Abstract
We compared allele sequences of two loci near the Arabidopsis lyrata self-incompatibility (S) loci with sequences of A. thaliana orthologs and found high numbers of shared polymorphisms, even excluding singletons and sites likely to be highly mutable. This suggests maintenance of entire S-haplotypes for long evolutionary times and extreme recombination suppression in the region.
BALANCING selection can sometimes maintain variants for very long evolutionary times, and it is often stated that the times can exceed the ages of related species, i.e., that variants arose before the split of related species (e.g., Ioerger et al. 1990; Klein et al. 1993; Richman et al. 1996; Clark 1997; Wu et al. 1998; Adams et al. 2000; Muirhead et al. 2002). Trans-specific polymorphism can provide strong evidence of long-term balancing selection, because it is highly unlikely to exist under neutrality, except between very closely related species that can share variants present in their common ancestor (Wiuf et al. 2004), and is expected only when the same alleles persist for long times, and not when alleles are regularly replaced by new alleles (“turnover”; see Muirhead et al. 2002).
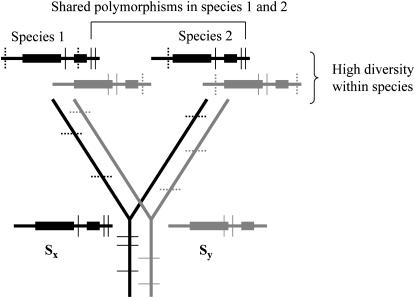
Recently, by examining human and chimpanzee gene sequences for trans-specific polymorphism, a search for evidence of long-term balancing selection concluded that it is infrequent in humans (Asthana et al. 2005). The principle of such tests depends on the fact that long-term balancing selection not only affects diversity at the sites that are under selection, but also leads to high diversity at nearby neutral sites. Within an ancestral species, a gene under balancing selection maintains different functional classes of alleles, and each allele class will acquire its own unique set of neutral mutations, causing variants to be associated with the allele in which they arose until recombination allows “migration” into a different allele (reviewed in Charlesworth et al. 2003a). Thus functionally different alleles will be differentiated at the amino acids that define those types and also at other sites within the region (linkage disequilibrium); i.e., there will be higher polymorphism over a region whose size depends on the local recombination frequency than in unlinked genome regions (Wiuf et al. 2004). When a species with such a balanced polymorphism splits into two, multiple different haplotypes will often pass to the daughter species (Figure 1 shows a hypothetical example). The resulting trans-specific polymorphism will initially maintain the same associations of variants as in the ancestor, but over evolutionary time this signal will become indistinct, as the sequences in each daughter species's copies of the functionally same allele recombine with other haplotypes of the locus, acquire new mutations, and evolve new, functionally different alleles, leading to allele turnover. Low recombination will lead to trans-specific polymorphism for longer evolutionary times.
Figure 1.
Lineages at an S-locus under long-term balancing selection and the effects of speciation. As an example, two haplotypes are shown with different S-alleles, x and y, which diverged before the common ancestor of two species (1 and 2). The different alleles at the pistil and pollen S-loci, SRK and SCR, are denoted by solid and shaded boxes. Mutations in the regions in and around the S-locus (thin horizontal lines in the tree) before the species became isolated remain associated with the haplotype in which they arose (shown as thin vertical lines in the haplotypes) until recombination occurs with a different haplotype. Species-specific differences (dotted lines) will also accumulate.
Here we show that the expected effect of balancing selection in leading to trans-specific polymorphism is detectable at loci near the self-incompatibility (SI) loci (S-loci) in species of the plant genus Arabidopsis. Because the formal population genetics theory shows that the region affected by a locus under balancing selection will be very small, in terms of the recombination distance (Wiuf et al. 2004), this result suggests very low recombination in the region. With recombination, the region of high diversity within the ancestral species is small (Takahata and Satta 1998), and only this region is likely to yield trans-specific polymorphisms. The results also suggest recent maintenance of multiple S-haplotypes in Arabidopsis thaliana, even though this species is now highly self-compatible. Balancing selection is well documented in self-incompatibility in several plant species (Richman et al. 1996; Takebayashi et al. 2003), with large numbers of functionally different S-alleles maintained for very long evolutionary times by the advantage of rare incompatibility alleles, i.e., very slow turnover, as theoretically predicted (Takahata 1990; Vekemans and Slatkin 1994). This should produce extremely high polymorphism at linked neutral sites (Nordborg et al. 1996; Takahata and Satta 1998; Takebayashi et al. 2004), and the considerable data now available document the expected high variability throughout the S-locus gene sequences. In all species where multiple S-alleles have been studied, their sequences differ greatly. Nucleotide diversity is extremely high in pistil recognition genes of gametophytic SI systems (e.g., Richman et al. 1996; Lu 2001, 2002) and in the pistil and pollen S-loci of species with sporophytic SI (Sato et al. 2002; Charlesworth et al. 2003c). Consistent with this evidence for long-term maintenance of S-alleles, several alleles with the same specificity are shared between Brassica oleracea and B. rapa (=campestris) (Kimura et al. 2002; Sato et al. 2003).
