Abstract
The homobasidiomycete Coprinus cinereus exhibits remarkable photomorphogenesis during fruiting-body development. Under proper light conditions, fruiting-body primordia proceed to the maturation phase in which basidia in the pileus undergo meiosis, producing sexual spores, followed by stipe elongation and pileus expansion for efficient dispersal of the spores. In the continuous darkness, however, the primordia do not proceed to the maturation phase but are etiolated: the pileus and stipe tissues at the upper part of the primordium remain rudimentary and the basal part of the primordium elongates, producing “dark stipe.” In this study we genetically analyzed five strains that produce dark stipes even if light conditions promoting the maturation are given and then characterized one of them, Uar801 (dst1-1). The dst1 gene was cloned as a DNA fragment that rescues the dst1-1 mutation. Dst1 is predicted to be a protein of 1175 amino acids that contains two PAS domains, a coiled-coil structure, and a putative, glutamine-rich, transcriptional activation domain (AD). One of the PAS domains exhibits significant similarity to the LOV domains of known blue-light receptors, suggesting that Dst1 is a blue-light receptor of C. cinereus. The dst1-1 mutation is predicted to truncate the putative AD in the C-terminal region.
BLUE light is known to be an important signal for many organisms, regulating their developmental and physiological processes. In plants, blue light has effects upon phototropism, chloroplast rearrangement, stomatal opening, hypocotyl elongation, and vegetative-to-floral transition, and several blue-light receptors, including phototropins and cryptochromes, have been identified (for reviews, see Lin 2000; Christie and Briggs 2001; Devlin 2002). In fungi, it has been found that the ascomycete Neurospora crassa has two blue-light receptors, White Collar-1 (WC-1) (Ballario et al. 1996; Froehlich et al. 2002; He et al. 2002) and Vivid (VVD) (Schwerdtfeger and Linden 2003). The former is an essential component for all known blue-light responses in this fungus, including circadian rhythms, conidium formation, and carotenoid biosynthesis, while the latter is crucial for responding to daily changes in light intensity. Homologs of WC-1 have recently been identified in two other ascomycetes, Tuber borchii (Ambra et al. 2004), Trichoderma atroviride (Casas-Flores et al. 2004), and in a heterobasidiomycete, Cryptococcus neoformans (Idnurm and Heitman 2005). Phototropins, WC-1, and VVD contain the light, oxygen, and voltage (LOV) domain, which forms a subgroup of the sensory domain superfamily Per-Arnt-Sim (PAS) (Taylor and Zhulin 1999) and is crucial for binding a flavin chromophore. Cryptochromes exhibit high sequence similarities to microbial DNA photolyases: they do not contain the LOV domain and have pterin in addition to flavin as chromophores (Lin 2000).
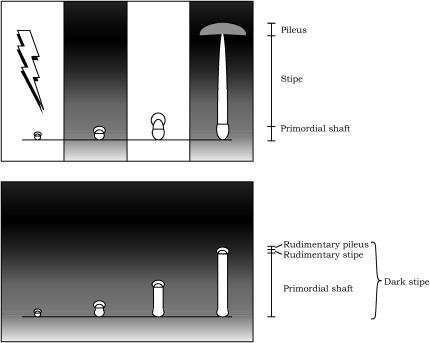
Fruiting body (mushroom) formation of the homobasidiomycete Coprinus cinereus is a most remarkable example of fungal development regulated by light (Tsusué 1969; Kamada et al. 1978; for a review, see Kües 2000). The development is initiated as a hyphal knot of 0.2 mm or less in diameter, which rapidly develops into a tiny primordium containing the rudimentary tissues of the future pileus and stipe. The primordium gradually enlarges and then proceeds to the fruiting-body maturation phase under proper light conditions, such as the 12 hr light/12 hr dark cycle (Kamada et al. 1978). In the maturation phase, basidia on the undersurface of the pileus undergo meiosis, resulting in the production of basidiospores, which is followed by stipe elongation and pileus expansion for efficient dispersal of the spores (Figure 1, top; Figure 2A). In the continuous darkness, however, the primordium does not proceed to the maturation phase but is etiolated: the pileus and stipe tissues at the upper part of the primordium remain rudimentary, and the primordial shaft, which is the basal part of the primordium, elongates, producing “dark stipe” (Figure 1, bottom; Figure 2B) (Tsusué 1969; Kamada et al. 1978). Under continuous light, the maturation starts but stops halfway: meiosis in the basidia is arrested at prophase I, the stipe elongates only slightly, and the pileus does not expand (Tsusué 1969; Kamada et al. 1978; Lu 2000). Action spectra have been determined for the initiation of the fruiting-body primordium, for the development of the primordium into the mature fruiting body, and for the inhibition of normal progress of meiosis in Coprinus congregatus, a close relative of C. cinereus, showing that UV-blue light is effective in all the processes examined (Durand and Jaques 1982; Durand and Furuya 1985). On the basis of the spectra determined, it has been hypothesized that a flavin-containing blue-light photoreceptor is responsible for mediating photomorphogenesis in Coprinus species (Durand and Jaques 1982; Durand and Furuya 1985). However, a photoreceptor(s) for blue light remains to be identified in homobasidiomycetes.
