Abstract
We report on the construction of a linkage map for brown trout (Salmo trutta) and its comparison with those of other tetraploid-derivative fish in the family Salmonidae, including Atlantic salmon (Salmo salar), rainbow trout (Oncorhynchus mykiss), and Arctic char (Salvelinus alpinus). Overall, we identified 37 linkage groups (2n = 80) from the analysis of 288 microsatellite polymorphisms, 13 allozyme markers, and phenotypic sex in four backcross families. Additionally, we used gene–centromere analysis to approximate the position of the centromere for 20 linkage groups and thus relate linkage arrangements to the physical morphology of chromosomes. Sex-specific maps derived from multiple parents were estimated to cover 346.4 and 912.5 cM of the male and female genomes, respectively. As previously observed in other salmonids, recombination rates showed large sex differences (average female-to-male ratio was 6.4), with male crossovers generally localized toward the distal end of linkage groups. Putative homeologous regions inherited from the salmonid tetraploid ancestor were identified for 10 pairs of linkage groups, including five chromosomes showing evidence of residual tetrasomy (pseudolinkage). Map alignments with orthologous regions in Atlantic salmon, rainbow trout, and Arctic char also revealed extensive conservation of syntenic blocks across species, which was generally consistent with chromosome divergence through Robertsonian translocations.
TELEOSTS of the family Salmonidae (e.g., trout and salmon) have long been suspected to be derived from polyploid evolution (Svärdson 1945). The current view, which is supported by genome size estimates (Ohno et al. 1968), chromosome counts (Phillips and Ràb 2001), and patterns of gene duplication (Allendorf et al. 1975), is that salmonid polyploidy is ancestral to the formation of extant taxa and presumably occurred as a result of whole-genome duplication in the early evolution of the family (Allendorf and Thorgaard 1984). While the age of the duplication event is still a matter of uncertainty (25–100 million years ago), residual signs of tetrasomy in modern salmonids (e.g., some chromosomes can form multivalents instead of bivalents at meiosis by pairing with more than one homolog) indicate that the contributing genomes probably shared extensive homology at the time of tetraploidization (Allendorf and Thorgaard 1984). Intraspecific genome duplication (autotetraploidy) is therefore the preferred hypothesis for the origin of polyploidy in this family (Allendorf and Thorgaard 1984), although tetraploidization following hybridization of closely related species with partially differentiated chromosomes (segmental allotetraploidy) cannot be excluded (Wright et al. 1983; Johnson et al. 1987).
Since the duplication event, the genome of salmonid fish has been reverting toward a diploid state (diploidization) through the differentiation of duplicated chromosome sets into distinct pairs of homeologs (Ohno et al. 1968). Independent segregation of homeologs (disomic inheritance) has been restored across most of the genome, with the exception of some chromosomes that still form multivalents and exchange chromatid segments with their ancestral counterpart during meiosis (reviewed in Allendorf and Thorgaard 1984). However, recombination between homeologous segments appears to be restricted to the subtelomeric or telomeric end of chromosomes as marker regions showing evidence of nondisomic segregations coincide with the distal end of linkage groups (Johnson et al. 1987; Sakamoto et al. 2000). This may be explained by a model of “secondary tetrasomy” in which homologous chromosomes first pair and recombine in regions proximal to the centromere followed by homeologous pairing and recombination toward the distal end of the chromosome (Allendorf and Thorgaard 1984; Allendorf and Danzmann 1997). Meiotic recombination between homeologs is therefore believed to retard the diploidization of distally located genes, while segments more proximal to the centromere are allowed to diverge at a faster rate (Allendorf et al. 1986).
Another consequence of the residual persistence of tetrasomy in salmonid species is an unusual gene segregation pattern (pseudolinkage) whereby recombinant progeny types are produced in excess of parental ones during meiosis, thus causing physically unlinked loci to appear in linkage disequilibrium when the phase of alleles is unknown (Morisson 1970; Davisson et al. 1973; Wright et al. 1983). Unlike recombination between homeologous DNA segments, pseudolinkage is not restricted to the distal end of chromosome arms and can also affect regions closer to the centromere (Wright et al. 1983; Johnson et al. 1987). Thus far, pseudolinkage, and in fact all forms of residual tetrasomy, has been observed only in male individuals and there is little evidence of similar patterns in females (although see Danzmann et al. 2005). The origin of these sex differences remains unclear, although it has been suggested that multivalent formation may be constrained during female meiosis due to greater specificity in the initiation of chromosome pairing relative to males (Allendorf and Thorgaard 1984).
The restoration of disomic inheritance across most of the salmonid genome presumably involved specific chromosome rearrangements that helped to differentiate homeologous regions and to promote the formation of meiotic bivalents (Allendorf and Thorgaard 1984). Although this aspect of the diploidization process is poorly understood, chromosome fusions appear to have played a predominant role in reshaping the salmonid karyotype (Hartley 1987). While extensive variation in chromosome number exists in the family (2n = 52–102), most salmonid species share a fairly similar number of chromosome arms (NF ∼100), which is approximately twice the number generally observed in other teleosts (Phillips and Ràb 2001). Extant salmonids are therefore believed to have evolved from an ancestral karyotype similar to that of most teleost fishes (2n = 48, NF = 48) and subsequently diverged through a series of centromere-to-centromere fusions (Robertsonian translocations) that maintained the overall number of chromosome arms (Allendorf and Thorgaard 1984).
Robertsonian translocations are also considered to be the primary mode of chromosome rearrangement both within and among salmonid genera (Phillips and Ràb 2001). According to Hartley (1987), two main categories may be distinguished among salmonid karyotypes, which essentially differ by the relative number of one-armed (acrocentric) and two-armed (metacentric) chromosomes, owing to differential rates of Robertsonian translocations. Type A salmonids, including brown trout (Salmo trutta) and Arctic char (Salvelinus alpinus), have diploid numbers close to 80, ∼100 chromosome arms, and more acrocentric than metacentric chromosomes. In contrast, species with ∼60 chromosomes, 104 chromosome arms, and higher numbers of metacentrics relative to acrocentrics are characterized as having B karyotypes (e.g., rainbow trout, Oncorhynchus mykiss). More recently, Phillips and Ràb (2001) proposed additional subcategories to account for unusual numbers of chromosome arms in lenoks (NF = 110–116), huchens (NF = 112–116), graylings (NF = 146–170), and Atlantic salmon (Salmo salar, NF = 72–74). Since Robertsonian translocations are not accompanied with a change in the number of chromosome arms, karyotype evolution in these species must have also involved other types of rearrangement, such as centromere-to-telomere (tandem) fusions and/or inversions that include the centromeric region (pericentric inversions).
In previous efforts to study the organization of the salmonid genome, we used microsatellite markers to develop linkage maps for rainbow trout (Sakamoto et al. 2000; Danzmann et al. 2005) and Arctic char (Woram et al. 2004). This allowed us to identify multiple chromosome regions presumably derived from whole-genome duplication on the basis of the detection of duplicate marker copies within species. Furthermore, we used a set of conserved markers across species to investigate interspecific patterns of chromosome evolution, also including mapping information from Atlantic salmon (Danzmann et al. 2005). In this study, we extended our analysis to another member of the salmonid family by developing a linkage map for brown trout. Since this species has more acrocentric than metacentric chromosomes (2n = 80, NF = 100–104), it is expected that its genome has experienced fewer chromosome rearrangements than most salmonids, which in turn should facilitate the identification of ancestral chromosome blocks and the reconstruction of karyotype evolution in the family.
MATERIALS AND METHODS
Biological material:
Reference families:
Source populations used to determine linkage arrangements among genetic markers were backcross pedigrees between distinct phylogenetic groups in the brown trout complex, including individuals from the Atlantic, Mediterranean, and marmoratus lineages (see Giuffra et al. 1996 for further details on phylogenetic relationships among lineages). Two Mediterranean × Atlantic F1 hybrid males were backcrossed to different Atlantic females to produce the families from which most of the data were collected (families 12 and 15). Two additional families (families 14 and 17) were also generated from the backcross of marmoratus × Atlantic F1 males to Atlantic females. Crosses were all performed in the winter of 1992 and raised at the Institut National de la Recherche Agronomique (INRA) fish experimental facilities. Fish were sacrificed 12–18 months posthatching in accordance with French regulations for animal care and stored at −20° for tissue and enzyme preservation. Marker analysis was performed on a subset of 45 progeny for families 12 and 15, and 48 progeny for families 14 and 17.
