Abstract
The identification of clones within bacterial populations is often taken as evidence for a low rate of recombination, but the validity of this inference is rarely examined. We have used statistical tests of congruence between gene trees to examine the extent and significance of recombination in six bacterial pathogens. For Neisseria meningitidis, Streptococcus pneumoniae, Streptococcus pyogenes, and Staphylococcus aureus, the congruence between the maximum likelihood trees reconstructed using seven house-keeping genes was in most cases no better than that between each tree and trees of random topology. The lack of congruence between gene trees in these four species, which include both naturally transformable and nontransformable species, is in three cases supported by high ratios of recombination to point mutation during clonal diversification (estimates of this parameter were not possible for Strep. pyogenes). In contrast, gene trees constructed for Hemophilus influenzae and pathogenic isolates of Escherichia coli showed a higher degree of congruence, suggesting lower rates of recombination. The impact of recombination therefore varies between bacterial species but in many species is sufficient to obliterate the phylogenetic signal in gene trees.
The extent and significance of recombination in bacterial species is unclear. Mechanisms that promote homologous recombination in the laboratory (transformation, transduction, or conjugation) have long been recognized and have the potential to mediate the replacement of regions of the bacterial chromosome with the corresponding regions from other members of the same or closely related species (1). The detection of high levels of linkage disequilibrium between alleles in many bacterial populations and the existence of clones or clonal complexes have led to a view that recombinational exchanges between bacterial lineages are rare in natural populations and that point mutation is the major source of the genetic variation observed within bacterial house-keeping genes (2).
In recent years this view has changed, because it has become apparent from nucleotide sequence data that recombinational exchanges are common in house-keeping genes from some bacterial species, and that the observed levels of linkage disequilibrium between alleles may sometimes be due to either ecological or geographical structuring within the population, or to poor sampling, rather than a low rate of recombination. Hence, linkage disequilibrium does not necessarily indicate a low rate of recombination, and conclusions drawn from the analysis of closely related isolates, which may reflect the recent emergence of adaptive clones, should not be extrapolated to more deep-rooted relationships within the population (3, 4).
Until recently, it was difficult to obtain empirical estimates of rates of recombination in natural populations of different bacteria. The application of multilocus sequence typing (MLST; ref. 5) for the unambiguous characterization of isolates of bacterial species is providing the sequences of ≈450-bp internal fragments of seven house-keeping genes from hundreds of isolates of several bacterial pathogens, and these data can be used to estimate the relative contributions of recombination and point mutation to clonal diversification. This approach uses the sequences to score the recombinational exchanges and point mutations that have occurred during the initial stages in the diversification of bacterial clones and thereby avoids the problems of distinguishing ancient recombinational exchanges from subsequent ones that occur when the sequences of distantly related isolates are analyzed (6–8). Because MLST focuses exclusively on house-keeping genes, the same arguments for selective neutrality of alleles that have previously been used for data from multilocus enzyme electrophoresis (MLEE; ref. 9) can also be applied to MLST data.
The significance of recombination can also be assessed through an examination of phylogenetic congruence among gene loci (10). In populations where recombination is rare, the genetic relationships inferred using the sequences of one house-keeping gene should be congruent with those obtained using other house-keeping genes, or with trees obtained using the pairwise differences between the allelic profiles assigned by MLEE or MLST. Congruent trees are therefore indicative of a relatively low contribution of recombination to clonal divergence and are likely to illustrate the true relationships between the major lineages of a bacterial species, and hence can be used as a framework to understand the evolutionary history of the species (2, 11). However, if recombination is the predominant mode of allelic change, deep phylogenies based on single gene loci will reflect the relationships between the alleles at the locus in question, which may be complex, but not the phylogenetic relationships between the isolates in which the alleles are found. We might therefore expect to observe very different gene trees for the same sample of isolates when different gene loci are examined. Because evolution is occurring by a net-like process, none of these trees can be used as a framework to understand the evolutionary history of the species (12).
In this study, we have used statistical tests to examine whether the relationships between distantly related isolates of six bacterial pathogens inferred from the sequences of one house-keeping gene show congruence with those obtained using the other house-keeping genes. In Hemophilus influenzae, and pathogenic isolates of Escherichia coli, there was some congruence between the different gene trees, indicating that recombination has not been sufficiently frequent to fully obscure the deep-rooted phylogenies in these species. However, in the remaining four species (Neisseria meningitidis, Streptococcus pneumoniae, Staphylococcus aureus, and Streptococcus pyogenes), we conclude that phylogenetic trees constructed using sequence data from one gene are in most cases no more congruent with the other gene trees than are trees of random topology. The extreme noncongruence between gene trees suggests that recombination has occurred with sufficient frequency to effectively disintegrate the “clonal frame” (13) of these species. This conclusion is supported by empirical estimates showing that alleles change 5- to 10-fold more frequently by recombination than by point mutation in N. meningitidis, Strep. pneumoniae, and Staph. aureus.
