CpG dinucleotide sites. Note that only four CpG's are newly created throughout this tree and that CpG is considerably underrepresented as in the human genome (Shioiri and Takahata 2001). In any event, when the ancestral Mkrn1-p1 was integrated into the mouse genome, there must have been n = 36 CpG's in the B and C regions. At these sites, k = 13 substitutions occurred during the stem lineage of Mudo Mkrn1-p1 and Musp Mkrn1-p1 that diverged from Mudo Mkrn1 t′ years ago. These substitutions reduced the number (n′ = 23) of CpG's at the common node of Mudo and Musp Mkrn1-p1 and accordingly increased the number of non-CpG's by 2k = 26. Since then, t years elapsed during which k′ = 7 and k′′ = 7 CpG substitutions occurred in the Mudo and Musp Mkrn1-p1 terminal branch, respectively.
We assume that the number of substitutions follows the Poisson distribution and that once a CpG site is mutated to a non-CpG site, the CpG is not restored for a short period of time. The probability of no substitutions during x years is then given by exp(−2rx), where r stands for the substitution rate at either C or G of a CpG site. The rate of r at CpG sites is thus estimated by the same correction formula as for amino acid substitutions (Kimura 1983):
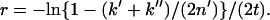 |
(1) |
Substituting the observed values of k′ = k′′ = 7 and n′ = 23 for Equation 1, we obtain r = 0.18/t. Similarly, if we compare the same set of sequences after excluding all the CpG's, we find 21 substitutions in the stem lineage of Mudo and Musp Mkrn1-p1 and 18 (or p = 18/1238) in both of the terminal branches. The rate at the non-CpG's becomes 0.0073/t. The ratio of these estimates is ∼25, implying that the CpG sites have evolved ∼25 times faster than the non-CpG sites. Nevertheless, since the number of CpG's is relatively small, the overall substitution rate may not be greatly enhanced. To compute the overall rate, one may also note that the heterogeneity of substitution rates among sites within individual sequences does not cause serious problems in phylogenetic and demographic analyses (Takahata and Satta 2002). We thus computed the overall distance between Mudo and Musp Mkrn1-p1 by taking the weighted average of the two 2rt values at the CpG's and non-CpG's. The overall distance (d = 2rt) then becomes 2.60%. This distance is substantially larger than the synonymous distance of 0.8% between the Mudo and Musp Mkrn1. If the divergence time (t) between the M. musculus complex and M. spretus is 1.5 MYA (Suzuki et al. 2004), the rate becomes 8.7 × 10−9/site/year.
With the above rate and a branch length of 2.65% for the stem lineage of Mudo and Musp Mkrn1-p1, we dated the emergence of Mkrn1-p1 as 4.5 MYA. This is slightly greater than the estimated time at which the Palearctic musculus and Southeast Asian cervicolor species groups began to diversify genetically. However, while this dating is consistent with the presence of the Mkrn1-p1 ortholog in M. caroli, it seems to be too recent for M. pahari or subgenus Coelomys to possess the ortholog (cf. Podlaha and Zhang 2004). Thus, our dating suggests that, like rats, Coelomys or any other mouse subgenera must have their own Mkrn1-derived processed pseudogenes.
Cotranscription of Mkrn1-derived processed pseudogenes:
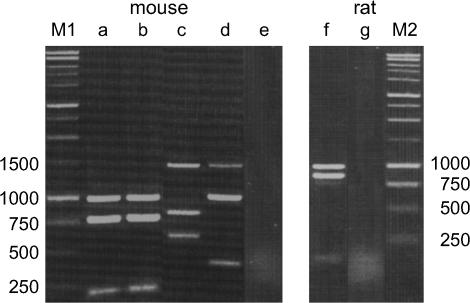
It is necessary to examine possible functional roles of the aforementioned processed pseudogenes in M. m. domesticus, M. spretus, M. caroli, and R. norvegicus. For this purpose, we carried out RT–PCR using total RNAs from testes of the above species (Figure 4). For each species, the designed primers can amplify both Mkrn1 and its processed pseudogene(s). Since the size of the expected PCR products of these sequences is almost the same, the products were digested by restriction enzymes to conveniently distinguish them from each other. The digestion pattern reveals that in each of the three mouse species, both Mkrn1 and Mkrn1-p1 are cotranscribed as in M. m. musculus (Hirotsune et al. 2003). To be thorough, we determined the nucleotide sequences of these PCR products. In M. m. domesticus and M. spretus, the TPPs are indeed Mkrn1-p1 orthologs. In M. caroli, Mkrn1-p2 is also transcribed together with Mkrn1-p1. Although the Mkrn1-p2 ortholog is also present in M. m. domesticus, it is 636 bp long and retains only exon 2 and exon 3 with an inserted LINE-1. It is unlikely that this sequence plays any functional role. Finally, we confirmed the cotranscription of Mkrnp1 in rats.
