Abstract
Heterodimers of structural maintenance of chromosomes (SMC) proteins form the core of several protein complexes involved in the organization of DNA, including condensation and cohesion of the chromosomes at metaphase. The functions of the complexes with a heterodimer of Smc5p and Smc6p are less clear. To better understand them, we created two S. cerevisiae strains bearing temperature-sensitive alleles of SMC5. When shifted to the restrictive temperature, both mutants lose viability gradually, concomitant with the appearance of nuclear abnormalities and phosphorylation of the Rad53p DNA damage checkpoint protein. Removal of Rad52p or overexpression of the SUMO ligase Mms21p partially suppresses the temperature sensitivity of smc5 strains and increases their survival at the restrictive temperature. At the permissive temperature, smc5-31 but not smc5-33 cells exhibit hypersensitivity to several DNA-damaging agents despite induction of the DNA damage checkpoint. Similarly, smc5-31 but not smc5-33 cells are killed by overexpression of the SUMO ligase-defective Mms21-SAp but not by overexpression of wild-type Mms21p. Both smc5 alleles are synthetically lethal with mms21-SA and exhibit Rad52p-independent chromosome fragmentation and loss at semipermissive temperatures. Our data indicate a critical role for the S. cerevisiae Smc5/6-containing complexes in both DNA repair and chromosome segregation.
STRUCTURAL maintenance of chromosomes (SMC) proteins have central roles in the organization, maintenance, and segregation of chromosomes. The single condensin complex in Saccharomyces cerevisiae and Schizosaccharomyces pombe compacts DNA, especially during cell division (Strunnikov et al. 1995). In vertebrate cells, one condensin complex has the same role, while a second condensin orders the compacted DNA into chromatids (Hirano 1998; Ono et al. 2003). During and after DNA replication and condensation, sister chromatids are linked together by the cohesin SMC complex throughout condensin-mediated compaction until the beginning of anaphase (Gruber et al. 2003). The cell has also exploited the SMC complexes for DNA manipulation outside of mitosis. In a variety of organisms, different combinations of SMC proteins and non-SMC subunits are used to assist meiotic division, recombinational DNA repair, and dosage compensation (Jessberger 2002). While these extramitotic functions of SMC complexes are generally organism specific, cell division in all eukaryotic cells requires a third class of SMC complex, the Smc5/6 complex, the normal cellular role of which has recently begun to emerge.
Radiation-sensitive mutants in the S. pombe smc6 gene (also known as rad18, see Table 1) were first isolated in 1975; the wild-type gene was cloned by complementation of the smc6-X UV sensitivity phenotype in 1995 (Nasim and Smith 1975; Lehmann et al. 1995). Cell division in the absence of wild-type Smc6 results in lethality concomitant with highly abnormal nuclear DNA morphology (Lehmann et al. 1995; Verkade et al. 1999; Fousteri and Lehmann 2000; Harvey et al. 2004). Smc6 mutants are unable to repair DNA damage caused by several exogenous agents and, despite normal initiation of the DNA damage checkpoint response, are unable to prevent mitosis in the presence of unrepaired damage (Verkade et al. 1999; Harvey et al. 2004). Various mutant alleles of smc6 have revealed that Smc6 has two roles in the cell: a nonessential role in recombinational DNA damage repair and a poorly understood essential role in maintaining genomic stability (Fousteri and Lehmann 2000).
TABLE 1.
Members of the Smc5/6 complexes
| S. cerevisiae name/alias | S. pombe name | Smc5/6 complex |
|---|---|---|
| SMC5 | spr18 | I, II |
| RHC18/SMC6 | rad18 | I, II |
| NSE1 | nse1 | I |
| MMS21 | nse2 | I |
| NSE3 | nse3 | I |
| NSE4/QRI2 | nse4/rad62 | I |
| NSE5 | ? | II |
| KRE29 | ? | II |
The number of Smc5/6 complexes is in dispute. Hazbun et al. described the two complexes shown here, while Zhao and Blobel isolated all eight proteins as one complex.
S. pombe Smc6 exists in a high-molecular-weight complex with Smc5 and several other proteins (Fousteri and Lehmann 2000). The other non-SMC elements (NSEs) (Nse1, Nse2, Nse3, and Rad62/Nse4) were identified by mass-spectrometric analysis of affinity-purified complexes (McDonald et al. 2003; Morikawa et al. 2004; Pebernard et al. 2004; Sergeant et al. 2005). Mutants of the NSEs also exhibit nuclear fragmentation and a sensitivity to DNA damage epistatic to the homologous recombination-dependent repair pathway (McDonald et al. 2003; Morikawa et al. 2004; Pebernard et al. 2004). Nse1, Nse2, and Nse3 are required for meiotic as well as mitotic division, but elimination of meiotic recombination by deletion of rec12 partially suppresses the requirement for Nse1 during meiotic division (Pebernard et al. 2004). A variety of other genetic interactions have been discovered between the SMC and NSE proteins and proteins involved in DNA replication and repair. S. pombe Rad60 is required for recombinational repair and is a target of the Cds1 checkpoint kinase that stabilizes stalled replication forks. Rad60 physically interacts with either Smc5 or Smc6, or perhaps with both proteins (Boddy et al. 2003). S. pombe nse1, nse2, nse3, and smc6-X mutant alleles are synthetically lethal with deletion of rad60 (Verkade et al. 1999; Boddy et al. 2003; Pebernard et al. 2004), and overexpression of Rad60 can partially suppress the DNA damage sensitivity phenotype of the smc6-X hypomorphic allele (Morishita et al. 2002). Overexpression of Brc1 suppresses the DNA damage sensitivity of smc6-74, and determination of genes required for this suppression has implicated the S. pombe Smc5/6 complex in postreplication DNA repair (Sheedy et al. 2005). Nse mutants have been implicated in the process of DNA replication, as they are also synthetically lethal with deletion of the mus81/eme1 Holliday junction endonuclease and the rqh1 and srs2 replication/recombination helicases (Morikawa et al. 2004; Pebernard et al. 2004). Additionally, the smc6-74 allele is synthetically lethal with the topoisomerase II hypomorphic allele top2-191 (Verkade et al. 1999).
S. pombe Nse2 was recently shown to have SUMO ligase activity and to bind to the coiled-coil region of Smc5 (Andrews et al. 2005; Sergeant et al. 2005). In addition to catalyzing its auto-sumoylation, Nse2 sumoylates Smc6 and Nse3, but not Smc5 or Nse1. In vivo, Smc6 is sumoylated in response to DNA damage. Mutation of the conserved PIAS-like zinc finger in Nse2 eliminates its SUMO ligase activity as well as the DNA damage-dependent sumoylation of Smc6. Interestingly, S. pombe cells bearing the catalytically inactive version of Nse2 can grow, but are very sensitive to DNA damage (Andrews et al. 2005).
In S. cerevisiae, a similarly large Smc5/6 complex has been identified (Fujioka et al. 2002). Large-scale affinity purification coupled with mass spectrometric analysis of uncharacterized, essential yeast genes revealed two S. cerevisiae Smc5/6 complexes, in which the Smc5/6 heterodimer is grouped with two different sets of NSEs (Hazbun et al. 2003). This view has recently been challenged by the isolation of all six NSE subunits in Mms21p immunoprecipitates (Zhao and Blobel 2005). If two different Smc5/6 complexes do indeed exist, their structure would be reminiscent of the vertebrate condensin I and condensin II complexes. Two-hybrid interactions between the non-SMC subunits defined a Nse1p/Nse3p/Nse4p subcomplex (Hazbun et al. 2003), an organization shared by the orthologous S. pombe proteins (Sergeant et al. 2005). All Smc5/6 complex I and complex II proteins are essential for the viability of S. cerevisiae cells. Mutant smc6 S. cerevisiae strains are hypersensitive to a broad spectrum of DNA-damaging agents and are defective in recombination induced by DNA damage (Onoda et al. 2004). Smc6p is enriched at telomeres and the rDNA and is required for their correct segregation (Torres-Rosell et al. 2005b). Nse1p, the first NSE of any Smc5/6 complex to be identified, was found to immunoprecipitate with the Smc5/6 heterodimer (Fujioka et al. 2002). Hypomorphic alleles of nse1 are sensitive to DNA damage; depletion of Nse1p or overexpression of mutant Nse1p results in the appearance of cells with aberrant morphology and extremely disordered nuclei (Fujioka et al. 2002). The S. cerevisiae ortholog of nse2 is MMS21. Mms21p is also a SUMO E3 ligase, the targets of which include Smc5p and Yku70p (Zhao and Blobel 2005). The catalytically inactive allele mms21-11 exhibits defects in maintaining nucleolar integrity, telomere clustering, and telomeric silencing (Zhao and Blobel 2005). Unlike the gross nuclear defects seen with Nse1p imbalances and with mms21-11, temperature-sensitive alleles of NSE4 (formerly known as QRI2) exhibit a fully reversible Rad24p-dependent arrest at the G2/M boundary when shifted to the restrictive temperature (Hu et al. 2005). In addition, nse4 alleles display DNA damage hypersensitivity and have high rates of plasmid loss (Hu et al. 2005).
We describe here the generation and characterization of two S. cerevisiae strains with distinctly different temperature-sensitive and hypomorphic alleles of SMC5. Smc5-31 and smc5-33 cells grown at the restrictive temperature accumulate as large-budded cells and have phosphorylated and activated the Rad53p DNA damage checkpoint protein. Cell-cycle arrest is activated less than once per cell division at high temperature and is accompanied by the generation of cells with disordered and occasionally fragmented nuclear DNA. At semipermissive temperatures, smc5-31 and smc5-33 strains lose and fragment chromosomes at high rates. Strikingly, smc5-31 but not smc5-33 cells are hypersensitive to DNA damage, suggesting at least two separable roles for Smc5p. Both overexpression of the Mms21p SUMO ligase and elimination of homologous recombination by deletion of rad52 partially suppress the temperature-sensitive phenotype of both alleles and dramatically increase the fraction of cells that survive inactivation of Smc5p. Our data indicate dual roles for Smc5/6 complex activity both during homologous recombination associated with DNA repair and in maintaining the physical integrity of chromosomes.