Figure 1.
Diagrams illustrating photomorphogenesis during fruiting-body development in C. cinereus. When the fungus is cultured under the 12 hr light/12 hr dark cycle, the mature fruiting body is formed. The first 5 min of light (500 lx) on the first day triggers fruiting-body maturation (top) (Kamada et al. 1978). When the fungus was cultured in continuous darkness, the primordium never proceeded to the fruiting-body maturation phase. Instead, the primordial shaft, which is a basal part of the primordium, elongated, producing the dark stipe (bottom).
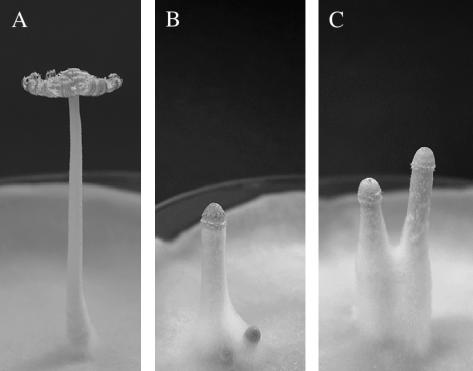
Figure 2.
Photographs showing a mature fruiting body developed on a culture of wild-type strain CopD5-12 under the 12 hr light/the 12 hr dark cycle (A), dark stipes formed on a culture of strain CopD5-12 grown in the continuous darkness (B), and dark stipes on a culture of the dark stipe1 mutant, Uar801, grown under the 12 hr light/12 hr dark cycle (C).
In this article, we first genetically analyze five photomorphogenetic mutants of C. cinereus, which produce dark stipes even if light conditions that normally promote the development of mature fruiting bodies are given (Figure 2C), resulting in identification of two putative gene loci, dst1 and dst2, responsible for the mutations. We then characterize one of the mutations, dark stipe1 (dst1-1). The dst1 gene is predicted to encode a protein of 1175 amino acid residues, which contains two PAS domains, a coiled-coil structure, and a putative, glutamine-rich, transcriptional activation domain (AD). One of the PAS domains has high similarity to the LOV domains of WC-1, VVD, and phototropins. These results strongly suggest that Dst1 is a blue-light receptor of C. cinereus.
MATERIALS AND METHODS
Strains, culture conditions, and genetic techniques:
Strains of C. cinereus listed in Table 1 were used. Malt extract/yeast extract/glucose (MY) medium (Rao and Niederpruem 1969) solidified by 2% agar in 9-cm petri dishes was used for routine mycelial cultures and for fruiting bodies for extraction of RNA. Slants of MY agar medium were used for examination of fruiting phenotypes. MY medium without agar in 9-cm petri dishes was used for mycelial cultures for extraction of DNA and RNA. MY medium was supplemented with 100 mg/liter of tryptophan for tryptophan-requiring strains. Cultures were maintained at 28° under a 12 hr light/12 hr dark regime unless otherwise stated. Crosses and isolation of basidiospore germlings were performed as described previously (Inada et al. 2001).
TABLE 1.
