Abstract
A quantitative trait locus (QTL), dth1.1, was associated with transgressive variation for days to heading in an advanced backcross population derived from the Oryza sativa variety Jefferson and an accession of the wild rice relative Oryza rufipogon. A series of near-isogenic lines (NILs) containing different O. rufipogon introgressions across the target region were constructed to dissect dth1.1 using substitution mapping. In contrast to the late-flowering O. rufipogon parent, O. rufipogon alleles in the substitution lines caused early flowering under both short- and long-day lengths and provided evidence for at least two distinct sub-QTL: dth1.1a and dth1.1b. Potential candidate genes underlying these sub-QTL include genes with sequence similarity to Arabidopsis GI, FT, SOC1, and EMF1, and Pharbitis nil PNZIP. Evidence from families with nontarget O. rufipogon introgressions in combination with dth1.1 alleles also detected an early flowering QTL on chromosome 4 and a late-flowering QTL on chromosome 6 and provided evidence for additional sub-QTL in the dth1.1 region. The availability of a series of near-isogenic lines with alleles introgressed from a wild relative of rice provides an opportunity to better understand the molecular basis of transgressive variation in a quantitative trait.
A wide range of natural variation for flowering time exists in wild and cultivated rice (Oryza sativa) varieties around the world. In contrast to Arabidopsis, which is a long-day plant, short days promote flowering in rice. Tropical rice varieties tend to be most sensitive to variations in photoperiod, with especially prolonged flowering under long days. As rice has been adapted to more temperate climates, it has been selected for photoperiod insensitivity to ensure normal flowering times under long days. Recent quantitative trait loci (QTL) studies have confirmed that multiple genes control the time to flowering, with multiple flowering-time loci, or heading-date QTL, segregating in any one population. Hundreds of heading-date QTL reported in >20 different studies in rice are documented at http://www.gramene.org. Strategies involving near-isogenic lines (NIL) development, high-resolution mapping, and QTL cloning have further characterized several heading-date QTL in rice (Yamamoto et al. 1998; Lin et al. 2000, 2003; Yano et al. 2000; Takahashi et al. 2001; Kojima et al. 2002; Monna et al. 2002).
The regulation of flowering time in plants has been most thoroughly studied in the model plant Arabidopsis where at least four distinct genetic pathways are involved in the transition from the vegetative to the reproductive stage. They are the photoperiod promotion pathway, the constitutive or autonomous pathway, the vernalization pathway, and the gibberellic acid promotion pathway (Mouradov et al. 2002; Simpson and Dean 2002; Yanovsky and Kay 2003; Putterill et al. 2004). Recent studies have identified putatively orthologous flowering-time genes in rice and Arabidopsis (Izawa et al. 2003). These studies confirm the presence of a conserved photoperiod pathway between Arabidopsis and rice, while at the same time providing clues to the reversal of gene function leading to the difference between short-day and long-day plants (Hayama and Coupland 2004; Putterill et al. 2004).
One of the unresolved questions in the study of quantitative traits concerns the molecular basis for transgressive variation. The occurrence of progeny displaying phenotypes more extreme than either parent has been observed for decades, and selection of offspring that are “better than the better parent” has long been practiced in the field of plant breeding. QTL analysis provides a way of identifying specific regions of chromosomes that contain genes associated with transgressive variation (deVicente and Tanksley 1993; Tanksley and McCouch 1997; Rieseberg et al. 2003). Transgressive variation for flowering time in rice has been detected in studies employing the wild relative Oryza rufipogon in crosses with four different cultivated varieties (Xiao et al. 1998; Moncada et al. 2001; Septiningsih et al. 2003; Thomson et al. 2003). In the study by Thomson et al. (2003), an O. rufipogon-derived QTL for days to heading, dth1.1, promoted early flowering in the recurrent parent (cv. Jefferson), despite the fact that the O. rufipogon parent flowers much later than the early flowering cultivar Jefferson. While there is only one report of a flowering-time QTL in this region associated with an intraspecies cross (Maheswaran et al. 2000), interspecific crosses are consistently associated with QTL for flowering time detected in the dth1.1-containing region on the short arm of chromosome 1 (Kohn et al. 1997; Doi et al. 1998; Xiao et al. 1998; Cai and Morishima 2002). This suggests that there may be genes for flowering time in wild Oryza relatives that did not pass through the genetic bottleneck(s) associated with domestication of O. sativa. These genes are likely to offer new possibilities for altering the flowering time of modern rice cultivars in ways that are inherently valuable for agriculture and not immediately obvious from the phenotype of the wild species.
To characterize the phenotypic effect of a specific QTL, it is helpful to separate it from other QTL associated with the same phenotype. One way to do this is through the creation of a set of NILs for the target QTL, thereby isolating a single donor introgression for the QTL in the background of the recurrent parent. By developing multiple NILs with introgressions covering different locations, substitution mapping can be employed to effectively dissect the QTL (Paterson et al. 1990). As suitable NILs are developed, progeny contrasts can be performed using heterozygous NILs to compare the phenotypic means of each genotypic class resulting from the segregation of a target introgression. Simultaneously, fixed homozygous NILs allow the QTL effect to be assayed in multiple environments and replicated trials. For this approach it is important to first remove all nontarget introgressions in the background that might confound the analysis of the QTL region. To further study the nature of transgressive QTL in rice, we have undertaken the molecular dissection of dth1.1 through near-isogenic line development, substitution mapping with heterozygous and homozygous NILs, and candidate gene analysis.
MATERIALS AND METHODS
NIL development:
The dth1.1 QTL was originally detected in an advanced backcross (BC2F2) QTL study with the O. sativa cultivar Jefferson as the recurrent parent and an O. rufipogon accession (IRGC105491) as the donor parent (Thomson et al. 2003). The molecular marker genotype data from the original QTL study, consisting of 153 SSR and RFLP markers across 258 BC2 families, were analyzed to identify the best families for NIL development. Pre-NIL families were chosen to contain different O. rufipogon introgressions at the targeted dth1.1 QTL region on the short arm of chromosome 1, as well as the fewest nontarget O. rufipogon segments in the rest of the genome. This was accomplished using the “NIL extraction” command in the QGene software (Nelson 1997). Five BC2 families were chosen (families 126, 131, 133, 323, and 342), and 16 BC2F2 individuals were backcrossed to the Jefferson recurrent parent in the summer of 1998 in Beaumont, Texas, resulting in 224 BC3 seeds. These five families can be traced back to 4 BC1 individuals (families 131 and 133 share the same BC1 ancestor). For continued NIL development, DNA was extracted from 131 BC3 individuals planted under greenhouse conditions (65 planted in Ithaca, NY, and 66 planted in Beaumont, TX, in 1999). Six SSR markers in the dth1.1 region (RM220, RM283, RM272, RM259, RM243, and RM23) were genotyped on the 131 BC3 plants (see Figure 1 for marker locations). To select against unwanted background introgressions, 32 additional SSR markers on other chromosomes were genotyped on the BC3 individuals predicted to have introgressions at these loci on the basis of the original BC2 genotype data. The SSR marker data were used for positive and negative selection to prioritize which BC3 individuals would be backcrossed to Jefferson. Of the original 131 BC3 plants, 42 were backcrossed to Jefferson in Ithaca, New York, resulting in 1923 BC4 seeds, while 15 BC3 plants were backcrossed in Beaumont, Texas, resulting in 349 BC4 seeds.
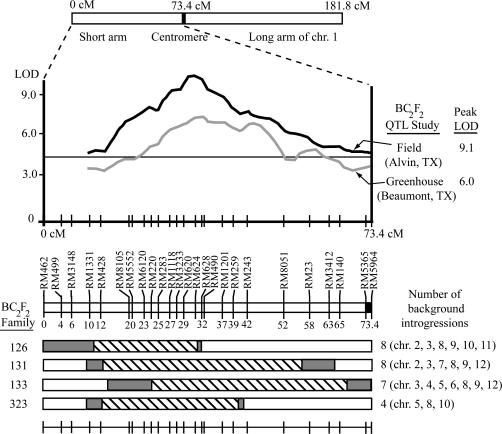
Figure 1.