MATERIALS AND METHODS
Generation of temperature-sensitive alleles:
All yeast strains are of the S288C background, are GAL2, and can be found in Table 2 (Brachmann et al. 1998); manipulation of strains and plasmids was done according to standard protocols (Ausubel et al. 2005) unless otherwise noted. A sequence-verified genomic clone of the S. cerevisiae SMC5 gene from nucleotide −173 to 4488 relative to the translation start site (AccI–BanI) was inserted into the SmaI site of pRS316 to make pGC233. pGC233 was transformed into a diploid heterozygous for a complete deletion of SMC5 to make yGC131 (Giaever et al. 2002). A haploid spore from yGC131 unable to grow on medium containing 5-fluoro-orotic acid (5-FOA) was isolated and named yGC137. A PmlI–HpaI fragment containing the CEN/ARS and LEU2 sequences of pRS415 was cloned into the EcoRV site of pACYC184 to create pGC250. A SalI–EagI fragment containing SMC5 was inserted into the SalI and EagI sites in pGC250 to give pGC251. PCR amplification of pGC251 was performed for 30 cycles under conditions designed to yield mutagenized products (1.5 mm MgCl2, 500 μm MnCl2, 200 μm dATP, 200 μm dCTP, 200 μm dTTP, 40 μm dGTP, 0.5 μm 5′-cgt tga tgc aat ttc tat gcg-3′, and 0.5 μm 5′-gga ggc aga caa ggt ata gg-3′). The purified PCR product was cotransformed with an EagI–SalI fragment of pGC251 into yGC137 by lithium acetate transformation. Cells with recombinant plasmids were selected by plating onto SC medium without leucine. After 3 days of growth at 22°, colonies were replica plated onto SC medium containing 5-FOA to select against cells carrying the wild-type SMC5 gene on the URA3 plasmid. After 4 days of growth at 22° or 36°, ∼10,000 transformants were visually screened for colonies able to grow at 22° but not at 36°. Multiple candidate strains were retested for conditional growth on SC 5-FOA and YPAD plates. Plasmid DNA was rescued from 21 strains and retransformed into yGC137. Leu+ transformants were grown on SC 5-FOA and retested for conditional growth at 22°, 30°, 32°, 34°, and 36°. Temperature-sensitive strains were assayed for growth on plates containing 0.005% methyl methanesulfonate. Of the 21 alleles, smc5-31 and smc5-33 (yGC231 and yGC233) were chosen for further analysis on the basis of their robust growth at 22° and 30°, the completeness of their temperature sensitivity at 36°, and their differential response to DNA damage. A wild-type control strain was generated by transformation of pGC251 into yGC137 and selection of transformants on SC −leu and then on SC 5-FOA to yield yGC141. Sequencing of the mutagenized alleles revealed the presence of the following mutations: smc5-31, K327I, K445T, A499V, H706L, K738R, A808V, and V1028D; and smc5-33, P39L, L253F, F331S, N338S, E351G, I374N, M471T, Y507C, T509A, and R712C.
TABLE 2.
Yeast strain genotypes
| Straina | Genotype |
|---|---|
| 100 | MATahis4 HIS3 LEU2 LYS2 MET15 URA3 SMC5 |
| 101 | MATα his4 HIS3 LEU2 LYS2 MET15 URA3 SMC5 |
| 131 | MATamet15Δ0 LYS2 smc5∷kanMX4 |
| MATα MET15 lys2Δ0 SMC5 [pGC233-URA3 SMC5] | |
| 135 | MATα [pGC233-URA3 SMC5] MET15 |
| 137 | MATa [pGC233-URA3 SMC5] |
| 141 | MATa [pGC251-LEU2 SMC5] |
| 231 | MATa [pGC251-LEU2 smc5-31] |
| 233 | MATa [pGC251-LEU2 smc5-33] |
| 159 | MATarad52∷kanMX4 SMC5 LYS2 |
| 160 | MATα rad52∷kanMX4 SMC5 MET15 |
| 177 | MATα rad52∷kanMX4 [pGC251-LEU2 SMC5] |
| 182 | MATα rad52∷kanMX4 [pGC251-LEU2 smc5-31] |
| 185 | MATα rad52∷kanMX4 [pGC251-LEU2 smc5-33] |
| 104 | MATamet15Δ0 LYS2 SMC5 |
| MATα MET15 lys2Δ0 SMC5 | |
| 170 | MATα MET15 |
| MATamet15Δ0 [pGC251-LEU2 SMC5] | |
| 172 | MATα MET15 |
| MATamet15Δ0 [pGC251-LEU2 smc5-31] | |
| 173 | MATα MET15 |
| MATamet15Δ0 [pGC251-LEU2 smc5-33] | |
| 187 | MATamet15Δ0 rad52∷kanMX4 LYS2 SMC5 |
| MATα MET15 rad52∷kanMX4 lys2Δ0 SMC5 | |
| 188 | MATaMET15 rad52∷kanMX4 [pGC251-LEU2 SMC5] |
| MATα met15Δ0 rad52∷kanMX4 [pGC251-LEU2 SMC5] | |
| 189 | MATaET15 rad52∷kanMX4 pGC251-LEU2 smc5-31] |
| MATα met15Δ0 rad52∷kanMX4 pGC251-LEU2 smc5-31] | |
| 190 | MATaET15 rad52∷kanMX4 [pGC251-LEU2 smc5-33] |
| MATα met15Δ0 rad52∷kanMX4 [pGC251-LEU2 smc5-33] | |
| 250 | MATα ade2Δ∷hisG his3 ARG1 [pGC251-LEU2 SMC5] |
| MATaADE2 his3Δ1 arg1∷MET15 | |
| 251 | MATα ade2Δ∷hisG his3 ARG1 [pGC251-LEU2 smc5-31] |
| MATaADE2 his3Δ1 arg1∷MET15 | |
| 252 | MATα ade2Δ∷hisG his3 ARG1 [pGC251-LEU2 smc5-33] |
| MATaADE2 his3Δ1 Arg1∷MET15 | |
| 280 | MATα ade2Δ∷hisG his3 ARG1 rad52Δ∷HIS3 [pGC251 SMC5] |
| MATaADE2 his3Δ1 arg1∷MET15 rad52Δ∷URA3 | |
| 281 | MATα ade2Δ∷hisG his3 ARG1 rad52Δ∷URA3 [pGC251 smc5-31] |
| MATaADE2 his3Δ1 arg1∷MET15 rad52Δ∷URA3 | |
| 282 | MATα ade2Δ∷hisG his3 ARG1 rad52Δ∷URA3 [pGC251 smc5-33] |
| MATaADE2 his3Δ1 arg1∷MET15 rad52Δ∷URA3 | |
| 301 | MATamms21∷pRS303 MMS21 [pGC233-URA3 SMC5] [pGC251 LEU2 SMC5] |
| 302 | MATamms21∷pRS303 MMS21 [pGC233-URA3 SMC5] [pGC251 LEU2 smc5-31] |
| 303 | MATamms21∷pRS303 MMS21 [pGC233-URA3 SMC5] [pGC251 LEU2 smc5-33] |
| 304 | MATamms21∷pRS303 mms21-SA [pGC233-URA3 SMC5] [pGC251 LEU2 SMC5] |
| 305 | MATamms21∷pRS303 mms21-SA [pGC233-URA3 SMC5] [pGC251 LEU2 smc5-31] |
| 306 | MATamms21∷pRS303 mms21-SA [pGC233-URA3 SMC5] [pGC251 LEU2 smc5-33] |
Unless otherwise noted, all strains are his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 smc5Δ∷kanMX4. Strains listed as his3 are either his3Δ1 or his3Δ200.
Strains heterozygous for both rad52Δ0 and smc5Δ0 were selected by streaking cells from mixtures of yGC141, yGC231, and yGC233 with yGC160 onto SC −leu −met plates. These heterozygous diploids were sporulated and DNA from both G418-resistant spores from 2:0 G418r:G418s-segregating tetrads was analyzed by Southern blot to confirm deletion of both SMC5 and RAD52 in the spore. This resulted in the isolation of yGC177, yGC182, and yGC185. Diploids doubly homozygous for rad52Δ0 and smc5Δ0 were generated by micromanipulation of schmoos formed from these strains and complementary MATα cells. yGC159 and yGC160 were crossed to create yGC187. RAD52 diploid strains were created by crossing yGC141, yGC231, and yGC233 with yGC135 and selecting on SC −ura −leu −met, followed by growth on SC 5-FOA. This cross yielded yGC170, yGC172, and yGC173. As diploid smc5-31 and smc5-33 strains proved to be genetically unstable, they were used for analysis and frozen as glycerol stocks immediately after generation.
ARG1 was replaced by MET15 in yGC141, yGC231, and yGC233 by integrative transformation and met+ transformants were screened for arginine auxotrophy; replacement of ARG1 in Met+ Arg− cells was confirmed by Southern blot. The ade2 and arg1∷MET15 markers were combined by mating to generate yGC250, yGC251, and yGC252. rad52 versions of these strains were made by disruption of RAD52 with either HIS3 or URA3 in the precursor haploids, followed by mating to create yGC280, yGC281, and yGC282. Disruption of RAD52 was confirmed by PCR.
Growth and viability assays:
Cells were grown in YPAD to saturation at 22°. The cultures were diluted in duplicate with fresh YPAD to OD600 = 0.1 and permitted to recover from stationary phase at 22° for 1 hr. Experimental cultures were then shifted to 36°; reference cultures were maintained at 22°. The cells were maintained in log phase throughout the experiment by dilution of the cultures with prewarmed YPAD as appropriate. At time 0 and every 2 hr thereafter, an aliquot of cells was removed, vortexed, and ∼1000 cells were plated onto 22° YPAD plates.
For extended viability experiments, dilutions of log-phase cells were plated onto prewarmed YPAD plates and incubated at 36° for the indicated number of days before return to 22°. Surviving cells were counted after 3 days of growth at 22°. The plating efficiency of yGC141, yGC231, and yGC233 cells is the same, both on YPAD and on SC medium (data not shown).
Fluorescence-activated cell sorting:
Cells were prepared for fluorescence-activated cell sorting (FACS) analysis as described (Nash et al. 1988) and analyzed on a Beckman–Coulter EPICS XL cytometer. Twenty-five thousand cells were assayed per sample.
DNA damage assays:
Early log-phase cells (OD600 < 0.4) were either 5-fold or 10-fold serially diluted, deposited on YPAD plates containing the drug indicated, and grown at 22° for 3 days. Methyl methanesulfonate (MMS), hydroxyurea (HU), and phleomycin [a bleomycin derivative, Sigma (St. Louis) P-9564] were mixed with molten 55° YPAD agar at the concentrations indicated. As the half-life of MMS in plates is very short, YPAD–MMS plates were always used exactly at 24 hr after they were poured. UV-C light was from a Mineralight UV S-11 source, and the intensity was quantitated using a UVX radiometer. For analysis of Rad53p phosphorylation, cells were grown in liquid culture at 22° to midlog phase, MMS was added to 0.03% or HU was added to 25 mm, they were grown for another 3 hr at 22°, and then samples were analyzed as described below.