Materials and Methods
Bacterial Isolates.
N. meningitidis.
A previously described MLST data set of 107 meningococcal isolates recovered predominantly from cases of invasive disease world-wide was used in the analysis (5).
Strep. pneumoniae.
The complete pneumococcal MLST dataset (as of May, 2000) was used in the analysis. These data included the 575 isolates previously analyzed (8) and 89 additional penicillin-resistant isolates from various countries.
Staph. aureus.
The MLST dataset of 240 isolates were recovered from community-acquired invasive disease and asymptomatic carriage from the same region (Oxfordshire, UK) over the same time period, 1997–1999 (ref. 14; N. P. J. Day, C. E. Moore, M. C. Enright, A. R. Berendt, J. Maynard Smith, M. D. Murphy, S. J. Peacock, B. G. Spratt & E. J. Feil, unpublished data).
Strep. pyogenes.
The MLST dataset of 212 isolates included multiple isolates of the most prevalent M serotypes, from world-wide sources, and a sample of 47 isolates, representing >70 emm-sequence types, recovered from invasive disease in 1995 and 1998 in Connecticut (M.C.E., B.G.S., A.K. & D.B., unpublished data).
H. influenzae.
The 37 isolates were selected from a large ribotyping study (15, 16), and from a previous MLEE study (17), to cover the diversity of typable and nontypable isolates. Twenty-nine of these isolates are encapsulated and represent multiple examples of all six serotypes; the other eight are diverse nonencapsulated isolates recovered from cases of otitis media in Finland as part of a study being undertaken by R. Moxon (Institute of Molecular Medicine, University of Oxford, Oxford, U.K.).
E. coli.
MLST has been developed for E. coli by T. Whittam and colleagues. The sequences of seven house-keeping loci from twenty-one pathogenic isolates of E. coli were analyzed. These include representatives of the enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC), and Shiga-toxin-producing E. coli (STEC) pathogenic groups from world-wide sources (11).
Neisseria species.
Sequence data were available for four loci (argF, recA, rho, 16S rRNA) from 14 named species of the genus Neisseria, which commonly colonize humans. A single randomly selected isolate of each of these species was extracted from the dataset described by Smith et al. (18).
Sequence Data.
For each of the isolates in the five MLST databases, the sequences of ≈450-bp internal fragments of seven house-keeping genes have been determined and are available at the MLST website (http://www.mlst.net/) or at T. Whittam's E. coli website (http://www.bio.psu.edu/people/faculty/whittam/lab/mlst). For the H. influenzae isolates, the sequences of ≈450-bp internal fragments of five loci (adk, pgi, recA, fucK, and mdh) were determined. These fragments were amplified by PCR and sequenced on both strands using the following primers: adkUP, GGTGCACCGGGTGCAGGTAA; adkDN, CCTAAGATTTTATCTAACTC; pgiUP, GGTGAAAAAATCAATCGTAC; pgiDN, ATTGAAAGACCAATAGCTGA; recAUP, ATGGCAACTCAAGAAGAAAA; recADN, TTACCAAACATCACGCCTAT; fucKUP, ACCACTTTCGGCGTGGATGG; fucKDN, AAGATTTCCCAGGTGCCAGA; mdhUP, TCATTGTATGATATTGCCCC; and mdhDN, ACTTCTGTACCTGCATTTTG. These sequences are also available on the MLST web site.
Phylogenetic Analysis.
A sample of diverse isolates of each species was used for the analysis of congruence. The H. influenzae and E. coli samples had been preselected for diversity, and all of the available isolates were used (corresponding to 37 and 21 isolates, respectively). Samples for N. meningitidis, Strep. pneumoniae, Staph. aureus, and Strep. pyogenes were obtained by constructing dendrograms using the percentage mismatches in the allelic profiles defined by MLST and subsequently truncating them (at linkage distances of 0.4–0.55) so that only 30–41 lineages were obtained. A single isolate from each of these lineages was selected at random to obtain a set of isolates of each species that are distantly related to each other. The 30 strains of N. meningitidis were those used in a previous analysis of congruence in this species (12). Single randomly selected isolates from each of 14 different Neisseria species (18) were also analyzed.
All phylogenetic trees used in the congruence analysis were reconstructed by using the maximum likelihood (ML) method available in the PAUP* package (version 4; Swofford 1999). The HKY85 model of DNA substitution was used in all cases, with the optimal ratio of transitions to transversions (Ts/Tv) and the α parameter, which describes the extent of rate variation among nucleotide sites assuming a discrete gamma distribution with eight categories, both estimated from the empirical data during tree reconstruction. These values, as well as the individual trees, are available from the authors on request.