(Top) The BC2F2 QTL interval plot for dth1.1 from the field and greenhouse environments, covering the short arm of chromosome 1 (Thomson et al. 2003). To facilitate comparisons between experiments, the SSR markers of the original QTL map were aligned with the genomic sequence from this region, and centimorgan distances corresponding to the Nipponbare/Kasalath map (RGP, Tsukuba, Japan) were used. In addition, the set of SSR markers used for the BC4F4 experiment are shown for all figures to enable comparisons, although only a subset of these markers were used at the BC2F2 generation. (Bottom) Genotypes of four selected BC2F2 families with O. rufipogon introgressions in the dth1.1 region. Confirmed heterozygous introgressions are shown as diagonal boxes, while shaded boxes represent potential introgression boundaries on the basis of BC2F2 genotype data. The number and chromosome locations for the background introgressions are listed for each family.
For the next round of NIL development, 960 BC4 seeds were planted in a greenhouse (Ithaca, NY; summer 1999), using deep plastic pots (2 in. in diameter, 7 in. deep) with one seed per pot. The BC4 individuals were genotyped with one to four SSR markers on chromosome 1 to identify the plants that were heterozygous (O. rufipogon/Jefferson) for dth1.1. Phenotypic selection was also applied, and selfed BC4F2 seed was harvested from selected individuals with the desired introgressions and early flowering time. The BC4F2 seed was used to select for recombinants in the dth1.1 region (see below). To complete the NIL development, the final round of negative selection against nontarget O. rufipogon segments was performed on BC4F4 families using 46 SSRs previously showing an O. rufipogon introgression in the original BC2F2 families. This resulted in the identification of 15 BC4F4 families with one to three remaining O. rufipogon segments in the background and nine NILs with no detected background segments, which were used to further dissect dth1.1. In this study, the term “NIL” is used to refer to lines that contain a single defined “target introgression” in the region of interest, with no remaining “background” introgressions in the rest of the genome, on the basis of the marker surveys described in each case. A “pre-NIL” may contain “background introgressions” whose positions are described by the molecular markers used to detect them.
DNA extraction:
Two different DNA extraction methods were used, depending on the planting design. For plants in the field and in 6-in. clay pots in the greenhouse, miniprep DNA extractions were performed using a chloroform extraction protocol. Approximately 1 × 2 cm of leaf tissue was harvested and folded into 1.5-ml microfuge tubes above a pool of liquid nitrogen to freeze the tissue. In the lab, the frozen tissue was crushed, 700 μl of DNA extraction buffer was added (100 mm Tris–HCl, 50 mm EDTA, 500 mm NaCl, 1.25% (w/v) SDS, 3.8 g/liter NaBisulfite), and the tubes were vortexed and incubated at 60° for 30 min. Subsequently, a chloroform extraction was performed with 24:1 chloroform:isoamyalcohol solution, followed by an ethanol precipitation and resuspension in 50 μl of dH2O. A 1:100 dilution of this solution was used in the PCR reaction.
For NIL populations planted in deep plastic pots, a high-throughput DNA extraction was used. Individuals were planted in sets of 96 deep pots in the greenhouse or growth chamber. These pots were arrayed in 8 × 12 matrices of 96 pots/matrix to facilitate subsequent DNA extraction and PCR in 96-well plates. Approximately 1 × 1 cm of leaf tissue was harvested directly into a 96-well flat-bottom plate above a layer of liquid nitrogen for freezing during the harvesting process. All 96 frozen tissue samples were then simultaneously crushed using a 96-prong tissue crusher (HyPure Seed Crusher HSC-200). The DNA was then extracted using the Matrix Mill apparatus (Harvester Technology; http://home.twcny.rr.com/htihome/) using the following protocol: alloy dowel pins were added to each sample well, to which 110 μl of 0.5 n NaOH was added, and the plate was covered with a Thermowell sealer and mixed in the Matrix Mill for 2 min. Ten microliters of the supernatant was transferred into a fresh 96-well plate containing 200 μl of a Tris/EDTA solution (0.05 Tris–HCl, pH 7.0, with 1 mm EDTA). Subsequently, 2 μl of the dilution was directly used in the PCR reactions. This protocol reduced the amount of labeling required, lessened the possibilities of errors due to handling individual samples, did not require chloroform or centrifugation, and increased the efficiency of extracting large numbers of samples.
SSR marker genotyping:
PCR was performed in 15-μl reactions containing 0.2 μm of each SSR primer, 200 μm dNTP mix, 50 mm KCl, 10 mm TRIS-Cl, pH 8.3, 1.5 mm MgCl, 0.01% gelatin, and 1 unit of taq polymerase. The PCR profile was: 94° for 5 min for initial denaturation, followed by 35 cycles of 94° for 30 sec, 55° for 30 sec, 72° for 30 sec, and finally by 5 min at 72° for final extension. The PCR reaction was performed in a PTC-225 tetrad thermocycler (MJ Research, Watertown, MA). The PCR products were mixed with 3× loading buffer (95% formamide, 10 mm NaOH, 0.05% bromophenol blue and 0.05% xylene cyanol) and run on 4% denaturing polyacrylamide gels using a manual sequencing gel apparatus followed by silver staining, as previously described (Panaud et al. 1996). The SSR markers were multiplexed three to seven times per gel, depending on the size of the polymorphic alleles for the Jefferson and O. rufipogon parents.
Development of novel SSR markers on chromosome 1:
For more precise mapping, new SSR markers were developed in the dth1.1 region on chromosome 1. To develop a new SSR marker for a specific region, the complete sequence of a P1-derived artificial chromosome (PAC) or BAC from the region of interest was entered into the online Simple Sequence Repeat Identification Tool (SSRIT) developed by the Cornell informatics group (Temnykh et al. 2001; http://www.gramene.org/db/searches/ssrtool). From the output of SSRIT (which identifies all perfect simple repeats in the sequence), the longer SSR motifs were prioritized for marker design to increase the chance of developing a polymorphic marker. To design PCR primers flanking the motif, several hundred bases surrounding the SSR motif were entered into the online primer design tool, Primer3 (Rozen and Skaletsky 2000; http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). Primers were then tested with Jefferson and O. rufipogon DNA to confirm single-copy products and to test for polymorphism. Ten new polymorphic markers were developed (Table 1). All of these primers were designed to have an annealing temperature of 55° in the PCR protocol.
TABLE 1.
Ten new SSR markers between 28.9 and 32.4 cM on chromosome 1
| Locus name | Marker reagenta | Forward and reverse primers (5′–3′) | Motif | Genomic cloneb |
|---|---|---|---|---|
| RM620 | MJT11 | F: GCAACTTCTGGAACTGGATG R: GCCTTCTCAGCGCAAAGTC | (GA)31 | AP001551 |
| RM621 | MJT40 | F: CGACAACTTTGAGTGCGAAG R: CCATGCATCAACACAACACA | (CG)10(AG)8 | AP002093 |
| RM622 | MJT43 | F: CAGCCTTGATCGGAAGTAGC R: TGCCGTGGTAGATCAGTCTCT | (CT)17 | AP003104 |
| RM623 | MJT44 | F: CATGTGGAAGCCAATCAGAG R: ACCAGCGGCACAGTACAAG | (CT)26 | AP003104 |
| RM624 | MJT13 | F: AGATGGTGCAAGCTAAGTTGG R: CGCATCAGTTGTTGTCAGTG | (GA)29 | AP001633 |
| RM625 | MJT46 | F: CCTAGCCAGTCCAACTCCTG R: GAGTGTCCGACGTGGAGTTC | (CCT)2 (CT)6 | AP002861 |
| RM626 | MJT47 | F: TGATGAGGCTCTAGCCGAGT R: CATGGACGAAGAAGCAAAGC | (GA)28 | AP002861 |
| RM627 | MJT48 | F: CGTGCGACAGTGGAGTAAAG R: AGCTGAGCTGATGGAGAGGA | (CCA)5 | AP002861 |
| RM628 | MJT50 | F: AGGCCATAAAGACCACGATG R: GATGTTCTCGCTAAGTCTTTCACTC | (GA)9 | AP002745 |
| RM629 | MJT52 | F: GTTCAGGTTTGCAGGTGGAC R: TAGCAGCTTGCTTGGATGTG | (CT)23 | AP002094 |
Laboratory reagent label for a specific primer pair that was used before conversion to the “RM” locus name.
Sequenced genomic PAC or BAC clone where the SSR marker is located.