Our previous work on diversity at loci linked to the A. lyrata S-locus suggests low recombination in the region on the basis of two kinds of evidence. First, we find high nucleotide diversity in the sequences of at least three of five such loci studied, even though they are not involved in incompatibility functions and show no evidence of themselves being under balancing selection; two of them have extremely high diversity (Kamau and Charlesworth 2005). Applying a recently developed model of gametophytic self-incompatibility (Takebayashi et al. 2004), we roughly estimated the recombination rate for the region to be <1 cM/6 Mb (Kamau and Charlesworth 2005), a value lower than most, if not all, other estimates for noncentromeric regions of plant genomes (e.g., Copenhaver et al. 1998; Thiel et al. 2003; Khrustaleva et al. 2005). Moreover, diversity is very low within S-haplotypes (on the basis of sequences of flanking genes carried in haplotypes with the same SRK sequence; J. Bechsgaard, E. Kamau and D. Charlesworth, unpublished results). Here we add an independent type of evidence from trans-specific polymorphism between A. lyrata and its self-compatible relative, A. thaliana, in the S-locus region, also suggesting low recombination in the region, but over a much longer evolutionary timescale and thus with an even lower recombination than our rough estimates from polymorphism within A. lyrata. Sequence diversity data from these species have recently become available for two genes, B80 and Aly8 (see below), allowing comparisons to be made. Shared polymorphisms were found in both genes, particularly in B80. These plants are far too distantly related for trans-specific polymorphism to be expected, unless very long-term balancing selection has acted at a locus very closely linked to the ones studied here, so that associations are maintained among variants of these loci over an extremely long timescale.
Sequence diversity in the A. thaliana genome region orthologous to the S-locus region of A. lyrata is low at the pollen S-locus, SCR1, but high at the pistil S-locus, SRK (Shimizu et al. 2004), and also at the linked orthologs of B80 (the U-box gene in Shimizu et al. 2004). Thus, despite having lost self-incompatibility, A. thaliana retains multiple different haplotypes in the S-locus region. This might happen if self-compatibility was lost in this species's ancestor by selection for a loss-of-function allele at a locus not in the S-locus region or one that recombines with SRK, rendering the SRK alleles neutral. If this occurred recently enough, there might not have been enough time for genetic drift to lead to fixation of one of the haplotypes present (see below). By comparing our B80 sequences with sequences of the orthologous gene of A. thaliana, we obtained evidence that recombination is indeed very infrequent.