A maximum likelihood method was also used to determine the extent of congruence among gene trees (12). First, for each gene in each species, the differences in log likelihood (Δ-ln L) were computed between the ML tree for that gene and the ML trees constructed on the other genes from that species, but with branch lengths optimized to maximize the likelihood of this topology on the reference data. Values for Ts/Tv and α were also reoptimized. To determine whether these differences in log likelihood are significantly different (they will not be if the gene trees are congruent), 200 random trees were created for each gene. The likelihoods of these trees were then estimated, again by optimizing branch lengths and Ts/Tv and α values, and the differences in log likelihood between these random trees and the ML tree for each gene were computed. These can then be considered as a null distribution of Δ-ln L values, as would be obtained when there is no more similarity in topology among gene trees than expected by chance. If the Δ-ln L values for the comparisons among the different ML trees fall within the 99th percentile of this null distribution, then we may say that they are significantly different and hence incongruent (Fig. 1; see also supplemental Figs. 2–7, which are published as supplemental data on the PNAS web site, www.pnas.org). Finally, to test whether the method is not adversely affected by the inclusion of very similar sequences, model congruent data sets containing little phylogenetic signal were created. Trees significantly more similar than random topologies were recovered (results not shown, available on request).
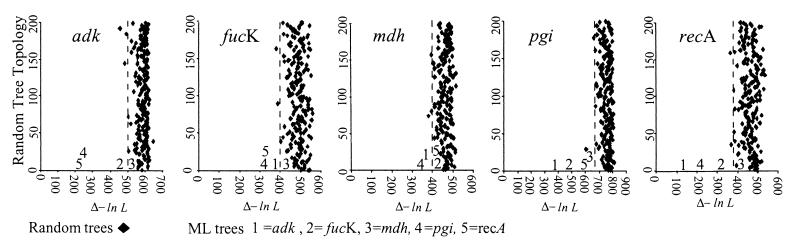
Figure 1.
Maximum likelihood analysis of congruence in H. influenzae. The ML tree of each locus is compared with the ML trees from the other four loci. The differences in likelihood (Δ-ln L) are shown between loci (squares) and between each locus and 200 trees of random topology (diamonds). The 99th percentile of the likelihood differences between the ML tree for each gene and the 200 random tree topologies is indicated by the dotted line.
Empirical Estimates of Recombination Rates.
The approach utilizes comparisons between very closely related isolates belonging to the same clonal complex. The large amount of data required is currently available for only three of the species, N. meningitidis, Strep. pneumoniae, and Staph. aureus. Although a large dataset for Strep. pyogenes is available, the sample was chosen to represent the diversity of this species and does not contain sufficient multiple examples of closely related isolates. The method for estimating recombinational parameters from MLST datasets has been described (6, 7) and was slightly modified as follows.
The MLST datasets were first divided into clonal complexes, defined as groups in which every isolate shares at least five identical alleles with at least one other isolate in the group. Within each clonal complex, pairwise comparisons between all MLST allelic profiles were made, and the profile that is associated with the highest number of single-locus variants (SLVs, isolates that differ at only one of the seven loci) was identified. This genotype is assumed to represent the most likely recent common ancestor from which all of the associated SLVs have descended (this assumption is justified below).
Comparisons were then made between the sequences of the alleles in each of the SLVs and those in the corresponding putative ancestral genotype. If the variant allele in an SLV differs at multiple nucleotide sites from the corresponding allele in its ancestral genotype, it is assigned as a recombinational event (multiple independent point mutations are unlikely because the SLV has retained identical sequences to the ancestral genotype at the other six loci). If, however, the difference involves only a single nucleotide site, the allele may have arisen by point mutation or by recombination between sequences that differ only at a single site. These possibilities are distinguished by examining the presence of the variant allele elsewhere in the dataset, because an allele introduced by recombination may, or may not, be present elsewhere in the dataset, whereas a point mutation will almost certainly result in a novel allele (8). If the majority (say, 80%) of imported alleles that have multiple changes are present in unrelated isolates in the dataset, then the majority of imported alleles that differ at a single site should also be found in the database, and the number of point mutations can be approximated to the number of novel variant alleles within SLVs that differ from their ancestral alleles at a single site. This approximation is most likely to be conservative with respect to the significance of recombination. Two different parameters are estimated: the ratios at which (i) alleles and (ii) an individual nucleotide site change by recombination as compared with mutation. The identification of SLVs and putative ancestral genotypes was carried out by using the BURST program (available at http://microbiology.ceid.ox.ac.uk/).
Results
Analysis of Congruence Between Gene Trees.