Substitution line mapping using NILs:
A BC4F2 population of 29 families with ∼60 plants/family (a total of 1831 plants) was grown in the greenhouse (Ithaca, NY; summer 2001). These families were planted in deep plastic pots (2 in. diameter and 7 in. deep) with 1 plant/pot. Of these, 846 individuals were genotyped with seven SSR markers (RM220, RM283, RM620, RM272, RM490, RM259, and RM243), and an additional 468 individuals were genotyped with RM620 alone (see Figure 1 for marker locations). Of the 846 individuals genotyped with the seven SSR markers, 801 represented segregating O. rufipogon introgressions between RM620 and RM490 and were used to select for new recombination events in that region. A pedigree of these materials shows that the BC4F2 families used for NIL evaluation arose from three different BC1 plants, four BC2 plants, 16 BC3 plants, and 25 BC4 plants. After selecting selfed seed from the desired recombinant individuals from the BC4F2 population, 42 BC4F3 families, totaling 1775 individuals, were grown in the greenhouse (Ithaca, NY; winter 2001–2002) in deep plastic pots. The entire population was genotyped with three SSR markers (RM283, RM628, and RM259) to select individuals with the desired O. rufipogon introgressions at dth1.1.
For the substitution line mapping, 53 BC4F4 families, totaling 1526 individuals, were grown in the greenhouse (Ithaca, NY; summer 2002) in deep plastic pots and phenotyped for flowering time. At the same time, phenotype data were also collected on 24 BC4F4 families (12 plants each family) planted in short (10 hr)- and long (16 hr)-day growth chambers in deep plastic pots at 30° day and 26° night temperatures. For these families, 32 SSR markers in the dth1.1 region and 46 markers at all other nontarget loci were genotyped to define the O. rufipogon introgressions. Subsequently, DNA extractions of several individual plants per family were performed to confirm the introgressions. Since most of the families had fixed O. rufipogon introgressions, bulk DNA extractions were performed by combining leaves from 10 individuals from each of the 53 families. Differences in days to flowering between the NILs and the Jefferson control were analyzed using Dunnett's multiple comparison statistic (family wide error rate P < 0.05; Minitab software).
Progeny contrasts using BC4F7 families:
To develop progeny contrasts for the final experiment, three segregating families (P9-84, P13-67, and P14-28) were selected at the BC4F5 generation and the BC4F6 seeds were grown and genotyped. Individuals with homozygous O. rufipogon introgressions at dth1.1 were selected for the R/R group, and individuals with homozygous Jefferson alleles at the target loci were selected for the J/J group. The J/J group served as internal controls in this analysis. The BC4F7 progeny representing both the R/R and J/J groups were grown in short (10 hr)- and long (14 hr)-day growth chambers at 30° day and 26° night temperatures. On average, 24 plants per family were grown in the long -day chamber and 9 plants per family were grown in the short-day chamber. The days to flowering for this experiment was measured as the days between germination and 50% anthesis for the first panicle of each plant. Statistical comparisons between the R/R and J/J groups were performed using t-tests (P < 0.05), while the comparisons between the NILs and the Jefferson control were performed using Dunnett's multiple-comparison statistic (experiment-wide P < 0.05; Minitab software).
Candidate gene analysis:
The protein sequences of 18 genes known to be involved in flowering-time pathways in Arabidopsis (CCA1, CO, CRY2, EMF1, FCA, FKF1, FLC, FRI, FT, FWA, GAI, GI, LFY, LHY, LD, SOC1, TOC1, and ZTL; Blazquez 2000) were used in protein–protein BLAST searches against the GenBank nonredundant database and in protein query-translated database BLAST searches against the high-throughput genomic sequence database to identify candidate genes in rice. The locations of the BLAST matches were identified using the BAC/PAC clone list for chromosome 1 from the Rice Genome Research Program in Japan (http://rgp.dna.affrc.go.jp/). The GenBank accession numbers for the predicted protein sequences of each rice candidate are BAB32917 (OsGI), BAB32999 (FT-L 8), BAB16494 and BAC00541 (FTL), BAB92226 and BAB32985 (MADS-like), BAA89564 and BAA87823 (PNZIP), AAK98529 (OsEMF1 by Aubert et al. 2001), and BAA94774 (OsEMF1 as predicted “unnamed” protein in GenBank).
RESULTS
Development of NILs containing O. rufipogon introgressions at dth1.1:
In the original QTL study, data from a field environment (Alvin, TX) showed a peak for dth1.1 with a LOD of 9.06 and R2 of 14.9%, while greenhouse (Beaumont, TX) data showed a LOD of 5.98 and R2 of 7.5% (Thomson et al. 2003). The shape of the interval plot for dth1.1, however, was very broad: for the field environment, the QTL plot was significant (LOD > 3.0) across ∼64 cM of the short arm of chromosome 1, while the plot for the greenhouse environment showed a significant QTL across 38 cM (Figure 1). While the original dth1.1 QTL was associated with transgressive variation, due to the O. rufipogon allele promoting earliness in comparison to the Jefferson allele in BC2F2 families, we sought to test whether O. rufipogon alleles at dth1.1 continued to promote early flowering in a near-isogenic background. To create NILs for dth1.1, repeated backcrossing to the recurrent parent Jefferson was combined with DNA marker genotyping, both at the QTL target for positive selection of O. rufipogon introgressions in this region and across the rest of the genome for negative selection against nontarget O. rufipogon introgressions. Four BC2F2 families (families 126, 131, 133, and 323) were chosen from the original study with O. rufipogon introgressions encompassing overlapping, but slightly different, sections of the dth1.1 QTL region, and between four and eight nontarget segments (Figure 1).
After backcrossing these families to Jefferson, positive and negative selection was applied on 131 BC3 individuals by genotyping 6 SSR markers across the dth1.1 region and 32 SSR markers across the rest of the genome. Subsequently, 57 BC3 individuals were backcrossed to Jefferson, and BC4 plants were genotyped at the dth1.1 region to identify individuals containing overlapping segments of the desired O. rufipogon introgressions. As previous QTL cloning studies found a single gene controlling a QTL located in the region under the QTL LOD peak (Frary et al. 2000; Fridman et al. 2000; Yano et al. 2000), we focused on the 4-cM region under the dth1.1 QTL peak to select new recombinants. Segregating O. rufipogon introgressions in BC4F2 families were used to identify recombinant individuals in this region. The flanking markers RM620 and RM490 were genotyped on 801 BC4F2 individuals, resulting in 42 recombinants identified. Subsequently, genotype data on 1775 BC4F3 individuals allowed plants with homozygous O. rufipogon introgressions to be selected for phenotyping in the BC4F4 generation. A comprehensive survey of the presence or absence of nontarget introgressions was then performed at the BC4F4 generation using 46 SSRs covering all locations previously showing an O. rufipogon introgression. This resulted in the identification of 15 BC4F4 families with one to three remaining O. rufipogon segments in the background and nine NILs with no detected background segments.
Substitution mapping with fixed O. rufipogon introgressions reveals at least two sub-QTL at dth1.1:
After creating near-isogenic lines with dth1.1 O. rufipogon alleles in the Jefferson background (and no detectable introgressions elsewhere in the genome), we sought to test whether individual O. rufipogon introgressions at different locations across dth1.1 continued to exhibit transgressive variation for earliness in comparison with the Jefferson parent. For the substitution line mapping, 53 BC4F4 families, totaling 1526 individuals, were grown in the greenhouse (∼12-hr day length), and 24 BC4F4 families of 12 plants for each family were planted in short (10 hr)- and long (16 hr)-day growth chambers and phenotyped for flowering time. For these families, 32 SSR markers in the dth1.1 region, and 46 markers at all other nontarget loci were genotyped to define the O. rufipogon introgressions. These families included nine NILs (with no detected background introgressions) as previously described and contained representatives for five different genotype groups with recombination breakpoints between RM620 and RM490 (Figure 2A). Unexpectedly, almost all of the families showed significantly early flowering associated with the O. rufipogon introgressions when compared to the Jefferson control (Figure 2B).