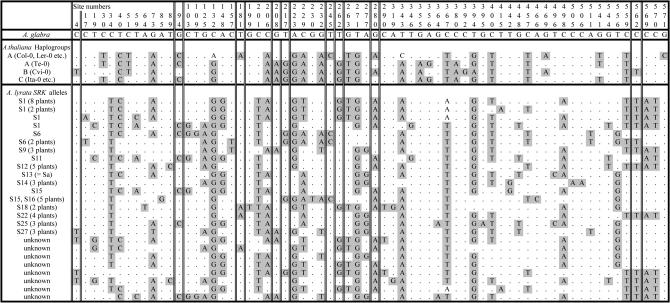
We studied two loci, B80 and ARK3 (called Aly8 in A. lyrata; see Charlesworth et al. 2003c), for which multiple sequences are available from A. thaliana and A. lyrata. These genes are located physically close to the functional self-incompatibility loci, SRK and SCR (Kusaba et al. 2001). We aligned our B80 sequences with sequences of the A. thaliana ortholog and with a sequence from the inbreeding species Arabis glabra (synonymous site divergence from A. thaliana averages 0.2; A. Kawabe, unpublished data). The B80 gene contains no introns and is a single exon of 1125 bp in A. thaliana; no alignment gaps were required. SRK diversity is very low in A. glabra, and the sequences from three different populations are all similar to SRK allele 31 of A. lyrata. We used the alignment of 577 nucleotides to infer the lineages in which the variants originated and to examine the data for polymorphisms shared between the 21 A. thaliana sequences obtained by Shimizu et al. (2004) and the sequences of 54 A. lyrata alleles, representing 25 different S-haplotypes (there are 6 different A. thaliana sequences, or 4 if singleton sites are ignored, and 28 different A. lyrata sequences, or 25 if ignoring singletons; see Figure 2). Of 104 variants in the B80 alignment, 11 were fixed differences between A. glabra and both the sister species, A. thaliana and A. lyrata, and there were no fixed differences between those two species. Of the 19 sites polymorphic within A. thaliana, 9 are apparently trans-specific polymorphisms, one of them a nonsynonymous variant (Figure 2). None of the shared variants involves CpNpG or CpG-prone sites (because of the known high mutation rate of such sites, these sites should be excluded, together with sites that might recently have mutated from such sites). None of the sites with shared polymorphisms is a singleton variant in A. lyrata, but 3 are singletons in A. thaliana, all of these being variants in the Cvi strain that were not seen in sequences of other strains. The average raw divergence of the sequences of this gene between these two species is close to the mean for other loci so there is no evidence for an unusually high mutation rate at this locus (Kamau and Charlesworth 2005).
Figure 2.
Variants in the B80 (U-box) gene within A. thaliana and A. lyrata, excluding fixed differences from A. glabra and singleton variants other than in the A. thaliana Cvi-0 strain. For A. thaliana and A. lyrata, a single sequence of each type seen within the species is shown. For A. thaliana, the haplogroups of the strains (defined by SRK sequences; see the U-box gene in Shimizu et al. 2004) are shown; for A. lyrata, the SRK allele, when known, is shown for each haplotype whose B80 sequence was used in the analysis (the numbers of each A. lyrata S-haplotype sequenced are also shown in the left column). The nine shared variants between the two species are boxed with thin double lines; the nonsynonymous one is at position 245.
Aly8, the ortholog of the A. thaliana ARK3 gene (Kusaba et al. 2001; Schierup et al. 2001), encodes an S-domain protein, resembling SRK. The results are less straightforward, because Aly8 is duplicated in at least some A. lyrata haplotypes (J. Hagenblad, J. Bechsgaard and D. Charlesworth, unpublished data); nevertheless, we can test for shared variants in the two species. Our sequences are from the first exon, slightly 5′ of the region where high polymorphism was found in A. thaliana (Shimizu et al. 2004) and just 3′ of the region where high diversity was previously found in A. lyrata (Charlesworth et al. 2003c). With a larger number of sequences (93 from A. thaliana and 34 from A. lyrata) in a sequence alignment (including both species) of only 318 bp after excluding gaps, three nonsingleton shared polymorphisms were found, plus four shared polymorphisms that are singletons in the A. thaliana sample; one other site is polymorphic in both species at a CpG-prone site, but not in the identical variants. There were 13 sites with fixed differences between the two species (4.1%), a strikingly low proportion for these species.
Trans-specific polymorphisms can arise in several ways. One possibility is chance occurrence of mutations at the same site since two species split. We calculated the probability that a polymorphism observed in one species will be found in a related species, using a recently derived analytical formula (equation 16 of Charlesworth et al. 2005). The split between A. thaliana and A. lyrata is estimated to have been ∼5 MYA on the basis of net silent-site divergence values of ∼12% (Wright et al. 2002; Schmid et al. 2005); this divergence estimates 2μT, where μ is the neutral mutation rate and T is the time of the speciation event. Within either species, silent-site diversity values are ∼10-fold lower than this, and these values are estimates of 4Neμ, where Ne is the species effective population size. Thus the observed divergence/diversity values provide an estimate of (2μT/4Neμ) = T, in units of 2Ne, of ∼10. With T = 10 we obtain a probability value of ∼4 × 10−5. A sequence of 577 bp, such as B80, is thus not expected to include as many as a single shared polymorphism. With T = 5, allowing for a generation time of >1 year, and only 318 bp, as for ARK3, two are expected. Moreover, if recurrent mutation caused the polymorphisms at the same sites in both Arabidopsis species, most of these sites should have polymorphisms of different nucleotides, whereas trans-specific polymorphisms due to long-term associations must be identical nucleotides. In the B80 gene, there are only two such polymorphic sites shared between A. thaliana and A. lyrata [at sites 343 and 367 (see Figure 2); these are, of course, not included in the count of shared polymorphisms]. T is also large enough that ancestral polymorphisms would be very unlikely to be retained in A. thaliana (Clark 1997), assuming that since this species lost functional self-incompatibility, the S-locus region was not maintained polymorphic by balancing selection.