The results of the maximum likelihood analysis of congruence are presented in Table 1. The most striking feature is that the differences between the likelihoods of trees (Δ-ln L) reconstructed on different genes are often very great, indicating that all are incongruent to some extent. The clearest examples of a lack of congruence between loci are provided by Strep. pneumoniae and Strep. pyogenes. Every comparison between gene trees in these two species produced likelihood differences that fell within the range seen for comparisons involving trees of random topology. Similar results were observed in N. meningitidis and Staph. aureus. For these species, only a small number of gene tree comparisons showed levels of topological similarity greater than the random expectation, and even in these cases the trees were very dissimilar, with likelihood differences falling only marginally outside the 99th percentile of the random distribution, as compared with the levels of congruence observed in H. influenzae and E. coli (Table 1; Suppl. Figs. 2–7).
Table 1.
Statistical tests of congruence between loci
| Bacterial species (no. isolates) | Gene | bp | π | Δ-ln L of ML tree | Δ-ln L of ML trees from other genes | 99th percentile Δ-ln L in random trees | Loci outside 99th percentile of random trees |
|---|---|---|---|---|---|---|---|
| E. coli (21) | arcA | 564 | 0.003 | 843.572 | 27.852–50.788 | 53.147 | aroE, icd, mdh, mtlD, pgi, rpoS |
| aroE | 456 | 0.018 | 860.313 | 69.726–149.340 | 139.995 | icd, mdh, mtlD, pgi, rpoS | |
| icd | 1,176 | 0.017 | 2,221.243 | 170.542–321.632 | 364.039 | arcA, aroE, mdh, mtlD, pgi, rpoS | |
| mdh | 846 | 0.013 | 1,529.923 | 73.672–210.526 | 210.922 | arcA, aroE, icd, mtlD, pgi, rpoS | |
| mtlD | 1,098 | 0.020 | 2,167.194 | 153.445–316.678 | 236.699 | aroE, icd, mdh, pgi, rpoS | |
| pgi | 978 | 0.014 | 1,817.052 | 94.717–257.440 | 178.269 | aroE, icd, mdh, mtlD, rpoS | |
| rpoS | 714 | 0.006 | 1,126.439 | 5.816–93.776 | 70.024 | aroE, icd, mdh, mtlD, pgi | |
| H. influenzae (37) | adk | 479 | 0.024 | 992.635 | 222.146–520.867 | 494.584 | fucK, pgi, rec |
| fucK | 451 | 0.024 | 837.845 | 310.062–440.454 | 399.627 | adk, pgi, rec | |
| mdh | 406 | 0.033 | 1,173.585 | 332.250–404.052 | 397.248 | adk, pgi | |
| pgi | 469 | 0.040 | 1,476.730 | 359.179–616.517 | 669.637 | adk, fucK, mdh, rec | |
| recA | 427 | 0.026 | 918.907 | 125.392–403.000 | 381.975 | adk, fucK, pgi | |
| N. meningitidis (30) | abcZ | 433 | 0.046 | 1,167.727 | 486.423–597.804 | 504.041 | fumC, gdh |
| adk | 465 | 0.009 | 764.176 | 139.803–156.868 | 134.151 | — | |
| aroE | 490 | 0.095 | 1,889.853 | 1,244.795–1,596.723 | 1,176.877 | — | |
| fumC | 465 | 0.017 | 974.543 | 172.168–231.833 | 186.717 | abcZ | |
| gdh | 501 | 0.017 | 947.074 | 240.369–312.717 | 229.519 | — | |
| pdhC | 480 | 0.054 | 1,455.959 | 687.918–845.640 | 676.075 | — | |
| pgm | 450 | 0.039 | 1,236.568 | 444.264–547.919 | 415.225 | — | |
| Strep. pneumoniae (40) | aroE | 405 | 0.007 | 707.481 | 162.162–171.607 | 143.932 | — |
| ddl | 441 | 0.016 | 1,089.352 | 237.612–266.662 | 202.116 | — | |
| gdh | 459 | 0.010 | 905.748 | 196.571–221.832 | 174.686 | — | |
| gki | 483 | 0.021 | 1,064.972 | 430.700–493.201 | 359.787 | — | |
| recP | 447 | 0.006 | 723.160 | 137.379–160.715 | 133.568 | — | |
| spi | 471 | 0.013 | 907.581 | 268.895–291.920 | 233.293 | — | |
| xpt | 486 | 0.008 | 867.223 | 148.206–157.570 | 134.643 | — | |
| Staph. aureus (38) | arcC | 456 | 0.008 | 756.869 | 197.330–260.448 | 202.504 | glpF |
| aroE | 456 | 0.011 | 839.810 | 207.142–288.318 | 242.507 | glpF, yqiL | |
| glpF | 465 | 0.004 | 731.576 | 78.188–149.162 | 116.081 | aroE, yqiL | |
| gmk | 429 | 0.008 | 658.350 | 153.494–195.390 | 148.037 | — | |
| pta | 474 | 0.006 | 765.706 | 195.291–232.350 | 182.386 | — | |
| tpi | 402 | 0.009 | 701.141 | 180.586–206.998 | 171.693 | — | |
| yqiL | 516 | 0.008 | 875.024 | 143.012–213.713 | 185.236 | aroE, glpF | |
| Strep. pyogenes (41) | gki | 498 | 0.013 | 958.175 | 323.073–351.450 | 281.164 | — |
| gtr | 450 | 0.008 | 829.388 | 118.240–142.246 | 117.052 | — | |
| murL | 438 | 0.005 | 716.867 | 144.926–167.725 | 129.570 | — | |
| mutS | 405 | 0.007 | 676.422 | 161.971–176.198 | 143.998 | — | |
| recP | 459 | 0.013 | 1,029.345 | 243.735–294.841 | 239.060 | — | |
| xpt | 471 | 0.004 | 819.777 | 76.926–83.659 | 63.367 | — | |
| yqiL | 434 | 0.005 | 748.693 | 117.028–133.732 | 103.659 | — | |
| Neisseria sp. (14) | argF | 696 | 0.146 | 3,504.516 | 58.675–287.783 | 291.965 | recA, rho, 16S rRNA |