Checkpoint activation:
Log-phase cultures were made 10% trichloroacetic acid and pelleted. The fixed cells were neutralized in 1 ml 1 m Tris pH 7.5, resuspended in 2× Laemmli buffer, 200 mg glass beads were added, and then they were vortexed six times for 30 sec. Lysates were electrophoresed on 8% acrylamide gels, and the gels were equilibrated and then transferred in Bjerrum buffer to PVDF membranes using a Bio-Rad (Hercules, CA) semidry transfer apparatus. Bjerrum buffer is (per liter): 5.8 g Tris, 2.9 g glycine, 375 mg SDS, and 200 ml methanol. Rad53p was detected with the yC-19 antibody (Santa Cruz Biochemicals) at a 1:500 dilution. In situ Rad53p kinase assays were performed according to Pellicioli et al. (1999).
Loss of heterozygosity assays:
Diploid cells were grown on SC −met plates, grown overnight at 22° in YPAD to saturation, and then diluted to OD600 = 0.1 in fresh YPAD. While the cells were in early log phase, the cultures were either shifted to 30° or left at 22°. After 3–4 hr (more than one full cell cycle at the assay temperature), the culture was diluted and plated onto lead plates. Lead plates contained 3 g peptone, 5 g yeast extract, 200 mg (NH4)2SO4 and 40 g glucose, 20 g Difco (Detroit) (low phosphate) agar, and 0.7 g/liter Pb(NO3)2 per liter (Brachmann et al. 1998; McMurray and Gottschling 2003). After 5–7 days of growth on lead plates, cells were visually assayed for brown sectoring indicative of the loss of the MET15 gene. In a MET15/met15Δ0 diploid, loss of MET15 will result in a brown sector on lead plates, whereas cells that have lost met15Δ0 will still be white and therefore not counted by this assay. The reported loss of heterozygosity (LOH) rates were multiplied by two to reflect this fact. To calculate the LOH rate, the number of sectored colonies was taken as the numerator and the total number of cell divisions analyzed as the denominator. If the colony was half sectored, one cell division was considered assayed; if quarter sectored, two divisions were considered assayed. No attempt was made to analyze eighth sectors, as the high LOH rate prevented reliable counting of them in smc5 strains. As LOH events that occur prior to plating will yield either pure white MET15 or pure brown met15Δ0 colonies, these colonies were subtracted from the denominator for strains smc5-31 and smc5-33. The relatively low LOH rate of BY4743, yGC170, and the chromosome XV MET15 insertion made it impossible to count the number of pure white colonies. For these strains the number of pure brown colonies was multiplied by two and subtracted from the denominator. LOH assays were done in duplicate, with 800–7000 cell divisions assayed per strain. LOH rates were calculated independently on the basis of the number of half-sectored colonies and on the basis of the number of quarter-sectored colonies; the resultant rates were quite similar and were averaged to give the reported rate.
For LOH measurement with yGC250, yGC251, yGC252, yGC280, yGC281, and yGC282, cells were grown in SD +his +lys +ura (−met −ade −arg) medium and then diluted into YPAD, grown to early log, and shifted to 30° for 4 hr before plating. For analysis of LOH at arg1Δ∷MET15, cells were plated on lead medium and black sectors were counted, picked, restruck on lead plates, and then assayed for the ADE2 color marker by streaking on YPD. The presence of any red colonies was scored as positive for chromosome loss. For analysis of LOH at ADE2, cells were plated on SC medium with one-quarter the normal concentration of adenine and red sectors were counted, picked, restruck on YPD plates, and then assayed for the MET15 color marker by streaking on lead plates. The presence of any black colonies was scored as positive for chromosome loss. The large majority of black sectors scored as red were uniformly red; all red sectors scored as black were uniformly black. Between 5000 and 30,000 colonies were screened per strain for LOH rate measurement with these strains.
Diploid mating rate measurement:
Measurement of the LOH rate at the MAT loci was performed using the method of the median (Lea and Coulson 1949). Strains were grown in YPAD at 22° to late-log phase and then diluted and split into five separate cultures. Log-phase cells from each culture were mixed with a 10-fold excess of yGC100 or yGC101, pelleted at 1000 × g, the medium was aspirated, and they were incubated at 22° for 6 hr. Cells were then resuspended in water, and dilutions plated onto SD medium. Colony formation was assayed after 5 days of growth on SD medium. Control experiments indicated that the mating efficiency in such experiments was ∼33%. The mating rate with yGC100 and yGC101 was averaged to give the reported value. The simple mean of the mating rate was very close to the rate determined by fluctuation analysis (see Figure 5 legend).
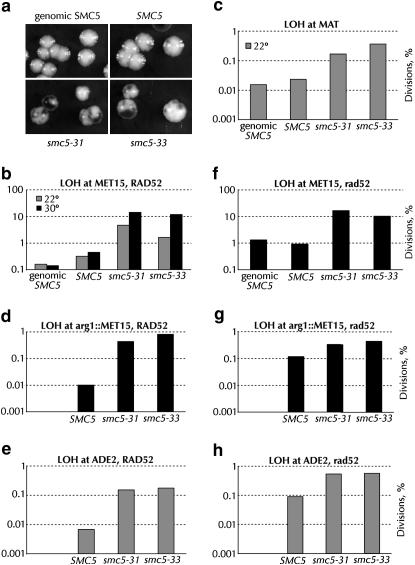
Figure 5.
Global, Rad52p-independent loss of heterozygosity in smc5 strains. All assays were performed at 30° unless noted. (a) Appearance of SMC5/SMC5 (BY4743), smc5Δ0/smc5Δ0 pGC251-SMC5 (yGC170), smc5Δ0/smc5Δ0 pGC251-smc5-31 (yGC172), and smc5Δ0/smc5Δ0 pGC251-smc5-33 (yGC173) colonies when plated on medium containing lead ions. (b) Percentage of cell divisions from the strains in a experiencing MET15 LOH at 22° and at 30°. In multiple experiments, 1000–5000 cell divisions were assayed per strain per temperature. For comparisons between wild type and smc5 at both temperatures, χ2-analysis returned values >50. The LOH assay is the only one we have performed to find any difference between episomal and genomic SMC5 (χ2 = 23). (c) LOH at 22° at the MAT loci on chromosome III in BY4743, yGC170, yGC172, and yGC173 cells. The rate of LOH at the MAT loci in wild-type cells is comparable to that found by Spencer and Hieter (Spencer et al. 1990). The simple mean of the data gave rates of 0.011, 0.015, 0.12, and 0.28% for BY4743, yGC170, yGC172, and yGC173, respectively. (d) LOH rates at 30° of rad52Δ0 versions of the strains in a (yGC187, yGC188, yGC189, and yGC190). χ2-analysis gave values >1000 for comparisons between wild type and smc5. The difference between yGC187 and yGC188 is not significant. (e) LOH in the arg1∷MET15 strains yGC250, yGC251, and yGC252. From 5000 to 33,000 cell divisions were analyzed. (f) LOH at arg1∷MET15 in rad52 cells. From 4000 to 20,000 cell divisions were analyzed. (g) LOH at ADE2 in rad52 cells. From 14,000 to 40,000 cell divisions were analyzed. (h) LOH at ADE2 in rad52 cells. From 8000 to 22,000 cell divisions were analyzed.
Overexpression of non-SMC subunits:
The NSE5, KRE29, NSE1, MMS21, NSE3, and NSE4 genes were cloned by PCR amplification of genomic DNA using the following oligonucleotides: NSE5, 5′-gaa ggg atc cgt aac cat gga tgg tgc gtt gat aaa ttc-3′ and 5′-tat gat ctc gag cct tgg cca tac ata cat tgc-3′; KRE29, 5′-gaa ggg atc cgt aac cat ggg aag cgt gaa ctc atc a-3′ and 5′-tat gat ctc gag tct att gcc ata aag gaa atg g-3′; NSE1, 5′-gaa ggg atc cgt aac cat gga ggt aca tga aga gca g-3′ and 5′-taa tca ctc gag cgt gtt caa tca cat gcc gtt t-3′; MMS21, 5′-gaa ggg atc cgt aac cat ggc ctt gaa cga taa tcc tat-3′ and 5′-tat gat ctc gag atg tgg tta tat acc aac ctt ttg-3′; NSE3, 5′-gaa ggg atc cgt aac cat gag ttc tat aga taa tga cag c-3′ and 5′-tat gat ctc gag gta acc cgt tta agt cca gtc t-3′; and NSE4, 5′-gaa ggg atc cgt aac cat gtc tag tac agt aat atc tag aa-3′ and 5′-tat gat ctc gag tcc aat tag gca atg gcc att g-3′. All amplified regions were sequenced in their entirety. All PCR products were Topo-TA cloned (Invitrogen, San Diego), and BamHI–XhoI fragments were inserted into BamHI–XhoI cut pJES19, a 2μ galactose-inducible URA3-marked vector (Stray and Lindsley 2003). Plasmids were introduced into BY4741, yGC141, yGC231, and yGC233 and transformants selected on SC −ura. Three colonies from each transformation were assayed for growth on SC −ura containing 2% dextrose or 2% galactose at 22° or 36°. All three transformants behaved identically in two different experiments; for clarity, only one transformant per strain is shown.
The mms21-SA presumptive SUMO ligase inactive version of Mms21p was made by making C200S and H202A mutations by site-directed mutagenesis. This allele is orthologous to the S. pombe nse2.SA allele (Andrews et al. 2005). To make mms21-SA strains, a fragment of MMS21 or mms21-SA was inserted into pRS303, linearized, and integrated into the MMS21 locus of yGC137. Correct integration was confirmed by PCR, and the strains were transformed with the SMC5, smc5-31, or smc5-33 plasmids.