Figure 4.
Restriction enzyme analyses of transcripts of Mkrn1 and its processed pseudogenes in Mus and Rattus. The RT–PCR products were digested by HindIII, EcoRI, or BglII and separated by electrophoresis on a 1.5% agarose gel. (Lanes a and b) In M. m. domesticus and M. spretus (SPRET/Ei), there is a HindIII site in Mkrn1, but not in Mkrn1-p1. (Lane c) In M. caroli, a HindIII site is present in Mkrn1-p1.a, but absent in Mkrn1-p1.b, Mkrn1, and Mkrn1-p2. (Lane d) In M. caroli, an EcoRI site is present in Mkrn1-p1.a/b and Mkrn1, but absent in Mkrn1-p2. (Lane f) In R. norvegicus, Mkrnp1 has a BglII site, but Mkrn1 does not. Lanes e and g represent negative controls in M. caroli and R. norvegicus, respectively. These control experiments were carried out using total RNA to confirm no contamination of genomic DNA in samples. The molecular weight standards are shown on the left- and rightmost lanes (M1 and M2).
Mkrn1-derived processed pseudogenes in mammals:
Human MKRN1 is orthologous to mouse Mkrn1. The gene is well conserved at the amino acid level, suggesting that the control of Makorin1 mRNA stability is important not only in mice but also in other animals. We surveyed the genome databases for humans, chimpanzees, dogs, and cows to examine the presence of Makorin1-derived processed pseudogenes. In humans, in addition to seven known processed pseudogenes (MKRN4, MKRN5, and MKRNP1–5), we found two more on chromosome 2 (MKRNP6) and chromosome 9 (MKRNP7). The orthologs of all these human sequences can also be found in the chimpanzee genome. In dogs, there are four processed pseudogenes that are annotated here as Cafa Mkrnp1–4 and, in cows, three that are annotated as Bota Mkrnp1–3. It should be kept in mind that none of these pseudogenes, with similar nomenclatures, have orthologous relationships with respect to different orders of Eutherian mammals. In humans and chimpanzees, the p-distances from the conspecific MKRN1 are 6–7%, suggesting that New World monkeys (NWMs) and prosimians may have different sets of processed pseudogenes. To test this possibility, we carried out genomic PCR with a primer set of 5F2 and 3R2 and obtained two pairs of distinct sequences from four NWMs and five from three prosimians, as mentioned earlier.
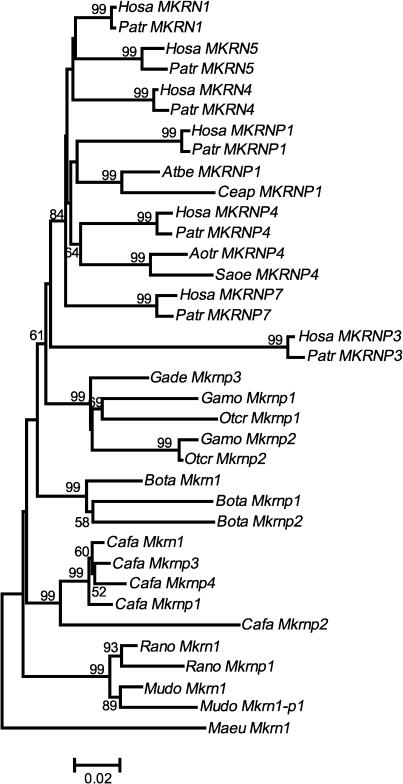
We constructed the phylogenetic tree of these mammalian sequences and rooted it by Maeu Mkrn1 in the wallaby (Figure 5). Two orthologous pairs of human and chimpanzee MKRN4 (∼1.5 kb) and MKRN5 (∼1.5 kb) cluster with Hosa MKRN1. In other mammals, Cafa Mkrnp1 and p2 for dogs or Bota Mkrnp1 and p2 for cows, respectively, cluster with their conspecific Mkrn1. Likewise, the prosimian sequences form a distinct cluster from the simian primate sequences. It is thus suggested that these prosimian sequences were also independently derived from their conspecific Mkrn1. The short branch leading to G. demidoff (Gade Mkrnp3) indicates its recent origin. This raises the possibility of a functional role, but the proof of this is beyond the present study. On the other hand, the four NWM sequences are closely related to either MKRNP1 or MKRNP4 