Another possibility is thus ancestral polymorphism. If we take as our null hypothesis that in A. thaliana no balancing selection has recently affected this region, because this species has lost self-incompatibility, A. thaliana alleles should have a common ancestor considerably more recent than the time of the split between A. thaliana and A. lyrata. We tested this null hypothesis further by calculating the probability of finding in A. thaliana nine or more shared polymorphisms at sites at which polymorphisms are observed in A. lyrata (81/577, or 14%, of sites in the B80 gene). We used the binomial theorem to approximate the hypergeometric probabilities of each possible number of A. thaliana polymorphisms occurring at these sites, assuming independence of the chances of a variant occurring at different sites in the sequence (since the proportion of polymorphic sites in A. lyrata is not close to zero). The approximation is valid since the number of sites examined is large (see Keeping 1962). The probability of finding nine or more shared polymorphic sites is 0.021. The probability that these will each have the same variants in both species is considerably lower, since if mutation occurs randomly, each site should have a chance of 1/3 of having the same mutation, assuming the same ancestral nucleotide for both species. For the ARK3/Aly8 genes, the chance of three or more shared polymorphic sites is high (53%), given the polymorphism level in A. lyrata (12.6% of all sites), but the chance that all three will have identical variants in both species is lower. Thus this locus also may have shared polymorphisms, although the conclusion is weaker than that for B80.
Our analysis underestimates the number of trans-specific polymorphisms, because our samples might not include rare variants, and the A. lyrata sample is from a limited sampling of populations (Schierup et al. 2001). However, many different alleles were included in the samples from both species, so this is not likely to be a large effect.
Because of their large divergence times, shared polymorphisms between A. thaliana and A. lyrata seem a priori highly unlikely. To check this, we examined loci that are probably not close to genes under balancing selection and whose alleles are thus not expected to be maintained for long evolutionary times. We found six reference loci for which sequence samples are available from both these species; the loci are Adh (Savolainen et al. 2000), Cauliflower (Purugganan and Suddith 1998; Wright et al. 2003), and Chi, FAH1, F3H, and MAM-L (see Ramos-Onsins et al. 2004). In an alignment of sequences of these six loci, with a total of 5172 bp, we found no shared polymorphisms, as expected for species that have diverged for a long evolutionary time [the expected life span of neutral alleles is 4Ne, although the range of values is wide, so that much longer times may occasionally be observed (Clark 1997)]. Thus the observed trans-specific polymorphisms in the S-locus region seem to require a selective explanation.
The shared polymorphisms are probably variants that differed between S-haplotypes in a self-incompatible ancestral species. Analysis of the A. lyrata B80 sequences does not suggest balancing selection acting at this locus itself, despite its high diversity, which seems to be attributable to linkage to the SRK locus (Kamau and Charlesworth 2005). Our results thus suggest that recombination between different S-alleles is rare enough across the region including the flanking genes (B80 and perhaps also ARK3) that entire haplotype sequences have been preserved between the species studied since their common ancestor. In the absence of recombination, each functionally distinct haplotype is expected to be almost uniform in sequence within the ancestral self-incompatible species, as is the case for the SRK sequence in A. lyrata (Charlesworth et al. 2003b). When daughter species become reproductively isolated, they will often have “trans-specific” allelic lineages with the same incompatibility type and, initially, similar sequences. If the regions flanking the S-locus also recombine very rarely, their sequences would also, like those of the same S-allele in related species, differ only by mutations that have substituted in one lineage or the other since the species split (Figure 1). There should thus be unusually few fixed differences between species, as observed, whereas raw divergence (uncorrected for polymorphism within the species) will be high, due to the long times to the common ancestors of the sequences, whether compared within or between species. It therefore appears that the species studied here must have shared S-allele lineages recently.