Meiotic gynogens:
Source material for centromere mapping were meiotic gynogens obtained through retention of the second polar body following protocols similar to those described in Chourrout (1980) with the modifications outlined in Quillet et al. (1991). Briefly, sperm pooled from several males and irradiated with ultraviolet light were used to activate eggs sampled from donor females. The extrusion of the second polar body was then inhibited by heat-shock treatment, thus blocking the second meiotic division and resulting in diploid, gynogenetic individuals whose chromosomes are derived from recombinant sister chromatids. For the purpose of this study, two sets of 47 gynogens were selected from the progeny of two Atlantic females (G1 and G2).
Genetic markers:
Protein systems:
Allozyme and other protein electrophoresis was performed in horizontal starch gels using liver, eye, blood, or muscle tissue according to methods described in Krieg and Guyomard (1985). Polymorphic systems included fumarate hydratase (EC 4.2.1.2, FH-1,2), malate dehydrogenase (EC 1.1.1.37, sMDH-A2, and sMDHB1,2), isocitrate dehydrogenase (EC 1.1.1.42, sIDHP-1, and sIDHP-2), l-lactate dehydrogenase (EC 1.1.1.27, LDH-C1), phospho-gluconate dehydrogenase (EC 1.1.1.44, PGDH), glycerol-3-phosphate dehydrogenase (EC 1.1.1.8, G3PDH-1), mannose-6-phosphate isomerase (EC 5.3.1.8, MPI), aspartate transaminase (EC 2.6.1.1, sAAT-1,2), carboxylic ester hydrolase (EC 3.1.1.-, EST-1), superoxide dismutase 4 (EC 1.15.1.1, sSOD-1), and transferrin (TF).
Microsatellites:
Genomic DNA samples were prepared from muscle or liver tissue preserved in ethanol following simplified phenol extraction and ethanol precipitation procedures (Estoup et al. 1993). Polymerase chain reactions (PCRs) were labeled with either radioactivity or fluorescence (see below and supplemental Table S1 at http://www.genetics.org/supplemental/ for specific PCR conditions for each marker). For radioactive PCR, the reaction mixture (10 μl) contained 80 ng of genomic DNA template, 1× reaction buffer (Promega, Madison, WI), 400 nm nonradioactive primer, 440 or 880 nm primer end-labeled with [γ-33P]dATP, 75 μm each dNTP (Amersham, Buckinghamshire, UK), 0.8–2.5 mm MgCl2 (Promega), 20 μg/ml bovine serum albumin, and 0.25 units Taq polymerase (Promega). Fluorescent reactions were amplified in 10-μl volumes with 40 ng of genomic DNA, 1× reaction buffer (Promega), 220 nm fluorescent primer, 800 nm nonfluorescent primer, 125 μm of each dNTP (Amersham), 0.8–2.0 mm MgCl2 (Promega), and 0.35 units Taq DNA polymerase (Promega). All amplifications were processed on MJ Research (Watertown, MA) PTC-100/200 or Hybaid MBS 0.2G thermocylcers for 5 min at 96°; 5 cycles of 1 min at 96°, 30 sec at 48°–60°, 30 sec at 72°; 19–25 cycles of 30 sec at 95°, 30 sec at 48°–60°, 30 sec at 72°; 5 min at 72°; and a final soak at 10°. After direct addition of loading buffer, the PCR products were denatured for 4 min at 95° and electrophoresed on 6% polyacrylamide denaturing gels for 1.5–6 hr, depending on fragment size. Fragments resolved by electrophoresis were visualized using either a fluorescence detection platform (Hitachi FMBIO II) or X-OMAT AR autoradiography films (Kodak).
Sex phenotype:
The phenotypic sex of progeny in the families 14 and 17 was determined by internal examination of the gonads after dissection and used as a male-specific dominant marker for linkage analysis as described in Woram et al. (2003).
Microsatellite nomenclature:
New microsatellite markers reported in this study were named in accordance with the convention outlined by Jackson et al. (1998), where the first three letters refer to the species of origin and the source laboratory is indicated as a suffix (see Sakamoto et al. 2000 and Woram et al. 2003 for a complete list of acronyms). Previously published markers were all designated in agreement with the original report (see supplemental Table S1 at http://www.genetics.org/supplemental/ for alternative designations used in other publications). Duplicated loci detected by a single pair of primers (i.e., markers amplifying more than two alleles in a single individual or apparent single markers mapping to distinct linkage groups across mapping parents) were identified with /i or /ii labels within the marker name. Type I microsatellite markers designed from functional sequences were named according to the gene product, including TF, myogenic factor (MYOD), trout red cell arrestin (TRCARR), and somatolactin (SL).
Linkage analysis:
Single-parent maps:
Segregation data were processed separately for each sex by analyzing male and female segregating alleles independently. Marker genotypes from each backcross family were initially analyzed with the LINKMFEX software package, version 1.7 (http://www.uoguelph.ca/∼rdanzman/software/LINKMFEX/). Conformity to the expected 1:1 segregation ratio was evaluated for each marker with <15% of missing data using a log-likelihood G-test (Sokal and Rohlf 1995) and linkage groups were assembled at a minimum LOD score of 4.0. In a second step, genotypes were converted into a backcross format and imported into CARTHAGENE (de Givry et al. 2005), version 0.99, which computes marker orders and map distances using a multipoint likelihood approach. Since algorithms implemented in CARTHAGENE require phase-known genotypes, the original allele complement of each marker was determined prior to conversion of the data. Parental phases in Mediterranean × Atlantic F1 hybrid males (families 12 and 15) were directly inferred by genotyping the grandparent males contributing the Mediterranean alleles, with a limited number of exceptions due to ambiguous transmission. The most likely phase of markers genotyped in the six other parents was inferred from allele combinations observed among linked markers under the assumption that females show only classical linkage and that the nature of linkage (i.e., classical vs. pseudolinkage) is consistent across males. Backcross-formatted genotypes were then checked for nonrecombinant markers, which were merged into a single composite marker for further analysis. Linkage groups with <10 recombinant markers were ordered following a comprehensive search of the likelihood space using the flips command with a window size equal to the number of markers. Larger linkage groups were ordered following a stepwise approach. The first step consisted of generating a seed map using the build command. In the following steps, the map was improved by local search methods, including simulated annealing, greedy, and genetic algorithms (see CARTHAGENE documentation for details). The optimality of the output was eventually verified using the flips (window size = 7) and polish options. In addition, a subset of markers ordered with high confidence was tentatively identified for each linkage group using the framework map function with a LOD threshold of 3.0. Graphic representations of linkage groups were generated with MAPCHART, version 2.1 (Voorrips 2002), using recombination fractions as estimates of map distances to account for the high levels of crossover interference previously reported in brown trout (Guyomard 1986).
Multi-parent maps:
Multi-parent maps were assembled by merging single-parent data sets by sex using the mergen option in CARTHAGENE. Ambiguous marker orders due to a lack of anchor markers among parents were treated as follows. In cases where ambiguous orders were caused by markers from families 14 and 17, we removed the corresponding genotypes from the merged data set and performed linkage analysis with the remaining data. However, when ambiguity arose from a lack of anchors between families 12 and 15, the multi-parent map was constructed with priority given to the total number of markers. Linkage groups, marker order, and map distances for each multi-parent map were computed as described above for single-parent maps.
Gene–centromere analysis:
Prior to analysis, each marker was assessed for deviation from Mendelian expectations using a log-likelihood ratio test (G-test). Marker–centromere distances (d) were estimated under a model of complete crossover interference with d = y/2, where y is the proportion of heterozygous genotypes in the progeny. When the same marker was informative in both donor females, a G-test was used to verify homogeneity of gene–centromere distances between females. At least two recombinant markers per linkage group were analyzed to determine chromosome orientation with respect to the centromere. The putative centromere region was approximated as the 95% confidence interval of the most proximal marker following the method outlined in Danzmann and Gharbi (2001).