in humans. Indeed, a shared 12-bp deletion supports that the A. belzebuth and C. apella sequences (Atbe MKRNP1 and Ceap MKRNP1) are orthologous to Hosa MKRNP1, and a shared 4-bp insertion supports that the S. oedipus and A. trivirgatus sequences (Saoe MKRNP4 and Aotr MKRNP4) are orthologous to Hosa MKRNP4 (supplement 2 at http://www.genetics.org/supplemental/). These processed pseudogenes were therefore retroposed prior to the divergence of simian primates, presumably during the time period in which LINE-1 was most active in the primate lineage (Batzer and Deininger 2002). By contrast, MKRN4 and MKRN5 appear to be of more recent origins and have been retroposed in the stem lineage of Catarrhini.
Figure 5.
The NJ tree of Makorin1 and its processed pseudogenes in primates and other mammals. The tree is based on the p-distances over 655 bp under complete deletion. The number close to each node represents the bootstrap value (≥50%) in 10,000 resamplings. The scale bar is represented at the bottom. The abbreviations of the genus and species names are: Hosa (Homo sapiens), Patr (Pan troglodytes), Saoe (Saguinus oedipus), Aotr (Aotus trivirgatus), Atbe (Ateles belzebuth), Ceap (Cebus apella), Gamo (Galago moholi), Otcr (Otolemur crassicaudatus), Gade (Galagoides demidoff), Cafa (Canis familiaris), Bota (Bos taurus), Rano (Rattus norvegicus), and Maeu (Macropus eugenii).
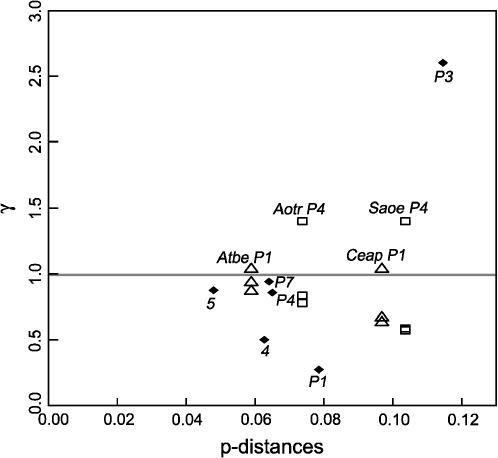
We examined if the B region of these processed pseudogenes in primates and other mammals has evolved without any selective constraint as in the mouse Mkrn1-p1. In a slightly different way from Figure 2, we plotted the ratio (γ) of the p-distances in the B region to the C region of orthologous pairs against the p-distances in the C region from Hosa MKRN1 (Figure 6). Unlike Mkrn1-p1, it is evident that the B region is more conserved than the C region for some processed pseudogenes (γ < 1) and that this trend becomes conspicuous as the p-distances in the C region increase. Most notable are MKRNP1 and MKRN4 in the comparison between the human and the chimpanzee. Their γ-values are 0.3 and 0.5, respectively. Although neither one is statistically different from 1 (P < 0.094 for MKRNP1 and P < 0.291 for MKRN4 in Fisher's exact test, 95% C.I. based on 1000 bootstrap samplings is 0–1.1 for MKRNP1 and 0–2.3 for MKRN4), both suggest the conservation of the B region relative to the C region after the species divergence 6 MYA. The MKRNP1 in NWMs also shows a similar trend, although less conspicuously (γ = 0.7). One reason for this discrepancy appears to be that the divergence between subfamilies A. trivirgatus and S. oedipus was as early as 20 MYA (Goodman et al. 1998), thereby including the early phase of the neutral evolution of MKRNP1. We also note that if we compare orthologous processed pseudogenes between distantly related species, the effect of enhanced substitution rates at CpG disappears. This rate slowdown, which may also be found in other processed pseudogenes, appears to stem from reduced CpG dinucleotides at equilibrium (cf. Nachman and Crowell 2000; Subramanian and Kumar 2003).