| recA | 711 | 0.130 | 3,175.912 | 29.021–243.857 | 312.017 | argF, rho, 16S rRNA | |
| rho | 1,026 | 0.126 | 4,549.233 | 48.935–431.534 | 434.414 | argF, recA, 16S rRNA | |
| 16S rRNA | 1,355 | 0.028 | 2,903.989 | 81.136–123.474 | 139.920 | argF, recA, rho |
In contrast, all of the gene trees in H. influenzae showed at least some congruence, and in some cases the differences in likelihoods fell well outside those of the random trees, revealing a relatively high degree of similarity in tree topology (Fig. 1). The congruence between gene trees was most apparent with adk, fucK, pgi, and recA, although far less congruence was seen in comparisons involving mdh, and pgi was the only locus in which every tree comparison fell outside of the 99th percentile of the random distribution. Even more congruence among loci was seen in the analysis of 21 E. coli isolates. In this species, all ML trees were more similar to each other than random topologies, and sometimes greatly so, as in the case of icd and mdh. The only exceptions were some comparisons involving arcA.
We also examined the level of congruence at a deeper phylogenetic level by comparing trees reconstructed for 14 different Neisseria species. The ML trees recovered using the argF, recA, and rho genes were clearly much more similar to each other than random trees and showed a higher degree of similarity than seen in most of the intraspecific comparisons, but, surprisingly, the differences in likelihood between the 16S rRNA tree and the other trees only just fell outside the 99th percentile of the random tree topologies.
The lowest levels of congruence were observed in Strep. pneumoniae and Strep. pyogenes, and these species also exhibited very low levels of average pairwise sequence divergence (π). However, we do not note a general correlation between congruence and sequence diversity. For example, although some of the loci from N. meningitidis are amongst the most divergent analyzed, there is little similarity between gene trees within this species. Thus, recombination occasionally takes place even between relatively diverged sequences in the meningococcus; recombinational imports of diverged sequences from other named Neisseria species have previously been noted in house-keeping genes of this species (19). Furthermore, E. coli exhibits the highest degree of congruence between loci despite the fact that some loci in this species are as conserved as those in Strep. pneumoniae.
Finally, it was also evident from our analysis that different pairs of loci vary in the extent to which they are congruent. For example, in H. influenzae, the pgi locus exhibits significant congruence with all of the other four loci, whereas mdh shows significant congruence with only two (Fig. 1). These differences are not explained in terms of the physical locations of the genes on the chromosome, because recA and fucK are the only closely linked genes, yet are not atypically congruent.
Estimates of Per Site and Per Allele Recombinational Parameters.
The estimate of recombinational parameters involves comparing isolates that are very closely related and differ by MLST at only one of the seven house-keeping loci. Analysis of the sequence differences between the variant allele of each SLV and the corresponding allele in the putative ancestral genotype allows the variant alleles to be assigned as the result of either recombination or mutation. In the current analysis, we assign alleles that differ at multiple sites as the result of recombination and, for those that differ at only a single site, we discriminate between those that are likely to have arisen by recombination rather than mutation (as described in Materials and Methods). This approach increased the estimate of the number of alleles changed by recombination compared with mutation [the per allele recombination/mutation (r/m) parameter] for N. meningitidis from the previously published lower-bound of 3.6:1 to 4.75:1 (7). The updated estimate for Strep. pneumoniae is very similar to the previous estimate (8.9:1), and the estimate for Staph. aureus is 6.5:1. These estimates are therefore strikingly similar for the three species. However, the relative probability that an individual nucleotide site will change by recombination or mutation (the per site r/m parameter) varies markedly, reflecting differences in sequence diversity and hence the average number of nucleotide sites changed per replacement. Hence, the per site r/m estimates are 100:1, 61:1, and 24:1 for N. meningitidis, Strep. pneumoniae, and Staph. aureus, respectively.