Recombination differences:
Recombination rates were compared among parents using the RECOMDIF module in LINKMFEX, which applies a two-way contingency G-test to the numbers of parental and recombinant genotypes inherited from each individual in the comparison. All analyses were performed with the option of comparing only adjacent marker intervals (see LINKMFEX documentation). Average recombination ratios over all adjacent marker intervals between two individuals were computed as the ratio between the proportion of recombinants across shared intervals between parents.
Map comparisons:
Map comparisons between brown trout and other salmonid species were performed using a combination of published reports and unpublished data for rainbow trout (Sakamoto et al. 2000; Nichols et al. 2003; Danzmann et al. 2005), Atlantic salmon (http://grasp.mbb.sfu.ca/), and Arctic char (Woram et al. 2004). Putative homeologies across species were inferred manually on the basis of the location of common markers between maps and visualized in a grid format using GRIDMAP, version 3.0a (http://cbr.jic.ac.uk/dicks/software/grid_map/index.html). Ambiguous homologies due to the comparison of syntenic blocks with duplicate homeologous copies were tentatively resolved on a case-by-case basis using both known homeologies within each species (Sakamoto et al. 2000; Nichols et al. 2003; O'Malley et al. 2003; Woram et al. 2004; Danzmann et al. 2005) and cross-species homologies previously identified among rainbow trout, Atlantic salmon, and Arctic char (Danzmann et al. 2005).
RESULTS
Map statistics:
Basic statistics calculated for single- and multi-parent maps are summarized in Table 1. Overall, we analyzed 302 genetic markers, most of which (288) were microsatellite repeats isolated from various salmonid species (see supplemental Table S1 at http://www.genetics.org/supplemental/ for details). In addition, we also used segregation data from 13 allozyme markers and the sex phenotype. Most map information was derived from families 12 and 15, with a smaller fraction of the markers genotyped in families 14 and 17. Also evident from Table 1, linkage maps derived from male parents contained more markers than their female counterparts due to higher levels of heterozygosity in hybrid males relative to pure-strain females (data not shown).
TABLE 1.
Summary of single- and multi-parent maps
| Source data | No. of markersa | No. of LGsb | Length (cM)c | |
|---|---|---|---|---|
| Family 12 | Male | 239 | 36 | 261.7 |
| Female | 159 | 36 | 427.2 | |
| Family 14 | Male | 113 | 28 | 112.9 |
| Female | 91 | 25 | 300.9 | |
| Family 15 | Male | 233 | 35 | 179.5 |
| Female | 182 | 42 | 604.5 | |
| Family 17 | Male | 63 | 14 | 44.6 |
| Female | 42 | 10 | 146.6 | |
| All parents | Male | 279 | 35 | 346.4 |
| Female | 242 | 43 | 912.5 | |
| Total | 302 | 37 | — |
Number of informative markers analyzed for map construction in each mapping population.
Number of linkage groups identified at a LOD threshold of 4.0.
Cumulative map distance calculated from summing across all marker intervals.
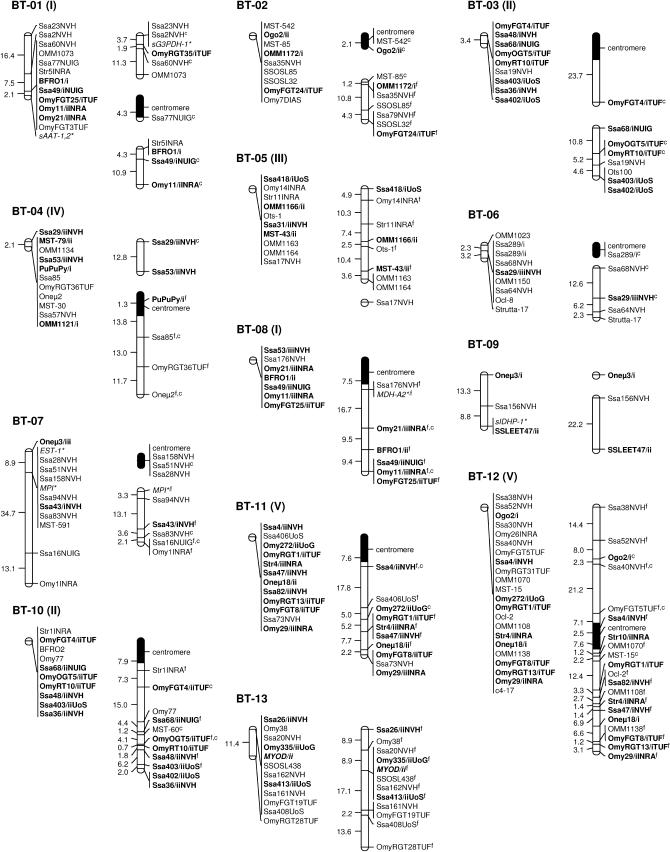
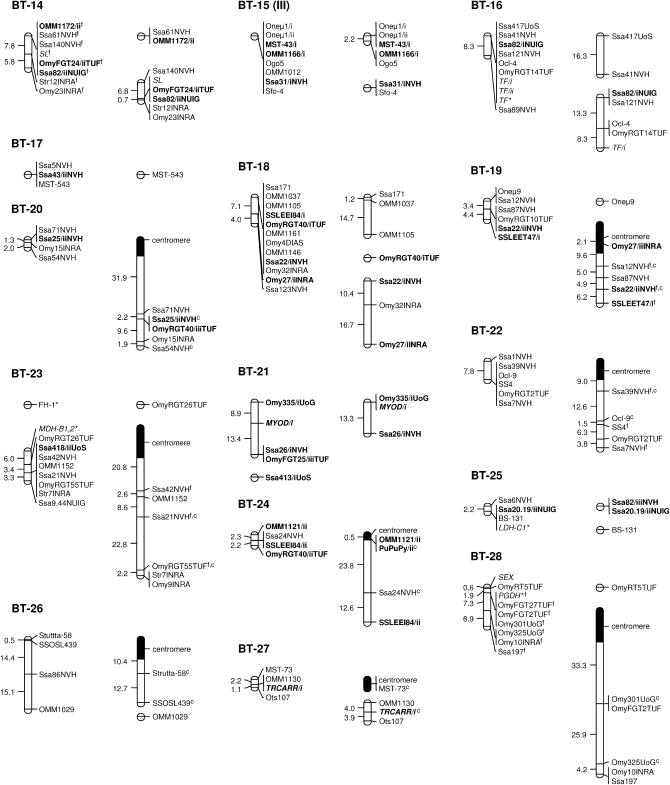
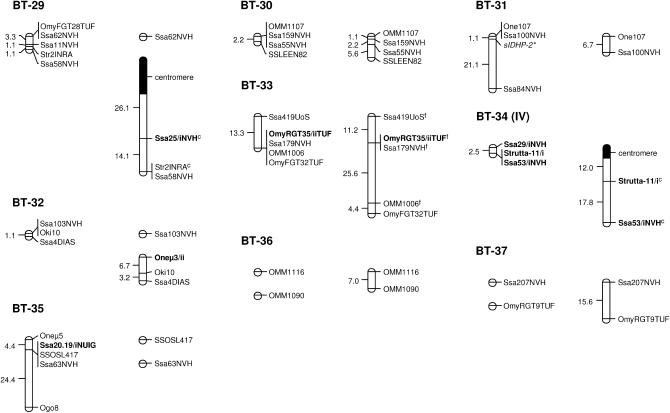
A total of 37 classical linkage groups (BT-01–BT-37) were identified across parents (LOD threshold = 4.0) with both individual and sex-specific variations (Table 1). Within each sex, there was generally good agreement between the number of informative markers and the number of linkage groups, which essentially reflected differences in genome coverage across individuals. Between sexes, however, variations in the number of linkage groups were primarily determined by differences in recombination rates. Eight linkage groups (BT-01, BT-02, BT-04, BT-07, BT-14, BT-15, BT-16, and BT-18) were thus found as multiple map segments in females, while occurring as a single block in males (Figure 1). Conversely, two small linkage groups of the female map (BT-36 and BT-37) corresponded to unlinked markers in the male parents. Cumulative map distances were also variable across parents with generally shorter maps in males (44.6–261.7 cM) and more extensive coverage in females (146.6–604.5 cM). The map derived from male parents (346.4 cM) was thus ∼2.5 times shorter than the female map (912.5 cM). Overall, a total of eight markers remained unassigned in both sexes (BFRO7, SSLEER15, Omy108INRA, Strutta-11/ii, sSOD-1*, Ssa85NVH, Ssa88/iNVH, and T3-13).