RESULTS
Generation of temperature-sensitive alleles of SMC5:
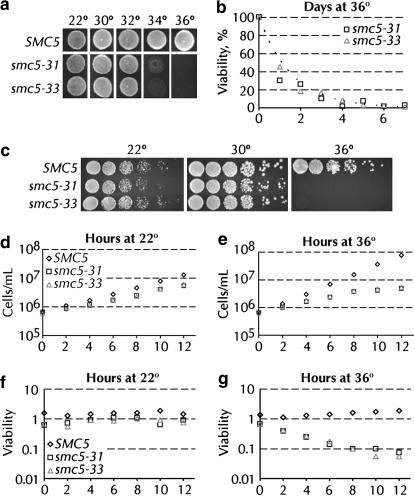
Temperature-sensitive alleles of SMC5 were obtained via plasmid shuffle between wild-type and mutagenized SMC5 genes covering a chromosomal deletion of SMC5. A population of PCR-mutagenized SMC5 genes was cotransformed with an episomal vector carrying SMC5 linearized to delete the coding sequence of the gene. After loss of the wild-type plasmid and screening for low temperature-dependent growth, 21 candidate alleles were obtained and assayed for growth at a variety of temperatures and on various media. Two alleles of SMC5 (smc5-31 and smc5-33) were chosen for further analysis on the basis of their near wild-type growth at 22° and 30°, the completeness of their temperature sensitivity at 36°, and their differential response to DNA damage. Gross observation of smc5-31 and smc5-33 cells grown at various temperatures revealed robust growth at temperatures up to 32°, but almost no growth at 36° (Figure 1, a and c). A SMC5 strain (yGC137) was transformed with SMC5, smc5-31, or smc5-33. These strains all grew equally well at 36° under selection for maintenance of both the wild-type and mutant plasmids, thus demonstrating that both smc5-31 and smc5-33 are recessive alleles (data not shown). The smc5-31 and smc5-33 alleles phenotypically revert approximately once per 5 × 108 and <1 × 108 cell divisions, respectively. The growth in liquid culture of SMC5, smc5-31, and smc5-33 strains was assayed quantitatively over a 12-hr period at 22° and 36°. At 22°, smc5-31 and smc5-33 cells grew slightly slower than wild type (Figure 1d). When shifted to 36°, the growth rates of these cultures slowly declined. At 10-hr postshift, cell division in the culture had largely ceased (Figure 1e). There were no significant differences in the viability of SMC5, smc5-31, and smc5-33 cells grown at 22° (Figure 1f). At 36°, however, the viability of SMC5 cells remained constant, whereas the viability of smc5 cells gradually decreased, reaching a low of 5% after 12 hr at 36° (Figure 1g). This decrease corresponds to a loss of approximately one-third of the culture's viability per cell division at 36°. Doubling the DNA content of smc5 cells roughly doubled the smc5-dependent increase in doubling time. At 22°, diploid smc5 cells grow slightly faster than haploids. At the semipermissive temperature of 30°, the increase in doubling times is 2.2- and 1.9-fold greater for smc5-31 and smc5-33 diploid cells, respectively, than for haploid smc5-31 and smc5-33 cells.
Figure 1.
Characterization of temperature-sensitive alleles of SMC5. (a) Thirty-five thousand cells from log-phase cultures were deposited onto YPAD plates prewarmed to the indicated temperature and grown at that temperature for 2 days. (b) Long-term viability of smc5-31 (yGC231) and smc5-33 (yGC233) cells at 36°. The dashed line is an exponential curve fit to the data. The half-life of both strains at 36° is 1.1 days, R2 = 0.74 and 0.97 for smc5-31 and smc5-33, respectively. (c) Twenty thousand cells and fivefold dilutions thereof were deposited onto prewarmed YPAD plates and grown at the indicated temperature for 2 days. (d and e) Liquid culture growth of SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells at 22° and 36°. (f and g) Viability of SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells grown at 22° and 36°. The experiment in d–g was repeated, and representative data are shown. The offset between viability data from liquid culture vs. that from solid medium arises from the fact that 100% of the cells plated in b are viable. The slow death of smc5 cells allows for some cell division, effectively increasing the number of cells assayed.
The slow decline of smc5-31 and smc5-33 cultures at the restrictive temperature prompted us to examine the ability of smc5 cells to survive for extended periods at 36°. Surprisingly, a small fraction of smc5-31 and smc5-33 cells remained viable after several days at 36°, growing into colonies when returned to 22° (Figure 1b). The loss of viability of cells in this experiment followed a simple exponential decay with a half-life of 1.1 days for both smc5-31 and smc5-33. Consistent with the slow loss of growth and viability in liquid culture, nonproductive microcolonies were observed on plates at 36° in this experiment. Both microcolony formation and the persistence of viable cells at 36° presumably arise from those cells that escaped the detrimental effects of Smc5p inactivation for several generations. There was no change in the steady-state level of Smc5p in wild-type and smc5 cells after 6 hr of exposure to 36° (data not shown).
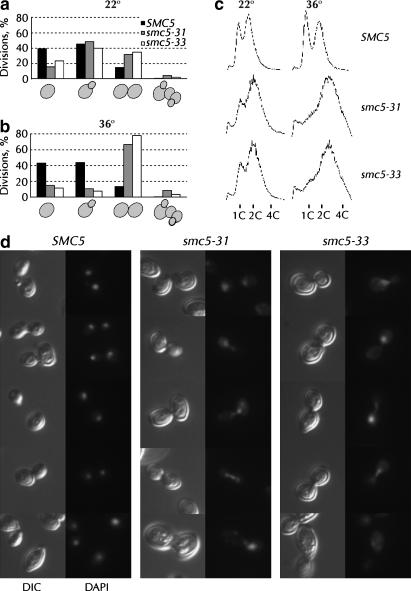
Cell-cycle profile and nuclear morphology of smc5 alleles:
The percentage of unbudded, small-budded, large-budded, or abnormal cells from the 12-hr time points at 22° and 36° in Figure 1, b and c, was determined by microscopic examination. A modest change in the steady-state cell-cycle distribution was observed at the permissive temperature with both smc5 alleles (Figure 2a). At the restrictive temperature, ∼70% of smc5-31 and smc5-33 cells accumulated as large-budded cells, consistent with a G2/M arrest (Figure 2b). Cells grown at both 22° and 36° were analyzed by FACS to monitor the progress of DNA replication. Consistent with the morphological data in Figure 2b, FACS analysis of smc5 cells after 6 hr at 36° revealed an accumulation of cells with ≥2C DNA content (Figure 2c). The DNA content of smc5 cells was slightly abnormal even at 22°. Microscopic inspection of smc5 cells grown at 36° indicated that they were much larger than SMC5 cells, indicating a block to cell division, not cell growth (Figure 2d). The DNA of these cells was stained with DAPI. In contrast to the compact, ordered nuclei of cells with wild-type SMC5, the nuclear morphology of smc5-31 and smc5-33 cells was abnormal (Figure 2d). Parts of the chromosomal DNA in smc5 cells were often pulled across the bud neck (Figure 2d, smc5-31 rows 2, 3, and 4 and smc5-33 rows 1, 3, and 4), asymmetrically segregated (Figure 2d, smc5-31 row 1), or (less frequently) fragmented and disordered (Figure 2d, smc5-33 row 5).
Figure 2.
Cell-cycle distribution and nuclear morphology of smc5 cells. The percentages of cells that were unbudded, single budded, large budded, and anomalous were assayed microscopically at 22° (a) and at 36° (b). More than 300–700 cells per strain at both time points were counted while blind with respect to the genotype and temperature. (c) Asynchronous cultures of SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells grown at 22° or at 36° for 6 hr were stained with propidium iodide and analyzed by FACS for DNA content. (d) DNA in cells from a were stained with 4′,6-diamidino-2-phenylindole (DAPI) and examined by fluorescence or differential interference contrast (DIC) microscopy.
Activation of the Rad53p DNA damage checkpoint in smc5 cells:
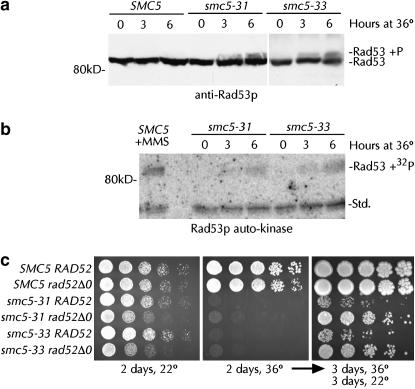
Given the accumulation of large-budded cells, the nuclear morphology of dying smc5-31 and smc5-33 cells, and the postulated involvement of the S. pombe Smc5/6 complex at the replication fork (Boddy et al. 2003; Morikawa et al. 2004; Pebernard et al. 2004), we investigated whether the DNA damage checkpoint had been activated in smc5-31 and smc5-33 cells at the restrictive temperature. Activation of the Rad53 protein via phosphorylation is common to all of the DNA damage-sensing pathways in S. cerevisiae (Melo and Toczyski 2002). Indicative of checkpoint activation, slower-migrating Rad53p phosphoforms were detected in smc5-31 and smc5-33 cells after 3 and 6 hr at 36° (Figure 3a). No Rad53p phosphorylation was observed in SMC5 cells at 22° or 36°. Phospho-Rad53p is a kinase that phosphorylates itself in addition to many proteins required for the response to DNA damage. To determine if the Rad53p phosphoforms observed in Figure 3a correspond to a genuine activation of Rad53p kinase activity and the DNA damage checkpoint, we assayed Rad53p autophosphorylation while bound to a membrane. When incubated with [γ-32P]ATP, a temperature- and time-dependent induction of Rad53p auto-kinase activity was observed in extracts from smc5 cells, demonstrating a genuine activation of the DNA-damage checkpoint (Figure 3b). The level of Rad53p activation seen in these cells after incubation at the restrictive temperature is about half of that seen when wild-type cells are treated with a high concentration of MMS (Figure 3b). We conclude that Smc5p is required for the normal maintenance and transmission of the genome. Lack of Smc5p function must result in the accumulation of the single-stranded DNA needed to activate the DNA-damage checkpoint and mediate cell-cycle arrest.
Figure 3.
DNA-damage checkpoint induction in smc5 cells and synthetic viability with rad52Δ0. (a) Lysates from SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells grown at 22° and at 36° for 0–6 hr were probed with anti-Rad53p antibody. (b) Lysates from a were assayed for Rad53p auto-kinase activity in situ after refolding on the membrane. SMC5 yGC141 cells exposed to 0.033% MMS for 2 hr served as a positive control for Rad53p activation. An unidentified lower-molecular-weight kinase (std) serves as a loading control. (c) Fifteen thousand, 3000, 600, 120, and 24 cells from log-phase SMC5 RAD52 (yGC141), SMC5 rad52Δ0 (yGC177), smc5-31 RAD52 (yGC231), smc5-31 rad52Δ0 (yGC182), smc5-33 RAD52 (yGC233), and smc5-33 rad52Δ0 (yGC185) cultures were grown on YPAD for 2 days at 22° or 36°. After 3 days at 36°, the plate was placed at 22° and allowed to grow for a further 3 days.