C. cinereus strains used in this study
| Strain | Genotype/description | Source |
|---|---|---|
| 5302 | A2B2 | This laboratory |
| 5337 | A8B7 | This laboratory |
| 292 | A3B1 trp1-1,1-6 | P. J. Pukkila |
| 326 | AmutBmut/homokaryotic fruiting | P. J. Pukkila |
| CopD5-12 | A12B12/homokaryotic fruiting | This laboratory |
| Uar801 | A12B12 dst1-1/homokaryotic fruiting | This laboratory |
| T140 | A3B1 trp1-1,1-6 dst1-1/a progeny of Uar801 × 292 | This study |
| R1428 | AmutBmut dst1-2/a REMI mutant from 326 | M. E. Zolan |
| R1428F1#72 | A8B7 dst1-2/ a progeny of R1428 × 5337 | This study |
| Uar290 | A12B12 dst2-3/a UV mutant from CopD5-12 | This laboratory |
| Uar351 | A12B12 dst2-2/a UV mutant from CopD5-12 | This laboratory |
| H1-1280 | AmutBmut dst2-1/a REMI mutant from 326 | M. E. Zolan |
Chromosome IX-specific cosmid library screening, the dst1 gene cloning, and sequencing:
A cosmid library of chromosome IX from the wild-type homokaryon, 5302, was constructed as described by Zolan et al. (1992). The vector (LLC5200) contains the trp1 gene of C. cinereus as a selectable marker (Pukkila and Casselton 1991). The library was composed of 672 clones (96 clones × 7 plates). Groups of 12 cosmid clones were cultured on a plate of Luria broth/ampicilline solid medium, mixed, and subjected to mini prep. Pooled DNAs were used to transform strain T140 (A3B1 trp1-1,1-6 dst1-1) as described by Binninger et al. (1987). Transformants were cultured on minimal medium to purify the transformed mycelium and then mated to R1428F1#72 (A8B7 dst1-2). The resulting dikaryons were cultured on slant medium to test for fruiting phenotypes. Of 140 trp+ transformants by a single pool of DNA, 1A, 6 produced wild-type fruiting bodies. Subsequent sib-selection revealed that a single cosmid clone, 1A1, has the rescuing activity.
Clone 1A1 was digested with EcoRI, EcoRV, ClaI, PstI, and BglII, and the digests were used to transform strain T140. We found that PstI and ClaI did not destroy the dst1 activity. Each fragment of the PstI digest was ligated into the PstI site of pGEM-7zf+ (Promega, Madison, WI) and examined for the dst1 activity by cotransformation of strain T140 with plasmid pCc1003 carrying the C. cinereus trp1 gene (Skrzynia et al. 1989). We found that a 10-kb fragment had the dst1 activity. Further digestion of the 10-kb PstI fragment with XbaI, followed by examination of the dst1 activity, revealed that a 6-kb PstI-XbaI fragment contained the whole dst1 gene. This 6-kb fragment was ligated into pBluescript II SK(+) and sequenced from both ends.
5′- and 3′-RACE experiments:
dst1 cDNA was amplified using an existing cDNA library synthesized with the Marathon cDNA amplification kit (CLONTECH, Palo Alto, CA) by Muraguchi and Kamada (1998) as a template. The gene-specific primers for the 5′ and 3′ RACE experiments are listed in Table 2 and their sites in the dst1 gene are shown in Figure 4A. PCR was performed with the Advantage cDNA PCR kit (CLONTECH) and the PCR products were cloned into pCR 2.1 vector with the TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced.
TABLE 2.
PCR primers used in this study
| Primer name | Sequence |
|---|---|
| a1 | GATGATGAACGCTGTTCTGG |
| a2 | ACCACCCCATCTCCCATT |
| b1 | ACGCAACAACGCCAGTTT |
| b2 | GGGTCGTAGATCTGCGTATGA |
| c1 | GGTGGAGCCCTTTTCTACCC |
| c2 | TTGGACCCCCACCTACTACC |
| d1 | GAAGGAGCGATGGGACTCTC |
| d2 | CTGCTCTTCCGCGCACGTACC |
| e1 | CGCCTACACAGCAGCATTT |
| e2 | GCAGCAGCAGATGTATGGAG |
| A1F1 | GATTACTACACGTCGACGCAGCAG |
| A5R2 | ACCCCACGGCCTCTTCAT |
| A4F3 | GAAGGAGCGATGGGACTCTC |
| 3DR3-2 | AATCGGGAAGGAGAGAGAGC |
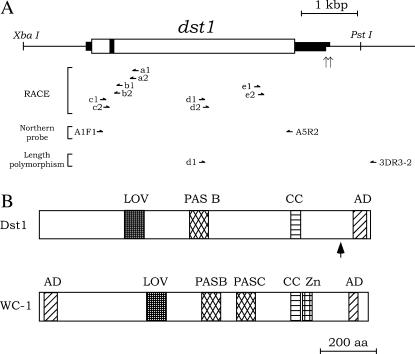
Figure 4.