The accuracy of the estimates depends on the correct assignments of ancestral genotypes, and hence the directionality of events, and these are supported by the finding that alleles assigned as having arisen by point mutation are significantly more likely to be novel than those of their predicted ancestors (P < 0.01; data not shown). Furthermore, although ancestral genotypes are assigned independently of the number of isolates with each genotype, the ancestral genotypes typically correspond to the most common allelic profile within the clonal complex. For example, Feil et al. (8) previously assigned ancestral genotypes within S. pneumoniae on the basis of numerical dominance, whereas in the current analysis we have used the parsimony-based approach outlined above. For all 21 of the previously identified clonal complexes that contain more than two different genotypes (the other minor clonal complexes are ignored in the current analysis), the two methods predict the same clonal ancestor.
Discussion
We have examined levels of congruence between housekeeping loci in six important species of pathogenic bacteria and also discussed empirical estimates of recombinational parameters for three of these species. In most cases, ML trees for different house-keeping genes of four of these species (N. meningitidis, Strep. pneumoniae, Strep. pyogenes, and Staph. aureus) are no more similar to each other than they are to trees of random topology. Even those comparisons of trees that gave a difference in likelihood that was outside the 99th percentile of the random trees were essentially incongruent because the differences in likelihood were still very great. The lack of congruence among gene trees is most easily explained as the consequence of a history of relatively frequent recombinational exchanges that over time have eliminated (or almost eliminated) the phylogenetic signal in each tree. In other words, it is not possible to detect a clonal frame in these species.
This view is supported by estimates of the ratio of recombinational exchanges to point mutations during the initial stages of clonal diversification. In the three species where these parameters could be estimated, an allele is approximately 5- to 10-fold more likely to change to a new allele by recombination than by point mutation. These are not relative rates of recombination and mutation per se, because some events, such as highly deleterious point mutations, or recombinational events between identical sequences, will not be observed. Any differences in the selective outcomes of a point mutation and a recombinational exchange are not considered because they are difficult to predict: a point mutation represents a small, untested, genetic change, whereas a recombinational exchange introduces single or multiple nucleotide changes that already exist in the population and so may have passed a selective filter. The parameters do, however, reflect the relative impact of recombination and point mutation on sequence variation within house-keeping loci in natural populations of these species.
Although these analyses mutually confirm the profound significance of recombination in the three pathogens where both approaches could be applied, ranking N. meningitidis, Staph. aureus, and Strep. pneumoniae in terms of their recombination rates is more difficult. Levels of congruence will depend not only on the frequency of recombination, but also on the precise sample of isolates examined, the average length of recombinant fragments, and evolutionary relationships between parental isolates; exchanges between closely related isolates will have less affect on overall tree topology than exchanges between distantly related isolates. Population genetic factors, such as the relative rates of population subdivision and gene flow, will also contribute, as discussed below.
Despite these caveats, there is some justification from both methods for concluding that recombination is slightly more significant in Strep. pneumoniae than in N. meningitidis or Staph. aureus. Strep. pneumoniae gives the highest per allele r/m parameter (8.9:1), and there are also no examples of congruence between loci in this species although some congruent pairs of loci are noted within N. meningitidis and Staph. aureus (Table 1). However, the highest per site r/m parameter is noted in N. meningitidis, which reflects the fact that there is a high degree of sequence divergence in this species (19), and it is unclear to what extent the per site and per allele r/m parameters contribute to levels of congruence among loci.
There was no obvious correlation between the degree of congruence and the transformability of the species. Both N. meningitidis and Strep. pneumoniae are naturally transformable and showed little congruence between gene trees. However, H. influenzae is also naturally transformable but showed far greater congruence between gene trees than the nontransformable Staph. aureus and Strep. pyogenes. In fact, there was an almost total absence of congruence between trees in Strep. pyogenes. Recombinational exchanges in Staph. aureus and Strep. pyogenes are presumably mediated by phage transduction, and it appears that the effects of phage-mediated transduction on population structure may in some cases be as great as, or even greater than, that of transformation.
Two of the six species, H. influenzae and E. coli, consistently showed statistically significant similarities between gene trees, although direct estimates of the r/m parameters have not yet been obtained for these species because large MLST data sets are currently unavailable. The relatively high level of congruence within the E. coli data set confirms the recent suggestion by Reid et al. (11), made by constructing compatibility matrices and using split decomposition analysis, that a phylogenetic signal common to all loci is present in these data. The interpretation of relatively low rates of recombination is in accord with extensive MLEE studies for both species (2, 20) but is curious for H. influenzae, given that the species is naturally transformable and contains large numbers of uptake sequences believed to promote the specific incorporation of “self” DNA into the cell (21). There is also clear mosaicism in some of the H. influenzae sequences used in the analysis (data not shown). Similarly, there is direct evidence from nucleotide sequences for recombination in E. coli (22–25), although there have also been several previous assertions of congruence between gene trees, and the existence of a clonal frame in this species (2, 13).
