Abstract
We report the results of a genetic screen designed to identify transcriptional coregulators of yeast heat-shock factor (HSF). This sequence-specific activator is required to stimulate both basal and induced transcription; however, the identity of factors that collaborate with HSF in governing noninduced heat-shock gene expression is unknown. In an effort to identify these factors, we isolated spontaneous extragenic suppressors of hsp82-ΔHSE1, an allele of HSP82 that bears a 32-bp deletion of its high-affinity HSF-binding site, yet retains its two low-affinity HSF sites. Nearly 200 suppressors of the null phenotype of hsp82-ΔHSE1 were isolated and characterized, and they sorted into six expression without heat-shock element (EWE) complementation groups. Strikingly, all six groups contain alleles of genes that encode subunits of Mediator. Three of the six subunits, Med7, Med10/Nut2, and Med21/Srb7, map to Mediator's middle domain; two subunits, Med14/Rgr1 and Med16/Sin4, to its tail domain; and one subunit, Med19/Rox3, to its head domain. Mutations in genes encoding these factors enhance not only the basal transcription of hsp82-ΔHSE1, but also that of wild-type heat-shock genes. In contrast to their effect on basal transcription, the more severe ewe mutations strongly reduce activated transcription, drastically diminishing the dynamic range of heat-shock gene expression. Notably, targeted deletion of other Mediator subunits, including the negative regulators Cdk8/Srb10, Med5/Nut1, and Med15/Gal11 fail to derepress hsp82-ΔHSE1. Taken together, our data suggest that the Ewe subunits constitute a distinct functional module within Mediator that modulates both basal and induced heat-shock gene transcription.
WHEN exposed to thermal or chemical stress, organisms respond by vigorously transcribing genes encoding heat-shock proteins (HSPs). HSPs function as molecular chaperones and protect the cell—along with ubiquitin, proteases, metallothioneins, and antioxidant enzymes—from damage caused by the expression of misfolded proteins. In the yeast Saccharomyces cerevisiae, the expression of heat-responsive genes is stimulated by the sequence-specific transcriptional activator heat-shock factor (HSF) Hsf1 (ScHSF) (Sorger and Pelham 1988; Nieto-Sotelo et al. 1990; Sorger 1990). In response to metabolic, oxidative, or osmotic stress, the transcription of a number of HSP genes is additionally enhanced by the gene-specific activators Msn2/Msn4 and Skn7 (Boy-Marcotte et al. 1998; Treger et al. 1998; Gasch et al. 2000; Raitt et al. 2000; Amoros and Estruch 2001; Kandror et al. 2004). Nonetheless, the only activator known to promote basal heat-shock gene transcription is HSF (McDaniel et al. 1989; Park and Craig 1989; Erkine et al. 1996). Whether this basal expression is an indirect consequence of HSF's role in establishing and maintaining a nucleosome-remodeled (“nucleosome-free”) structure over the transcription start site (Gross et al. 1993; Erkine et al. 1996), or whether HSF plays a more direct role in recruiting transcriptional coactivators under noninducing conditions, is unknown.
HSF is of additional interest, given that it can activate its target genes in the absence of several key general transcription factors (GTFs). These include Taf9 (TAFII17, constituent of both SAGA and TFIID), Med17/Srb4 and Med22/Srb6 (both subunits of Mediator), TFIIA, Kin28 (TFIIH kinase), and even the C-terminal domain (CTD) of the large subunit of RNA polymerase II (Apone et al. 1998; Lee and Lis 1998; McNeil et al. 1998; Moqtaderi et al. 1998; Chou et al. 1999). Moreover, activated HSF has been shown to mediate gene-wide histone displacement and can do so in the absence of prominent chromatin remodeling (Swi/Snf), histone modification (Set1, Gcn5), and transcriptional elongation (Paf1) complexes (Zhao et al. 2005). These observations suggest the possibility that HSF uses a novel route for transcriptional activation of its target genes, a notion supported by artificial recruitment experiments. The latter has led to the suggestion that HSF's C-terminal activation domain can activate transcription via the Rgr1 subcomplex of Mediator (Lee et al. 1999).
Mediator is an evolutionarily conserved transcriptional coregulator, composed of 25 subunits in yeast, that integrates signals from sequence-specific activators and repressors to the general transcriptional machinery (GTM) (reviewed in Malik and Roeder 2000; Myers and Kornberg 2000; Rachez and Freedman 2001; Boube et al. 2002). Originally described as an activity in fractionated yeast extracts that stimulated both basal and activator-dependent transcription (Kelleher et al. 1990; Flanagan et al. 1991), Mediator physically interacts with the pol II CTD and under certain conditions copurifies with the 12-subunit core pol II as a holoenzyme (Koleske and Young 1994). Its association with pol II is reversible: it binds tightly to the hypophosphorylated, recruitment-competent pol IIa isoform, but very weakly (if at all) to hyperphosphorylated, elongation-competent pol IIo (Svejstrup et al. 1997). This is consistent with findings, both in vivo and in vitro, that Mediator remains at the promoter following escape of pol II (Pokholok et al. 2002), where it may serve to facilitate reinitiation (Yudkovsky et al. 2000). In addition, at several yeast promoters, recruitment of Mediator has been found to precede that of pol II (Bhoite et al. 2001; Cosma et al. 2001; Bryant and Ptashne 2003), demonstrating that it can exist free of RNA polymerase in vivo. Mediator has been shown to associate exclusively with the UAS of GAL genes and does so independently of pol II and GTFs (Kuras et al. 2003).
Biochemical and electron microscopic analysis of yeast Mediator have indicated the presence of three discrete domains: the “head,” composed of Med6, Med8, Med11, Med17/Srb4, Med18/Srb5, Med19/Rox3, Med20/Srb2, and Med22/Srb6; the “middle,” composed of Med1, Med4, Med7, Med9, Med10/Nut2, Med21/Srb7, and Med31/Soh1; and the “tail,” composed of Med2, Med3/Hrs1, Med5/Nut1, Med14/Rgr1, Med15/Gal11, and Med16/Sin4 (Boube et al. 2002; Beve et al. 2005). A kinase module, composed of Cdk8/Srb10, CycC/Srb11, Med12/Srb8, and Med13/Srb9, loosely associates with the 21-subunit core Mediator complex (Liu et al. 2001). Biochemical isolation of a stable Rgr1 subcomplex, consisting of subunits of the middle and tail domains, has also been described (Lee and Kim 1998).
Each domain of Mediator appears to be directly targeted by sequence-specific regulators. For example, the MED17/TRAP80 subunit of Drosophila Mediator, ortholog of the head domain subunit Srb4, has been shown to engage in direct interaction with Drosophila HSF (Park et al. 2001). Likewise, mammalian MED17/TRAP80 is specifically targeted by p53 and VP16 (Ito et al. 1999). On the other hand, the mammalian middle domain subunit, MED1/TRAP220, is targeted by nuclear receptors (Ito et al. 2000). Three subunits of the yeast tail domain, Gal11, Med2, and Hrs1 (constituting the “Gal11 module”), physically interact with the Gcn4 activation domain in vitro and contribute to the recruitment of Mediator at Gcn4-regulated promoters (Zhang et al. 2004). Other subunits may be targets of corepressors. Srb7 binds the global yeast repressor Tup1, both in vivo and in vitro, and an Srb7 mutation that obviates Srb7–Tup1 interactions derepresses Tup1-regulated genes in S. cerevisiae (Gromoller and Lehming 2000). Specific subunits within Mediator also may interact with GTFs such as TBP, TFIIB, TFIIE, or TFIIH (Sakurai and Fukasawa 2000; Kang et al. 2001).
Consistent with their physical interactions, genetic analysis suggests that individual Mediator subunits can act as either positive or negative regulators of transcription (reviewed in Carlson 1997). For example, the yeast Gal11 and Rox3 subunits are required for Gal4 activation of galactose-inducible genes (Suzuki et al. 1988; Brown et al. 1995), Nut2 for Gcn4-mediated activation of amino acid biosynthetic genes (Han et al. 1999), and Med11 for MFα2 transcriptional activation (Han et al. 1999). Similar activator-specific functions of individual subunits have been described for Drosophila Mediator, including a role for MED23 in heat-shock gene expression and MED16 in lipopolysaccharide gene expression (Kim et al. 2004). In addition, the tail subunits of yeast Mediator have been shown to repress transcription of specific genes, including HO, SUC2, PHO5, GAL1, and IME1 (Stillman et al. 1994; Piruat et al. 1997; Tabtiang and Herskowitz 1998; Han et al. 2001; Nishizawa 2001). The kinase module has also been shown to negatively regulate transcription, either via Cdk8/Srb10 phosphorylating the pol II CTD prior to formation of the preinitiation complex (PIC) (Hengartner et al. 1998) or by its directly phosphorylating DNA-bound activators, thereby leading to their ubiquitin-mediated proteolysis or export from the nucleus (Chi et al. 2001). Subsequent to PIC formation, Mediator enhances Kin28 phosphorylation of the pol II CTD at Ser5 (Guidi et al. 2004), suggesting a way in which Mediator may positively regulate gene transcription. These and other observations point to a complex role for Mediator as an integrator of intracellular signals.
We report evidence that subunits found in all three structural domains of Mediator govern yeast heat-shock gene transcription. Genes encoding these subunits, MED7, NUT2, RGR1, ROX3, SIN4, and SRB7, were identified in a genetic screen for extragenic suppressors of a crippled heat-shock gene, hsp82-ΔHSE1, that retains only a pair of low-affinity HSF sites. Recessive mutations in each of these genes enhance the basal transcription not only of hsp82-ΔHSE1, but also of wild-type heat-shock genes. Furthermore, the more severe mutations diminish heat-shock-induced transcription, thereby revealing a functional module within Mediator that collaborates with HSF in governing the dynamic range of heat-shock gene expression.