Figure 6.
The ratio (γ) of the p-distances in the B region to those in the C region in orthologous pairs of MKRN1-derived processed pseudogenes in simian primates (ordinate) and the p-distances in the C region of these processed pseudogenes measured from Hosa MKRN1 (abscissa). As in Figure 5, there are 16 such pairs, of which 6 are for MKRNP1 (open triangles); 6 are for MKRNP4 (open squares); and 1 each is for MKRN4, MKRN5, MKRNP3, and MKRNP7. These last four comparisons are indicated by solid diamonds, but distinguished by different numbers. The six comparisons of MKRNP1 or MKRNP4 are divided into two according to the p-distances on the abscissa. These four trios are indicated by Atbe P1 and Ceap P1 or Aotr P4 and Saoe P4. The lowest γ-value among the MKRNP1s is realized by the comparison between the human and the chimpanzee.
DISCUSSION
In this analysis of mouse Mkrn1-p1's, we have isolated the bona fide orthologs in several Mus species and used them to argue that the B and C regions have been evolving in a strictly neutral fashion (Kimura 1968, 1983). Despite the essential role of the B region of Mkrn1-p1 (Hirotsune et al. 2003), there is little or no evidence for the sequence conservation relative to the C region. This conclusion is different from that in Podlaha and Zhang (2004), which resulted from the erroneous sequences that were amplified for M. caroli and M. cervicolor in the Southeast Asian species group. We have pointed out that the PCR primer design used in Podlaha and Zhang (2004) does not guarantee the amplification of orthologous sequences owing to the presence of multiple copies of Mkrn1-derived processed pseudogenes in the Mus genome. Specifically, the three B region sequences of M. caroli, M. cervicolor, and M. pahari may not come from the species specified and the C region sequence of M. caroli may be that of paralogous Muca Mkrn1-p2.
However, if Mkrn1-p1 constantly decays, there will be a stage at which the transcript no longer titrates out the destabilizing factor of Mkrn1 mRNAs. There are then two ways of overcoming this situation. One way is the continuous production of Mkrn1-derived processed pseudogenes before the fatal decay of old ones. The alternative way is that in the absence of a new cotranscribed processed pseudogene, further decay of Mkrn1-p1 becomes detrimental to the carriers, thereby being subjected to negative selection. This is exactly what Podlaha and Zhang (2004) assumed for the B region sequences in the evolution of mouse Mkrn1-p1's. However, this alternative has not been applied to mice simply because of recurrent occurrences of Mkrn1-derived processed pseudogenes in the genome. We have shown that Mkrn1-p2 found in M. m. domesticus and M. caroli is older than Mkrn1-p1 and suggested that Mkrn1-p2 once played the same role as Mkrn1-p1, which emerged ∼4.5 MYA. The remnant of Mkrn1-p2 can still be seen in the Mus genome and is even transcribed in M. caroli. The eventual fate in the presence of a newcomer is, however, to be free from negative selection. This is most conspicuously exemplified by the fragmented Mkrn1-p2 in the M. m. domesticus genome. In addition, there are two other Makorin1-derived pseudogenes in the M. m. domesticus genome that are truncated and substantially different from the conspecific Makorin1 (p = 5.5 and 8.5%). Under this recurrent production of processed pseudogenes, the nucleotide substitution rate is considerably enhanced by changes at the CpG dinucleotide sites.
Podlaha and Zhang (2004) found a Mkrn1-p1-like partial sequence in M. pahari, but excluded it from their phylogenetic analysis. M. pahari belongs to subgenus Coelomys, which diverged from the subgenus Mus >6 MYA (Suzuki et al. 2004) and occupies an interesting phylogenetic position in the present context. Our dating of the origin of Mkrn1-p1, however, suggests that M. pahari does not possess the Mkrn1-p1 ortholog, but instead possesses other Mkrn1-derived processed pseudogenes of different origins. The proof of the absence of Mkrn1-p1 in M. pahari requires the same approach as that taken for the isolation of Muca Mkrn1-p1 and is currently under way. In any event, the rapid turnover of Mkrn1-p1 and Mkrn1-p2 in mice implies that M. pahari, rats, and any other mammal must possess their own Mkrn1-derived processed pseudogenes. Indeed, we have found species-specific processed pseudogenes in rats, dogs, cows, and primates. In rats, we have also demonstrated that Rano Mkrnp1 on chromosome 9 is of almost full length and transcribed. Although evidence for the function similar to Mkrn1-p1 is only circumstantial, our finding of Rano Mkrnp1 transcription suggests that the same stabilizing mechanism of Mkrn1 mRNAs exists in individual mammals. In dogs, we found candidate processed pseudogenes: Cafa Mkrnp1–2 with such small p-distances as 1.2–1.5% from Cafa Mkrn1. These are almost intact and likely function if they are transcribed.