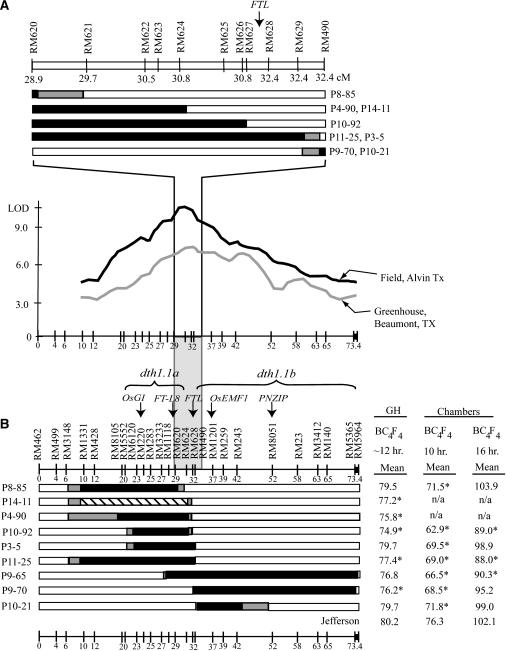
Figure 2.
(A) The region between RM620 and RM490 is expanded to show the newly developed SSRs and the precise recombinant breakpoints in this region. The relative marker distances were estimated from the continuous genomic sequence for this region. (B) BC4F4 NILs with O. rufipogon introgressions in the dth1.1 region grown in the greenhouse (∼12-hr day length) and short (10 hr)- and long-day (16 hr) growth chambers. Solid boxes represent known homozygous O. rufipogon introgressions, diagonal boxes represent heterozygous introgressions, and shaded boxes represent regions of recombination. An average of 24 individuals/line were analyzed in the greenhouse experiment, 12 individuals/line in the short-day growth chamber, and 11 individuals/line in the long-day growth chamber. Family averages were compared to the Jefferson control, and families flowering significantly earlier than Jefferson are indicated by an asterisk (Dunnett's test, family error rate P < 0.05, individual error rate P < 0.0009). The locations of the sub-QTLs and candidate genes are shown above the markers. Not shown is the MADS-box candidate, which is tightly linked to FT-L 8 in the dth1.1a region.
If a single gene controlled dth1.1, about half of the introgressions would be significant for early flowering, while the other half would not be significant. Since a number of the early flowering introgressions are not overlapping, these data clearly showed that at least two sub-QTL, dth1.1a and dth1.1b, control dth1.1 (Figure 2B). For example, in examining the NILs grown in the short-day growth chamber, the significant early flowering families P8-85 (4.8 days early) and P10-92 (13.4 days early) have O. rufipogon introgressions telomeric to RM628, while the early flowering families P9-70 (7.8 days early) and P10-21 (4.5 days early) have O. rufipogon introgressions centromeric to RM628, with no overlap between these two groups (Figure 2A). Although the short-day growth chamber data are the most consistent in supporting multiple early flowering sub-QTL at dth1.1, the greenhouse data generally agree with the short-day data. For example, in the greenhouse experiment, the early flowering families P4-90 and P10-92 (telomeric to RM628) and P9-70 (centromeric to RM628) also support the presence of at least two sub-QTL (Figure 2B). On the other hand, other early flowering families, such as P11-25, P3-5, and P9-65, have overlapping introgressions that cannot be used to strictly delimit dth1.1. The data from the NILs supported the presence of at least two sub-QTL, both of which exhibit transgressive variation with the O. rufipogon allele, causing early flowering in a near-isogenic Jefferson background in comparison to the Jefferson parent.
Identification of candidate genes for the dth1.1 sub-QTL:
To identify candidate genes for the multiple sub-QTL in the dth1.1 region, 18 proteins known to effect flowering time in Arabidopsis were used in BLAST searches against the Nipponbare rice genomic sequence. As of December 2005, the dth1.1 region on the short arm of chromosome 1 was completely covered with contiguous sequence with the exception of three physical gaps (at 52.7, 62.5, and 73.1 cM). Six BLAST hits were located in the dth1.1 region, with amino acid similarity to Arabidopsis proteins GIGANTEA (GI), FT, SOC1, EMF1, and the Pharbitis nil protein PNZIP. Three of the matches were located in the dth1.1a region. The first of these, with 73% amino acid similarity to GI, appears to be the only strong match to GI in the rice genome and has been named OsGI (Fowler et al. 1999; Hayama et al. 2002). OsGI is located on the PAC clone P0666G04 near marker RM220 on chromosome 1 (Figure 2B). Another candidate in the dth1.1a region, located on clone P0489A05 near RM1118, has 54% amino acid similarity to Arabidopsis FT and was previously referred to as FT-L 8 by Izawa et al. (2002). A third candidate in this region, on the overlap of clones B1015E06 and P0489A05 near RM1118, shows 62% similarity to the MADS-box protein SOC1. Between the dth1.1a and dth1.1b regions was another match to FT (on the overlap of clones P0665D10 and P0489G09) with 87% similarity to Arabidopsis FT, previously referred to as FTL by Izawa et al. (2002). In the dth1.1b region, two candidate genes were identified, the first being a match with 37% similarity to EMF1, previously referred to as OsEMF1 (Aubert et al. 2001). OsEMF1 appears to be the only rice homolog to EMF1 and is located on clone P0485D09 near RM1201 (Figure 2B). In addition, a keyword search of the GenBank database for genes involved in flowering identified a gene from Japanese morning glory (P. nil) that is phytochrome-regulated and possibly involved in photoperiodic flower induction in short-day plants, originally named PNIL34 (GenBank accession no. U37437) and later published as PNZIP (Zheng et al. 1998). A BLAST search of this gene identified a single putative homolog in the rice genome, a predicted gene with 93% amino acid similarity to PNZIP located in the dth1.1b region on the overlap of clones P0025D05 and P0003H10 near RM8051 (Figure 2B). Although the two sub-QTL still cover large genomic regions containing dozens of predicted genes, these genes, on the basis of their similarity to known flowering-time genes, represent the most promising candidates underlying the sub-QTL regions.
O. rufipogon introgressions in pre-NILs also affect flowering time:
In addition to the nine NILs used to dissect dth1.1, 15 BC4F4 families with one or two background introgressions were grown to test the effect on flowering time of nontarget O. rufipogon introgressions, either separately or in combination with O. rufipogon alleles at dth1.1. For example, family P2-53 has no detected O. rufipogon segments in the dth1.1 region and yet flowers significantly earlier than the Jefferson control (Figure 3B). The most likely explanation for early flowering in this family is associated with an introgression on the top of chromosome 4 that contains QTL dth4.1, a previously reported QTL where the O. rufipogon allele confers earliness in this population (Thomson et al. 2003). Family P14-28 provides evidence that the O. rufipogon introgression at RM3–RM3353 on chromosome 6 delays flowering under long days; this family is fixed for the chromosome 6 segment, and while it is segregating for an O. rufipogon introgression at dth1.1, it consistently flowers significantly later than Jefferson under long days, with no significant difference under short days. Families P21-53, P2-17, P2-40, and P2-53 provide additional support for the hypothesis that the chromosome 6 introgression delays flowering under long days; all contain the chromosome 6 introgression (as well as one or more additional introgressions, including dth1.1 for all but P2-53) and all flower early under short days, but not under long days (Figure 3B). The only family to flower significantly later under both short and long days was family P9-84. This family is unique in that it contains two nontarget O. rufipogon segments on chromosomes 6 and 9, as well as a small O. rufipogon introgression covering part of the dth1.1 region on chromosome 1 (Figure 3B).
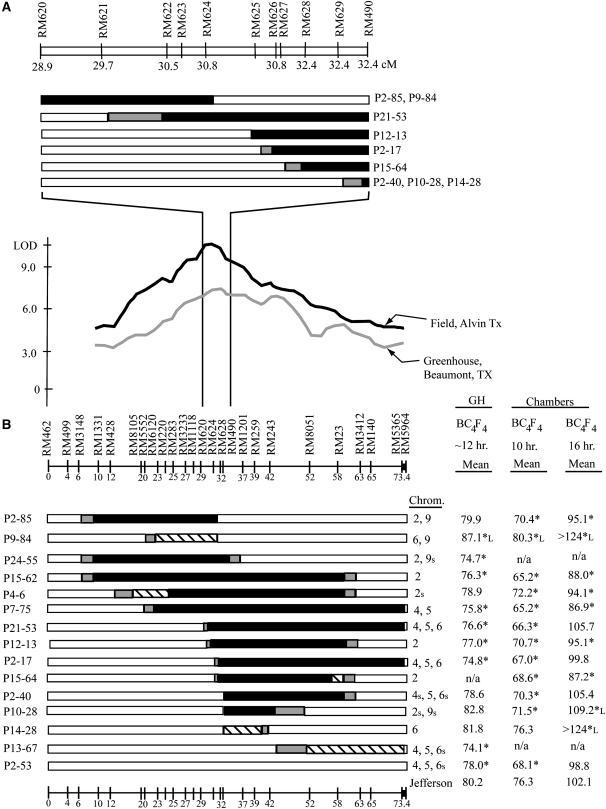
Figure 3.