MATERIALS AND METHODS
Yeast strain construction:
Strain employed in genetic screen:
To place HIS3 under the regulation of the hsp82-ΔHSE1 promoter, we used oligonucleotide-directed mutagenesis (Norrander et al. 1983; Gross et al. 1993) to introduce a synthetic XhoI site at position −6 relative to the ATG initiation codon of hsp82. The template was an EcoRI fragment of hsp82-ΔHSE1 [spanning −1362–+1543 (wild-type coordinates)] cloned into M13mp18. The mutation was confirmed by sequencing and the EcoRI fragment was subcloned into the URA3 integrating vector, pRS306. The HIS3 ORF (+1–+663), PCR amplified and appended with XhoI sites, was then ligated into the XhoI-opened pRS306 construct, creating pHD102. The construct was linearized at the unique StuI site within the URA3 gene and targeted to the ura3-1 locus of the hsp82-ΔHSE1 recipient strain, KEY102, creating HDY1002 (see Table 1).
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| SLY101 | MATα ade- can1-100 cyh2r his3-11,15 leu2-3,112 trp1-1 ura3 | Lee and Gross (1993) |
| KEY102 | SLY101; hsp82-ΔHSE1 | Gross et al. (1993) |
| HDY1002 | KEY102; hsp82-ΔHSE1/HIS3∷URA3, TRP1+ | This study |
| HS3000 | HDY1002; hsp82-ΔHSE1/lacZ∷ LEU2 | This study |
| HS1001 | HS3000; hsp82-ΔHSE1-HIS3∷ura3Δ (KanMX excised by Cre recombinase) | This study |
| HS1002 | HS1001; MATa | This study |
| HS1003 | HS3000; leu2∷hsp82- ΔHSE1-lacZ∷leu2∷KanMX | This study |
| HS1004 | HS1003; MATa | This study |
| HS1005 | HS1001; sin4Δ∷KanMX | This study |
| J79 | HS1004; sin4-1001 | This study |
| J1 | HS1004; rgr1-1001 | This study |
| S5 | HS1004; med7-1001 | This study |
| J121 | HS1004; med7-1002 | This study |
| S2 | HS1004; srb7-1001 | This study |
| J20 | HS1004; srb7-1002 | This study |
| J15 | HS1004; nut2-1001 | This study |
| J84 | HS1004; nut2-1002 | This study |
| J34 | HS1004; rox3-1001 | This study |
| B20 | HS1004; rox3-1002 | This study |
| RRG2 | HS1001; srb10Δ∷KanMX | This study |
| DAD2 | HS1001; srb2Δ∷KanMX | This study |
| DAD3 | HS1001; gal11Δ∷KanMX | This study |
| JHD1 | HS1001; nut1Δ∷KanMX | This study |
| SBK2000 | J20; LEU2+ | This study |
| SBK2001 | HS1001; SRB7- KanMX | This study |
| SBK2002 | HS1001; met16Δ∷KanMX | This study |
| SBK2003 | HS1001; pfk27Δ∷KanMX | This study |
| SBK2004 | HS1001; ybl094CΔ∷KanMX | This study |
| SBK2005 | J121; LEU2+ | This study |
| SBK2006 | S5; LEU2+ | This study |
| SBK2007 | J15; LEU2+ | This study |
| SBK2008 | J84; LEU2+ | This study |
| SBK2009 | B20; LEU2+ | This study |
| SBK2010 | J34; LEU2+ | This study |
| DY150 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | D. J. Stillman |
| DY2694 | DY150; MATα rgr1-Δ2∷LEU2 | D. J. Stillman |
| SBK501 | MATα leu2∷hsp82-ΔHSE1-lacZ∷leu2∷KanMX trp1 ura3 rgr1-Δ2∷LEU2 | This study |
| S288C | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| HSF1-GFP | S288C; HSF1-GFP | Invitrogen |
To fuse the hsp82-ΔHSE1 promoter to lacZ, hsp82-ΔHSE1 spanning nucleotides −806–+309 (relative to the initiating AUG codon) was PCR amplified and appended with SmaI and PstI sites. This fragment was cloned into the integrating vector YIp366, fusing hsp82-ΔHSE1 with the lacZ ORF at codon 8. The resultant hsp82/lacZ fusion was thus composed of 103 amino acids encoded by HSP82, 13 amino acids encoded by the multicloning site, and 1016 amino acids encoded by lacZ. The hsp82/lacZ construct, linearized at the unique AflII site within the LEU2 gene, was then transformed into HDY1002. Integration into the leu2-3,112 locus was confirmed by genomic PCR, creating HS3000. Strains HS1001 and HS1003 were constructed by one-step gene disruptions in HS3000 of URA3 and LEU2, respectively, using KanMX-loxP as described (Guldener et al. 1996). The KanMX (Kanr) gene was excised in HS1001 by ectopically expressed Cre recombinase (Guldener et al. 1996). HS1002 and HS1004, MATa derivatives of HS1001 and HS1003, respectively, were generated by galactose-induced expression of HO endonuclease of parental strains harboring pJH132 (GAL1/HO-URA3-CEN; gift of James Haber, Brandeis University).
Other strains:
To construct an hsp82-ΔHSE1/lacZ rgr1-Δ2 double-mutant strain, we crossed DY2694 (generously provided by David Stillman, University of Utah) with HS1004. Diploids were selected on media lacking uracil and leucine and then sporulated. Tetrads were dissected and three spores harboring both rgr1-Δ2 and hsp82-ΔHSE1/lacZ were identified. All three were assayed for lacZ expression using a liquid β-galactosidase (β-gal) assay (see below) and found to exhibit comparable expression levels. One of them, SBK501, was employed in this study. To construct a sin4Δ knockout of HS1001, we used the KanMX-loxP gene disruption method as described above, creating HS1005.
Selection of His+ revertants:
Independent 5-ml cultures of strain HS1004 were grown overnight at 30° to saturation in 1% yeast extract, 2% peptone, and 2% glucose supplemented with 0.04 mg/ml adenine (YPDA). A total of 100–200 μl of the saturated culture were spread onto synthetic medium lacking histidine and containing 1.5 mm 3-aminotriazole (3-AT) and incubated for 6–10 days at 30°. Use of 3-AT, a competitive inhibitor of IGP dehydratase (HIS3 gene product), was necessary to prevent growth of the HS1004 parental strain in the absence of a suppressing mutation. Mutagens were avoided to minimize the possibility of generating multiple mutations. On some plates, a large number of colonies of varying sizes were observed while on other plates no colonies were present. Colonies of varying sizes were picked and restreaked on 1.5 mm 3-AT plates lacking histidine to confirm growth of revertants. Using this method, >1000 His+ revertants were generated.
Screening for lacZ expression:
Each of the revertants was screened for expression of the hsp82-ΔHSE1/lacZ gene to confirm that the mutation affected hsp82-ΔHSE1 transcription and that it was acting in trans. Master plates of His+ revertants were constructed and used in XGAL assays. On average, only one in five revertants turned blue relative to the parental strain [XGALBlue (XGALB) phenotype] and thus, by definition, harbored an extragenic suppressor mutation.
Scoring of secondary growth phenotypes:
To facilitate cloning of the suppressor gene by plasmid complementation (see below), His+ XGALB suppressors were evaluated for the presence of growth phenotypes. The following phenotypes were tested: slow growth at 30° on YPDA (Slg−); inability to grow on YPDA at 37° (temperature sensitivity, Ts−); inability to grow on YPDA at 15° (cold sensitivity); inability to grow on synthetic media lacking either inositol (Ino−) or inorganic phosphate (Pho−); sensitivity to 300 mm urea (Us−), 6% ethanol (Es−) 3% formamide (Fs−), or 5 mm caffeine (Caf−) (all added to YPDA); and inability to grow on YPG (1% yeast extract, 2% peptone, 3% glycerol) (Gly−).
Determination of recessivity:
His+ XGALB mutants were backcrossed to HS1001, a MATα Leu+ Ura− counterpart of HS1004 (see Table 1). Diploids were selected on −His, −Leu synthetic medium and scored for their β-gal (XGAL) and secondary growth phenotypes. In all cases, both primary and secondary phenotypes were complemented, demonstrating recessivity.
Sorting of suppressors into complementation groups:
Suppressors with selectable growth phenotypes were crossed to MATα Leu+ Ura− spores of each complementation group (obtained by dissection of sporulated diploids derived from HS1001 backcrosses of a representative member). Ura+ Leu+ diploids were scored for retention of the XGALB phenotype (and, when ambiguous, the growth phenotype as well); those that did were considered members of that particular complementation group.
Cloning of suppressors by plasmid complementation and confirmation by genetic linkage:
EWE1:
Spontaneous His+ XGALB mutant J79, exhibiting a strong Ino− phenotype, was streaked on medium containing 1.0 mg/ml 5-fluoroorotic acid (FOA) to select a spontaneous FOAR (ura3−) mutant. The resultant strain, termed J79u, was transformed with a YCp50-based S. cerevisiae genomic DNA library (Rose et al. 1987). Ura+, Ino+ transformants were selected. Plasmids were then isolated from two transformants, and their inserts were sequenced (Iowa State University DNA Sequencing and Synthesis Facility) using YCp50-specific primers straddling the unique BamHI site (forward primer: CTGCTCGCTTCGCTACTTGG; reverse primer: CGATATAGGCGCCAGCAACC). The genomic inserts consisted of overlapping fragments (5 kb in length) derived from chromosome XIV (spanning coordinates 204,917–210,464 and 204,993–210,339); both encompassed SIN4. To demonstrate that the complementing activity was conferred by SIN4, we PCR amplified the SIN4 ORF and its flanking upstream region and cloned the resultant fragment into pRS316. The SIN4 construct was then transformed into J79u and shown to complement both Ino− and XGALB phenotypes. The XGALB phenotype of two other members of this complementation group, J73 and G128, was also fully complemented by the SIN4 plasmid. To confirm that EWE1 is SIN4, we deleted SIN4 and assayed the β-gal phenotype of the resultant strain, HS1005. It exhibited robust XGALB and liquid β-gal phenotypes (see Figure 3 below), thereby demonstrating that EWE1 is SIN4.
Figure 3.