Amelioration of the smc5 phenotype by removal of Rad52p:
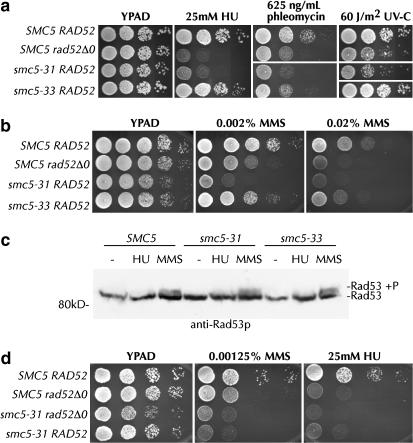
Stalled replication forks may be inappropriately processed by the recombination machinery into lethal products (Seigneur et al. 1998; Lopes et al. 2001; Cha and Kleckner 2002; Sogo et al. 2002). If Smc5p functions at the replication fork, then such toxic recombination products may be formed when Smc5p is incapacitated and may contribute to the death of the cell. We reasoned that elimination of recombination would prevent the formation of such structures and thereby allow the growth or prevent the death of smc5-31 and smc5-33 cells at the restrictive temperature. When grown at the restrictive temperature, rad52Δ0 smc5-31 and rad52Δ0 smc5-33 cells indeed grew very slightly at 36° (Figure 3c, middle, rows 4 and 6). Upon shifting down to 22° after 3 days at 36°, 25-fold more smc5 rad52Δ0 cells were found to have survived exposure to the restrictive temperature than smc5 RAD52 cells (Figure 3c, right, compare rows 3 and 5 with rows 4 and 6). Surprisingly, at 22° deletion of RAD52 caused a synthetic sick phenotype with both smc5-31 and smc5-33 (Figure 3c, left). Quantitative measurement of growth revealed a 55% increase in the doubling time of smc5 rad52Δ0 strains relative to smc5 RAD52 strains (Table 3). Viability analysis showed no difference between smc5 RAD52 and smc5 rad52Δ0 strains (data not shown). We conclude that smc5 cells undergo Rad52p-dependent lethality at 36°, but that growth is aided by Rad52p at 22°.
TABLE 3.
Epistasis analysis of rad52Δ and smc5-31
| Doubling time (min) | Relative to SMC5 RAD52 | |
|---|---|---|
| YPAD, 22° | ||
| SMC5 RAD52 | 147 | 1 |
| SMC5 rad52Δ0 | 169 | 1.15 |
| smc5-31 RAD52 | 156 | 1.6 |
| smc5-31 rad52Δ0 | 226 | 1.55 |
| Doubling time (min) | Relative to without HU | |
| YPAD + 25 mm HU, 22° | ||
| SMC5 RAD52 | 170 | 1.16 |
| SMC5 rad52Δ0 | 247 | 1.46 |
| smc5-31 RAD52 | 231 | 1.48 |
| smc5-31 rad52Δ0 | 341 | 1.50 |
Smc5-31 but not smc5-33 cells are hypersensitive to DNA damage:
S. cerevisiae strains with mutations of SMC6, NSE1, and MMS21; S. pombe smc6, nse1, nse2, and nse3 mutants; and human cells depleted for hMms21 exhibit defects in their response to DNA damage (Prakash and Prakash 1977; Lehmann et al. 1995; Verkade et al. 1999; Fujioka et al. 2002; McDonald et al. 2003; Harvey et al. 2004; Morikawa et al. 2004; Onoda et al. 2004; Pebernard et al. 2004; Hu et al. 2005; Potts and Yu 2005; Torres-Rosell et al. 2005b). Alleles of S. pombe smc6 and nse2 have been isolated that support the normal growth of cells, but are sensitive to DNA-damaging agents, suggesting the existence of more than one function for the S. pombe Smc5/6 complex (Fousteri and Lehmann 2000; Andrews et al. 2005). SMC5 is an essential gene in yeast. As the growth rates of smc5-31 and smc5-33 cells are almost identical, the two alleles must have the same ability to complement the essential role of Smc5p. Smc5-31 and smc5-33 are differently sensitive to DNA damage, however. The smc5-31 allele is hypersensitive to even very low doses of HU, phleomycin, and UV-C (Figure 4a). Notably, smc5-31 is about as sensitive as a rad52Δ0 strain to a variety of MMS concentrations and to 25 mm HU (Figure 4b, Table 3). In sharp contrast, smc5-33 strains are only slightly more sensitive than wild type to phleomycin and MMS (Figure 4, a and b). No difference in DNA-damage sensitivity was observed between yGC141 cells with episomal SMC5 and cells with a chromosomal copy of SMC5 (data not shown). We conclude that S. cerevisiae Smc5p, like S. pombe Smc6 and Nse2, has one essential function and a second nonessential role in DNA repair.
Figure 4.
The DNA-damage response is different in smc5-31 and smc5-33. (a) Ten thousand, 1000, 100, and 10 cells from log-phase SMC5 RAD52 (yGC141), SMC5 rad52Δ0 (yGC177), smc5-31 RAD52 (yGC231), and smc5-33 RAD52 (yGC233) cultures were plated onto YPAD medium containing the indicated concentrations of drug or exposed to UV-C at 60 J/m2 and allowed to grow for 3 days at 22°. (b) One-hundred-twenty thousand, 12,000, 1200, 120, and 12 cells were plated on YPAD medium containing the indicated concentration of MMS. Similar results were obtained with plates containing 0.04% MMS, but growth of even wild-type strains on this medium is poor. (c) Lysates from SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells grown for 3 hr at 22° in YPAD, YPAD + 25 mm HU, or YPAD + 0.03% MMS were probed with anti-Rad53p antibody. (d) As in a, but with SMC5 RAD52 (yGC141), SMC5 rad52Δ0 (yGC177), smc5-31 rad52Δ0 (yGC182), and smc5-31 RAD52 (yGC231). The above assays were performed multiple times.
Smc5-31 cells may be hypersensitive to DNA-damaging agents because they are unable to sense DNA damage or because they are unable to effect DNA repair. Activation of the Rad53 protein kinase is an event that occurs after DNA damage has been detected, but before it is repaired. To distinguish between a failure to sense and a failure to repair exogenous DNA damage in smc5-31 cells, we assayed the activation of Rad53p in cells exposed to 0.03% MMS and 25 mm HU for 3 hr at 22°. Wild-type, smc5-31, and smc5-33 cells all produce approximately equal levels of phosphorylated Rad53p in response to MMS exposure (Figure 4c). Longer exposures of this gel indicated lower but equal levels of HU-induced Rad53p phosphorylation across all three strains (data not shown). We conclude that the hypersensitivity of smc5-31 cells is caused by a failure downstream from the detection of DNA damage and checkpoint activation.
DNA damage is repaired by four main pathways: recombinational repair, nucleotide or base excision repair, and nonhomologous end joining. All recombination in S. cerevisiae is thought to be dependent on the Rad52 protein (Symington 2002). We found that smc5-31 was epistatic with rad52Δ0 with respect to DNA damage sensitivity (Figure 4d). Measurement of the doubling time of these strains confirmed this observation, as the HU-dependent increase in doubling time was nearly the same in rad52Δ0, smc5-31, and rad52Δ0 smc5-31 cells (Table 3).
Rad52p-independent chromosome loss and fragmentation in smc5 cells:
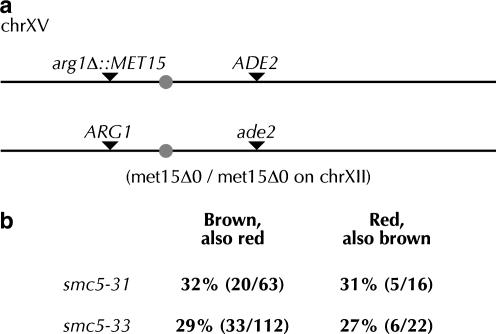
S. pombe rad18 alleles have an unusually high rate of minichromosome loss (Verkade et al. 1999); nse4 alleles in S. cerevisiae lose plasmids at elevated rates (Hu et al. 2005). To understand the consequences of loss of Smc5p function on the transmission of normal chromosomes, we examined the rate of LOH at the MET15 locus in smc5-31 and smc5-33 strains. When deposited on plates containing Pb2+ ions, met15 cells turn dark brown because hydrosulfide ions leaking from the cell react with Pb2+ to form a brown lead sulfide precipitate (Ono et al. 1991; Cost and Boeke 1996). In a MET15/met15 heterozygous diploid, the rate of loss of MET15 can be scored as the frequency of half-white, half-brown colonies. Quantitation of the rate of LOH at both 22° and 30° revealed substantial, temperature-dependent LOH increases in smc5-31 and smc5-33 cells (Figure 5b). At 30°, both smc5-31 and smc5-33 cells lost heterozygosity ∼30-fold more often than yGC170 wild-type cells and ∼100-fold more often than cells with chromosomal SMC5.
We investigated whether LOH in smc5 cells was specific for the MET15 locus or was a more general phenomenon. When diploid cells lose a MAT locus, they become competent to mate. If loss of MAT in diploids is random, then half of the cells that have lost a MAT locus should be functionally a and half functionally α. We measured the mating rate of diploid smc5-31 and smc5-33 cells with MATa and MATα tester strains to form prototrophic triploid cells. Smc5-31 and smc5-33 cells mated to both MATa and MATα cells equally well at rates ∼10-fold greater than those of wild-type cells (Figure 5c). Furthermore, we measured LOH rates in strains heterozygous for the MET15 gene inserted into ARG1 on the left arm of chromosome XV and in strains heterozygous for the ADE2 gene in its normal location on the right arm of the same chromosome (ade2 cells are red). Loss of these markers was elevated from 22- to 81-fold in smc5 cells (Figure 5, d and e). We therefore conclude that a reduction in Smc5p function causes a global chromosomal instability.
Several molecular mechanisms could be responsible for the LOH observed in smc5-31 and smc5-33 strains. Gene conversion, break-induced replication, internal deletion (such as between Ty1 or δ elements), chromosome loss, or chromosome fragmentation could all result in the conversion of MET15 to met15. The first three processes listed require homologous recombination. To determine if smc5-dependent LOH was recombinational, we constructed smc5 diploids homozygous for rad52. At MET15 on chromosome XII, LOH rates in smc5 rad52 cells were about the same as those in smc5 RAD52 cells (Figure 5f). LOH in SMC5 cells increased ∼10-fold in the absence of Rad52p (Figure 5f).
MET15 is distal to the rDNA loci on chromosome XII. The Smc5/6 complex is known to be enriched at the rDNA (Torres-Rosell et al. 2005b). DNA damage repaired by recombination in the rDNA is known to occur at a relatively high rate and is likely responsible for most of the observed MET15 LOH. Recombination in the rDNA can occur via a conventional double-strand-break repair mechanism, but single-strand annealing can also occur because the rDNA consists of tandem repeats. While the consensus is that single-strand annealing is Rad52p dependent (Symington 2002), there is a report of Rad52p-independent single-strand annealing at this locus (Ozenberger and Roeder 1991). To confirm that LOH in smc5 strains is Rad52p independent, we also measured LOH rates at the arg1Δ∷MET15 insertion and at the ADE2 locus in rad52 cells. In smc5 cells without Rad52p, LOH at arg1Δ∷MET15 decreased only slightly; LOH at ADE2 increased slightly (Figure 5, g and h). In contrast, LOH in SMC5 rad52Δ cells increased ∼10-fold (Figure 5, g and h). At all loci tested, LOH in smc5 cells is clearly not dependent on Rad52p and must therefore be a result of chromosome loss or chromosome fragmentation.