Figure 1.
Sex-specific linkage maps of the brown trout genome. Each linkage group (numbered BT-01–BT-37) is represented by a male (left) and a female (right) map derived from the joint analysis of segregation data from families 12, 14, 15, and 17. Roman letters (I–V) in parentheses with linkage group numbers designate pseudolinkage groups. Map distances are indicated in centimorgans to the left of each marker interval (markers within nonrecombinant intervals are shown along the same vertical line). Italicized marker names indicate gene markers, including type I microsatellites and allozymes (shown with an asterisk). A superscript “f” indicates framework markers ordered with high confidence (LOD threshold = 3.0). Markers with multiple copies (denoted as “/i,” “/ii,” and “/iii”) across linkage groups (excluding putative tandem duplications) are shown in boldface type. Markers used for the orientation of linkage groups with respect to the centromere are indicated with a superscript “c” and the position of centromeres is represented by 95% confidence intervals (solid segments) in the female map.
Sex linkage:
The sex-determining locus (SEX) was previously localized to linkage group BT-28, along with seven microsatellite markers (Woram et al. 2003). The current map contains one additional sex-linked marker (PGDH*) located within 1.9 cM of SEX (Figure 1).
Centromere mapping:
Gene–centromere distances were estimated for 49 microsatellite markers assigned to the female map (see supplemental Table S2 at http://www.genetics.org/supplemental/) and used to infer chromosome morphology for 20 of 37 linkage groups (Figure 1). In most cases, the putative centromere region was approximated to the distal end of the linkage group, thus suggesting acrocentric chromosomes for BT-02, BT-03, BT-06, BT-07, BT-08, BT-10, BT-11, BT-19, BT-20, BT-22–BT-24, BT-26–BT-29, and BT-34. In three cases (BT-01, BT-04, and BT-12), the centromere was internal to the linkage group and supported a metacentric architecture for each of these chromosomes.
Recombination rates:
Significant differences in recombination rates were detected both within and between sexes (Table 2). Among female parents, recombination rates were significantly higher (P < 0.05) in families 12 and 17 relative to families 14 and 15, respectively. However, these findings may not be representative of genomewide differences as only a limited number of map intervals (n < 20) were sampled in each comparison. Indeed, the female parents in families 12 and 15 showed a wide range of recombination ratios across linkage groups (data not shown), although the overall proportions of recombinants were similar in both individuals. In males, four of six comparisons were statistically significant at the 5% level with the family 15 male consistently showing lower recombination than the three other males (Table 2). In addition, recombination rates were generally higher in the family 12 male than in the family 17 male, although this difference was only slightly significant.
TABLE 2.
Pairwise comparisons of recombination rates between mapping parents
| F12 (f) | F14 (f) | F15 (f) | F17 (f) | F12 (m) | F14 (m) | F15 (m) | F17 (m) | |
|---|---|---|---|---|---|---|---|---|
| F12 (f) | — | 18 | 57 | 19 | 80 | 16 | 78 | 14 |
| F14 (f) | 0.6a | — | 19 | 9 | 27 | 35 | 26 | 9 |
| F15 (f) | 0.9 | 1.0 | — | 19 | 89 | 22 | 86 | 19 |
| F17 (f) | 0.8 | 0.9 | 1.6a | — | 21 | 8 | 27 | 22 |
| F12 (m) | 4.7a | 5.0a | 4.9a | defa | — | 47 | 160 | 24 |
| F14 (m) | 2.9a | 3.0a | 4.4a | 4.2a | 1.0 | — | 45 | 19 |
| F15 (m) | 9.2a | 6.5a | 6.4a | defa | 2.2a | 2.5a | — | 24 |
| F17 (m) | 13.2a | 4.8a | 8.7a | 16.0a | 0.2a | 0.9 | 0.3a | — |
Parent designations refer to family (F) and sex (f, female; m, male). The number of chromosome segments and the average recombination ratio for each pairwise comparison are given above and below the diagonal, respectively (def, undefined ratio). Recombination ratios were calculated with the column and row individuals in the numerator and denominator, respectively.
Significant differences between individuals (P < 0.05).
All 16 pairwise comparisons between male and female parents were highly significant (P < 0.001) and reflected large differences in recombination rates between the sexes (Table 2). In all cases, the male parent showed reduced recombination relative to the female parent. Across all pairwise comparisons (n = 579), recombination rates in females were 6.4 times greater than those in males. However, sex-specific differences were not consistent across the genome and in fact male recombination exceeded female recombination in 49 of 579 comparisons (although not always significantly). Further investigations indicated that, whenever gene–centromere distances were available (22 comparisons), marker intervals showing increased male recombination were localized in the putative telomeric regions (>40 cM from the centromere; data not shown). For example, Ssa2NVH, which was located 48.7 cM from the centromere on linkage group BT-01, showed no recombination with Ssa23NVH in the female map, while the two markers occurred 16.4 cM apart in males (Figure 1). Similarly, higher male recombination at the distal ends of linkage groups BT-07 (Ssa16NUIG-Omy1INRA) and BT-28 (Omy10INRA-Ssa197) was associated with large gene–centromere distances for Ssa16NUIG (49.0 cM) and Ssa197 (45.7 cM), respectively.
Segregation distortion:
Possible evidence for segregation distortion (P < 0.05 in single G-tests) was observed for a total of 82 markers (22 linkage groups) across parents (see supplemental Figure S1 at http://www.genetics.org/supplemental/ for detailed results for each individual). When P-values were adjusted for multiple tests using a Bonferroni-type correction to take into account the number of linkage groups identified in each individual, segregation distortion remained statistically significant for 12 markers. Seven of them were detected in the family 14 male and localized to short map regions on linkage groups BT-01 (Ssa2NVH and sAAT-1,2*) and BT-24 (PuPuPy/ii, Ssa24NVH, SSLEEI84/ii, Ssa88/iiNVH, and Str10/iiINRA). Another four (OmyOGT5/iiTUF/ii, Ssa68/iiNUIG, BFRO2, and Str1INRA) were clustered within 2.1 cM on linkage group BT-10 in the family 17 male. Finally, high segregation distortion was also detected at OMM1116 in the family 12 male, which might explain why this marker remained unlinked at a LOD threshold of 4.0.
Duplicated markers:
Overall, 56 microsatellite markers (24.8%) detected a pair of polymorphic loci in the brown trout genome. Duplicated markers were found on a total of 31 linkage groups (Table 3). Duplicate copies for Ssa289 (BT-06), Oneμ1 (BT-15), and TF (BT-16) were tightly linked on three separate linkage groups and were therefore interpreted as internal homologies due to local events of tandem duplication. Other markers, however, pointed to possible regions of ancestral homology between linkage groups. Most linkage groups shared putative homology with one (N = 18) or two (N = 10) other linkage groups in the current map. Exceptions to this general pattern were BT-12 and BT-24, which showed marker homology to three different linkage groups. Ten conserved syntenic blocks were identified between putative homeologous regions on the basis of the colocalization of multiple duplicates (Table 3). In addition, 10 more pairs of linkage groups showed homologous affinities on the basis of a single pair of duplicates, which may represent additional duplicated segments inherited from the tetraploid ancestor.
TABLE 3.