(A) The region between RM620 and RM490 is expanded to show the newly developed SSRs and the precise recombinant breakpoints in this region. The relative marker distances were estimated from the continuous genomic sequence for this region. (B) BC4F4 families with O. rufipogon introgressions in the dth1.1 region and nontarget introgressions grown in the greenhouse and short- and long-day growth chambers. Nontarget introgressions are indicated by the chromosome numbers to the right of each graphical genotype, with segregating introgressions labeled with an “s” (chromosome 2: RM174, RM29, RM5812; chromosome 4: RM8213, RM307; chromosome 5: RM334, RM3170; chromosome 6: RM170, RM3, RM3353; chromosome 9: RM6839, RM5535, RM257). For these families, an average of 26 individuals/line were analyzed in the greenhouse experiment, 12 individuals/line in the short-day growth chamber, and 9 individuals/line in the long-day growth chamber. Family averages were compared to the Jefferson control, and significantly early flowering families are indicated with an asterisk, while significantly late-flowering families have an asterisk followed by “L” (Dunnett's test, family error rate P < 0.05, individual error rate P < 0.0009). Individuals in families P9-84 and P14-28 had not yet flowered when the long-day chamber experiment ended and therefore are shown to have >124 days flowering time.
A photoperiod sensitivity effect, calculated as the days to flowering under short days subtracted from the days to flowering under long days, can be seen for all materials tested; however, some families clearly showed a greater photoperiod effect than others. The Jefferson parent flowered 26 days later under long days when compared to short days, indicating that this variety is moderately photoperiod sensitive. The range of photoperiod differences seen across the nine NILs (none of which have any detectable background introgressions), which flowered between 19 and 32 days later under long days than under short days, is similar to that of Jefferson. In the BC4F4 families with background introgressions, however, six families showed larger photoperiod effects: family P2-17 with 33 days, family P2-40 with 35 days, family P10-28 with 38 days, family P21-53 with 39 days, and families P9-84 and P14-28 with >43 days difference between the short- and long-day growth chambers. In the case of family P10-28, the photoperiod effect was strong enough to cause an opposite effect between short and long days: this family had significantly early flowering under short days, but flowered significantly later than the Jefferson control under long days (Figure 3). Notably, five of these six families shared the same background introgression covering the region including RM3353, RM170, and RM3 on chromosome 6, possibly indicating an O. rufipogon allele in this region contributing a strong photoperiod effect in the Jefferson background. While most of these families also contained two other background introgressions on chromosomes 4 and 5, the early flowering of family P7-75 under long days suggests that O. rufipogon alleles on chromosomes 4 and 5 do not contribute to the late-flowering long-day effect, since family P7-75 contains just the chromosome 4 and 5 segments, but not the introgression on chromosome 6 (Figure 3B).
Because three of the BC4F4 families had a segregating O. rufipogon allele at dth1.1 and fixed background introgressions (P9-84, P14-28, and P13-67), we took the opportunity to employ progeny contrasts to measure the O. rufipogon allele effect at dth1.1 in combination with the fixed nontarget introgressions. The BC4F6 progeny from these three segregating families were genotyped to select individuals homozygous for O. rufipogon in the dth1.1 region (the R/R allele individuals), as well as individuals homozygous for Jefferson (the J/J allele individuals). The BC4F7 progeny were then tested in both short- and long-day growth chambers. A comparison between the J/J groups and the Jefferson control provides data concerning the effect of the background O. rufipogon introgressions. In this case, the J/J groups for families P9-84 and P14-28 flowered significantly later than Jefferson under both short- and long-day conditions—at 18.1 and 17.8 days later in short days and at 23.9 and 18.4 days later in long days, respectively (Figure 4). This provides strong evidence that the O. rufipogon allele at the background introgression on chromosome 6 contributes to late flowering. Likewise, the photoperiod effect of the J/J P9-84 group was 39 days, while the Jefferson control was 33 days, again supporting the presence of an O. rufipogon allele in the background, providing an increased photoperiod effect. Ironically, the O. rufipogon parent in this experiment had only a 5-day difference between the short- and long-day flowering times (Figure 4).
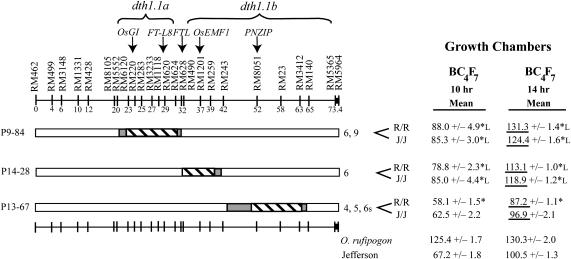
Figure 4.
BC4F7 families with homozygous O. rufipogon alleles at dth1.1 compared to homozygous Jefferson alleles, grown under short (10 hr)- and long (14 hr)-day growth chambers. An average of 8 individuals/line were analyzed in the short-day growth chamber, and an average of 23 individuals/line were analyzed in the long-day chamber. Family averages were compared to the Jefferson control and significantly early flowering families are indicated by an asterisk, while late-flowering families have an asterisk followed by “L” (Dunnett's test, family error rate P < 0.05, individual error rate P < 0.0009). In addition, the families with a fixed O. rufipogon introgression (R/R) were compared to those lacking the O. rufipogon allele at that locus (J/J) using a t-test (P < 0.05) and significantly different pairs are underlined. The families with significant differences under long days were P13-67 and P14-28 with an early flowering effect due to the O. rufipogon allele at dth1.1 and P9-84 with a late-flowering effect due to the O. rufipogon allele at dth1.1.
Evidence for additional sub-QTL at dth1.1:
While a strict interpretation of the NIL data can distinguish only two sub-QTL, dth1.1a and dth1.1b, additional evidence from the pre-NIL BC4F7 progeny contrasts suggests additional sub-QTL in the dth1.1 region. In the long-day chamber, the homozygous O. rufipogon (R/R) lines flowered significantly earlier than the homozygous Jefferson (J/J) lines for two of the families: P14-28 at 5.8 days earlier (P < 0.005) and P13-67 at 9.7 days earlier (P < 0.001; Figure 4). Since the introgressions in these two families do not overlap, these results provide evidence for another early flowering O. rufipogon sub-QTL within the dth1.1b region. The short-day data support the same trend toward earliness from the O. rufipogon alleles: family P14-28 at 6.2 days earlier and P13-67 at 4.4 days earlier, although these are not statistically significant due to the large phenotypic variance within the families.
There is also evidence for a late-flowering O. rufipogon allele at dth1.1 from family P9-84, as seen by the significant 6.9-day difference between the R/R and J/J groups for this family under long days (P < 0.02; Figure 4). Since the O. rufipogon introgression at dth1.1 in family P9-84 is in a similar region to that of several of the early flowering NILs (such as P10-92), it is possible that epistasis between dth1.1 and the nontarget alleles causes this late-flowering effect that cannot be explained by a single introgression alone.
DISCUSSION
Multiple sub-QTL at dth1.1:
The recent advances in QTL cloning have begun to unravel the molecular nature of quantitative traits, providing essential information concerning the number of genes underlying QTL and the relationship between QTL and major genes. Many of the first QTL to be cloned were those of relatively large effect, such as Hd1 with up to 67% of the total genetic variance explained by this QTL, EDI with up to 56%, Ovate with 48–67%, and fw2.2 with up to 30% of the variance explained (Frary et al. 2000; Yano et al. 2000; El-Assal et al. 2001; Liu et al. 2002). After map-based cloning, a single gene was found to control each of these QTL. In the case of Hd1, this QTL was also found to be allelic to the major gene Se1; likewise, Ovate had also been identified as both a major gene and a QTL. Similarly, map-based cloning determined that a large-effect plant height QTL on chromosome 1, ph1.1, was allelic to the major semidwarf gene Sd1 (Septiningsih 2002). Although the number of cloned QTL is still low, it appears that QTL of large effect are often controlled by single genes, and in some cases are allelic to known “major” genes. In contrast, other QTL have proven more complex. Tightly linked QTL controlling the same trait have been described in tomato in the cases of Brix9-2-5 and PW9-2-5, in rice with the heading-date QTL Hd3a and Hd3b, and in Arabidopsis with two tightly linked growth-rate QTL (Fridman et al. 2002; Monna et al. 2002; Kroymann and Mitchell-Olds 2005). As seen by our results, it now appears that the flowering-time QTL dth1.1 also presents a complex locus with multiple, linked genes controlling the QTL. As more QTL are investigated in detail, these data will provide valuable information on whether the majority of QTL are controlled by a single gene or by a group of multiple linked genes and a better understanding of whether functionally linked genes are independently or coordinately regulated. It is possible that the earliest examples of cloned QTL tend to be biased toward the simple model due to the length of time needed to clone extremely complex QTL.