There are two classes of explanation to reconcile direct evidence for recombination within nucleotide sequences with congruence between gene trees. The first is simply that recombination occurs occasionally but not at a sufficient frequency to disrupt the clonal frame. The second explanation is the possibility of ecological substructuring within the population; for E. coli, this possibility is discussed elsewhere (4). The sequence diversity among H. influenzae is considerable (Table 1), and this species could well encompass distinct biological or ecological clusters. The greater levels of congruence within this species may therefore reflect barriers to gene flow between different subgroups within the population; if so, the results tell us little about the extent of recombination within subgroups. Included within this data set are representatives of all of the major disease-causing serotypes (a–f), and these provide the most likely biological basis for such substructuring; these serotypes are clearly resolved in the MLST data, and also by using data generated by ribotyping (15, 16) and MLEE (17). A more extensive MLST data set containing large numbers of isolates from each of the different serotypes should allow estimates of recombination parameters in this species. Such a dataset would clarify whether the significant congruence between loci reflects relatively low rates of recombination, both within and between subgroups, or is a consequence of the lumping together of isolates from different subgroups.
Finally, the analysis of 14 named Neisseria species revealed good congruence for three house-keeping loci, presumably because of a combination of partial ecological separation and/or a reduction in transformation efficiency between species, compared with within species, because of high levels of sequence divergence between species. In contrast, the 16S rRNA tree exhibited far lower levels of congruence between the species. One possible explanation for this difference is a suggestion made by Cohan (26) as follows. Very conserved loci within a freely recombining population may become subject to a positive feedback loop, whereby low levels of divergence facilitate recombination, which in turn tempers the divergence between taxa. This effect has been discussed previously with respect to a comparison of adk sequences between different Neisseria species (27). Whatever the explanation, the observation of low levels of congruence in 16S rRNA is ironic because this locus is frequently used in bacterial systematics and is assumed to represent the true phylogeny between taxa.
Concluding Remarks.
Here we compare levels of congruence between housekeeping loci in six bacterial pathogens and note that in four of these species (N. meningitidis, Staph. aureus, Strep. pyogenes, and Strep. pneumoniae) there is little or no congruence. High rates of recombination in N. meningitidis have been proposed previously, from estimates of linkage disequilibrium (3), the presence of frequent mosaic structure in house-keeping genes (19), and the lack of congruence between gene trees (12, 27). There is much less information about levels of recombination in Strep. pneumoniae, Staph. aureus, and Strep. pyogenes. MLST and/or MLEE has revealed that populations of each of these species (particularly those recovered from disease) principally consist of a limited number of widespread clonal lineages. The ability to identify clusters of isolates with identical allelic profiles (clones), from different countries, over a period of decades, should not therefore be taken as evidence for low rates of recombination.
With the possible exception of Salmonella enterica (28), few bacterial species appear to be truly clonal, such that recombinational exchanges are absent, or so rare that they are observed only in genes under strong selection for diversity. In these species, clones should be stable because diversification at house-keeping loci depends almost entirely on the accumulation of point mutations, but, with increasing ratios of recombination to point mutation, bacterial clones should become increasingly transient. Except in a few species where rates of recombination are so high that new adaptive genotypes diversify so rapidly that clones cannot become established in the population [e.g., Neisseria gonorrhoeae (3, 29) or Helicobacter pylori (30)], the diversification process in most species appears to be sufficiently slow that clones (or clonal complexes) can still be recognized over the short term (tens or hundreds of years). However, over the long term, the impact of relatively frequent recombination is to obliterate the phylogenetic signal in gene trees such that the relationships between major lineages of many bacterial species should be depicted as a network rather than a tree. For these bacterial species the practice of mapping phenotypic characters, or life history traits, onto trees produced from sequence data, or from MLEE or MLST data, is likely to lead to erroneous assumptions about the evolutionary origins of these traits. No deep phylogenetic structure (clonal frame) is present in these populations, and reliable information about the relatedness of isolates will be evident only within the clusters of very similar genotypes belonging to the same clonal complex that have diversified from a recent common ancestor. However, phylogenetic signal is clearly present in trees of some species, and it is unclear why the impact of recombination appears to vary widely among species.