Putative homologies between linkage groups on the basis of shared marker assignments
| Linkage group | Putative homologies | ||||
|---|---|---|---|---|---|
| BT-01 | BT-08 (5) | BT-21 (1)a | BT-33 (1) | ||
| BT-02 | BT-12 (1) | BT-14 (2) | |||
| BT-03 | BT-10 (8) | ||||
| BT-04 | BT-06 (1)a | BT-08 (1)a | BT-24 (2) | BT-34 (2) | |
| BT-05 | BT-15 (3) | BT-23 (1) | |||
| BT-06 | BT-04 (1)a | BT-06 (1) | BT-34 (1)a | ||
| BT-07 | BT-09 (1)a | BT-17 (1) | BT-32 (1)a | ||
| BT-08 | BT-01 (5) | BT-04 (1)a | BT-21 (1)a | BT-34 (1)a | |
| BT-09 | BT-07 (1)a | BT-19 (1) | BT-32 (1) | ||
| BT-10 | BT-03 (8) | ||||
| BT-11 | BT-12 (10) | BT-25 (1)a | |||
| BT-12 | BT-02 (1) | BT-11 (10) | BT-24 (1) | BT-25 (1)a | |
| BT-13 | BT-21 (4) | ||||
| BT-14 | BT-02 (2) | BT-16 (1) | |||
| BT-15 | BT-05 (3) | BT-15 (1) | |||
| BT-16 | BT-14 (1) | BT-16 (1) | |||
| BT-17 | BT-07 (1) | ||||
| BT-18 | BT-19 (2) | BT-20 (1)a | BT-24 (2) | ||
| BT-19 | BT-09 (1) | BT-18 (2) | |||
| BT-20 | BT-18 (1)a | BT-24 (1)a | BT-29 (1) | ||
| BT-21 | BT-01 (1)a | BT-08 (1)a | BT-13 (4) | BT-21 (1) | |
| BT-22 | |||||
| BT-23 | BT-05 (1) | ||||
| BT-24 | BT-04 (2) | BT-12 (1) | BT-18 (2) | BT-20 (1)a | UNA |
| BT-25 | BT-11 (1)a | BT-12 (1)a | BT-35 (1) | ||
| BT-26 | |||||
| BT-27 | BT-30 (1) | ||||
| BT-28 | |||||
| BT-29 | BT-20 (1) | ||||
| BT-30 | BT-27 (1) | ||||
| BT-31 | |||||
| BT-32 | BT-07 (1)a | BT-09 (1) | |||
| BT-33 | BT-01 (1) | ||||
| BT-34 | BT-04 (2) | BT-06 (1)a | BT-08 (1)a | UNA | |
| BT-35 | BT-25 (1) | ||||
| BT-36 | |||||
| BT-37 | |||||
For each linkage group, the number in parentheses indicates the number of markers shared with the linkage group shown on the left. UNA, markers currently unassigned to a linkage group at a LOD threshold of 4.0.
Secondary homologies due to the amplification of a third marker copy in addition to the primary duplicates presumably derived from whole-genome duplication (see text).
Interestingly, six microsatellite markers (2.7%) detected a third polymorphic locus in addition to the putative homeologs (OmyFGT25TUF, OmyRGT40TUF, Oneμ3, Ssa29NVH, Ssa53NVH, and Ssa82NVH). In most cases, the additional locus showed distinctive allele sizes and amplification patterns, which allowed discrimination of primary duplicates (i.e., duplicates presumably derived from tetraploidization in the salmonid ancestor) from the supernumerary copy (thereafter denoted by “/iii”). Alternatively, primary duplicates were distinguished from the supernumerary copy on the basis of putative homeologies supported by other markers (Table 3).
Pseudolinkage:
Direct evidence for pseudolinkage as indicated by a significant excess of recombinant genotypes was obtained from male recombination data in families 12 and 15 (Table 4). Linkage groups BT-04/BT-34, BT-05/BT-15, and BT-11/12 showed pseudolinkage affinities (LOD threshold = 4.0) in both males, while BT-03 and BT-10 were pseudolinked in the family 15 male only. Similar results were obtained in the family 14 and 17 males, although in this case pseudolinkage associations were not directly inferred from phase-known genotypes but tentatively deduced from marker assignments in the males of families 12 and 15 (see materials and methods). Furthermore, we also found suggestive pseudolinkage (3.0 < LOD < 4.0) between BT-01 and BT-08 in the family 17 male. Although relatively weak, we believe this association to be real because BT-01 contains two allozyme markers (sG3PDH-1* and AAT-1,2*) previously assigned to pseudolinkage group I in the composite map compiled by May and Johnson (1990).
TABLE 4.
Pseudolinkage associations detected in the four males used for map construction
| Male parent
|
||||
|---|---|---|---|---|
| Linkage groups | Family 12 | Family 14 | Family 15 | Family 17 |
| BT-01/BT-08 | NS | NS | NS | b |
| BT-03/BT-10 | NS | a | a | a |
| BT-04/BT-34 | a | a | a | — |
| BT-05/BT-15 | a | NS | a | — |
| BT-11/BT-12 | a | a | a | a |
NS, nonsignificant test. —, tests could not be performed due to lack of marker information.
A significant excess of recombinant genotypes between markers assigned to linkage groups in the first column, LOD > 4.0.
A significant excess of recombinant genotypes between markers assigned to linkage groups in the first column, LOD > 3.0.
Cross-species homologies:
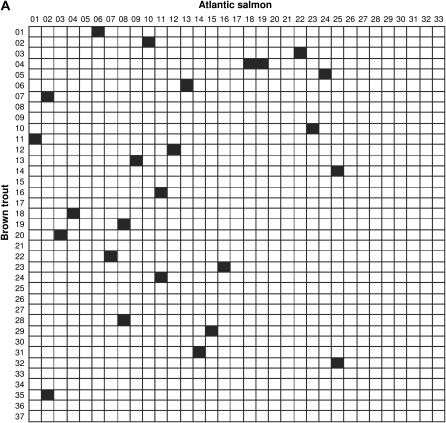
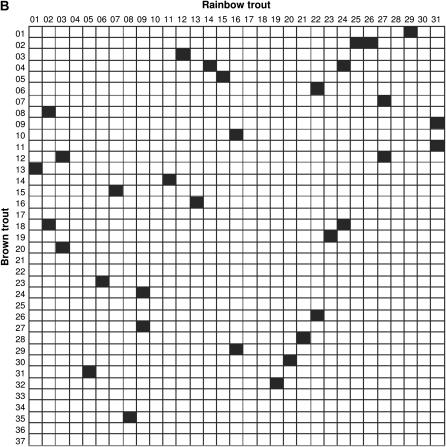
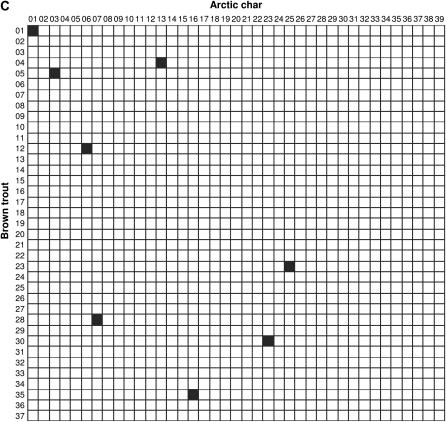
Comparisons of linkage arrangements with other salmonid species were performed on the basis of variable numbers of anchor markers identified in the current maps for Atlantic salmon (N = 120), rainbow trout (N = 140), and Arctic char (N = 42). Overall, the compilation of map information across species (see supplemental Table S3 at http://www.genetics.org/supplemental/) pointed to 38, 47, and 30 possible homologies between linkage groups in brown trout and each comparison species, respectively. However, since a large number of putative homologies were supported by a single marker, we restricted our analysis to the fraction of linkage groups sharing at least two common markers (syntenic blocks) to minimize spurious results due to misallocation of homology. In Atlantic salmon, a total of 25 syntenic blocks were identified with cross-homology in brown trout (Figure 2A). In all cases but one, linkage groups in brown trout shared homologous segments with a single linkage group in the Atlantic salmon map. The one exception was linkage group BT-04, which showed syntenic homology to both AS-18 and AS-19. Conversely, linkage groups in Atlantic salmon (N = 21) shared a syntenic block with either one (N = 17) or two (N = 4) linkage groups in brown trout. Syntenic blocks detected in rainbow trout (N = 33) revealed a generally similar pattern, with the identification of single-chromosome homologies for 25 of 29 linkage groups examined in brown trout (Figure 2B). Linkage groups with multiple homologies were BT-02, BT-04, BT-12, and BT-18, which contained homologous segments from RT-25/RT-26, RT-14/RT-24, RT-03/RT-27, and RT-02/RT-24, respectively. In turn, eight rainbow trout linkage groups (RT-02, RT-03, RT-09, RT-16, RT-22, RT-24, RT-27, and RT-31) were homologous to more than one linkage group in brown trout. In Arctic char, the limited number of anchor markers in the current map restricted the comparison to only eight syntenic blocks, which all involved single linkage groups (Figure 2C).