The situation is different in primates and cows in that even the most closely related processed pseudogenes to the functional ones exhibit p-distances as large as 6–7% or are as old as ∼35 MY. The rather old ages of these processed pseudogenes may be associated with the reduced LINE-1 activity and/or with the lowered “hidden” promoter activity in the genome (Cheng et al. 2005; FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group 2005). Whatever the reason, we have hypothesized that under lowered production rates of TPPs, organisms must rely on the alternate strategy mentioned above. We cannot directly prove or disprove this hypothesis by experiments, but our evolutionary approach has revealed that the B region of MKRN4 and MKRNP1 in humans and chimpanzees tends to evolve more slowly than the C region (Figure 6). Since it is known that MKRN4 is transcribed, we were curious whether or not MKRNP1 is also transcribed. The RT–PCR experiment for human testis confirmed the transcription of MKRNP1 as well (data not shown). Thus, both MKRN4 and MKRNP1 are transcribed in humans so that they may play the same role as Mkrn1-p1 in mice. However, although it is true that no newcomer appeared during the past 35 MY or so, it is not immediately clear why these TPPs must be simultaneously conserved. Alternatively, despite the apparent older age of MKRNP1 than MKRN4, one may ask which one is more functionally important. To address these questions, it is interesting to compare orthologous pairs of MKRNP1 and MKRN4 in more closely related species of Old World monkeys (OWMs) or NWMs than at the subfamily level. If we can observe lowered γ-values as in the comparison between humans and chimpanzees, we may generalize the conservation and assess the relative importance of MKRN4 and MKRNP1. Comparisons of DNA polymorphism between the two regions may help to reveal differential selective constraints. Also, measuring relative amounts of these transcripts may give us some clue, by noting that the Makorin1 transcription is more uniform over tissues in humans than over those in mice (Hirotsune et al. 2003; Kim et al. 2005). In these contexts, we agree with Podlaha and Zhang (2004): The B region of Makorin1-derived TPPs tends to be subjected to negative selection without newcomers in the genome. However, a question remains as to the biological significance about such a devious and fortuitous mechanism for regulating mRNA stability of such a developmentally important gene as mammalian Makorin1. It also remains to be answered whether this coevolving system is found in other phyla.
Acknowledgments
We are grateful to Jianzhi Zhang for providing us with the then unpublished manuscript. We thank Toshihiko Shiroishi, the National Institute of Genetics, for providing us with the BLG2/MsRbrc and MSM/MsRbrc mice strains. We also thank Michael Kryshak for his editorial assistance. This work was supported in part by the Japan Society for Promotion of Science grant 12304046 to N.T.