Supplementary Material
Acknowledgments
We are grateful to Valerie Bouchet for providing invaluable insights by a ribotype restriction fragment length polymorphism (RFLP)-based phylogenetic analysis of more than 400 H. influenzae isolates, and to Richard Moxon for help in selecting and providing H. influenzae isolates. We also gratefully acknowledge Anthony Berendt, Michael Murphy, and Sharon Peacock for the provision of Staph. aureus isolates. We thank the members of the Finnish Otitis Media Study Group at the National Public Health Institute in Finland for the provision of NTHi strains from the inner ear fluid, obtained as part of the Finnish Otitis Media Cohort Study. E.J.F. is supported by the Wellcome Trust. E.C.H. is supported by The Royal Society. B.G.S. is a Wellcome Trust Principal Research Fellow. D.E.B. is an Established Investigator of the American Heart Association and is supported by the National Institutes of Health (GM60793) and the American Heart Association (Grant-in-Aid). N.P.J.D. is a Wellcome Trust Career Development Fellow. R.G. was supported in part by research grant awards from the National Institutes of Health (DK-50838) and the Cystic Fibrosis Foundation (USA). M.C.E. is supported by The Royal Society. C.E.M., M.S.C., and J.Z. are supported by the Wellcome Trust. D.W.H. is supported by a Medical Research Council program grant.
Abbreviations
- SLV
single-locus variant
- MLST
multilocus sequence typing
- MLEE
multilocus enzyme electrophoresis
- ML
maximum likelihood
- r/m
recombination/mutation
Footnotes
References
- 1.Levin B R. In: The Evolution of Sex. Michod R E, Levin B R, editors. Sunderland, MA: Sinauer; 1988. pp. 194–211. [Google Scholar]
- 2.Selander R K, Musser J M. In: Molecular Basis of Bacterial Infections. Iglewski B H, Clark V L, editors. San Diego: Academic; 1990. pp. 11–36. [Google Scholar]
- 3.Maynard Smith J, Smith N H, O'Rourke M, Spratt B G. Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman D S. Trends Ecol Evol. 1997;12:16–22. doi: 10.1016/s0169-5347(96)10057-4. [DOI] [PubMed] [Google Scholar]
- 5.Maiden M C, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, et al. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guttman D S, Dykhuizen D E. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 7.Feil E J, Maiden M C J, Achtman M, Spratt B G. Mol Biol Evol. 1999;16:1496–1502. doi: 10.1093/oxfordjournals.molbev.a026061. [DOI] [PubMed] [Google Scholar]
- 8.Feil E J, Maynard Smith J, Enright M C, Spratt B G. Genetics. 2000;154:1439–1450. doi: 10.1093/genetics/154.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selander R K, Caugant D A, Ochman H, Musser J M, Gilmour M N, Whittam T S. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykhuizen D E, Green L. J Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reid S D, Herbelin C J, Bumbaugh A C, Selander R K, Whittam T S. Nature (London) 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 12.Holmes E C, Urwin R, Maiden M C J. Mol Biol Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- 13.Milkman R, McKane Bridges M. Genetics. 1990;126:505–517. doi: 10.1093/genetics/126.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright M C, Day N P J, Davies C E, Peacock S J, Spratt B G. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolduc G, Bouchet V, Jiang R-Z, Geisselsoder J, Truong Q, Rice P A, Pelton S I, Goldstein R. Infect Immun. 2000;68:4505–4517. doi: 10.1128/iai.68.8.4505-4517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolduc G. Ph.D. Thesis. Boston: Boston University School of Medicine; 1999. [Google Scholar]
- 17.Musser J M, Kroll J S, Granoff D M, Moxon E R, Brodeur B R, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G, et al. Rev Infect Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- 18.Smith N H, Holmes E C, Donovan G M, Carpenter G A, Spratt B G. Mol Biol Evol. 1999;16:773–783. doi: 10.1093/oxfordjournals.molbev.a026162. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Bowler L D, Spratt B G. Mol Microbiol. 1997;23:799–812. doi: 10.1046/j.1365-2958.1997.2681633.x. [DOI] [PubMed] [Google Scholar]
- 20.Musser J M, Kroll J S, Moxon E R, Selander R K. Proc Natl Acad Sci USA. 1988;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith H O, Gwinn M L, Salzberg S L. Res Microbiol. 1999;150:603–616. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- 22.Nelson K, Selander R K. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd E F, Nelson K, Wang F, Whittam T S, Selander R K. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milkman R, McKane Bridges M. Genetics. 1993;133:455–468. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson K, Selander R K. Proc Natl Acad Sci USA. 1994;91:10227–10231. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohan F M. Evolution. 1995;49:164–175. doi: 10.1111/j.1558-5646.1995.tb05968.x. [DOI] [PubMed] [Google Scholar]
- 27.Feil E, Zhou J, Maynard Smith J, Spratt B G. J Mol Evol. 1996;43:631–640. doi: 10.1007/BF02202111. [DOI] [PubMed] [Google Scholar]
- 28.Selander R K, Smith N H. Med Rev Microbiol. 1990;1:219–228. [Google Scholar]
- 29.O'Rourke M, Stevens E. J Gen Microbiol. 1993;139:2603–2611. doi: 10.1099/00221287-139-11-2603. [DOI] [PubMed] [Google Scholar]
- 30.Suerbaum S, Maynard Smith J, Bapumia K, Morelli G, Smith N H, Kunstmann E, Dyrek I, Achtman M. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.