While many of the first QTL to be cloned were of large effect, our dth1.1 target explained only 8–15% of the total variance for flowering time in the original QTL population. We chose this QTL, in part, to test whether a QTL of moderate effect could be efficiently cloned using a positional strategy and to compare the underlying genetic structure of this locus to the other large-effect QTL that had already been cloned. In pursuing the standard fine-mapping strategy for cloning a single gene underlying a QTL, we identified 42 recombinants in the 4-cM region under the dth1.1 peak in the BC4F2 generation. After designing 10 new SSR markers in this 950-kb region, we should have had a resolution of ∼100 kb to map a single flowering-time gene in this region, given the number of recombinants and the marker density. The BC4F4 data, however, reversed our assumption of a single gene model and at the same time forced an abrupt change in the mapping paradigm that we had been using. Whereas a single gene/QTL model can be mapped by selecting for recombination events within a large introgression, a multiple gene/QTL model requires two subsequent recombinant screens to identify new recombination events at both ends of the target segment. Therefore, to fine map multiple linked genes controlling a common trait requires the development of sub-NILs containing small segments of donor introgressions in the background of the recurrent parent that allow each sub-QTL to be isolated independently. As seen in the case of dth1.1, the presence of multiple sub-QTL contributing to earliness prevented almost any portion of the large dth1.1 region to be excluded on the basis of nonsignificance for early flowering. Due to the complexity of the locus, the sub-QTL dth1.1a and dth1.1b still cover relatively large regions of 10 and 32 cM, respectively (Figure 2). With additional rounds of screening for recombinants using the new sub-QTL NILs, however, it should be possible to fine map each sub-QTL to more precisely delimit the gene locations.
Candidate genes for dth1.1:
Since each sub-QTL still covers a region containing dozens of predicted genes, we chose to employ a positional candidate gene strategy to identify high-priority candidates in our sub-QTL target regions. Although we cannot rule out any of the predicted genes in the target regions, the high-priority candidates allow for a more focused effort to gather evidence to support or reject the possible roles of these candidates as causal agents for each sub-QTL. Sequence similarity searches identified five promising candidate genes in the original dth1.1 QTL region. Although these candidates are linked on the short arm of rice chromosome 1, there is no evidence of linkage of their respective homologs in Arabidopsis. Three of five of these candidates are putative homologs to genes known to function in the photoperiod pathway. An early gene in the photoperiod pathway is GI, which encodes a novel protein predicted to be a membrane protein (Fowler et al. 1999; Park et al. 1999) and was determined to be a nuclear protein involved in phytochrome signaling (Huq et al. 2000). In rice, a partial cDNA sequence to GIGANTEA was noted by Fowler et al. (1999) and was subsequently isolated in a differential display experiment and named OsGI by Hayama et al. (2002). Overexpression and RNAi silencing experiments with OsGI have shown that OsGI inhibits flowering in rice under long days, suggesting a reversal in the regulatory function of GI between Arabidopsis and rice (Hayama et al. 2003). OsGI, which appears to be the only copy of a GI homolog in the rice genome, is located on the short arm of chromosome 1 in the region underlying the sub-QTL dth1.1a. In our data, the O. rufipogon allele at dth1.1a promotes flowering under both short and long days; however, a late-flowering effect was seen in the family P9-84. The O. rufipogon allele may confer a different phenotypic effect than was seen in the overexpression or RNAi silencing experiments. Gene expression analysis of OsGI among the different NILs may provide clues to the function of the O. rufipogon allele at this locus.
Downstream of GI in the photoperiod pathway is CONSTANS (CO), followed by two early target genes, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1), which have been shown to be required for CO to promote flowering (Samach et al. 2000). SOC1 encodes a MADS-box transcription factor and may play a role in activating floral meristem identity genes such as LFY (Samach et al. 2000). FT encodes a putative phosphatidylethanolamine-binding protein that shares significant similarity with TERMINAL FLOWER 1 (TFL1); while FT promotes flowering, TFL1 inhibits flowering (Araki et al. 1998; Kardailsky et al. 1999; Kobayashi et al. 1999). Approximately 70 MADS-box genes are found in rice (Nam et al. 2004). One MADS-box gene was found in our dth1.1a region; however, since there are ∼70 MADS-box genes in rice (Nam et al. 2004), the likelihood of this colocation happening by chance precludes this from being a high-priority candidate gene. At least nine putative FT homologs have been identified in rice (Izawa et al. 2002; Kojima et al. 2002). Kojima et al. (2002) has identified the rice photoperiod-sensitivity QTL Hd3a on chromosome 6, which promotes flowering under short days, as encoding a protein with high similarity to Arabidopsis FT. Under long days, HD1 represses the expression of FT orthologs in rice, in contrast to Arabidopsis, where CO promotes the expression of FT (Izawa et al. 2002; Hayama et al. 2003). Another putative FT homolog in rice, FTL, has been shown to promote flowering in rice when overexpressed (Izawa et al. 2002). Although FTL is located between the sub-QTL dth1.1a and dth1.1b on chromosome 1, its potential role in the flowering-time QTL dth1.1 cannot be ruled out; there is still the possibility of more than two sub-QTL controlling dth1.1. In addition, a second putative FT homolog, FT-L 8, is located in the dth1.1a region. FT-L 8 is closely linked to the MADS-box candidate and within the same sub-QTL as OsGI. Additional recombinants are needed to more precisely map dth1.1a and to isolate these three candidates into separate NILs.
A key repressor of flowering time in Arabidopsis is EMF1, since emf1 knockout mutants bypass the vegetative stage and flower directly upon germination (Bai and Sung 1995). There appears to be a single homolog to EMF1 in the rice genome: OsEMF1 (Aubert et al. 2001), located in the dth1.1b sub-QTL region. Another model plant for studying flowering time is the short-day Japanese morning glory, P. nil (also referred to as Ipomoea nil). Several light-regulated genes have been isolated from P. nil, including the CONSTANS homolog PnCO, the floral induction response gene INRPK1, and the light-regulated PNZIP (Zheng et al. 1998; Bassett et al. 2000; Liu et al. 2001). PNZIP encodes a protein with a leucine zipper motif and has been shown to be regulated by phytochrome and to follow a circadian pattern of gene expression (Zheng et al. 1998). There is only one putative homolog to PNZIP in the rice genome, which is located in dth1.1b. Although a putative Arabidopsis homolog to PNZIP, AT103, has been identified, the function in Arabidopsis is still unknown. Likewise, more evidence is needed to confirm the role of the rice PNZIP homolog in effecting flowering time in rice.
Transgressive variation for flowering time in rice:
Given that the dth1.1 QTL confers transgressive variation for days to flowering in the Jefferson background, we proceeded to explore the underlying cause of the transgressive variation by genetically dissecting this QTL-containing region. Our work aimed to test the hypothesis that transgressive variation for flowering time associated with dth1.1 was the result of a single gene derived from O. rufipogon in the dth1.1 region that interacted epistatically with another genetic factor(s) in the Jefferson genetic background. By developing NILs and dissecting the dth1.1 QTL through a substitution mapping approach, we were able to simultaneously test the alternative hypothesis, namely that several genes in the dth1.1 region contributed to the transgressive phenotype in rice. If this alternative hypothesis were true, we designed our approach to provide material that would enable us to examine whether the genes underlying the dth1.1 QTL interacted with each other and/or with other genetic factor(s) in the Jefferson background to produce the transgressive phenotype. Two NILs from our study provide clear evidence for transgressive variation at both sub-QTL and provide the genetic materials for further dissection of this QTL: at dth1.1a NIL P10-92 flowered 13 days earlier than Jefferson and at dht1.1b NIL P9-70 flowered 7 days earlier than the Jefferson control under short days.