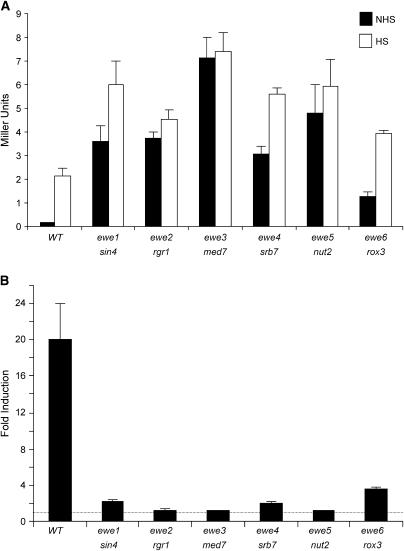
ewe suppressors strongly activate noninduced transcription of the hsp82/lacZ reporter. (A) A representative suppressor of each complementation group was grown in rich medium at 30° to early log phase and then split into two aliquots, noninduced (NHS) and heat shock induced (HS). The latter culture was subjected to a 30° → 39° thermal upshift for 45 min and allowed to recover at 30° for 20 min; then both cultures were harvested, whole-cell extracts were isolated, and β-gal levels were determined as described in materials and methods. Depicted are means ±SEM (n = 3). Strains used were HS1004 (WT), HS1005 (ewe1/sin4), J1 (ewe2/rgr1), S5 (ewe3/med7), S2 (ewe4/srb7), J15 (ewe5/nut2), and J34 (ewe6/rox3). (B) Fold inducibility of hsp82-ΔHSE1 in the wild-type strain and spontaneous ewe mutants (derived from the data of A). Dashed line indicates a level of 1.0 (no induction).
EWE2:
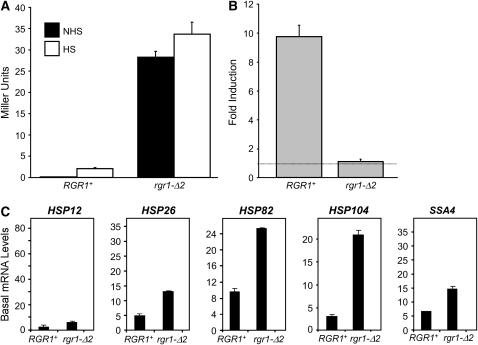
Spontaneous suppressor J1 was transformed with the YCp50-based genomic library as above and selected for Ts+ transformants on −Ura. Three different plasmids, containing overlapping inserts (spanning coordinates 274,710–286,266, 274,104–∼282,500, and 273,213–278,613 of chromosome XII) and encompassing RGR1, were isolated. To further strengthen the possibility that the mutation was in fact in RGR1, J1 and two other members of the EWE2 complementation group, J14 and J22, were crossed to an rgr1-Δ2 strain (Li et al. 1995) (DY2694, gift of David J. Stillman, University of Utah). The resultant diploids retained the XGALB and Ts− phenotypes, indicating noncomplementation. In contrast, crossing of DY2694 to S5, S2, and J15, members of the complementation groups EWE3, EWE4, and EWE5 (see below), as well as to the parental strain HS1004, resulted in complementation (XGALW). Confirmation that the EWE2 is RGR1 came from the observation that an engineered deletion of the C-terminal domain of Rgr1 (creating strain SBK501 as described above) conferred a strong β-gal+ phenotype (see Figure 6A).
Figure 6.
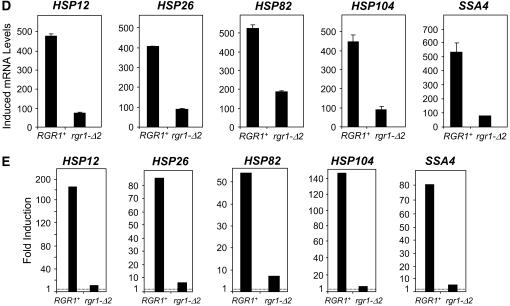
The C-terminal truncation mutant rgr1-Δ2 strongly enhances the basal transcription of both hsp82-ΔHSE1 and wild-type HSP genes, while drastically reducing their induced transcription. (A) hsp82-ΔHSE1/lacZ (β-gal) expression levels in RGR1+ (HS1004) and rgr1-Δ2 (SBK501) strains under noninducing and heat-shock-inducing conditions were determined as in Figure 3. Shown are means ±SEM (n = 4). (B) Dynamic range of expression of the strains analyzed in A. Dashed line indicates a level of 1.0 (no induction). (C and D). Northern analyses of RGR1+ (DY150) and rgr1-Δ2 (DY2694) strains maintained at 30° (C) or subjected to a 15-min 39° heat shock (D) using HSP-specific probes as in Figure 5. Illustrated are means ±SD (n = 2 or 3). (E) Dynamic range of heat-shock gene expression of RGR1+ and rgr1-Δ2 strains (data from C and D).
EWE3:
Spontaneous suppressor S5 was transformed with the YCp50-based genomic library as above and selected for growth at 30° on synthetic complete medium lacking uracil and containing 300 mm urea (Us+ phenotype). Transformants were screened for complementation of the XGAL phenotype (XGALW), and five candidates were identified. Restriction analysis of plasmids isolated from these indicated the presence of two distinct, but overlapping inserts. They were both sequenced and found to span coordinates 65,248–70,216 and 64,636–71,484 of chromosome XV, a locus containing MED7. MED7 was therefore PCR amplified, cloned into pRS316, and transformed into S5. It was found to fully complement both XGALB and Us− phenotypes. To further strengthen the possibility that EWE3 is MED7, we used homologous recombination to replace PFK27 (located 622 bp away from MED7), with the KanMX marker in strain HS1001, creating strain SBK2003. SBK2003 was then crossed to SBK2005 and SBK2006, isogenic LEU2+ (KanS) derivatives of spontaneous suppressors J121 and S5, respectively. KanR, Ura+ diploids were selected, sporulated, and dissected. In all cases (of 44 four-spore viable tetrads assayed), kanamycin resistance segregated opposite Slg− and XGALB phenotypes, thereby demonstrating that the mutation is tightly linked to MED7.
EWE4:
Spontaneous suppressor S2 was transformed with YCp50 library as described above, and Us+ transformants selected. Seven were screened for their XGAL phenotype, and four showed complementation (XGALW). Three plasmids were isolated; restriction analysis indicated the inserts were related although not identical. One was sequenced and spanned coordinates 1,072,800–1,080,059 on chromosome IV. As this region encompassed SRB7, we amplified the gene and its regulatory region by PCR, cloned the product into pRS315, and transformed the resultant SRB7 construct into S2. We observed that it fully complemented the Us− phenotype and partially complemented the XGALB phenotype. To strengthen the possibility that EWE4 is SRB7, we used homologous recombination to target the KanMX marker 100 bp downstream of SRB7 in strain HS1001, creating strain SBK2001. SBK2001 was then crossed to SBK2000, an isogenic LEU2+ derivative of the spontaneous suppressor J20, and KanR, Ura+ diploids were selected, sporulated and dissected. In all cases (47 four-spore viable tetrads assayed), kanamycin resistance segregated opposite Slg− and XGALB phenotypes, thereby demonstrating that the mutation is tightly linked to SRB7.
EWE5:
As for EWE3 and EWE4, spontaneous suppressor J15, bearing a Us− growth phenotype, was transformed with the YCp50 genomic library and complementation of its Us− phenotype was tested at 30°. A single transformant that could grow on synthetic complete medium lacking uracil and containing 300 mm urea was identified; its lacZ phenotype was similarly complemented (XGALW). Curing of the plasmid on FOA restored Us− and XGALB phenotypes. The plasmid was isolated and sequenced and found to contain an insert of 10.8 kb (coordinates 869,847–880,543 on chromosome XVI). As this region encompassed NUT2, we PCR amplified the gene and its 5′-flank and cloned it into pRS315. Leu+ transformants were found to be Us+ and XGALW, consistent with EWE5 being NUT2. To further strengthen this possibility, we used homologous recombination to replace MET16 (located 444 bp away from NUT2) with the KanMX marker in strain HS1001, creating strain SBK2002, and linkage analysis was conducted as described for EWE3 (44 four-spore viable tetrads assayed). Kanamycin resistance segregated opposite Slg− and XGALB phenotypes, thereby demonstrating that the mutation is tightly linked to NUT2.
EWE6:
Spontaneous His+ XGALB suppressor J34 (representative of a complementation group of 33 members) was transformed as above. Transformants capable of complementing the Ts− phenotype of J34 were selected. Plasmids were isolated from four Ts+ transformants; two distinct plasmids were isolated on the basis of their restriction digestion pattern. Their inserts were sequenced and found to be ∼15 kb in size, overlapping and derived from chromosome II (spanning coordinates 32,706–47,972 and 31,877–46,717). Both encompassed ROX3. ROX3 was therefore PCR amplified and cloned into pRS316, and the resultant construct was transformed into J34. It complemented both Ts− and XGALB phenotypes, implicating EWE6 as being ROX3. To strengthen this possibility, YBL094C (located 491 bp away from ROX3) was replaced with KanMX, creating strain SBK2004, and this strain was crossed to SBK2009 and SBK2010, LEU2+ derivatives of the spontaneous suppressors B20 and J34. Resultant diploids were selected, sporulated, and dissected (45 four-spore viable tetrads assayed). Kanamycin resistance segregated opposite Slg− and XGALB phenotypes, thereby demonstrating that the mutation is tightly linked to ROX3.
β-Galactosidase measurements:
XGAL plate assays:
To screen for suppressors and score dissected spores for lacZ expression, we used either a highly sensitive, nonlethal β-gal plate assay as described (Duttweiler 1996) or a nitrocellulose filter lift assay (Vojtek et al. 1993).
ONPG liquid assays:
Cultures were grown at 30° in YPDA to an A600 of 0.3–0.7 and then split into two portions. One was maintained at 30° (noninduced) and the other was heat-shocked at 39° for 45 min. The latter was then returned to 30° for 20 min to permit efficient export and translation of lacZ mRNA. At this point, cells were harvested and the activity of β-gal was determined from clarified extracts using this equation: units = (1000 × A420)/(t × V × A600), where A420 is a measure of the absorbance of the ONPG reaction, A600 is a measure of cell density, t is time in min, and v is volume in milliliters (Ausubel et al. 1995).
Microscopy:
GFP images were collected with an Olympus AX70 microscope using a 41001 filter set (Chroma Technology, Brattleboro, VT), a ×100 numerical aperture 1.25 Olympus objective equipped with a Bioptechs (Butler, PA) objective heater, and a CoolSNAP HQ charge-coupled device camera (Roper Scientific, Duluth, GA).