As already mentioned, A. thaliana is self-compatible. Using A. thaliana in our comparisons of sequence variants therefore does not correspond precisely to the situation in which the same alleles are maintained by balancing selection in two related self-incompatible species. If loss of SI in A. thaliana was due to a selective sweep at one of the S-loci, it should have caused low diversity across this region, so at most a few variants that arose after the event are expected in the species, since loss of SI was probably recent (Shimizu et al. 2004). Trans-specific polymorphisms would then be highly unlikely. Even if self-compatibility evolved through a mutation at an unlinked locus (which would not cause such rapid diversity loss), there has probably been enough time for genetic drift to have led to loss of all but one lineage at the S-locus. Applying the standard population genetics formula, a reduction in diversity to 40% as reported for the SRK pseudogene in a population with the Ne value of ∼400,000 estimated for A. thaliana (Shimizu et al. 2004) is expected to take 600,000 generations (Charlesworth and Vekemans 2005). Yet in A. thaliana diversity is very high in both SRK and the closely linked ortholog of B80 (Shimizu et al. 2004). A. thaliana therefore probably retained incompatibility and shared functional S-allele lineages with the ancestor of A. lyrata during most of the much longer time (see above) since the species's common ancestor, and balancing selection may thus have maintained S-allele variation in both species until quite recently. This is supported by our finding of trans-specific variants, which should not be present if A. thaliana evolved self-compatibility very long ago. If so, our comparison is essentially equivalent to one between two self-incompatible species (the self-incompatible ancestor of A. thaliana and A. lyrata). Unless there is another nearby locus at which alleles are maintained by long-term balancing selection in A. thaliana and A. lyrata, we can account for our findings at the B80 locus only if A. thaliana lost functional self-incompatibility recently enough that multiple haplotypes have remained present.
The results suggest that the region, including at least the S-loci and B80, has recombined very rarely since the species split. Low recombination is predicted in the S-locus region, because recombination generates self-compatible combinations of the pollen and pistil incompatibility loci (Casselman et al. 2000). This conclusion is similar to that for a part of the human MHC region containing three class II genes, where very high nucleotide diversity and strong linkage disequilibrium were found for sites in the intergenic regions, suggesting that entire haplotypes across the region have been maintained since before humans evolved and that recombination may have been rare since before the common ancestor with other species such as chimpanzee (Raymond et al. 2005). However, as explained above, balancing selection maintaining many alleles can affect diversity at nearby neutral sites, even without the evolution of a low recombination rate (Takahata and Satta 1998). Given the generally low recombination rate in humans, it is thus unclear whether the findings for MHC could be explained by a model in which many alleles are maintained by balancing selection under average recombination rates.
The size of the region of low recombination around the Arabidopsis S-locus cannot yet be accurately estimated, because different S-haplotypes are rearranged with respect to gene order and distances between genes. Physical distance information is currently available for three haplotypes, two functional S-haplotypes in A. lyrata, and one nonfunctional haplotype from A. thaliana (Kusaba et al. 2001). The gene order is the same in all three haplotypes, with B80 and ARK3 (or Aly8 in A. lyrata) on opposite sides of the S-loci, spanning a region of several tens of kilobases. The low recombination suggested by our results thus seems to extend into the nonrearranged regions flanking the S-loci at least as far as B80. Given the apparent extreme suppression of recombination, and likelihood that a quite large genome region is affected, family studies may be able to test for linkage between genes at large physical distances (which could potentially be estimated with data from the planned A. lyrata genome sequence). Such independent tests for recombination suppression would be very valuable, particularly as there is a puzzle about the results from A. thaliana. Shimizu et al. (2004) suggested that the loss-of-function mutation that causes loss of incompatibility in A. thaliana occurred at the SCR1 locus. This requires the assumption that recombination occurs frequently enough that the resulting selective sweep as the compatible haplotype spread affected only the SCR1 locus (which suffered a severe loss of sequence diversity), but not the flanking ARK3 and U-box genes. If recombination occurred very infrequently, this interpretation would be less plausible (Charlesworth and Vekemans 2005). Thus, either the low SCR1 diversity must be due to some cause other than the proposed selective sweep or this region of the genome must have had much higher recombination in A. thaliana's ancestor at the time when self-compatibility evolved. This puzzle should be resolved in the future.