One of the major causes of transgressive segregation in plants is the creation of novel combinations of complementary alleles from two parents, resulting in progeny with extreme phenotypes (Rick 1976; deVicente and Tanksley 1993). In the case of dth1.1, the combination of O. rufipogon alleles at dth1.1 with the background of Jefferson alleles at all other loci results in transgressive segregation for early flowering time. A comparative QTL analysis of rice heading-date QTL on the short arm of chromosome 1 reveals several other published QTL in the same region as dth1.1. Of 17 rice QTL studies examined (Li et al. 1995; Xiao et al. 1995, 1996, 1998; Kohn et al. 1997; Lu et al. 1997; Yano et al. 1997; Doi et al. 1998; Lin et al. 1998; Xiong et al. 1999; Maheswaran et al. 2000; Bres-Patry et al. 2001; Moncada et al. 2001; Cai and Morishima 2002; Yu et al. 2002; Hittalmani et al. 2003; Septiningsih et al. 2003), five heading-date QTL were identified in the dth1.1 region, four of which were detected in interspecific crosses in rice (Kohn et al. 1997; Doi et al. 1998; Xiao et al. 1998; Cai and Morishima 2002) and one in an intraspecific cross (Maheswaran et al. 2000). Interestingly, in the thoroughly studied intraspecific Nipponbare/Kasalath population, 14 heading-date QTL have been identified, none of which are located on chromosome 1 (Yano 2001). These results suggest that across different O. sativa varieties the alleles at dth1.1 are largely the same, while the natural variation present in the wild species O. rufipogon provides a novel source of allelic diversity. This supports the hypothesis presented by Tanksley and McCouch (1997) that there are many favorable alleles that were “left behind” by the domestication process and that these alleles can be efficiently “recovered” using advanced backcross QTL analysis.
The O. rufipogon alleles at the multiple loci underlying dth1.1 also present the opportunity to better understand the genetics and the molecular mechanism(s) underlying transgressive variation for flowering time in rice. For example, the presence of O. rufipogon introgressions in the same region that leads to both early and late flowering, as seen by the early flowering P10-92 compared to the late-flowering P9-84, presents the possibility of linked alleles with opposite effects in the dth1.1a region. In that situation, additional transgressive variation could be gained through recombination events between the linked loci. There is also the possibility of an epistatic interaction between a single gene at dth1.1a with different O. rufipogon background introgressions. For example, P9-84 is the only family that combines dth1.1a with an O. rufipogon introgression on the top of chromosome 6. This introgression on chromosome 6 is found in six BC4F4 families and appears to have a strong photoperiod effect in all of these families. While the chromosome 6 introgression may delay flowering under long days independently of chromosome 1 loci, a possible epistatic interaction between dth1.1a and the chromosome 6 allele may lead to late flowering under short days as well. It is noteworthy that the chromosome 6 region identified in this study overlaps the location of two flowering-time QTL identified by Monna et al. (2002), Hd3a and Hd3b, where the Kasalath allele at Hd3b causes late heading under long days but not under short days in the Nipponbare background. It will be of interest to evaluate the precise effects of O. rufipogon alleles at Hd3a and Hd3b in different combinations with the dth1.1 sub-QTL in the materials generated in this study.
Conclusion:
Our substitution mapping results have revealed at least two sub-QTL at the flowering-time QTL dth1.1, with the O. rufipogon alleles promoting early flowering in this region on the short arm of chromosome 1. Furthermore, additional evidence suggests a third early flowering sub-QTL in the dth1.1b region, as well as the possibility of a late-flowering sub-QTL in the dth1.1a region. In addition, the presence of an early flowering O. rufipogon introgression on chromosome 4 and a late-flowering introgression with a strong photoperiod effect on chromosome 6 was also detected in combination with several of the dth1.1 sub-QTL. In comparing these results to other QTL studies, it appears that this complex locus was revealed primarily due to the use of an interspecific population, indicating the value of employing the natural variation inherent in O. rufipogon alleles to dissect the control of flowering time in rice.
Acknowledgments
We gratefully acknowledge the following students for contributing to this project in the greenhouse and laboratory: graduate student Suzy Strickler; undergraduate students Christina Hoffman, Lisa Polewczak, Jennifer Lee, Danielle Ameen, Kevin Hong, and Vincent Lee; and the high school students involved in this project through our summer internship program: Juliet Girard, Roshan Prabhu, and Joshua Esnard. We are also thankful for the help of our greenhouse, laboratory, and administrative staff, especially Fumio Onishi, Jason Coburn, and Lois Swales. In addition, we acknowledge X. H. Lai and Anna McClung (U. S. Department of Agriculture (USDA)–Agricultural Research Service, Beaumont, TX) for backcrossing the 16 BC2F2 individuals that formed the basis of the NILs and Josefina Alcala, John Mann, and Mark Walton at RiceTec (Alvin, TX) for supporting field trials of our substitution lines. This material is based upon work supported by the National Science Foundation (NSF) under grant no. 0110004. Graduate student fellowships to M.J.T. and J.D.E. were funded by a Cornell University Plant Cell and Molecular Biology interagency grant (NSF, USDA, U. S. Department of Energy). A graduate research assistantship to E.M.S. was funded by the Rockefeller Foundation.
References
- Araki, T., N. Kobayashi, H. Kaya and A. Iwabuchi, 1998. The flowering-time gene FT and regulation of flowering in Arabidopsis. J. Plant Res. 111: 277–281. [Google Scholar]
- Aubert, D., L. Chen, Y. H. Moon, D. Martin, L. A. Castle et al., 2001. EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13: 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, S., and Z. R. Sung, 1995. The role of Emf1 in regulating the vegetative and reproductive transition in Arabidopsis-Thaliana (Brassicaceae). Am. J. Bot. 82: 1095–1103. [Google Scholar]
- Bassett, C. L., M. L. Nickerson, R. A. Cohen and M. S. Rajeevan, 2000. Alternative transcript initiation and novel post-transcriptional processing of a leucine-rich repeat receptor-like protein kinase gene that responds to short-day photoperiodic floral induction in morning glory (Ipomoea nil). Plant Mol. Biol. 43: 43–58. [DOI] [PubMed] [Google Scholar]
- Blazquez, M. A., 2000. Flower development pathways. J. Cell Sci. 113: 3547–3548. [DOI] [PubMed] [Google Scholar]
- Bres-Patry, C., M. Lorieux, G. Clement, M. Bangratz and A. Ghesquiere, 2001. Heredity and genetic mapping of domestication-related traits in a temperate japonica weedy rice. Theor. Appl. Genet. 102: 118–126. [Google Scholar]
- Cai, H. W., and H. Morishima, 2002. QTL clusters reflect character associations in wild and cultivated rice. Theor. Appl. Genet. 104: 1217–1228. [DOI] [PubMed] [Google Scholar]
- deVicente, M. C., and S. D. Tanksley, 1993. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi, K., A. Yoshimura and N. Iwata, 1998. RFLP mapping and QTL analysis of heading date and pollen sterility using backcross populations between Oryza sativa L. and Oryza glaberrima Steud. Breed. Sci. 48: 395–399. [Google Scholar]
- El-Assal, S., C. Alonso-Blanco, A. J. M. Peeters, V. Raz and M. Koornneef, 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29: 435–440. [DOI] [PubMed] [Google Scholar]
- Fowler, S., K. Lee, H. Onouchi, A. Samach, K. Richardson et al., 1999. GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frary, A., T. C. Nesbitt, S. Grandillo, E. Knaap, B. Cong et al., 2000. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. [DOI] [PubMed] [Google Scholar]
- Fridman, E., T. Pleban and D. Zamir, 2000. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. USA 97: 4718–4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E., Y. S. Liu, L. Carmel-Goren, A. Gur, M. Shoresh et al., 2002. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genomics 266: 821–826. [DOI] [PubMed] [Google Scholar]
- Hayama, R., and G. Coupland, 2004. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135: 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., T. Izawa and K. Shimamoto, 2002. Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43: 494–504. [DOI] [PubMed] [Google Scholar]