LOH can be a result from loss of all or part of a chromosome. To distinguish between these two mechanisms, we employed the strains heterozygous for ADE2/ade2 on the left arm of chromosome XV and heterozygous for arg1Δ∷MET15 on the right arm of chromosome XV (Figure 6a). We purified cells from brown sectors and assayed for the presence or absence of ADE2. Similarly, we purified cells from red sectors and assayed for the presence of MET15. As smc5 LOH is Rad52p independent, loss of one marker indicates chromosome fragmentation, and loss of both indicates chromosome loss. In both cases, about one-third of the cells that had lost one marker also lost the other (Figure 6b). LOH in smc5 strains therefore results mostly from chromosome fragmentation. As the expected rates of arg1Δ∷MET15 and ADE2 loss are 0.6 and 0.2%, respectively (Figure 5, d and e), loss of one marker is linked to loss of the other far more than would be expected by chance. smc5 strains therefore also suffer a high rate of chromosome loss. We conclude that Smc5p normally functions to protect the physical integrity of chromosomes and to promote their accurate segregation.
Figure 6.
Loss of heterozygosity in smc5 cells is a result of chromosome fragmentation and loss. (a) Diagram of colorimetric markers on chromosome XV in yGC250, -251, and -252 and yGC280, -281, and -282. The diagram is to scale. (b) Frequency of loss of both the MET15 and ADE2 markers in yGC251 and -252.
Mms21p overproduction suppresses smc5 temperature sensitivity but not the DNA damage sensitivity of smc5-31 cells:
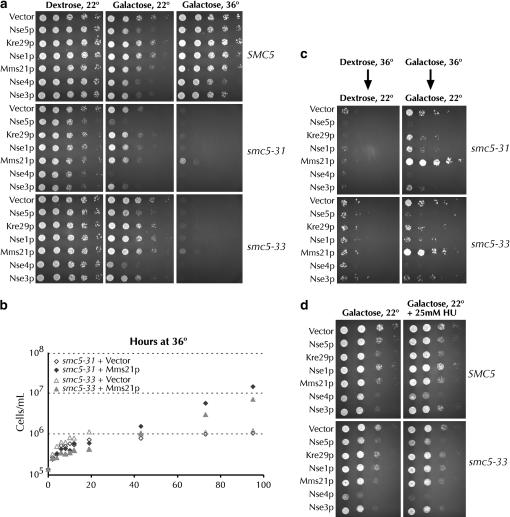
We investigated whether overexpression of the NSE components of the Smc5/6 complexes could alleviate the temperature sensitivity of smc5-31 and smc5-33 strains. When cells with galactose-inducible versions of NSE1, MMS21, NSE3, NSE4, NSE5, and KRE29 were plated on galactose, Mms21p overproduction was found to somewhat suppress the temperature sensitivity of both the smc5-31 and the smc5-33 alleles (Figure 7a). No suppression was seen when the cells were grown on dextrose at 36° (data not shown). When analyzed in liquid galactose medium at 36°, strains overproducing Mms21p initially grew slightly more slowly than the control, but ultimately outgrew control strains with kinetics similar to those seen on solid medium (Figure 7b). To ensure that growth of smc5 cells at the nonpermissive temperature was dependent on overexpression of MMS21 and not due to spontaneous or Mms21p-promoted reversion of the smc5 alleles, we obtained 10 independent isolates of both smc5-31 and smc5-33 strains with the GAL-MMS21 plasmid at 22°. These 20 strains were verified as temperature sensitive when grown on dextrose-containing medium at 36°. As expected, the cells grew at 36° when plated on galactose-containing medium. After 3 days of growth at 36°, the cells were transferred to 5-FOA medium at 22° to select for loss of the MMS21 plasmid. The cells were then reassayed for growth on galactose medium at 36°. All 20 strains regained their temperature sensitivity upon loss of the MMS21 plasmid, demonstrating that Mms21p overproduction was responsible for their growth at 36°. In addition to allowing growth at the restrictive temperature, overproduction of Mms21p permitted a large number of smc5 cells to survive exposure to 36° (Figure 7c). Overexpression of Nse5p, Nse3p, and Nse4p was actually toxic to wild-type cells and to cells with either smc5 allele (Figure 7, a and c). While overexpression of Mms21p suppressed the temperature-dependent death of smc5-31 cells, it neither alleviated nor exacerbated smc5-31 cells' HU sensitivity (data not shown). In contrast, the toxicity of Nse4p overexpression to SMC5 or smc5-33 cells was enhanced when these cells were grown on medium containing 25 mm HU (Figure 7d).
Figure 7.
Overexpression of MMS21 suppresses the temperature sensitivity of smc5 cells. (a) SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells were transformed with either pRS316 or a 2μ vector with the indicated gene driven by the GAL1 promoter. Fifteen thousand, 3000, 600, 120, and 24 cells were plated on prewarmed SC–ura–leu medium containing 2% dextrose or 2% galactose as indicated. Cells were allowed to grow for 3.5 days. Equal growth was seen on dextrose at 36° (data not shown). Transformants of strains with an intact chromosomal SMC5 locus performed identically to yGC141 transformants under all conditions in these assays (data not shown). Three transformants per strain were tested and the experiment was performed twice. Overexpression of the relevant protein was assayed by Coomassie stain (data not shown). (b) Mms21p suppression of smc5 in liquid culture. smc5-31 and smc5-33 strains with either pRS316 or GAL1-inducible MMS21 were grown in dextrose until midlog and then shifted to 36° SC–ura Gal medium. The doubling times of smc5-31 + Mms21p and smc5-33 + Mms21p strains are 16.3 and 17.7 hr, respectively. (c) Plates from a were grown at 22° for 3 days following 3.5 days of growth at 36°. (d) Overexpression of Nse4p causes DNA damage hypersensitivity. This was done as in a, except dilutions were spotted on SC–ura–leu plates with 25 mm hydroxyurea.
Suppression of smc5 by Mms21p requires Mms21p's SUMO ligase activity:
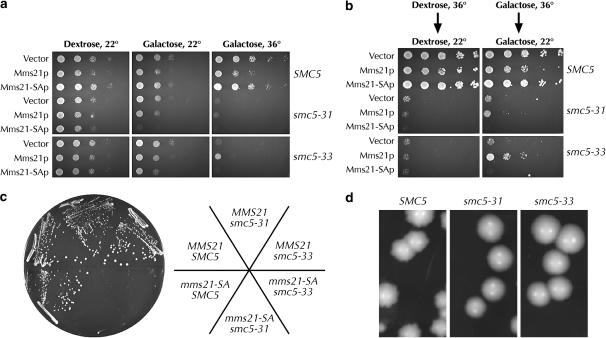
Mms21p has an essential role in the cell and is also a SUMO E3 ligase (Andrews et al. 2005; Zhao and Blobel 2005). To determine whether Mms21p-dependent suppression of smc5 temperature sensitivity requires its SUMO ligase activity, we created an allele of MMS21 lacking the conserved cysteine and histidine residues within the E3 ligase SP-RING finger. We refer to this allele as mms21-SA, as it is exactly orthologous to the S. pombe Nse2.sa allele (Andrews et al. 2005). When Mms21-SAp expression was induced with galactose, smc5-31 cells failed to grow even at the permissive temperature (Figure 8a). Unlike wild-type Mms21p, Mms21-SAp expression was unable to suppress the temperature sensitivity of smc5-33 or to allow smc5-33 cells to survive exposure to the nonpermissive temperature (Figure 8, a and b). Cells expressing mms21-SA via its endogenous promoter in the absence of wild-type MMS21 were unable to grow at 22° in the absence of a URA3 plasmid containing wild-type SMC5 (Figure 8c). We conclude that the smc5 alleles are synthetically lethal with mms21-SA and infer that control of sumoylation is one critical function of Smc5p.
Figure 8.
mms21-SA cannot suppress smc5 and is synthetically lethal with it. (a) Suppression of smc5 temperature sensitivity by Mms21p requires the SUMO ligase activity of Mms21p. SMC5 (yGC141), smc5-31 (yGC231), and smc5-33 (yGC233) cells were transformed with either pRS316 or a 2μ vector with the indicated gene driven by the GAL1 promoter. Twenty-five thousand, 5000, 1000, 200, and 40 cells were plated. The overall level of suppression was lower than that in Figure 7a, as cells were grown for 2.5 days. (b) Mms21p promotes survival of smc5 cells at the restrictive temperature. This was done as in Figure 7c. (c) The smc5 and mms21-SA alleles are synthetically lethal. MMS21 or mms21-SA cells containing SMC5 on a URA3 plasmid and SMC5, smc5-31, or smc5-33 on a LEU2 plasmid (yGC301–306) were grown on 5-FOA medium at 22° for 5 days. This assay was done in triplicate with independent transformants. On one FOA plate there was slight growth of some smc5-33 mms21-SA cells. (d) Hypomorphism of smc5 alleles rescues the nibbled-colony morphology of “wild-type” yGC141. yGC141 (SMC5), yGC231 (smc5-31), and yGC233 (smc5-33) cells were grown for 5 days on YPAD at 22°.
The “wild-type” strain used in creating the yeast knock-out collection produces nibbled colonies when grown for several days at 22°, but not when grown at 30°. This phenotype has also been observed in sumoylation mutants, arising from overreplication of normally harmless 2μ circle DNA due to undersumoylation of 2μ replication factors (Zhao et al. 2004; Chen et al. 2005; Dobson et al. 2005). Our smc5-31 and smc5-33 alleles suppress the nibbled colony phenotype of SMC5 cells, further suggesting a misregulation of sumoylation in these cells (Figure 8d).
Nse1p contains a RING-finger domain common to ubiquitin E3 ligases (Fujioka et al. 2002). SUMO and ubiquitin modification often change protein function, sometimes in an antagonistic manner (Seeler and Dejean 2003; Papouli et al. 2005; Pfander et al. 2005). Since Mms21p's SUMO ligase activity is required for growth of smc5 cells at the restrictive temperature, and if Nse1p is a ubiquitin ligase, then co-overexpression of Nse1p with Mms21p might eliminate the thermoprotective effect of Mms21p overproduction. It did not (data not shown).