Figure 2.
Conservation of syntenic blocks in brown trout and other salmonid species. Each cell represents a pairwise comparison of linkage groups between brown trout (rows) and one of three salmonid species (columns), namely Atlantic salmon (A), rainbow trout (B), and Arctic char (C). Linkage groups are numbered according to the nomenclature currently in use for each species (see materials and methods). Solid cells indicate comparisons with at least two common markers between linkage groups (syntenic homologies).
DISCUSSION
Genome coverage:
The linkage map that we assembled contains a total of 37 linkage groups. According to most reports, brown trout has 80 chromosomes in its complement (Phillips and Ràb 2001), which suggests that we have identified three linkage groups fewer than expected. However, we cannot exclude the possibility that some chromosomes are represented by more than one linkage group. In particular, BT-36 and BT-37 involve a single pair of markers (OMM1116/OMM1090 and Ssa207NVH/OmyRGT9TUF, respectively), which may coincide with the distal end of larger linkage groups. Indeed, since male recombination appears to be higher in telomeric regions relative to female recombination, the lack of linkage for the two pairs of markers in the male map may be indicative of terminal marker intervals within the female map. Because female linkage groups are often fragmented into multiple map segments, it is therefore possible that BT-36 and BT-37 represent the telomeric regions of chromosomes partially covered by other linkage groups. Notwithstanding, we believe that the vast majority of current linkage groups are representative of distinct chromosomes due to the general lack of recombination in male parents and the complementary nature of recombination dynamics between sexes (i.e., regions of high recombination in males tend to show lower recombination rates in females and vice versa). In addition, we are also confident that the identification of linkage groups in males was not confounded by pseudolinkage, as previously suggested for similar studies in other salmonid species where phase information was not available (Sakamoto et al. 2000; Woram et al. 2004).
On the basis of the observation that salmonid species exhibit complete or near-complete crossover interference during female meiosis, Young et al. (1998) estimated that the genetic length of the female genome in rainbow trout should approximate a total of 2600 cM (i.e., 50 cM × 52 chromosome arms, assuming that recombination occurs only once per chromosome arm). Since brown trout has 100–104 chromosome arms (Phillips and Ràb 2001), the predicted total length for the female genome in this species is therefore 2500–2600 cM, of which 35–37% is currently covered by the linkage map (912.5 cM). Since interference levels during male meiosis are currently unknown, a tentative estimate of the length of the male genome may be obtained from a recent map update for rainbow trout (Nichols et al. 2003), which represents the most extensive coverage of a salmonid genome to date (4590 cM). If a similar size is assumed in brown trout, the current map (346.4 cM) might therefore cover <10% of the male genome.
Sex differences in recombination rates:
Quantitative differences in the amount of meiotic recombination between males and females are known to occur in a wide range of organisms and tend to follow a general pattern whereby recombination is reduced in the heterogametic sex (Sakamoto et al. 2000). This is consistent with our observation of higher recombination rates in female brown trout (overall ratio is 6.4) since sex is determined by male heterogamety in this species (Quillet et al. 1991). In fact, male heterogamety is the general mechanism of sex determination in salmonids (reviewed in Woram et al. 2003) and all species studied thus far show a marked reduction of recombination in male gametes (Johnson et al. 1987; Sakamoto et al. 2000; Gilbey et al. 2004; Moen et al. 2004; Woram et al. 2004; Danzmann et al. 2005). Interestingly, however, female recombination does not always exceed male recombination and sex-specific ratios have been showed to reverse toward telomeric regions in both rainbow trout (Sakamoto et al. 2000) and brown trout (this study). These findings are reminiscent of recombination patterns along human chromosomes, where pericentromeric regions generally show higher female-to-male genetic-distance ratios, while telomeric ends exhibit equal or greater male recombination (Broman et al. 1998; Lynn et al. 2000; Kong et al. 2002). In salmonids, however, this pattern is much more pronounced than in any other vertebrate studies so far, which is believed to reflect structural constraints imposed by the formation of multivalents during male meiosis (Johnson et al. 1987; Sakamoto et al. 2000).
The fluctuations of sex-specific recombination ratios along the chromosome in brown trout and other salmonid species also raise the question of whether current estimates reflect actual differences in the total number of crossovers or mere variations in chiasma localization. As discussed above, female recombination is thought to occur at an average frequency of a single crossover per chromosome arm. In males, it is reasonable to assume that at least one crossover per chromosome pair is required to ensure proper segregation (Hawley et al. 1994). Under these assumptions, sex-specific recombination rates are expected to depend on both the number of chromosome arms and the number of chromosomes. In brown trout, the genomewide female-to-male recombination ratio should therefore be on the order of 1.2:1 (50–52 chromosome arms/40 chromosomes), or less if homologous pairs experience more than one crossover in males. Slightly higher ratios (1.6–1.8:1) should be observed in the rainbow trout but still not as high as current estimates. One possible explanation for this apparent contradiction may be differential marker coverage of the male and female genomes. If male recombination is indeed confined to the telomeric regions of chromosomes, it is possible that incomplete linkage maps may reflect a higher proportion of female crossovers than male crossovers. This, in turn, would cause an underestimation of crossover frequency in males relative to females and therefore artificially inflated recombination ratios.
Evolution of homeologous chromosomes:
Nearly 25% of the microsatellite markers used for map construction coamplified two polymorphic loci. However, this is likely an underestimate of the duplication rate as potential duplicates with only one polymorphic locus were scored as single markers due to the difficulty of distinguishing monomorphic loci from PCR artifacts. The cross-amplification of duplicate loci with a single primer set from salmonid DNA is generally attributed to residual homology between ancestrally duplicated chromosome regions (e.g., Sakamoto et al. 2000). In fact, other scenarios, such as tandem duplication, transposition, and segmental duplication, may account for the occurrence of multiple-copy sequences. For example, duplicate copies for Ssa289, One1μ, and TF more likely originated from local events of tandem duplication rather than polyploidy since no recombination was observed among them. The origin of triplicate markers is more equivocal in that no clear pattern was evident in their distribution and we are aware of only a few precedents in other salmonid species (e.g., OMM1197 in rainbow trout; Danzmann et al. 2005). Perhaps most intriguing is the lack of linkage between Ssa29/iiiNVH and Ssa53/iiiNVH, whereas primary duplicates for these markers mapped to the same pair of linkage groups (BT-04/BT-34). This suggests that Ssa29/iiiNVH and Ssa53/iiiNVH arose either from independent duplication events or a single block duplication followed by chromosome rearrangement. Alternatively, each locus may occur on different copies of a DNA region that has been duplicated multiple times in the course of salmonid evolution.
In addition to the above cases, we found 53 pairs of duplicate microsatellites that presumably reflect homeologous relationships between linkage groups. Overall, homeology patterns thus established were generally consistent with the conservation of whole homeologous arms as multiple duplicated markers were observed to map to the same pair of chromosome arms (see, for example, BT-03/BT-10 and BT-11/BT-12). However, we also uncovered linkage arrangements that may provide evidence for interarm exchanges. For example, BT-12 appears to represent a metacentric chromosome containing homeologous markers from at least three linkage groups instead of the expected two following Robertsonian translocation (one per chromosome arm). Upon closer inspection, this finding is most likely explained by the homeologous affinity conferred by Str10INRA, which resides in the proximal region of the chromosome arm sharing extensive homeology with BT-11 (Figure 1). The second copy of Str10INRA was assigned to linkage group BT-24 (although not shown in Figure 1 due to ambiguous marker order), which otherwise incorporates homeologous regions from BT-04 and BT-18. Taken together, these results indicate that homeologous copies of a relatively small chromosome segment surrounding Str10INRA may have been translocated to both BT-12 and BT-24 from other linkage groups in the genome. Suggestive evidence for interarm translocations may also be obtained from the acrocentric chromosome assigned to BT-19, which appears to be a mosaic of at least two ancestral arms. Although further confirmation is needed, current marker arrangements indeed suggest that the proximal region of BT-19 (Omy27INRA-Ssa22NVH) is homeologous to linkage group BT-18, while its distal end (SSLEET47) may share a short homeologous segment with BT-09.