Acknowledgments
We thank Brian Charlesworth, Magnus Nordborg (University of Southern California), and Xavier Vekemans (University of Lille I) for helpful discussions. This work was funded by a grant to D.C. from the Environmental Genomics Programme of the Natural Environment Research Council of the United Kingdom.
References
- Adams, E. J., S. Cooper, G. Thomson and P. Parham, 2000. Common chimpanzees have greater diversity than humans at two of the three highly polymorphic MHC class I genes. Immunogenetics 51: 410–424. [DOI] [PubMed] [Google Scholar]
- Asthana, S., S. Schmidt and S. Sunyaev, 2005. A limited role for balancing selection. Trends Genet. 21: 30–32. [DOI] [PubMed] [Google Scholar]
- Casselman, A. L., J. Vrebalov, J. A. Conner, A. Singhal, J. Giovanni et al., 2000. Determining the physical limits of the Brassica S-locus by recombinational analysis. Plant Cell 12: 23–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, D., and X. Vekemans, 2005. How and when did Arabidopsis thaliana become highly self-fertilising? BioEssays 27: 472–476. [DOI] [PubMed] [Google Scholar]
- Charlesworth, B., D. Charlesworth and N. H. Barton, 2003. a The effects of genetic and geographic structure on neutral variation. Annu. Rev. Ecol. Evol. Syst. 34: 99–125. [Google Scholar]
- Charlesworth, D., C. Bartolomé, M. H. Schierup and B. K. Mable, 2003. b Haplotype structure of the stigmatic self-incompatibility gene in natural populations of Arabidopsis lyrata. Mol. Biol. Evol. 20: 1741–1753. [DOI] [PubMed] [Google Scholar]
- Charlesworth, D., B. K. Mable, M. H. Schierup, C. Bartolomé and P. Awadalla, 2003. c Diversity and linkage of genes in the self-incompatibility gene family in Arabidopsis lyrata. Genetics 164: 1519–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., C. Bartolomé and V. Noel, 2005. The detection of shared and ancestral polymorphisms. Genet. Res. 86: 149–157. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., 1997. Neutral behavior of shared polymorphism. Proc. Natl. Acad. Sci. USA 94: 7730–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver, G. P., W. E. Browne and D. Preuss, 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger, T. R., A. G. Clark and T.-H. Kao, 1990. Polymorphism at the self-incompatibility locus in Solanaceae predates speciation. Proc. Natl. Acad. Sci. USA 87: 9732–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamau, E., and D. Charlesworth, 2005. Balancing selection and low recombination affect diversity near the self-incompatibility loci of the plant Arabidopsis lyrata. Curr. Biol. 15: 1773–1778. [DOI] [PubMed] [Google Scholar]
- Keeping, E. S., 1962. Introduction to Statistical Inference. Van Nostrand, Princeton, NJ.
- Khrustaleva, L. I., P. E. deMelo, A. W. vanHeusden and C. Kik, 2005. The integration of recombination and physical maps in a large-genome monocot using haploid genome analysis in a trihybrid Allium population. Genetics 169: 1673–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, R., K. Sato, R. Fujimoto and T. Nishio, 2002. Recognition specificity of self-incompatibility maintained after the divergence of Brassica oleracea and Brassica rapa. Plant J. 29: 215–223. [DOI] [PubMed] [Google Scholar]
- Klein, J., N. Takahata and C. O'hUigin, 1993. Trans-specific Mhc polymorphism and the origin of species in primates. J. Med. Primatol. 22: 57–64. [PubMed] [Google Scholar]
- Kusaba, M., K. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative, A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., 2001. Roles of lineage sorting and phylogenetic relationship in the genetic diversity at the self-incompatibility locus of Solanaceae. Heredity 86: 195–205. [DOI] [PubMed] [Google Scholar]
- Lu, Y., 2002. Molecular evolution at the self-incompatibility locus of Physalis longifolia (Solanaceae). J. Mol. Evol. 54: 784–793. [DOI] [PubMed] [Google Scholar]
- Muirhead, C. A., N. L. Glass and M. Slatkin, 2002. Multilocus self-recognition systems in fungi as a cause of trans-species polymorphism. Genetics 161: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg, M., B. Charlesworth and D. Charlesworth, 1996. Increased levels of polymorphism surrounding selectively maintained sites in highly selfing species. Proc. R. Soc. Lond. Ser. B 163: 1033–1039. [Google Scholar]