DISCUSSION
We isolated S. cerevisiae strains with alleles of SMC5 that require low temperature for viability and exploited this conditionality to examine the consequences of loss of Smc5p function. Our data imply that Smc5p has at least two separable functions in the cell, one in the cell's recombinational response to DNA damage and a second function in recombination that is essential. Removal of Smc5p by deletion of the SMC5 gene or depletion of the protein is lethal (Giaever et al. 2002; Torres-Rosell et al. 2005a). It is unclear whether our SMC5 alleles are completely null for the essential function of Smc5p at the restrictive temperature. The observation that diploid cells are more severely affected by loss of Smc5p function can be interpreted as evidence against a null allele; however, it is also conceivable that normal Smc5p function is required simply less than once per cell cycle per genome or that the presence of a homologous chromosome exacerbates the smc5 phenotype. Given the extraordinary sensitivity of the smc5-31 strain to DNA damage, we suspect that the smc5-31 allele is null or nearly so for Smc5p's function in the DNA damage response.
Smc5-31 and smc5-33 cells activate the Rad53p-dependent DNA damage checkpoint at the restrictive temperature, suggesting that lack of Smc5p function causes DNA damage or prevents the repair of spontaneous DNA damage. Similar results have recently been obtained with a temperature-sensitive allele of SMC6 and with another allele of SMC5 (Torres-Rosell et al. 2005b). Like S. pombe smc6 mutants, smc5-31 and smc5-33 cells may be defective in maintaining G2/M arrest (and perhaps repairing the damage), transiting through a lethal anaphase (Harvey et al. 2004). Alternately, irreparable DNA damage may result from the absence of functional Smc5p, such as the irreversible replication fork collapse seen in mec1 and rad53 cells (Cha and Kleckner 2002; Sogo et al. 2002). Whatever the type of DNA damage, it is clear that smc5 cells do not die at the restrictive temperature from a failure to sense it. Similarly, Rad53p phosphorylation is also induced when wild-type and smc5 cells are exposed to exogenous DNA-damaging agents. Smc5 cells appear to be generally competent at DNA damage detection and checkpoint induction. In contrast to these results, temperature-sensitive alleles of S. cerevisiae NSE4 show activation of Rad53p at both the permissive and the restrictive temperatures, perhaps reflecting a hypomorphism of these alleles (Hu et al. 2005). Like smc5-31, all four nse4 alleles confer hypersensitivity to DNA damage despite Rad53p activation (Hu et al. 2005). We do not know if the differential response of smc5-31 and smc5-33 to exogenous DNA-damaging agents reflects only a partial incapacitation of Smc5p in smc5-33 (a classic separation-of-function mutation) or whether the smc5-33 allele contains an intragenic suppressor mutation that restores DNA damage tolerance to an otherwise null-like allele. As Smc5-31p lacks a consensus sumoylation site, the former is more likely (see below).
Onoda and colleagues have isolated S. cerevisiae strains with alleles of SMC6 (smc6-56) that permit growth at 25° but not at 37° (Onoda et al. 2004). These authors measured the level of inappropriate interchromosomal recombination stimulated by DNA-damaging agents at 30° and found that it was lower in an smc6-56 strain. The inference is that Smc6p is normally involved in the recombinational repair of DNA, as the error rate of this process went down in its absence. Recombination related to endogenous levels of DNA damage was unaffected. Our observation of a synthetic-sick phenotype in rad52Δ0 smc5-31 and rad52Δ0 smc5-33 cells at 22° complements the somewhat indirect evidence obtained with the smc6-56 strain for an essential role of the S. cerevisiae Smc5/6 complex in recombination (Figure 3c). At 22°, smc5-31 and smc5-33 cells must be either hypomorphic for an essential recombination-related function that is suppressed by normal levels of Rad52p or produce elevated amounts of recombination intermediates, toxic unless resolved by homologous recombination. Conversely, the more severe defect in Smc5p function found at 36° leads to a recombination-dependent lethality (Figure 4c). The exact mechanism underlying this complex phenotype is unclear. Perhaps at the restrictive temperature Smc5p fails to associate with DNA, revealing an otherwise masked substrate for the recombination machinery. We cannot exclude the possibility that the synthetic viability of smc5 rad52 strains arises merely from the slower growth of these cells (and therefore fewer cell cycles at the restrictive temperature). As with smc6-56, we find that the DNA-damage hypersensitivity of smc5-31 is epistatic with rad52Δ0 (Figure 4b). In contrast to the smc6-56 allele, the wild-type DNA-damage response of the smc5-33 allele allows the Smc5/6 complex's role in this pathway to be unambiguously separated from its essential role in recombination.
Smc5 strains exhibit a Rad52p-independent loss of heterozygosity on at least three different chromosomes at both 22° and 30°. Analysis of LOH on chromosome XV revealed that smc5-dependent LOH is predominantly due to chromosome fragmentation, as ∼70% of the brown cells that had lost the MET15 marker retained ADE2 and remained white. We believe that our analysis may in fact underestimate the amount of LOH due to chromosome fragmentation. Although presumably centric, a remaining broken chromosome may be generally unstable due to inefficient telomere addition or because recombination or single-strand annealing with another chromosome creates an unstable dicentric molecule. Subsequent loss of this chromosome would score as complete chromosome loss, yet would have occurred via a two-step fragmentation-based pathway. We cannot be certain that the 30% of cells that have lost both markers represents cells that have lost chromosome XV in one event. Given the genetic interactions that we have found between Smc5p and Rad52p, and those between members of the Smc5/6 complex and proteins involved in DNA replication (Boddy et al. 2003; Morikawa et al. 2004; Pebernard et al. 2004; Torres-Rosell et al. 2005b), it may be that the inability of smc5-31 and smc5-33 cells to faithfully transmit their chromosomes is related to a failure of DNA metabolism and subsequent chromosome fragmentation. Alternately, one-step chromosome loss may in fact occur in a minority of cases in smc5 cells. The Nse5p subunit of the Smc5/6 complex has two-hybrid interactions with Mcm21p and Pac1p, two proteins involved in chromosome segregation (Hazbun et al. 2003). Mcm21p is part of the COMA kinetochore complex that serves as a bridge between the DNA-binding kinetochore components and those bound to microtubules and is required for the complete assembly of the outer kinetochore (Ortiz et al. 1999; De Wulf et al. 2003). Pac1p is required for the correct positioning of the mitotic spindle (Lee et al. 2003). Perhaps these interactions reflect a role for the Smc5/6 complex in chromosome transmission outside of preventing chromosome fragmentation.
Removal of Rad52p consistently increased LOH in SMC5 strains by ∼10-fold (Figure 5, c, f, and h). While there is no large increase in LOH with smc5 strains, we are somewhat hesitant to infer an epistatic relationship between SMC5 and RAD52. Seeing such an increase with MET15 on chromosome XII would not be possible, as >10% of cells are affected in RAD52 strains. Assuming smc5 LOH is in fact epistatic with rad52, it is likely to be caused by a failure between the commitment to recombination and its execution. As rad52 cells are synthetically viable with smc5 (Figure 3c) (Torres-Rosell et al. 2005b), the lethal event in smc5 cells must not be chromosome breakage or loss.
For the smc5-31 strains at 22°, the LOH rate was ∼65-fold higher at the MET15 marker on chromosome XII than at MAT on chromosome III, while the magnitude of the Smc5p-dependent defect was similar (4.7% of cell divisions vs. 0.07%; 15-fold greater than wild type at MET15 vs. ∼8-fold greater at MAT). Similar results were obtained with the smc5-33 allele and when assaying LOH at the arg1Δ∷MET15 insertion and at ADE2. The normal position of the MET15 locus distal from the rDNA repeats on chromosome XII likely accounts for this difference. This highly repetitive and fragile region of the S. cerevisiae genome is known to experience recombination-dependent LOH several times more often than other loci, to require the Rrm3p helicase to prevent chromosomal breakage and hyperrecombination caused by replication fork stalling, and to be very enriched for Smc6p (Ivessa et al. 2000; McMurray and Gottschling 2003; Torres-Rosell et al. 2005b). Significantly, the normal crescent-shaped morphology of the nucleolus is disturbed in smc6-9 and mms21-11 cells (Torres-Rosell et al. 2005b; Zhao and Blobel 2005). Missegregation of this region in smc6 cells was observed microscopically by following several fluorescently marked loci on chromosome XII (Torres-Rosell et al. 2005b). Missegregation was seen in up to 50% of cell divisions at the restrictive temperature. The lower frequency we observe in our marker-based assay likely results from the use of a semipermissive temperature (which is required to enable colony formation). In contrast, the higher sensitivity of our approach allows us to measure the effect of Smc5p's removal at loci not assayable microscopically. Although defects in Smc5p are very visible at the rDNA-distal MET15, the consequence of loss of Smc5p function is not proportionately larger for chromosome XII than for elsewhere in the genome.
Nse2 in S. pombe, Mms21p in S. cerevisiae, and hMms21 in humans are all SUMO E3 ligases that bind to Smc5p and are essential for growth (Andrews et al. 2005; Potts and Yu 2005; Sergeant et al. 2005; Zhao and Blobel 2005). Elimination of the SUMO-ligase activity by mutation of Nse2p's zinc finger or truncation of this portion of Mms21p resulted in cells that were extremely sensitive to DNA damage, yet viable. We find that overexpression of Mms21p is able to prevent death of smc5-31 and smc5-33 cells at the restrictive temperature but cannot suppress the DNA damage hypersensitivity of smc5-31 cells. The SUMO ligase activity of Mms21p is required for this suppression. While Mms21 overexpression might be a bypass suppressor of smc5, another possible model to explain these results follows. Smc5p has four possible sumoylation sites, all in the coiled-coil region [ψKX(E/D); lysines 327, 667, 733, and 919]. Mutations in Smc5-31p and Smc5-33p cluster in the coiled-coil region that binds Nse2 in S. pombe and presumably Mms21p in S. cerevisiae. At the restrictive temperature, the Smc5p-Mms21p interaction might be disfavored, leading to loss of Smc5p sumoylation. Overexpression of Mms21p may simply skew the equilibrium toward binding and therefore sumoylation of Smc5p. Significantly, Smc5-31p contains a K327I mutation and is extremely sensitive to DNA damage in a manner not suppressible by Mms21p overexpression. Elimination of Mms21p/Nse2 SUMO ligase activity yields a similarly severe DNA-damage phenotype (Andrews et al. 2005; Potts and Yu 2005; Zhao and Blobel 2005). Perhaps general sumoylation of Smc5p is required for viability and sumoylation of K327 in particular is required for DNA-damage tolerance. Overexpression of Mms21-SAp may titrate endogenous Mms21p and prevent sufficient sumoylation of Smc5-31p, but not of Smc5-33p. Consistent with this, overexpression of Mms21-SAp creates a modest sensitivity to hydroxyurea in smc5-33 cells (data not shown). If this model is true, then Mms21p SUMO ligase activity should be essential, yet it is not. In the absence of Mms21p SUMO ligase activity Smc5p sumoylation is greatly reduced but not eliminated (Zhao and Blobel 2005). It is not clear, however, if Mms21p-independent sumoylation of Smc5p occurs in MMS21 strains or whether this is an adaptation that allows cells to survive in the absence of functional Mms21p. Clearly many features of this model remain to be tested.