Northern blot hybridization:
Northerns were performed as described (Erkine and Gross 2003). The SSA4-specific hybridization probe was generated by linear amplification of a PCR fragment generated using forward primer +1827–+1848 (relative to ATG) and reverse primer +1922–+1901. All other probes were as previously described (Erkine and Gross 2003; Zhao et al. 2005). Hybridization signals were detected on a Storm 860 PhosphorImager and quantified using ImageQuant 1.11 software.
Chromatin immunoprecipitation:
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Sekinger and Gross 2001), employing a polyclonal (rabbit) antibody raised against recombinant GST–ScHSF (Erkine et al. 1996). Promoter-specific primers for SSA4 (coordinates relative to ATG) were forward, −450–−425, and reverse, −65–−93. All other primer pairs were as previously described (Sekinger and Gross 2001; Erkine and Gross 2003).
RESULTS
Experimental rationale:
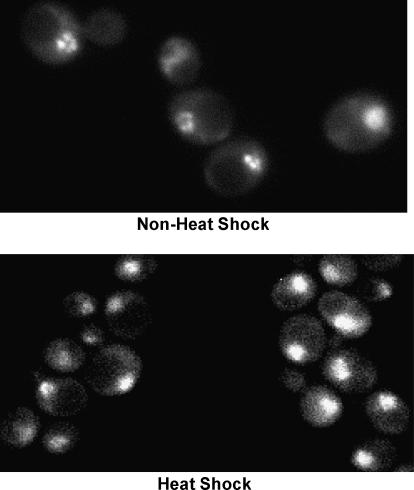
The ability of yeast HSF to activate transcription in response to thermal stress, and the role of its activation and regulatory domains in this process, have been extensively investigated (e.g., Nieto-Sotelo et al. 1990; Sorger 1990; Bonner et al. 1992; Bulman et al. 2001; Chen and Parker 2002; Erkine and Gross 2003; Hashikawa and Sakurai 2004). However, a poorly understood function of HSF is its ability to stimulate basal transcription. That this activity is central to its physiological role is suggested by several observations. First, the HSF1 gene is essential for viability, even at low temperatures (Sorger and Pelham 1988). Second, as shown in Figure 1, HSF localizes to the yeast nucleus under non-heat-shock conditions, consonant with it having a nuclear function even in the absence of stress. This pattern of intracellular localization resembles that of Drosophila HSF (Westwood et al. 1991), but contrasts with mammalian HSFs, which are principally located in the cytoplasmic compartment in the absence of stress (Sarge et al. 1993; Sheldon and Kingston 1993). And third, yeast HSF binds high-affinity HSEs under noninducing conditions, both in vitro and in vivo (Sorger et al. 1987; Jakobsen and Pelham 1988; Gross et al. 1990; Erkine et al. 1995, 1999; Giardina and Lis 1995; Sekinger and Gross 2001; Hahn et al. 2004) (see Table 3 below). In line with this, targeted mutagenesis of high-affinity HSEs severely reduces the basal transcription of HSP82, HSC82, and SSA1 (McDaniel et al. 1989; Park and Craig 1989; Gross et al. 1993; Erkine et al. 1996).
Figure 1.
HSF localizes to the yeast nucleus under both noninducing and heat-shock-activating conditions. Strain HSF1-GFP was cultivated and photographed under non-heat-shock (30°) or heat-shock (37° for 5 min) conditions as indicated.
TABLE 3.
HSF occupancy levels
| Gene | Noninduced | Induced |
|---|---|---|
| HSP12 | 0.7 | 0.7 |
| HSP26 | 0.4 | 0.95 |
| HSP82 | 1.0 | 2.1 |
| HSC82 | 1.0 | 1.0 |
| SSA3 | 0.2 | 0.65 |
| SSA4 | 1.3 | 3.4 |
HSF ChIP signals at the indicated gene promoters were normalized to those of noninduced HSP82 and represent the means of at least two independent experiments. Cells were heat-shock induced at 39° for 15 min. For these assays, PHO5 served as a nonspecific ChIP control as previously described (Sekinger and Gross 2001).
Demonstration of the constitutive nuclear localization and DNA binding of yeast HSFs raises the question of what represses its potent N- and C-terminal activation domains in the absence of stress. Several “masking” domains have been identified. These include an N-terminal repressor domain (Sorger 1990), a region that overlaps the DNA-binding and trimerization domains (Nieto-Sotelo et al. 1990; Bonner et al. 1992; Chen and Parker 2002), and a heptapeptide sequence termed CE2 located adjacent to the C-terminal activation domain (Jakobsen and Pelham 1991). While these have been traditionally thought to physically interact with one or both activation domains, the possibility of alternative or additional mechanisms of repression, including the recruitment of corepressors, has not been ruled out. Recently, a role for protein kinase A in maintaining HSF in a repressed state, at least with respect to its regulation of HSP12 and HSP26, has been reported (Ferguson et al. 2005).
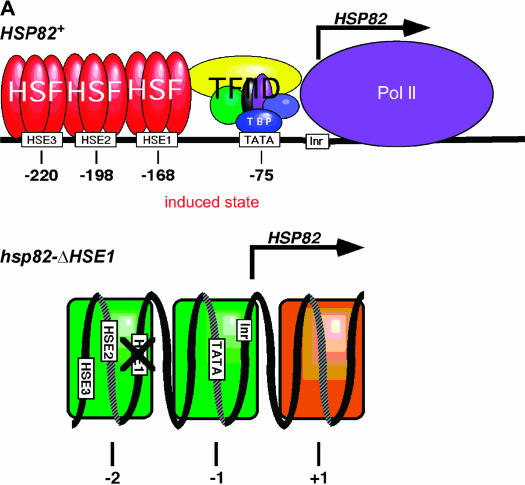
To identify additional proteins that might collaborate with HSF in regulating noninduced heat-shock gene transcription, we devised a genetic screen to isolate extragenic suppressors that restored expression to an hsp82 allele bearing a deletion of its high-affinity HSF site. This 32-bp mutation, termed ΔHSE1 (Gross et al. 1993), markedly diminishes affinity of the hsp82 promoter for HSF both in vitro and in vivo (Erkine et al. 1999; Sekinger and Gross 2001). Notably, two low-affinity HSEs are retained at this allele that are weakly occupied by HSF (Sekinger and Gross 2001). Deletion of HSE1 also obviates formation of the DNase I hypersensitive (DH), nucleosome-free region characteristic of the wild-type promoter (Gross et al. 1993). In place of the DH site—which, despite its accessibility, is occupied by histones under noninducing conditions (Zhao et al. 2005)—are two stably positioned nucleosomes, one centered over the mutated UAS and the other centered over the core promoter (Nuc-2 and Nuc-1, respectively) (Gross et al. 1993; Venturi et al. 2000). Accompanying this structural transformation is a 100-fold drop in noninduced transcription (Gross et al. 1993). Promoter chromatin architecture of wild-type and mutant hsp82 alleles is illustrated in Figure 2A.
Figure 2.
Structural phenotypes of hsp82 alleles and suppressor screen strategy. (A) Chromatin structure of the wild type and mutant hsp82 promoter. At the wild-type allele, HSF occupies HSE1 prior to heat shock, and all three HSEs following it. Its promoter nucleosomes are highly remodeled (DNase I hypersensitive) under non-heat-shock conditions and displaced, along with ORF nucleosomes, upon heat shock (depicted) (Zhao et al. 2005). At the hsp82-ΔHSE1 allele, HSF occupancy is reduced 20-fold relative to HSP82+ and its promoter nucleosomes (shaded green) are stable and precisely positioned. They are not detectably altered by heat shock. Nucleosome +1, located downstream of the initiation site, is also detectable at HSP82+ under noninducing conditions. (B) Selection/screen strategy employed in this study. Depicted are the three hsp82 alleles (all chromosomal integrants) present in strain HS1004 and the primary phenotypes of the parental strain and suppressor mutants.
Experimental strategy:
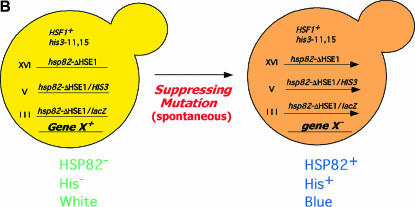
To select for suppressors of the null expression phenotype of hsp82-ΔHSE1, we fused the mutant promoter to the HIS3 coding region (in a nonrevertible his3 background), integrated the chimeric gene into the genome, and selected for spontaneous His+ revertants. In addition, to ensure the presence of an extragenic mutation, we integrated a second chromosomal reporter gene, lacZ, regulated by the same hsp82-ΔHSE1 promoter, and screened His+ revertants for β-gal expression. We also targeted the ΔHSE1 mutation to the gene's native locus using a similar integration strategy (summarized in Figure 2B), creating the strain termed HS1004 (see Table 1).
We selected >1000 independent His+ revertants. Roughly 1 in 5 of these restored expression of the integrated hsp82/lacZ reporter (as indicated by an XGALB phenotype in plate assays), consistent with the presence of an extragenic suppressing mutation. We scored suppressors for conditional growth phenotypes, including temperature sensitivity; cold sensitivity; inositol or phosphate auxotrophy; and sensitivity to ethanol, caffeine, and formamide. We also tested suppressors for enhanced sensitivity to urea (Us−), which to our knowledge has not been previously examined (S. cerevisiae growth phenotypes reviewed in Hampsey 1997). We reasoned that protein-folding defects arising from spontaneous mutations might be exacerbated by exposure to low concentrations of urea, a protein denaturant that should readily enter cells, given its small size. Indeed, a number of suppressors displayed enhanced sensitivity to 300 mm urea. Interestingly, Us− suppressors typically did not exhibit impaired growth on 3% formamide, sensitivity to which is thought to also result from folding defects (Aguilera 1994).
Suppressors were backcrossed to an isogenic strain of the opposite mating type (HS1001); all were found to be recessive. They were then sorted into complementation groups, and a representative of each, chosen on the basis of the strength of its primary (β-gal) and secondary (growth) phenotypes, was used to clone the wild-type allele from a yeast genomic library by plasmid complementation. To confirm the assignment of each suppressor gene, we performed genetic linkage analysis or, alternatively, engineered a mutation directly into it and assayed the β-gal phenotype of the resultant strain (see materials and methods).