- Purugganan, M. D., and J. I. Suddith, 1998. Molecular population genetics of the Arabidopsis CAULIFLOWER regulatory gene: nonneutral evolution and naturally occurring variation in floral homeotic function. Proc. Natl. Acad. Sci. USA 95: 8130–8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., B. E. Stranger, T. Mitchell-Olds and M. Aguadé, 2004. Multilocus analysis of variation and speciation in the closely related species Arabidopsis halleri and A. lyrata. Genetics 166: 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C. K., A. Kas, M. Paddock, R. Qiu, Y. Zhou et al., 2005. Ancient haplotypes of the HLA class II region. Genome Res. 15: 1250–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman, A. D., M. K. Uyenoyama and J. R. Kohn, 1996. Allelic diversity and gene genealogy at the self-incompatibility locus in the Solanaceae. Science 273: 1212–1216. [DOI] [PubMed] [Google Scholar]
- Sato, T., T. Nishio, R. Kimura, M. Kusaba, G. Suzuki et al., 2002. Coevolution of the S-locus genes SRK, SLG and SP11/SCR in Brassica oleracea and B. rapa. Genetics 162: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., R. Fujimoto, K. Toriyama and T. Nishio, 2003. Commonality of self-recognition specificity of S haplotypes between Brassica oleracea and Brassica rapa. Plant Mol. Biol. 52: 617–626. [DOI] [PubMed] [Google Scholar]
- Savolainen, O., C. H. Langley, B. P. Lazzaro and H. Freville, 2000. Contrasting patterns of nucleotide polymorphism at the alcohol dehydrogenase locus in the outcrossing Arabidopsis lyrata and the selfing Arabidopsis thaliana. Mol. Biol. Evol. 17: 645–655. [DOI] [PubMed] [Google Scholar]
- Schierup, M. H., B. K. Mable, P. Awadalla and D. Charlesworth, 2001. Identification and characterization of a polymorphic receptor kinase gene linked to the self-incompatibility locus of Arabidopsis lyrata. Genetics 158: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, K. J., S. Ramos-Onsins, H. Ringys-Beckstein, D. Weisshaar and T. Mitchell-Olds, 2005. A multilocus sequence survey in Arabidopsis thaliana reveals a genome-wide departure from a neutral model of DNA sequence polymorphism. Genetics 169: 1601–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K. K., J. M. Cork, A. L. Caicedo, C. A. Mays, R. C. Moore et al., 2004. Darwinian selection on a selfing locus. Science 306: 2081–2084. [DOI] [PubMed] [Google Scholar]
- Takahata, N., 1990. A simple genealogical structure of strongly balanced allelic lines and trans-species polymorphism. Proc. Natl. Acad. Sci. USA 87: 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., and Y. Satta, 1998. Footprints of intragenic recombination at HLA loci. Immunogenetics 47: 430–441. [DOI] [PubMed] [Google Scholar]
- Takebayashi, N., P. B. Brewer, E. Newbigin and M. K. Uyenoyama, 2003. Patterns of variation within self-incompatibility loci. Mol. Biol. Evol. 20: 1778–1794. [DOI] [PubMed] [Google Scholar]
- Takebayashi, N., E. Newbigin and M. K. Uyenoyama, 2004. Maximum-likelihood estimation of rates of recombination within mating-type regions. Genetics 167: 2097–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel, T., W. Michalek, R. Varshney and A. Graner, 2003. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106: 411–422. [DOI] [PubMed] [Google Scholar]
- Vekemans, X., and M. Slatkin, 1994. Gene and allelic genealogies at a gametophytic self-incompatibility locus. Genetics 137: 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., K. Zhao, H. Innan and M. Nordborg, 2004. The probability and chromosomal extent of trans-specific polymorphism. Genetics 168: 2363–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2003. Subdivision and haplotype structure in natural populations of Arabidopsis lyrata. Mol. Ecol. 12: 1247–1263. [DOI] [PubMed] [Google Scholar]
- Wu, J., S. J. Saupe and N. L. Glass, 1998. Evidence for balancing selection operating at the het-c heterokaryon incompatibility locus in filamentous fungi. Proc. Natl. Acad. Sci. USA 95: 12398–12402. [DOI] [PMC free article] [PubMed] [Google Scholar]