Why is Mms21p overproduction a relatively weak suppressor of smc5-31 and smc5-33? It may very well be that the somewhat complex phenotype of smc5-31 and smc5-33 reflects both a loss of Smc5p function and a gain of Mms21p function. The hypomorphism of the smc5 strains at 22° may result from a reduction in the affinity of Mms21p for Smc5p. At 36°, the interaction may be eliminated. Mms21p unbound from Smc5p may be free to catalyze the sumoylation of proteins not normally its substrates (e.g., 2μ replication proteins and recombination factors). Overproduction of Mms21p may therefore restore the Smc5p loss-of-function portion of the smc5-31 and smc5-33 phenotype, but exacerbate inappropriate sumoylation by Mms21p. In addition to assaying the NSE components of the Smc5/6 complex for the ability to suppress smc5-31 and smc5-33 temperature sensitivity, we have selected ∼75,000 transformants of a 2μ genomic library for this phenotype, but recovered only SMC5. If smc5-31 and smc5-33 temperature sensitivity reflects both a loss and a gain of function, it may be difficult to find a single protein capable of suppressing both.
Finally, our observation that Nse4p overproduction confers hydroxyurea sensitivity to wild-type and smc5-33 cells nicely complements the observation of a defect in the DNA damage response when Nse4p is hypomorphic (Hu et al. 2005; Zhao and Blobel 2005). Three of the four S. cerevisiae Smc5/6 complex I NSEs (all but Nse3p) have therefore been implicated in the cell's response to DNA damage.
Acknowledgments
We thank James Stray for providing expert technical advice and pJES19, Kenji Kohno for providing reagents, and members of Jasper Rine's laboratory for use of equipment and for providing strains. Hector Nolla of the University of California (UC) Berkeley FACS Facility and Steven Ruzin and Denise Schichnes of the UC Berkeley Biological Imaging Facility provided generous technical instruction. Members of the Cozzarelli and Rine labs and Kristin Robrock provided helpful suggestions and comments on the manuscript. This work was funded by grant GM31657 to N.R.C. from the National Institutes of Health (NIH). G.J.C. was the Amgen Fellow of the Damon Runyon Cancer Research Foundation (grant 1702-02) and is currently funded via a Kirschstein National Research Service Award from the NIH (F32 GM075645-01).
Note added in proof: Miyabe et al. (I. Miyabe, T. Morishita, T. Hishida, S. Yonei, H. Shinagawa, 2006, Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell Biol. 26: 343–353) recently provided evidence for the ability of the homologous recombination machinery to convert rad60-1-dependent DNA damage into a form unrecognizable by DNA damage checkpoints. Similarly, it is possible that the DNA damage sensed in smc5 cells via Rad53p activation is secondary to an initial lesion not sensed by this checkpoint.
References
- Andrews, E. A., J. Palecek, J. Sergeant, E. Taylor, A. R. Lehmann et al., 2005. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25: 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 2005. Current Protocols in Molecular Biology. Wiley, New York.
- Boddy, M. N., P. Shanahan, W. H. Mcdonald, A. Lopez-Girona, E. Noguchi et al., 2003. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 23: 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- Cha, R. S., and N. Kleckner, 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606. [DOI] [PubMed] [Google Scholar]
- Chen, X. L., A. Reindle and E. S. Johnson, 2005. Misregulation of 2 micron circle copy number in a SUMO pathway mutant. Mol. Cell. Biol. 25: 4311–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost, G. J., and J. D. Boeke, 1996. A useful colony colour phenotype associated with the yeast selectable/counter-selectable marker MET15. Yeast 12: 939–941. [DOI] [PubMed] [Google Scholar]
- De Wulf, P., A. D. Mcainsh and P. K. Sorger, 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17: 2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, M. J., A. J. Pickett, S. Velmurugan, J. B. Pinder, L. A. Barrett et al., 2005. The 2 micron plasmid causes cell death in Saccharomyces cerevisiae with a mutation in Ulp1 protease. Mol. Cell. Biol. 25: 4299–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri, M. I., and A. R. Lehmann, 2000. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 19: 1691–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka, Y., Y. Kimata, K. Nomaguchi, K. Watanabe and K. Kohno, 2002. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5–SMC6 complex involved in DNA repair. J. Biol. Chem. 277: 21585–21591. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gruber, S., C. H. Haering and K. Nasmyth, 2003. Chromosomal cohesin forms a ring. Cell 112: 765–777. [DOI] [PubMed] [Google Scholar]
- Harvey, S. H., D. M. Sheedy, A. R. Cuddihy and M. J. O'Connell, 2004. Coordination of DNA damage responses via the Smc5/Smc6 complex. Mol. Cell. Biol. 24: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun, T. R., L. Malmstrom, S. Anderson, B. J. Graczyk, B. Fox et al., 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Hirano, T., 1998. SMC protein complexes and higher-order chromosome dynamics. Curr. Opin. Cell Biol. 10: 317–322. [DOI] [PubMed] [Google Scholar]
- Hu, B., C. Liao, S. H. Millson, M. Mollapour, C. Prodromou et al., 2005. Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol. Microbiol. 55: 1735–1750. [DOI] [PubMed] [Google Scholar]
- Ivessa, A. S., J. Q. Zhou and V. A. Zakian, 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489. [DOI] [PubMed] [Google Scholar]
- Jessberger, R., 2002. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell. Biol. 3: 767–778. [DOI] [PubMed] [Google Scholar]
- Lea, D. E., and C. A. Coulson, 1949. The distribution of numbers of mutants in bacterial populations. J. Genet. 49: 264–285. [DOI] [PubMed] [Google Scholar]
- Lee, W. L., J. R. Oberle and J. A. Cooper, 2003. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, A. R., M. Walicka, D. J. Griffiths, J. M. Murray, F. Z. Watts et al., 1995. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 15: 7067–7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani et al., 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561. [DOI] [PubMed] [Google Scholar]
- McDonald, W. H., Y. Pavlova, J. R. Yates, 3rd and M. N. Boddy, 2003. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 278: 45460–45467. [DOI] [PubMed] [Google Scholar]
- McMurray, M. A., and D. E. Gottschling, 2003. An age-induced switch to a hyper-recombinational state. Science 301: 1908–1911. [DOI] [PubMed] [Google Scholar]
- Melo, J., and D. Toczyski, 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14: 237–245. [DOI] [PubMed] [Google Scholar]
- Morikawa, H., T. Morishita, S. Kawane, H. Iwasaki, A. M. Carr et al., 2004. Rad62 protein functionally and physically associates with the smc5/smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 24: 9401–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita, T., Y. Tsutsui, H. Iwasaki and H. Shinagawa, 2002. The Schizosaccharomyces pombe rad60 gene is essential for repairing double-strand DNA breaks spontaneously occurring during replication and induced by DNA-damaging agents. Mol. Cell. Biol. 22: 3537–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash, R., G. Tokiwa, S. Anand, K. Erickson and A. B. Futcher, 1988. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 7: 4335–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim, A., and B. P. Smith, 1975. Genetic control of radiation sensitivity in Schizosaccharomyces pombe. Genetics 79: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, B., N. Ishii, S. Fujino and I. Aoyama, 1991. Role of hydrosulfide ions in methylmercury resistance in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 57: 3183–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, T., A. Losada, M. Hirano, M. P. Myers, A. F. Neuwald et al., 2003. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121. [DOI] [PubMed] [Google Scholar]
- Onoda, F., M. Takeda, M. Seki, D. Maeda, J. Tajima et al., 2004. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in Saccharomyces cerevisiae. DNA Repair 3: 429–439. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank and J. Lechner, 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13: 1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger, B. A., and G. S. Roeder, 1991. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol. Cell. Biol. 11: 1222–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papouli, E., S. Chen, A. A. Davies, D. Huttner, L. Krejci et al., 2005. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19: 123–133. [DOI] [PubMed] [Google Scholar]
- Pebernard, S., W. H. McDonald, Y. Pavlova, J. R. Yates, 3rd and M. N. Boddy, 2004. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell 15: 4866–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes et al., 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18: 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander, B., G. L. Moldovan, M. Sacher, C. Hoege and S. Jentsch, 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433. [DOI] [PubMed] [Google Scholar]
- Potts, P. R., and H. Yu, 2005. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25: 7021–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash, L., and S. Prakash, 1977. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86: 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler, J. S., and A. Dejean, 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4: 690–699. [DOI] [PubMed] [Google Scholar]
- Seigneur, M., V. Bidnenko, S. D. Ehrlich and B. Michel, 1998. RuvAB acts at arrested replication forks. Cell 95: 419–430. [DOI] [PubMed] [Google Scholar]
- Sergeant, J., E. Taylor, J. Palecek, M. Fousteri, E. A. Andrews et al., 2005. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol. Cell. Biol. 25: 172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy, D. M., D. Dimitrova, J. K. Rankin, K. L. Bass, K. M. Lee et al., 2005. Brc1-mediated DNA repair and damage tolerance. Genetics 171: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo, J. M., M. Lopes and M. Foiani, 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. [DOI] [PubMed] [Google Scholar]
- Spencer, F., S. L. Gerring, C. Connelly and P. Hieter, 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray, J. E., and J. E. Lindsley, 2003. Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J. Biol. Chem. 278: 26238–26248. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A. V., E. Hogan and D. Koshland, 1995. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev. 9: 587–599. [DOI] [PubMed] [Google Scholar]
- Symington, L. S., 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66: 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell, J., F. Machin and L. Aragon, 2005. a Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle 4: 868–872. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell, J., F. Machin, S. Farmer, A. Jarmuz, T. Eydmann et al., 2005. b SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7: 412–419. [DOI] [PubMed] [Google Scholar]
- Verkade, H. M., S. J. Bugg, H. D. Lindsay, A. M. Carr and M. J. O'Connell, 1999. Rad18 is required for DNA repair and checkpoint responses in fission yeast. Mol. Biol. Cell 10: 2905–2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., and G. Blobel, 2005. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 102: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., C. Y. Wu and G. Blobel, 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167: 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]