Suppressors of the hsp82-ΔHSE1 null transcription phenotype encode components of Mediator:
All suppressors sorted into one of six complementation groups, termed EWE1-6 (for expression without heat-shock element). Strikingly, the genes represented by these complementation groups encode subunits of a single protein complex, pol II Mediator. Two of the six genes, EWE1/SIN4 and EWE2/RGR1, encode subunits of Mediator's tail domain; three others, EWE3/MED7, EWE4/SRB7, and EWE5/NUT2, encode subunits of its middle domain; and the sixth, EWE6/ROX3, encodes a subunit in the head domain (summarized in Table 2). To quantify the effect of the suppressor mutations on hsp82-ΔHSE1 expression, we measured β-gal levels of a representative allele from each complementation group. As expected, the suppressing mutations enhanced noninduced hsp82-ΔHSE1 expression, and in most cases quite substantially (30- to 70-fold; Figure 3A, solid bars; summarized in Table 2). The mutations also increased heat-shock-induced β-gal levels, but far less markedly (1.8- to 3.5-fold; open bars). As a consequence, the suppressors reduced the dynamic range of expression of hsp82-ΔHSE1 from 20-fold to, in most cases, <2-fold (Figure 3B).
TABLE 2.
Summary of EWE suppressor screen
| Complementation group
|
No. of members
|
Noninduced expression (β-gal or mRNA levels)
|
||||
|---|---|---|---|---|---|---|
| Gene | Growth phenotypes | hsp82-ΔHSE1 | HSP12 | HSP104 | ||
| Wild type | — | None | 1 | 1 | 1 | |
| EWE1 | MED16/SIN4 | Ts−, Ino−, Pho−, Us−, Fs−, Gly− | ≥100 | 35 | 1 | 1 |
| EWE2 | MED14/RGR1 | Ts−, Slg− | 38 | 38 | 2 | 1.8 |
| EWE3 | MED7 | Us−, Slg− | 7 | 71 | 3.3 | 3.4 |
| EWE4 | MED21/SRB7 | Us−, Slg− | 7 | 31 | 4 | 1.4 |
| EWE5 | MED10/NUT2 | Us−, Ts−, Slg− | 2 | 49 | 9 | 4.2 |
| EWE6 | MED19/ROX3 | Ts−, Us−, Slg− | 33 | 13 | 21 | 1.5 |
His+ revertants were selected on −His medium containing 1.5 mm 3-aminotriazole. Expression levels are provided for a representative member of each complementation group (see legend to Figure 5); for EWE1, strain HS1005 was used. mRNA levels are indicated for HSP12 and HSP104; β-gal levels for hsp82-ΔHSE1/lacZ. Expression levels are relative to wild type and are summaries of data presented in Figures 3 and 5. Growth phenotypes are listed in approximate order of severity and are defined in materials and methods.
Mutations in other subunits of Mediator fail to rescue the hsp82-ΔHSE1 null transcription phenotype:
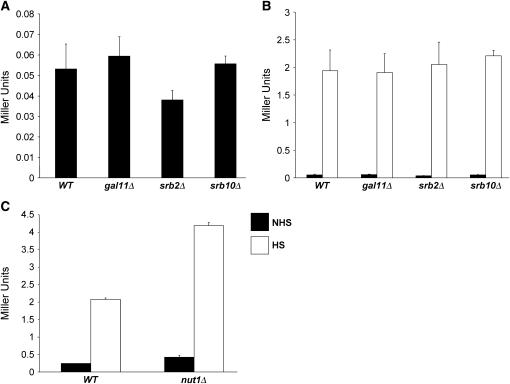
Given the scope of the selection and the large size of several complementation groups (three containing >30 members each; Table 2), it seemed possible that we had saturated the screen. Yet our selection failed to pull out either SRB10 or GAL11, which encode well-characterized negative regulatory subunits of Mediator. Indeed, Cdk8/Srb10, located in the loosely associated kinase module, negatively regulates transcription via its ability to phosphorylate both the CTD (Hengartner et al. 1998) and gene-specific activators (Chi et al. 2001), as discussed above. Additionally, Cdk8/Srb10 may play a role in Tup1-mediated repression (Kuchin and Carlson 1998). Gal11, located in the tail module, has been shown to negatively regulate transcription of genes such as SUC2 and SOL4 (Han et al. 2001), as well as of artificial promoter fusions (Nishizawa 2001). Therefore, as Cdk8/Srb10 and Gal11 are encoded by nonessential genes, we constructed isogenic srb10Δ and gal11Δ derivatives of the parental strain. We also deleted SRB2, which encodes a nonessential subunit of the head domain, and NUT1, which encodes a tail domain subunit that negatively regulates the expression of oxidative phosphorylation genes (Beve et al. 2005). However, as shown in Figure 4A, deletion of SRB10, GAL11, or SRB2 fails to enhance the low basal expression of hsp82-ΔHSE1; its induced expression is likewise unaffected (Figure 4B). The nut1Δ mutation was found to have a modest (approximately twofold) stimulatory effect on hsp82-ΔHSE1 transcription (Figure 4C), considerably less than the spontaneous suppressors. Therefore, Gal11, Srb10, and Nut1, despite their ability to negatively regulate the transcription of many genes, are functionally distinct from Ewe subunits, as is the head subunit Srb2.
Figure 4.
Disruption of GAL11, SRB2, SRB10, or NUT1 has little or no affect on hsp82-ΔHSE1 expression. Wild-type (WT) and mutant strains were cultivated, split into NHS and HS aliquots, and assayed for β-gal activity as in Figure 3. (A) Noninduced hsp82-ΔHSE1/lacZ expression in WT (HS1001), gal11Δ (DAD3), srb2Δ (DAD2), and srb10Δ (RRG2) strains. (B) Heat-shock-induced expression of hsp82-ΔHSE1/lacZ (open bars) compared with nonshocked expression (solid bars) in the same strains. (C) hsp82-ΔHSE1/lacZ expression in WT and nut1Δ strains (HS1001 and JHD1, respectively) under NHS and HS conditions as indicated. (A–C) means ±SEM (n = 3).
Mediator mutations derepress the basal transcription of wild-type HSP genes:
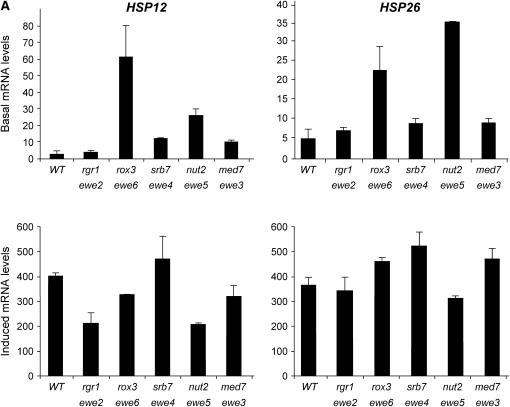
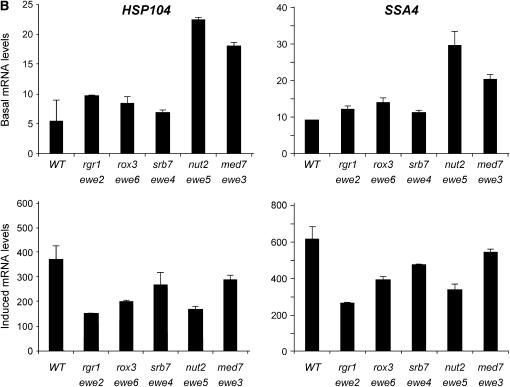
We next investigated the effects of the suppressors on wild-type heat-shock gene expression. In addition to HSP82, the chaperone-encoding genes HSP12, HSP26, HSP104, and SSA4 are regulated by HSF (Halladay and Craig 1995; Lee et al. 2002; Ferguson et al. 2005). And, as summarized in Table 3, the promoter regions of these genes are occupied by HSF even under noninducing conditions (see also Hahn et al. 2004). Nonetheless, they exhibit interesting differences in their regulation (e.g., see Ferguson et al. 2005), and their transcription might be affected by the ewe suppressors in distinct ways. As shown in Figure 5, basal HSP mRNA levels generally increased in the presence of each ewe mutation. For instance, suppressor J34, harboring a spontaneous mutation in rox3, strongly enhanced the basal transcript levels of HSP12 and HSP26 (21-fold and 4.5-fold, respectively). J15, bearing a spontaneous nut2 mutation, enhanced the noninduced expression of all four genes (3- to 9-fold), as did the med7 suppressor S5 (2- to 3-fold). Furthermore, the srb7 suppressor S2 derepressed HSP12 basal transcription 4-fold.
Figure 5.
HSP gene expression of ewe suppressors under control and heat-shock-inducing conditions. Northern analysis of wild-type (WT; HS1004), rgr1 (J1), rox3 (J34), srb7 (S2), nut2 (J15), and med7 (S5) strains cultivated in rich medium to early log phase in 30°. Cultures were either maintained at 30° (basal) or shifted to 39° for 15 min (induced) prior to isolation of total cellular RNA. Steady-state HSP mRNA levels were detected using gene-specific probes; signals were normalized to those of ACT1 (y-axis values are quotients: HSP mRNA/ACT1 mRNA). ACT1 expression, in turn, was independently quantified in each suppressor mutant relative to the abundance of the pol III transcript, SCR1, and found to be unaffected (S. B. Kremer and D. S. Gross, data not shown). For all panels, means of two or three independent experiments ±SD are illustrated.