Residual tetrasomy:
According to May and Johnson (1990), at least six groups of pseudolinkage have been documented in salmonids, although it is unclear whether all six combinations occur in every species. Therefore, we can only suggest that the current map for brown trout may have one less pseudolinkage group than expected. However, with the exception of pseudolinkage group I (see results), further investigation of our results in relation to the compilation of May and Johnson (1990) was unfortunately not possible due to a lack of common markers between the two studies. More recently, Sakamoto et al. (2000) interpreted the apparent linkage of OmyRT10/iTUF to OmyRT10/iiTUF in the rainbow trout as evidence for pseudolinkage between RT-12 and RT-16 (formerly known as linkage groups Fi and Fii, respectively). This is consistent with the assignment of OmyRT10TUF duplicates to the pseudolinkage group consisting of BT-03 and BT-10 in brown trout, along with other syntenic markers from RT-12 and RT-16 (Figure 2). Similarly, the fact that Gilbey et al. (2004) detected pseudolinkage between a microsatellite marker (SSLEEN17) and SEX in Atlantic salmon may correlate with evidence of cross-homology between the sex linkage group in this species (AS-01) and BT-11 in brown trout (Figure 2). Since linkage group AS-01 shares extensive homeology with AS-12 (Danzmann et al. 2005), which itself contains a syntenic block from BT-12 (Figure 2), it is indeed tempting to speculate that the pseudolinkage association identified by Gilbey et al. (2004) represents the Atlantic salmon counterpart of the affinity between BT-11 and BT-12.
Wright et al. (1983) proposed a general model for pseudolinkage in salmonid species, where the production of excess recombinants is determined by preferential pairing and alternate disjunction of homeologous chromosomes in males of hybrid origin. Specifically, the model postulates that acrocentric–metacentric pairs of chromosomes sharing homeologous arms form rod tetravalents in which homeologs derived from the same parent show greater pairing affinity and go to opposite poles, thus causing recombinant progeny types to be transmitted in excess of parentals. In an extension of this model, Allendorf and Thorgaard (1984) modified previous assumptions to account for other categories of multivalents with no restriction on chromosome architecture. In brown trout, rod tetravalents are reportedly the only type of homeologous pairing observed at meiosis (Wright et al. 1983) and pseudolinkage arrangements identified in this species are remarkably consistent with Wright et al.'s (1983) predictions. The most illustrative example is that of BT-11 and BT-12, which clearly represents a pair of acrocentric and metacentric chromosomes sharing extensive arm homeology (Figure 1). Pseudolinkage affinities exhibited by BT-01/BT-08 and BT-04/BT-34 appear to follow the same pattern, although putative metacentrics (BT-01 and BT-04) are still broken up into multiple fragments. A similar conclusion may also be speculated for BT-05/BT-15 until the morphology of these chromosomes is resolved. The case of BT-03/BT-10, however, may not conform to Wright et al.'s (1983) model because both linkage groups are currently classified as acrocentrics.
Chromosome evolution across species:
Cross-species comparisons showed that syntenic regions in brown trout are generally conserved in Atlantic salmon, rainbow trout, and to a lesser extent, Arctic char. If Robertsonian translocations have been the predominant mode of chromosome rearrangement associated with species divergence (see Introduction), then one would expect that most chromosomes contain syntenic blocks from either one or two homologs, depending on chromosome architecture (i.e., acrocentric and metacentric, respectively). In addition, since chromosomes in brown trout are prevalently acrocentric in structure, it is expected that cross-homologies with this species primarily involve single chromosome arms. Indeed, we found only four linkage groups (BT-02, BT-04, BT-12, and BT-18) with multiple homologous segments across species. As previously discussed, BT-04 and BT-12 have been assigned to metacentric chromosomes and we also found preliminary evidence that BT-18 has two chromosome arms (our unpublished data). The case of BT-02, however, is unusual in that two distinct genomic regions in rainbow trout map to the same chromosome arm in brown trout. Upon closer examination of comparative maps, we determined that most of BT-02 from the centromere region (MST-542) to Ssa35NVH is homologous to RT-25, while the distal end incorporates three markers currently assigned to RT-26 in rainbow trout (Ssa79NVH, SSOSL32, and OmyFGT24/iiTUF). If, as we believe, RT-25 and RT-26 do represent distinct chromosomes, at least two scenarios may account for this pattern. In the first scenario, Ssa79NVH, SSOSL32, and OmyFGT24/iiTUF may have been relocated to the telomeric end of BT-02 following the translocation of a short chromosome segment from the as-yet-unidentified homolog of RT-26 in brown trout. The second scenario comes from the observation that brown trout has two larger chromosome arms in its karyotype that presumably evolved through tandem fusions or pericentric inversions after divergence from the rainbow trout lineage (Hartley 1987). Therefore, it is possible that cross-homology patterns detected with BT-02 reflect one of these compound chromosome arms, although we recognize that additional markers are needed to confirm this hypothesis.
Since rainbow trout has more metacentric than acrocentric chromosomes, we expected that most linkage groups in this species would share syntenic homologies with two distinct chromosomes in brown trout. In practice, we found only a minority of such linkage groups (8/24 comparisons), which is most likely explained by an insufficient number of common markers and a conservative assessment of cross-species homologies. Nevertheless, the fact that these linkage groups are currently recognized as metacentric chromosomes in rainbow trout (Danzmann et al. 2005) is consistent with the occurrence of Robertsonian translocations following species divergence. Similarly, four linkage groups in Atlantic salmon contained homologous segments from more than one chromosome region in brown trout (AS-02, AS-08, AS-11, and AS-25) and may therefore provide additional examples of Robertsonian fusions. However, since centromeres have yet to be localized in this species, it cannot be excluded that one or more of these linkage groups are in fact representative of the larger chromosome arms known to occur in this species (Hartley 1987).
Despite the possibility of Robertsonian translocations and other forms of chromosome rearrangement, we also found evidence that some arm combinations may be conserved across species. For example, the fact that markers lying across the centromere of BT-01 are syntenic on AS-06 in Atlantic salmon, RT-29 in rainbow trout, and AC-01 in Arctic char (see details in supplemental Table S3 at http://www.genetics.org/supplemental/) indicates that all four species may have inherited the same metacentric chromosome from a common ancestor before the divergence of the genera Oncorhynchus, Salmo, and Salvelinus. Another case of linkage conservation across the centromere involves BT-12 in brown trout and AS-12 in Atlantic salmon, which apparently represent the same pair of chromosome arms (see supplemental Table S3 at http://www.genetics.org/supplemental/). Interestingly, we also found that microsatellite markers close to the centromere in brown trout have small gene–centromere distances in Atlantic salmon (our unpublished results), thus indicating that we most likely identified a conserved metacentric chromosome. However, the fact that chromosome arms combined into BT-12 have been assigned to distinct linkage groups in rainbow trout (RT-03 and RT-27, respectively) suggests either a Robertsonian translocation specific to the Salmo species or secondary rearrangement in the Oncorhynchus lineage. Since only one of the two chromosome arms is currently identified in Arctic char (AC-06), additional anchor markers will be needed to distinguish between these two scenarios.
Acknowledgments
We thank Martine Andriamanga, Sandrine Anne, and Vanessa Rouaud for technical assistance; Stéphane Mauger for testing fluorescent primers in brown trout; Thomas Schiex, Patrick Chabrier, and Martin Bouchez for help with CARTHAGENE; Matthew Couchman for assistance with GRIDMAP; as well as two anonymous reviewers and Barbara Mable for useful comments on previous versions of the manuscript. This research was funded by the European Union Fisheries, Agriculture, and Agro-Industrial Research program (CT96-1591: SALMAP) and the Institut National de la Recherche Agronomique.