- Hayama, R., S. Yokoi, S. Tamaki, M. Yano and K. Shimamoto, 2003. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422: 719–722. [DOI] [PubMed] [Google Scholar]
- Hittalmani, S., N. Huang, B. Courtois, R. Venuprasad, H. E. Shashidhar et al., 2003. Identification of QTL for growth- and grain yield-related traits in rice across nine locations of Asia. Theor. Appl. Genet. 107: 679–690. [DOI] [PubMed] [Google Scholar]
- Huq, E., J. M. Tepperman and P. H. Quail, 2000. GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 97: 9789–9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., T. Oikawa, N. Sugiyama, T. Tanisaka, M. Yano et al., 2002. Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16: 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Y. Takahashi and M. Yano, 2003. Comparative biology comes into bloom: genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 6: 113–120. [DOI] [PubMed] [Google Scholar]
- Kardailsky, I., V. K. Shukla, J. H. Ahn, N. Dagenais, S. K. Christensen et al., 1999. Activation tagging of the floral inducer FT. Science 286: 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., H. Kaya, K. Goto, M. Iwabuchi and T. Araki, 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kohn, J. R., N. Leyva, R. Dossey, B. Sobral and H. Morishima, 1997. Quantitative trait locus analysis of trait variation among annual and perennial ecotypes of Oryza rufipogon. Intl. Rice Res. Notes 22: 4–5. [Google Scholar]
- Kojima, S., Y. Takahashi, Y. Kobayashi, L. Monna, T. Sasaki et al., 2002. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43: 1096–1105. [DOI] [PubMed] [Google Scholar]
- Kroymann, J., and T. Mitchell-Olds, 2005. Epistasis and balanced polymorphism influencing complex trait variation. Nature 435: 95–98. [DOI] [PubMed] [Google Scholar]
- Li, Z., S. R. M. Pinson, J. W. Stansel and W. D. Park, 1995. Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 91: 374–381. [DOI] [PubMed] [Google Scholar]
- Lin, H. X., T. Yamamoto, T. Sasaki and M. Yano, 2000. Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor. Appl. Genet. 101: 1021–1028. [Google Scholar]
- Lin, H. X., Z. W. Liang, T. Sasaki and M. Yano, 2003. Fine mapping and characterization of quantitative trait loci Hd4 and Hd5 controlling heading date in rice. Breed. Sci. 53: 51–59. [Google Scholar]
- Lin, S. Y., T. Sasaki and M. Yano, 1998. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor. Appl. Genet. 96: 997–1003. [Google Scholar]
- Liu, J., J. Yu, L. McIntosh, H. Kende and J. A. Zeevaart, 2001. Isolation of a CONSTANS ortholog from Pharbitis nil and its role in flowering. Plant Physiol. 125: 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., J. Van Eck, B. Cong and S. D. Tanksley, 2002. A new class of regulatory genes underlying the cause of pear-shaped tomato fruit. Proc. Natl. Acad. Sci. USA 99: 13302–13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., L. Shen, Z. Tan, Y. Xu, P. He et al., 1997. Comparative mapping of QTLs for agronomic traits of rice across environments by using a doubled-haploid population. Theor. Appl. Genet. 94: 145–150. [DOI] [PubMed] [Google Scholar]
- Maheswaran, M., N. Huang, S. Sreerangasamy and S. McCouch, 2000. Mapping quantitative trait loci associated with days to flowering and photoperiod sensitivity in rice (Oryza sativa L.). Mol. Breed. 6: 145–155. [Google Scholar]
- Moncada, P., C.-P. Martinez, J. Borrero, M. Chatel, H. Gauch, Jr. et al., 2001. Quantitative trait loci for yield and yield components in an Oryza sativa × Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102: 41–52. [Google Scholar]
- Monna, L., X. Lin, S. Kojima, T. Sasaki and M. Yano, 2002. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling heading date in rice. Theor. Appl. Genet. 104: 772–778. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., F. Cremer and G. Coupland, 2002. Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14(Suppl.): S111–S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam, J., J. Kim, S. Lee, G. An, H. Ma et al., 2004. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc. Natl. Acad. Sci. USA 101: 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.-C., 1997. QGENE: software for marker-based genomic analysis and breeding. Mol. Breed. 3: 239–245. [Google Scholar]
- Panaud, O., X. Chen and S. R. McCouch, 1996. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597–607. [DOI] [PubMed] [Google Scholar]
- Park, D. H., D. E. Somers, Y. S. Kim, Y. H. Choy, H. K. Lim et al., 1999. Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582. [DOI] [PubMed] [Google Scholar]
- Paterson, A. H., J. W. DeVerna, B. Lanini and S. D. Tanksley, 1990. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes in an interspecies cross of tomato. Genetics 124: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., R. Laurie and R. Macknight, 2004. It's time to flower: the genetic control of flowering time. BioEssays 26: 363–373. [DOI] [PubMed] [Google Scholar]
- Rick, C. M., 1976. Natural variability in wild species of Lycopersicon and its bearing on tomato breeding. Agraria 30: 249–259. [Google Scholar]
- Rieseberg, L. H., A. Widmer, A. M. Arntz and J. M. Burke, 2003. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Samach, A., H. Onouchi, S. E. Gold, G. S. Ditta, Z. Schwarz-Sommer et al., 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616. [DOI] [PubMed] [Google Scholar]
- Septiningsih, E. M., 2002. Identification, near-isogenic line development and fine mapping of quantitative trait loci from the rice cultivar IR64 and its wild relative Oryza rufipogon. Ph.D. Thesis, Cornell University, Ithaca, NY.
- Septiningsih, E. M., J. Prasetiyono, E. Lubis, T. H. Tai, T. Tjubaryat et al., 2003. Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 107: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Simpson, G. G., and C. Dean, 2002. Arabidopsis, the Rosetta stone of flowering time? Science 296: 285–289. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., A. Shomura, T. Sasaki and M. Yano, 2001. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 98: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanksley, S. D., and S. R. McCouch, 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277: 1063–1066. [DOI] [PubMed] [Google Scholar]
- Temnykh, S., G. DeClerck, A. Lukashova, L. Lipovich, S. Cartinhour et al., 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, M. J., T. H. Tai, A. M. McClung, M. E. Hinga, K. B. Lobos et al., 2003. Mapping quantitative trait loci for yield, yield components, and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 107: 479–493. [DOI] [PubMed] [Google Scholar]
- Xiao, J., J. Li, L. Yuan and S. D. Tanksley, 1995. Dominance is the major genetic basis of heterosis in rice as revealed by QTL analysis using molecular markers. Genetics 140: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J., J. Li, L. Yuan and S. D. Tanksley, 1996. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific cross. Theor. Appl. Genet. 92: 230–244. [DOI] [PubMed] [Google Scholar]
- Xiao, J., J. Li, S. Grandillo, S. N. Ahn, L. Yuan et al., 1998. Identification of trait-improving quantitative trait loci alleles from a wild rice relative, Oryza rufipogon. Genetics 150: 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L. Z., K. D. Liu, X. K. Dai, C. G. Xu and Q. Zhang, 1999. Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor. Appl. Genet. 98: 243–251. [Google Scholar]
- Yamamoto, T., Y. Kuboki, S. Y. Lin, T. Sasaki and M. Yano, 1998. Fine mapping of quantitative trait loci Hd-1, Hd-2, and Hd-3 controlling heading date in rice, as single Mendelian factors. Theor. Appl. Genet. 97: 37–44. [Google Scholar]
- Yano, M., 2001. Genetic and molecular dissection of naturally occurring variation. Curr. Opin. Plant Biol. 4: 130–135. [DOI] [PubMed] [Google Scholar]
- Yano, M., Y. Harushima, Y. Nagamura, N. Kurata, Y. Minobe et al., 1997. Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor. Appl. Genet. 95: 1025–1032. [Google Scholar]
- Yano, M., Y. Katayose, M. Ashikari, U. Yamanouchi, L. Monna et al., 2000. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M. J., and S. A. Kay, 2003. Living by the calendar: how plants know when to flower. Nat. Rev. Mol. Cell Biol. 4: 265–275. [DOI] [PubMed] [Google Scholar]
- Yu, S. B., J. X. Li, C. G. Xu, Y. F. Tan, X. H. Li et al., 2002. Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor. Appl. Genet. 104: 619–625. [DOI] [PubMed] [Google Scholar]
- Zheng, C. C., R. Porat, P. Lu and S. D. O'Neill, 1998. PNZIP is a novel mesophyll-specific cDNA that is regulated by phytochrome and the circadian rhythm and encodes a protein with a leucine zipper motif. Plant Physiol. 116: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]