Other suppressor/gene combinations were not as striking, such as those involving the sin4 suppressor HS1005 and the rgr1 suppressor J1 (Figure 5, Table 2, and data not shown). Nonetheless, rgr1-Δ2, encoding a C-terminal deletion of Rgr1 (Jiang et al. 1995; Li et al. 1995), not only strongly derepressed the hsp82-ΔHSE1/lacZ reporter (Figure 6A, solid bars), thereby demonstrating that rgr1-Δ2 is a bona fide ewe suppressor, but also significantly enhanced the basal transcription of wild-type heat-shock genes (Figure 6C). As was true for the spontaneous ewe mutants, individual heat-shock genes respond somewhat differently to the rgr1-Δ2 mutation: HSP12, moderately occupied by HSF (Table 3) and coregulated by Msn2/Msn4 (Gasch et al. 2000), is only weakly derepressed, whereas HSP82, strongly occupied by HSF, is strongly derepressed, as is HSP104, whose transcription is coregulated by HSF and Msn2/Msn4 (Treger et al. 1998; Grably et al. 2002). Coupled with a dramatic reduction in induced transcription (Figure 6D), HSP genes exhibit a severely compromised dynamic range of expression in context of the rgr1-Δ2 mutation (Figure 6E). This defect is also evident for the hsp82-ΔHSE1/lacZ reporter (Figure 6B). We note that the spontaneous ewe mutants have a comparatively weak effect on induced HSP mRNA levels (e.g., compare rgr1-1001 in Figure 5 with rgr1-Δ2 in Figure 6).
ewe suppressors also derepress non-heat-shock gene expression:
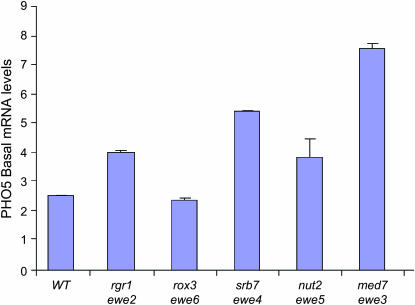
Finally, to investigate whether the ewe mutations might also derepress the basal transcription of non-HSP genes, we assayed PHO5 transcription under repressing (phosphate-rich medium) conditions. Like that of hsp82-ΔHSE1, the PHO5 promoter is assembled into an array of sequence-positioned nucleosomes (Almer and Horz 1986). Under repressing conditions, its principal activator, Pho4, is phosphorylated by the Pho80/Pho85 cyclin–CDK complex, thereby sequestering it in the cytoplasm (Kaffman et al. 1998). Under these same conditions, the Pho2 transcriptional activator weakly associates with the PHO5 promoter, although its presence does not lead to significant transcription (Nourani et al. 2004). Thus, noninduced PHO5 resembles hsp82-ΔHSE1 in two ways: the nucleosomal state of its promoter and the constitutive presence of a gene-specific activator. And, as shown in Figure 7, spontaneous mutations in srb7 and med7 modestly derepress PHO5 basal transcription (2.2- and 3.0-fold, respectively). The other suppressors had less of an effect. In other experiments, we observed that none of the suppressors detectably derepressed ACT1 (data not shown; see Figure 5 legend). Thus, although the derepressing effect of individual ewe mutations is not limited to heat-shock genes, the Ewe module does not globally restrict pol II transcription.
Figure 7.
Basal transcription of PHO5 is elevated in the context of certain ewe mutations. Northern analysis of wild-type (WT; HS1004), rgr1 (J1), rox3 (J34), srb7 (S2), nut2 (J15), and med7 (S5) strains cultivated in rich YPDA medium to early log phase in 30°. PHO5 transcript levels were normalized to those of ACT1. Depicted are means of two independent experiments ±SD.
DISCUSSION
EWE genes encode subunits of the pol II Mediator:
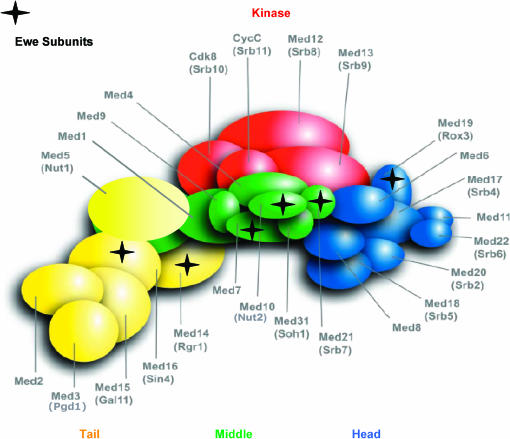
In this study, we employed a genetic screen to identify coregulators of yeast heat-shock gene transcription. The screen, which pulled out nearly 200 extragenic suppressors, proved to be remarkably specific. All suppressors sorted into six complementation groups, each of which contains alleles of genes encoding subunits of the pol II Mediator. Mediator is a modular and dynamic complex that connects gene-specific activators to the GTM by acting as signal sensor, integrator, and processor (reviewed in Boube et al. 2002). We have termed these suppressors EWE1-6. Interestingly, three EWE complementation groups encode subunits—Med7, Srb7, and Nut2—that map to the middle module of Mediator, as defined by electron microscopy and in vitro reconstitution experiments (Dotson et al. 2000; Kang et al. 2001). Two others encode subunits that map to its tail module (Rgr1 and Sin4), and one of the complementation groups encodes Rox3, which maps to its head module (Boube et al. 2002). All six proteins are highly conserved and have orthologs in human Mediator (Sato et al. 2004), and all but Sin4 are essential for viability. Relative locations of the Ewe subunits within Mediator are illustrated in Figure 8. Notably, Med7 and Srb7 physically contact Nut2 but not each other, and Sin4 contacts Rgr1. Rox3 does not appear to contact any one of the other five.
Figure 8.
Location of Ewe subunits within yeast pol II Mediator as depicted in an integrated interaction map. Subunit locations were deduced from the outcome of pairwise two-hybrid assays (Guglielmi et al. 2004 and references therein). Modified from Gugliemi et al. (2004) and presented here with permission.
While other genetic screens have implicated Sin4 and Rgr1 (Jiang et al. 1995; Wang and Michels 2004); Nut2, Sin4, and Rox3 (Tabtiang and Herskowitz 1998); or Sin4, Srb7, and Rox3 (Li et al. 2005) as participants in common regulatory pathways, to our knowledge, none has identified a unified regulatory role for all six proteins. This raises the possibility of a functional role for the Ewe subunits distinct from those previously ascribed to the Rgr1 or Gal11/Sin4 modules (reviewed in Bjorklund and Gustafsson 2004). Notably, we were aided in our identification of three of the complementation groups—EWE3/MED7, EWE4/SRB7, and EWE5/NUT2—by use of a novel conditional growth phenotype, enhanced sensitivity to 300 mm urea.
It is surprising that our screen failed to pull out genes encoding chromatin-associated proteins, especially since a previous screen, using a similar strategy (selection of bypass suppressors of a UAS deletion, in the earlier case of SUC2), isolated recessive mutations in several chromatin-associated proteins, including H2A, H2B, H3, Spt6, Spt10, and Spt16, in addition to three others: Bur1 and Bur2, which compose a cyclin/cyclin-dependent kinase heterodimer, and Bur6, a subunit of the heterodimeric NC2 negative general transcription factor (Prelich and Winston 1993). The EWE screen isolated none of these. One explanation is that the repressive chromatin structure associated with the repressed hsp82-ΔHSE1 promoter, illustrated in Figure 2A, is more stable than the chromatin assembling the suc2ΔUAS core promoter and thus not perturbed by recessive mutations in chromatin proteins. In addition, the hsp82-ΔHSE1 promoter still retains binding sites for a gene-specific activator, whereas the suc2ΔUAS promoter retains none. It may be for this reason that the BUR screen failed to isolate any Mediator subunits, or it may be simply that SUC2 and HSP82 are regulated in a distinct manner.
Sin4 may be functionally distinct from the other five Ewe subunits:
It is possible that, despite representing the largest complementation group, Sin4 functions differently from the other five suppressors. We suggest this for three reasons. First, recessive mutations in sin4 frequently act as bypass suppressors of activator defects, particularly in the context of promoter/gene fusions such as used here (Jiang et al. 1995; Tabtiang and Herskowitz 1998; Mizuno and Harashima 2000; Wang and Michels 2004; Li et al. 2005). Second, despite strongly derepressing the hsp82-ΔHSE1/HIS3 and hsp82-ΔHSE1/lacZ fusion genes, loss-of-function sin4 mutants fail to measurably enhance the basal transcript levels of nonchimeric HSP genes, including HSP12, HSP26, HSP104, SSA4, and hsp82-ΔHSE1 itself (Table 2 ; H. Singh, S. B. Kremer and D. S. Gross, unpublished observations). This is consistent with previous findings that a sin4 null mutation derepresses an HO/lacZ fusion gene but not HO itself (Tabtiang and Herskowitz 1998); sin4Δ also derepresses PHO5/lacZ but not PHO5 itself (discussed in Tabtiang and Herskowitz 1998). Third, deletion of SIN4 has little effect on induced HSP12 and HSP26 transcript levels (S. B. Kremer and D. S. Gross, unpublished observations), in marked contrast to the dramatic reductions observed in the rgr1-Δ2 mutant and also seen to some degree in the other spontaneous ewe mutants. Taken together, these observations argue against Sin4 being functionally equivalent to the other five Ewe subunits in regulating heat-shock gene expression.
Inactivating mutations in Ewe subunits likely increase the synthesis, rather than the stability, of HSP mRNAs:
By the criteria of both liquid β-galactosidase and Northern hybridization assays, ewe mutations increase heat-shock gene expression. While it is formally possible that these mutations act by increasing HSP transcript stability, three lines of evidence argue that they act principally by enhancing HSP transcription. First, the abundance of both HSP gene transcripts and those of hsp82/lacZ increases in ewe mutants. As HSP82 mRNA stability determinants map to its 3′-UTR (S. Lindquist, personal communication), and the 3′-UTR of the hsp82/lacZ transcript consists exclusively of lacZ sequence, the increase in hsp82/lacZ abundance is unlikely to stem from an HSP gene-specific enhancement of transcript stability. Second, a general increase in pol II transcript levels, while providing a possible explanation, is also unlikely, given that we measured abundance of HSP mRNAs relative to that of ACT1 mRNA; such an increase would probably be incompatible with cell viability as well. Third, heat-shock-induced HSP mRNA levels are not increased, and indeed are generally diminished, in ewe mutants. Thus, if ewe suppressors act to increase HSP mRNA stability, they would do so only under nonstressful conditions, which we believe is unlikely.
How might ewe suppressors act to negatively regulate heat-shock gene transcription?