References
- Balakirev, E. S., and F. J. Ayala, 2003. Pseudogenes: Are they “junk” or functional DNA? Annu. Rev. Genet. 37: 123–151. [DOI] [PubMed] [Google Scholar]
- Batzer, M. A., and P. L. Deininger, 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3: 370–380. [DOI] [PubMed] [Google Scholar]
- Chen, M.-J., T. Shimada, A. D. Moulton, M. Harrison and A. W. Nienhuis, 1982. Intronless human dihydrofolate reductase genes are derived from processed RNA molecules. Proc. Natl. Acad. Sci. USA 79: 7435–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J., P. Kapranov, J. Drenkow, S. Dike, S. Brubaker et al., 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154. [DOI] [PubMed] [Google Scholar]
- Dorus, S., S. L. Gilbert, M. L. Foster, R. J. Barndt and B. T. Lahn, 2003. The CDY-related gene family: coordinated evolution in copy number, expression profile and protein sequence. Hum. Mol. Genet. 12: 1643–1650. [DOI] [PubMed] [Google Scholar]
- FANTOM Consortium and RIKEN Genome Exploration Research Group and Genome Science Group, 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Fitch, W. M., 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20: 406–416. [Google Scholar]
- Gojobori, T., W. H. Li and D. Graur, 1982. Patterns of nucleotide substitution in pseudogenes and functional genes. J. Mol. Evol. 18: 360–369. [DOI] [PubMed] [Google Scholar]
- Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider et al., 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9: 585–598. [DOI] [PubMed] [Google Scholar]
- Gray, T. A., L. Hernandez, A. H. Carey, M. A. Schaldach, M. J. Smithwick et al., 2000. The ancient source of a distinct gene family encoding proteins featuring RING and C3H zinc-finger motifs with abundant expression in developing brain and nervous system. Genomics 66: 76–86. [DOI] [PubMed] [Google Scholar]
- Harrison, P., M. D. Zheng, Z. Zhang, N. Carriero and M. Gersten, 2005. Transcribed processed pseudogenes in the human genome: an intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res. 33: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune, S., N. Yoshida, A. Chen, L. Garret, F. Sugiyama et al., 2003. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 423: 91–100. [DOI] [PubMed] [Google Scholar]
- Jukes, T. H., and C. R. Cantor, 1969. Evolution of protein molecules, pp. 21–132 in Mammalian Protein Metabolism, edited by H. N. Munro. Academic Press, New York.
- Kim, J. H., S.-M. Park, M. R. Kang, S.-Y. Oh, T. H. Lee et al., 2005. Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19: 776–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1968. Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge, UK.
- Korneev, S., J. H. Park and M. O'Shea, 1999. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 19: 7711–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura and M. Nei, 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5: 150–163. [DOI] [PubMed] [Google Scholar]
- Li, W. H., T. Gojobori and M. Nei, 1981. Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239. [DOI] [PubMed] [Google Scholar]
- Long, M., and C. H. Langley, 1993. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260: 91–95. [DOI] [PubMed] [Google Scholar]
- Long, M., E. Betrán, K. Thornton and W. Wang, 2003. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 4: 865–874. [DOI] [PubMed] [Google Scholar]
- Nachman, M. W., and S. L. Crowell, 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and S. Kumar, 2000. Molecular Evolution and Phylogenetics. Oxford University Press, New York.
- Podlaha, O., and J. Zhang, 2004. Non-neutral evolution of the transcribed pseudogene Makorin1-p1 in mice. Mol. Biol. Evol. 21: 2202–2209. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Shioiri, C., and N. Takahata, 2001. Skew of mononucleotide frequencies, relative abundance of dinucleotides, and DNA strand asymmetry. J. Mol. Evol. 53: 364–376. [DOI] [PubMed] [Google Scholar]
- Skowronski, J., and M. F. Singer, 1986. The abundant LINE-1 family of repeated DNA sequences in mammals: genes and pseudogenes. Cold Spring Harbor Symp. Quant. Biol. 51: 457–464. [DOI] [PubMed] [Google Scholar]
- Subramanian, S., and S. Kumar, 2003. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 13: 838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., T. Shimada, M. Terashima, K. Tsuchiya and K. Aplin, 2004. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 33: 626–646. [DOI] [PubMed] [Google Scholar]
- Takahata, N., and Y. Satta, 2002. Pre-speciation coalescence and the effective size of ancestral populations, pp. 52–71 in Modern Developments in Theoretical Population Genetics, edited by M. Slatkin and M. Veuille. Oxford University Press, Oxford.
- Thompson, J., T. Gibson, F. Plewniak, F. Jeanmougin and D. Higgins, 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin, E. F., 1985. Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet. 19: 253–272. [DOI] [PubMed] [Google Scholar]
- Yano, Y., R. Saito, N. Yoshida, A. Yoshiki, A. Wynshaw-Boris et al., 2004. A new role for expressed pseudogenes as ncRNA: regulation of mRNA stability of its homologous coding gene. J. Mol. Med. 82: 414–422. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., and M. Gerstein, 2004. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 14: 328–335. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., P. Harrison, Y. Liu and M. Gerstein, 2003. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13: 2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., N. Carriero and M. Gerstein, 2004. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 20: 62–67. [DOI] [PubMed] [Google Scholar]