References
- Allendorf, F. W., and R. G. Danzmann, 1997. Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 145: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allendorf, F. W., and G. H. Thorgaard, 1984. Tetraploidy and the evolution of salmonid fishes, pp. 1–46 in Evolutionary Genetics of Fishes, edited by B. J. Turner. Plenum Press, New York.
- Allendorf, F. W., F. M. Utter and B. P. May, 1975. Gene duplication within the family Salmonidae: detection and determination of the genetic control of duplicate loci through inheritance studies and examination of populations, pp. 415–431 in Isozymes, Vol. IV: Genetics and Evolution, edited by C. L. Markert. Academic Press, New York.
- Allendorf, F. W., J. E. Seeb, K. L. Knudsen, G. H. Thorgaard and R. F. Leary, 1986. Gene-centromere mapping of 25 loci in rainbow trout. J. Hered. 77: 307–312. [Google Scholar]
- Broman, K. W., J. C. Murray, V. C. Sheffield, R. L. White and J. L. Weber, 1998. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 63: 861–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourrout, D., 1980. Thermal induction of diploid gynogenesis and triploidy in the eggs of the rainbow trout (Salmo gairdneri Richardson). Reprod. Nutr. Dev. 20: 727–733. [DOI] [PubMed] [Google Scholar]
- Danzmann, R. G., and K. Gharbi, 2001. Gene mapping in fishes: a means to an end. Genetica 111: 3–23. [DOI] [PubMed] [Google Scholar]
- Danzmann, R. G., M. Cairney, M. M. Ferguson, K. Gharbi, R. Guyomard et al., 2005. A comparative analysis of the rainbow trout genome with two other species of fish (Arctic char and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48: 1037–1051. [DOI] [PubMed] [Google Scholar]
- Davisson, M. T., J. E. Wright, Jr. and L. M. Atherton, 1973. Cytogenetic analysis of pseudolinkage of LDH loci in the teleost genus Salvelinus. Genetics 73: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Givry, S., M. Bouchez, P. Chabrier, D. Milan and T. Schiex, 2005. CARTHAGENE: multi-population integrated genetic and radiation hybrid mapping. Bioinformatics 21: 1703–1704. [DOI] [PubMed] [Google Scholar]
- Estoup, A., P. Presa, F. Krieg, D. Vaiman and R. Guyomard, 1993. (CT)n and (GT)n microsatellites: a new class of genetic markers for Salmo trutta L. (brown trout). Heredity 71: 488–496. [DOI] [PubMed] [Google Scholar]
- Gilbey, J., E. Verspoor, A. McLay and D. Houlihan, 2004. A microsatellite linkage map for Atlantic salmon (Salmo salar). Anim. Genet. 35: 98–105. [DOI] [PubMed] [Google Scholar]
- Giuffra, E., R. Guyomard and G. Forneris, 1996. Phylogenetic relationships and introgression patterns between incipient parapatric species of Italian brown trout, Salmo trutta L. Mol. Ecol. 5: 207–220. [Google Scholar]
- Guyomard, R., 1986. Gene segregation in gynogenetic brown trout (Salmo trutta L.): systematically high frequencies of post-reduction. Genet. Sel. Evol. 18: 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley, S. E., 1987. The chromosomes of salmonid fishes. Biol. Rev. 62: 197–214. [Google Scholar]
- Hawley, R. S., J. A. Frazier and R. Rasooly, 1994. Separation anxiety: the etiology of nondisjonction in flies and people. Hum. Mol. Genet. 3: 1521–1528. [DOI] [PubMed] [Google Scholar]
- Jackson, T. R., M. M. Ferguson, R. G. Danzmann, A. G. Fishback, P. E. Ihssen et al., 1998. Identification of two QTL influencing upper temperature tolerance in three rainbow trout (Oncorhynchus mykiss) half-sib families. Heredity 80: 143–151. [Google Scholar]
- Johnson, K. R., J. E. Wright, Jr. and B. May, 1987. Linkage relationships reflecting ancestral tetraploidy in salmonid fish. Genetics 116: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31: 241–247. [DOI] [PubMed] [Google Scholar]
- Krieg, F., and R. Guyomard, 1985. Population genetics of French brown trout (Salmo trutta L.): large geographical differentiation of wild populations and high similarity of domesticated stocks. Genet. Sel. Evol. 17: 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, A., C. Kashuk, M. B. Petersen, J. A. Bailey, D. R. Cox et al., 2000. Patterns of meiotic recombination on the long arm of human chromosome 21. Genome Res. 10: 1319–1332. [DOI] [PubMed] [Google Scholar]
- May, B., and K. R. Johnson, 1990. Synthetic linkage map of salmonid fishes, pp. 4.151–4.159 in Genetic Maps of Complex Genomes, Vol. 5, edited by S. J. O'Brien. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Moen, T., B. Høyheim, H. Munck and L. Gomez-Raya, 2004. A linkage map of Atlantic salmon (Salmo salar) reveals an uncommonly large difference in recombination rate between the sexes. Anim. Genet. 35: 81–92. [DOI] [PubMed] [Google Scholar]
- Morisson, W. J., 1970. Nonrandom segregation of two lactate dehydrogenase subunit loci in trout. Trans. Am. Fish. Soc. 1: 193–206. [Google Scholar]
- Nichols, K. M., W. P. Young, R. G. Danzmann, B. D. Robison, C. Rexroad et al., 2003. A consolidated linkage map for rainbow trout (Oncorhynchus mykiss). Anim. Genet. 34: 102–115. [DOI] [PubMed] [Google Scholar]
- Ohno, S., U. Wolf and N. B. Atkin, 1968. Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. [DOI] [PubMed] [Google Scholar]
- O'Malley, K. G., T. Sakamoto, R. G. Danzmann and M. M. Ferguson, 2003. Quantitative trait loci for spawning time and body weight in rainbow trout: testing for conserved effects across ancestrally duplicated chromosomes. J. Hered. 94: 273–284. [DOI] [PubMed] [Google Scholar]
- Phillips, R., and P. Ràb, 2001. Chromosome evolution in the Salmonidae (Pisces): an update. Biol. Rev. Camb. Philos. Soc. 76: 1–25. [DOI] [PubMed] [Google Scholar]
- Quillet, E., L. Foisil, B. Chevassus, D. Chourrout and F. G. Liu, 1991. Production of all triploid and all female brown trout for aquaculture. Aquat. Living Resour. 4: 27–32. [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal, R. R., and J. F. Rohlf, 1995. Biometry, Ed. 3. W. H. Freeman, New York.
- Svärdson, G., 1945. Chromosome studies of Salmonidae. Rep. Swedish State Inst. Freshwater Fish. Res. 23: 1–151. [Google Scholar]
- Voorrips, R. E., 2002. Mapchart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Woram, R. A., C. McGowan, J. A. Stout, K. Gharbi, M. M. Ferguson et al., 2004. A genetic linkage map for Arctic char (Salvelinus alpinus): evidence for higher recombination rates and segregation distortion in hybrid versus pure strain mapping parents. Genome 47: 304–315. [DOI] [PubMed] [Google Scholar]
- Woram, R. A., K. Gharbi, T. Sakamoto, B. Høyheim, L.-E. Holm et al., 2003. Comparative genome analysis of the primary sex-determining locus in salmonid fishes. Genome Res. 13: 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, Jr., J. E., K. Johnson, A. Hollister and B. May, 1983. Meiotic models to explain classical linkage, pseudolinkage, and chromosome pairing in tetraploid derivative salmonid genomes. Isozymes Curr. Top. Biol. Med. Res. 10: 239–260. [PubMed] [Google Scholar]
- Young, W. P., P. A. Wheeler, V. H. Coryell, P. Keim and G. H. Thorgaard, 1998. A detailed linkage map of rainbow trout produced using doubled haploids. Genetics 148: 839–850. [DOI] [PMC free article] [PubMed] [Google Scholar]