Repeated isolation of the Ewe subunits as suppressors of hsp82-ΔHSE1 suggests that they compose a functionally distinct module within Mediator. Indeed, targeted deletion of SRB2, SRB10, or GAL11 fails to suppress, even slightly, the null transcription phenotype of hsp82-ΔHSE1, and a nut1Δ mutation only weakly suppresses. Our findings are consistent with previous work showing that an srb10Δ mutation has little effect on the constitutive expression of HSP82, HSC82, and SSA1 (Holstege et al. 1998). Therefore, an important functional role for the Ewe subunits is to negatively regulate noninduced HSP gene transcription. How might they accomplish this?
One possibility is that ewe mutations act indirectly to derepress HSP (and other) gene promoters. That is, the ewe mutations may impair the expression of gene(s) that themselves negatively regulate constitutive HSP transcription. However, it seems unlikely that an unbiased selection such as the one described here would isolate six subunits of Mediator yet fail to isolate a single gene whose protein product directly regulates the hsp82-ΔHSE1 promoter, as well as the promoters of wild-type HSP genes. Indeed, a direct role for Mediator in governing the noninduced expression of heat-shock genes is suggested by the fact that Mediator subunits have been detected within the upstream regions of several heat-shock genes, including HSP12, HSP82, HSP104, and SSA4, in cells grown under nonstressful conditions (30°, midlog growth phase) in a genomewide analysis (J.-C. Andrau and F. Holstege, personal communication). In this regard, Mediator, through its Ewe module, may act directly to repress basal heat-shock gene transcription. This could be achieved by impairing the recruitment of GTFs (including RNA pol II itself) and/or impairing their assembly into a functionally competent PIC. Other scenarios are also possible and are the subject of current investigation.
It is intriguing to consider that in repressing basal transcription, the Ewe module may serve a role similar to that of the TFIIB recognition element (BRE), mutation of which has been shown to result in a similar phenotype in mammalian systems: loss of the dynamic range of expression due to a loss of repression of basal transcription (Evans et al. 2001). Thus, both the Mediator components identified here and the BRE may function to regulate the transition of the PIC from an initiation-competent to an elongation-competent complex.
ewe mutations can severely impair induced transcription:
Evidence for a positive role of the Ewe module in activated transcription comes from expression assays of heat-shock-induced cells. These reveal a drastic reduction in induced transcript levels of wild-type heat-shock genes in the rgr1-Δ2 mutant and, to a lesser extent, in the spontaneous rgr1-1001 and nut2-1001 mutants. Our data thus lend support to earlier speculation that HSF activates transcription of its target genes via a Mediator subcomplex containing Rgr1, Nut2, and Srb7 (Lee et al. 1999). Of note, the strong requirement for Rgr1 in HSF activation contrasts dramatically with the dispensability of other GTFs, including the Srb4 and Srb6 subunits of Mediator, the TFIIH kinase, and the pol II CTD (see the Introduction).
A special role for Mediator at promoters with constitutively bound activators:
It is tempting to speculate that Mediator evolved to negatively regulate noninduced heat-shock gene transcription since HSF, unlike many inducible yeast activators, constitutively resides in the nucleus where it binds high-affinity target sequences. Thus, Mediator may be needed to prevent promiscuous heat-shock gene transcription. In the ewe mutants identified here, Mediator's negative regulatory role is compromised and basal HSP gene transcription is significantly elevated. It might be anticipated that other constitutively bound activators will exhibit a similar requirement, and indeed we find that PHO5, bearing a constitutively bound activator, is derepressed by ewe mutants. On the other hand, the nucleosomal nature of the noninduced HSP82+ promoter (Sekinger and Gross 2001; Erkine and Gross 2003; Zhao et al. 2005), shared by other HSP gene promoters (Erkine et al. 1996) and greatly pronounced in mutants such as hsp82-ΔHSE1 (Gross et al. 1993), might elicit a requirement for Mediator, although the reason for this is unclear. Clearly, additional work will be required to distinguish between these and other potential mechanisms by which the Ewe module of Mediator governs gene transcription.
Acknowledgments
We thank Xin Liu, Elena V. Sambuk, Geraldine Alba, Jorge Herrera-Diaz, Christina Venturi, Beth Zweig, Staci Deaton, Jana K. Shirey, and Lindsey Garner for technical assistance; Mike Hampsey for helpful suggestions during the early stages of this project; Kelly Tatchell for assistance with microscopy; David Stillman and James Haber for providing strains and plasmids; Michel Werner for permission to reproduce the interaction map of yeast Mediator; and Sri Balakrishnan and Susan Gross for helpful comments on the manuscript. This work was supported by grants awarded to D.S.G. from the National Institutes of Health (GM45842), the National Science Foundation (MCB-0450419), and the Biomedical Research Institute and Feist-Weiller Cancer Center of the Louisiana State University Health Sciences Center.
References
- Aguilera, A., 1994. Formamide sensitivity: a novel conditional phenotype in yeast. Genetics 136: 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almer, A., and W. Horz, 1986. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 5: 2681–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoros, M., and F. Estruch, 2001. Hsf1p and Msn2/4p cooperate in the expression of the Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 39: 1523–1532. [DOI] [PubMed] [Google Scholar]
- Apone, L. M., C. A. Virbasius, F. C. P. Holstege, J. Wang, R. A. Young et al., 1998. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2: 653–661. [DOI] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1995. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Beve, J., G. Z. Hu, L. C. Myers, D. Balciunas, O. Werngren et al., 2005. The structural and functional role of med5 in the yeast mediator tail module. J. Biol. Chem. 280: 41366–41372. [DOI] [PubMed] [Google Scholar]
- Bhoite, L. T., Y. Yu and D. J. Stillman, 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15: 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund, S., and C. M. Gustafsson, 2004. The mediator complex. Adv. Protein Chem. 67: 43–65. [DOI] [PubMed] [Google Scholar]
- Bonner, J. J., S. Heyward and D. L. Fackenthal, 1992. Temperature-dependent regulation of a heterologous activation domain fused to yeast heat shock transcription factor. Mol. Cell. Biol. 12: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boube, M., L. Joulia, D. L. Cribbs and H.-M. Bourbon, 2002. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110: 143–151. [DOI] [PubMed] [Google Scholar]
- Boy-Marcotte, E., M. Perrot, F. Bussereau, H. Boucherie and M. Jacquet, 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180: 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T. A., C. Evangelista and B. L. Trumpower, 1995. Regulation of nuclear genes encoding mitochondrial proteins in Saccharomyces cerevisiae. J. Bacteriol. 177: 6836–6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, G. O., and M. Ptashne, 2003. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Cell 11: 1301–1309. [DOI] [PubMed] [Google Scholar]
- Bulman, A. L., S. T. Hubl and H. C. Nelson, 2001. The DNA-binding domain of yeast heat shock transcription factor independently regulates both the N- and C-terminal activation domains. J. Biol. Chem. 276: 40254–40262. [DOI] [PubMed] [Google Scholar]
- Carlson, M., 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13: 1–23. [DOI] [PubMed] [Google Scholar]
- Chen, T., and C. S. Parker, 2002. Dynamic association of transcriptional activation domains and regulatory regions in Saccharomyces cerevisiae heat shock factor. Proc. Natl. Acad. Sci. USA 99: 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan et al., 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15: 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, S., S. Chatterjee, M. Lee and K. Struhl, 1999. Transcriptional activation in yeast cells lacking transcription factor IIA. Genetics 153: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma, M. P., S. Panizza and K. Nasmyth, 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7: 1213–1220. [DOI] [PubMed] [Google Scholar]
- Dotson, M. R., C. X. Yuan, R. G. Roeder, L. C. Myers, C. M. Gustafsson et al., 2000. Structural organization of yeast and mammalian mediator complexes. Proc. Natl. Acad. Sci. USA 97: 14307–14310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duttweiler, H. M., 1996. A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends Genet. 12: 340–341. [DOI] [PubMed] [Google Scholar]
- Erkine, A. M., and D. S. Gross, 2003. Dynamic chromatin alterations triggered by natural and synthetic activation domains. J. Biol. Chem. 278: 7755–7764. [DOI] [PubMed] [Google Scholar]
- Erkine, A. M., C. C. Adams, M. Gao and D. S. Gross, 1995. Multiple protein-DNA interactions over the yeast HSC82 heat shock gene promoter. Nucleic Acids Res. 23: 1822–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine, A. M., C. C. Adams, T. Diken and D. S. Gross, 1996. Heat shock factor gains access to the yeast HSC82 promoter independently of other sequence-specific factors and antagonizes nucleosomal repression of basal and induced transcription. Mol. Cell. Biol. 16: 7004–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkine, A. M., S. F. Magrogan, E. A. Sekinger and D. S. Gross, 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol. Cell. Biol. 19: 1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, R., J. A. Fairley and S. G. E. Roberts, 2001. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 15: 2945–2949.11711430 [Google Scholar]
- Ferguson, S. B., E. S. Anderson, R. B. Harshaw, T. Thate, N. L. Craig et al., 2005. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics 169: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan, P. M., R. J. Kelleher, M. H. Sayre, H. Tschochner and R. D. Kornberg, 1991. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen et al., 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina, C., and J. T. Lis, 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol. Cell. Biol. 15: 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grably, M. R., A. Stanhill, O. Tell and D. Engelberg, 2002. HSF and Msn2/4p can exclusively or cooperatively activate the yeast HSP104 gene. Mol. Microbiol. 44: 21–35. [DOI] [PubMed] [Google Scholar]
- Gromoller, A., and N. Lehming, 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J. 19: 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, D. S., K. E. English, K. W. Collins and S. Lee, 1990. Genomic footprinting of the yeast HSP82 promoter reveals marked distortion of the DNA helix and constitutive occupancy of heat shock and TATA elements. J. Mol. Biol. 216: 611–631. [DOI] [PubMed] [Google Scholar]
- Gross, D. S., C. C. Adams, S. Lee and B. Stentz, 1993. A critical role for heat shock transcription factor in establishing a nucleosome-free region over the TATA-initiation site of the yeast HSP82 heat shock gene. EMBO J. 12: 3931–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi, B., N. L. van Berkum, B. Klapholz, T. Bijma, M. Boube et al., 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32: 5379–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi, B. W., G. Bjornsdottir, D. C. Hopkins, L. Lacomis, H. Erdjument-Bromage et al., 2004. Mutual targeting of mediator and the TFIIH kinase Kin28. J. Biol. Chem. 279: 29114–29120. [DOI] [PubMed] [Google Scholar]
- Guldener, U., S. Heck, T. Fiedler, J. Beinhauer and J. H. Hegemann, 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24: 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, J. S., Z. Hu, D. J. Thiele and V. R. Iyer, 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24: 5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halladay, J. T., and E. A. Craig, 1995. A heat shock transcription factor with reduced activity suppresses a yeast HSP70 mutant. Mol. Cell. Biol. 15: 4890–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey, M., 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133. [DOI] [PubMed] [Google Scholar]
- Han, S. J., Y. C. Lee, B. S. Gim, G. H. Ryu, S. J. Park et al., 1999. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol 19: 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, S. J., J. S. Lee, J. S. Kang and Y. J. Kim, 2001. Med9/Cse2 and Gal11 modules are required for transcriptional repression of a distinct group of genes. J. Biol. Chem. 276: 37020–37026. [DOI] [PubMed] [Google Scholar]
- Hashikawa, N., and H. Sakurai, 2004. Phosphorylation of the yeast heat shock transcription factor is implicated in gene-specific activation dependent on the architecture of the heat shock element. Mol. Cell. Biol. 24: 3648–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner, C. J., V. E. Myer, S.-M. Liao, C. J. Wilson, S. S. Koh et al., 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2: 43–53. [DOI] [PubMed] [Google Scholar]
- Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner et al., 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Ito, M., C. X. Yuan, S. Malik, W. Gu, J. D. Fondell et al., 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 3: 361–370. [DOI] [PubMed] [Google Scholar]
- Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell and R. G. Roeder, 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5: 683–693. [DOI] [PubMed] [Google Scholar]
- Jakobsen, B. K., and H. R. B. Pelham, 1988. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol. Cell. Biol. 8: 5040–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen, B. K., and H. R. B. Pelham, 1991. A conserved heptapeptide restrains the activity of the yeast heat shock transcription factor. EMBO J. 10: 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. W., P. R. Dohrmann and D. J. Stillman, 1995. Genetic and physical interactions between yeast RGR1 and SIN4 in chromatin organization and transcriptional regulation. Genetics 140: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman, A., N. M. Rank, E. M. O'Neill, L. S. Huang and E. K. O'Shea, 1998. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature 396: 482–486. [DOI] [PubMed] [Google Scholar]
- Kandror, O., N. Bretschneider, E. Kreydin, D. Cavalieri and A. L. Goldberg, 2004. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol. Cell 13: 771–781. [DOI] [PubMed] [Google Scholar]
- Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee et al., 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003–42010. [DOI] [PubMed] [Google Scholar]
- Kelleher, R. J., P. M. Flanagan and R. D. Kornberg, 1990. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215. [DOI] [PubMed] [Google Scholar]
- Kim, T. W., Y. J. Kwon, J. M. Kim, Y. H. Song, S. N. Kim et al., 2004. MED16 and MED23 of Mediator are coactivators of lipopolysaccharide- and heat-shock-induced transcriptional activators. Proc. Natl. Acad. Sci. USA 101: 12153–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske, A. J., and R. A. Young, 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368: 466–469. [DOI] [PubMed] [Google Scholar]
- Kuchin, S., and M. Carlson, 1998. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol. Cell. Biol. 18: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., T. Borggrefe and R. D. Kornberg, 2003. Association of the Mediator complex with enhancers of active genes. Proc. Natl. Acad. Sci. USA 100: 13887–13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D., and J. T. Lis, 1998. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature 393: 389–392. [DOI] [PubMed] [Google Scholar]
- Lee, D.-K., S. Kim and J. T. Lis, 1999. Different upstream transcriptional activators have distinct co-activator requirements. Genes Dev. 13: 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., and D. S. Gross, 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13: 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph et al., 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Lee, Y. C., and Y. J. Kim, 1998. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18: 5364–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., T. Quinton, S. Miles and L. Breeden, 2005. Genetic interactions between mediator and the late G1-specific transcription factor Swi6 in Saccharomyces cerevisiae. Genetics 171: 477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., S. Bjorklund, Y. W. Jiang, Y. J. Kim, W. S. Lane et al., 1995. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 92: 10864–10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., J. A. Ranish, R. Aebersold and S. Hahn, 2001. Yeast nuclear extract contains two major forms of RNA polymerase II Mediator complexes. J. Biol. Chem. 276: 7169–7175. [PubMed] [Google Scholar]
- Malik, S., and R. G. Roeder, 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25: 277–283. [DOI] [PubMed] [Google Scholar]
- McDaniel, D., A. J. Caplan, M. S. Lee, C. C. Adams, B. R. Fishel et al., 1989. Basal-level expression of the yeast HSP82 gene requires a heat shock regulatory element. Mol. Cell. Biol. 9: 4789–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, J. B., H. Agah and D. Bentley, 1998. Activated transcription independent of the RNA polymerase II holoenzyme in budding yeast. Genes Dev. 12: 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, T., and S. Harashima, 2000. Activation of basal transcription by a mutation in SIN4, a yeast global repressor, occurs through a mechanism different from activator-mediated transcriptional enhancement. Mol. Gen. Genet. 263: 48–59. [DOI] [PubMed] [Google Scholar]
- Moqtaderi, Z., M. Keaveney and K. Struhl, 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2: 675–682. [DOI] [PubMed] [Google Scholar]
- Myers, L. C., and R. D. Kornberg, 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69: 729–749. [DOI] [PubMed] [Google Scholar]
- Nieto-Sotelo, J., G. Wiederrecht, A. Okuda and C. S. Parker, 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62: 807–817. [DOI] [PubMed] [Google Scholar]
- Nishizawa, M., 2001. Negative regulation of transcription by the yeast global transcription factors, Gal11 and Sin4. Yeast 18: 1099–1110. [DOI] [PubMed] [Google Scholar]
- Norrander, J., T. Kempe and J. Messing, 1983. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26: 101–106. [DOI] [PubMed] [Google Scholar]
- Nourani, A., R. T. Utley, S. Allard and J. Cote, 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23: 2597–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H.-O., and E. A. Craig, 1989. Positive and negative regulation of basal expression of a yeast HSP70 gene. Mol. Cell. Biol. 9: 2025–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J. M., J. Werner, J. M. Kim, J. T. Lis and Y.-J. Kim, 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8: 9–19. [DOI] [PubMed] [Google Scholar]
- Piruat, J. I., S. Chavez and A. Aguilera, 1997. The yeast HRS1 gene is involved in positive and negative regulation of transcription and shows genetic characteristics similar to SIN4 and GAL11. Genetics 147: 1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok, D. K., N. M. Hannett and R. A. Young, 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9: 799–809. [DOI] [PubMed] [Google Scholar]
- Prelich, G., and F. Winston, 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez, C., and L. P. Freedman, 2001. Mediator complexes and transcription. Curr. Opin. Cell Biol. 13: 274–280. [DOI] [PubMed] [Google Scholar]
- Raitt, D. C., A. L. Johnson, A. M. Erkine, K. Makino, B. Morgan et al., 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell. 11: 2335–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M. D., P. Novick, J. H. Thomas, D. Botstein and G. R. Fink, 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237–243. [DOI] [PubMed] [Google Scholar]
- Sakurai, H., and T. Fukasawa, 2000. Functional connections between mediator components and general transcription factors of Saccharomyces cerevisiae. J. Biol. Chem. 275: 37251–37256. [DOI] [PubMed] [Google Scholar]
- Sarge, K. D., S. P. Murphy and R. I. Morimoto, 1993. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 13: 1392–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., C. Tomomori-Sato, T. J. Parmely, L. Florens, B. Zybailov et al., 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14: 685–691. [DOI] [PubMed] [Google Scholar]
- Sekinger, E. A., and D. S. Gross, 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105: 403–414. [DOI] [PubMed] [Google Scholar]
- Sheldon, L. A., and R. E. Kingston, 1993. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock factor 2. Genes Dev. 7: 1549–1558. [DOI] [PubMed] [Google Scholar]
- Sorger, P. K., 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62: 793–805. [DOI] [PubMed] [Google Scholar]
- Sorger, P. K., and H. R. B. Pelham, 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54: 855–864. [DOI] [PubMed] [Google Scholar]
- Sorger, P. K., M. J. Lewis and H. R. B. Pelham, 1987. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329: 81–84. [DOI] [PubMed] [Google Scholar]
- Stillman, D. J., S. Dorland and Y. Yu, 1994. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast Swi5 transcriptional activator. Genetics 136: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Y. Nogi, A. Abe and T. Fukasawa, 1988. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4991–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svejstrup, J. Q., Y. Li, J. Fellows, A. Gnatt, S. Bjorklund et al., 1997. Evidence for a mediator cycle at the initiation of transcription. Proc. Natl. Acad. Sci. USA 94: 6075–6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabtiang, R. K., and I. Herskowitz, 1998. Nuclear proteins Nut1p and Nut2p cooperate to negatively regulate a Swi4p-dependent lacZ reporter gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 4707–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treger, J. M., A. P. Schmitt, J. R. Simon and K. McEntee, 1998. Transcriptional factor mutations reveal regulatory complexities of heat shock and newly identified genes in Saccharomyces cerevisiae. J. Biol. Chem. 273: 26875–26879. [DOI] [PubMed] [Google Scholar]
- Venturi, C. B., A. M. Erkine and D. S. Gross, 2000. Cell cycle-dependent binding of yeast heat shock factor to nucleosomes. Mol. Cell. Biol. 20: 6435–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek, A. B., S. M. Hollenberg and J. A. Cooper, 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214. [DOI] [PubMed] [Google Scholar]
- Wang, X., and C. A. Michels, 2004. Mutations in SIN4 and RGR1 cause constitutive expression of MAL structural genes in Saccharomyces cerevisiae. Genetics 168: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood, J. T., J. Clos and C. Wu, 1991. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature 353: 822–827. [DOI] [PubMed] [Google Scholar]
- Yudkovsky, N., J. A. Ranish and S. Hahn, 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229. [DOI] [PubMed] [Google Scholar]
- Zhang, F., L. Sumibcay, A. G. Hinnebusch and M. J. Swanson, 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 24: 6871–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., J. Herrera-Diaz and D. S. Gross, 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25: 8985–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]