(A) Structure of the dst1 gene. The thick lines, open box, and solid box indicate the regions transcribed, a deduced ORF, and the intron, respectively. Two dst1 transcripts with distinct poly(A) binding sites were identified by 3′-RACE experiments. Vertical arrows indicate the 3′-ends of the dst1 transcripts. The primers for 5′- and 3′-RACE experiments, for the Northern probe, and for length polymorphism are indicated by horizontal half-arrows. (B) Schematic alignment of domains in Dst1 and WC-1. WC-1 is a blue-light receptor in N. crassa (Ballario et al. 1996). The vertical arrow beneath the Dst1 protein indicates the site of the dst1-1 mutation, which truncates AD in the C-terminal region.
Southern analysis:
DNA was transferred to Hybond N (Amersham, Arlington Heights, IL) according to Sambrook et al. (1989). The Gene Images system (Amersham) was used for probe labeling and detection.
Northern analysis:
Total RNAs were prepared from vegetative mycelia, fruiting-body tissues at various developmental stages, and dark stipes. Vegetative mycelia in MY liquid medium were harvested on a nylon filter, washed thoroughly with distilled water, and squashed between sheets of filter paper. Fruiting body tissues were harvested from cultures grown under a 12 hr light/12 hr dark regime. Dark stipes were harvested from cultures grown in the continuous darkness for 2 days after preincubation for 8 days under a 12 hr light/12 hr dark regime. Samples harvested were frozen in liquid nitrogen and ground into fine powder with a mortar and pestle. RNA was extracted from the powder using Isogen (Nippon Gene, Toyama, Japan). About 15 μg of total RNA was fractionated by electrophoresis in 1.0% agarose formaldehyde gel, transferred to a Hybond-N+ membrane (Amersham) according to Sambrook et al. (1989), and then fixed by UV crosslinking using a UV Stratalinker 1800 (Stratagene, La Jolla, CA). A part (2640 bp) of the dst1 ORF was amplified using the 6-kb PstI/XbaI fragment as a template and a pair of primers, A1F1 and A5R2 (see Table 2; Figure 4A), labeled using the Gene Images system (Amersham), and used as the probe.
DNA isolations:
Cosmid and plasmid DNAs were isolated with the Plasmid Miniprep kit (Bio-Rad, Hercules, CA). Genomic DNA from C. cinereus was prepared as described by Zolan and Pukkila (1986).
RESULTS
Genetic analysis of the dark stipe mutations:
We have obtained five dark stipe mutants, which produce the dark-stipe structure under the light conditions that promote the development of mature fruiting bodies in C. cinereus wild-type strains. Three of the mutants, Uar801, Uad290, and Uad351, were isolated from the homokaryotic fruiting strain, CopD5-12, after UV mutagenesis (Muraguchi et al. 1999), and the remaining two strains, R1428 and H1-1280, were found among hygromycin-resistant transformants of the AmutBmut strain after restriction enzyme-mediated integration (REMI) mutagenesis (Cummings et al. 1999). We first carried out dominance tests on these mutants by mating them with wild-type homokaryons and leaving the resulting heterozygous dikaryons to the conditions that promote the development of mature fruiting bodies. The dikaryons all produced mature fruiting bodies, showing that the five mutations are all recessive. We then performed complementation tests in dikaryons among the five mutations, which revealed that the mutants compose two complementation groups (Table 3). The presumptive gene responsible for the mutations in one group containing strains Uar801 and R1428 was designated dst1 (dark stipe1), and that in the other group containing strains Uad290, Uad351, and H1-1280 was designated dst2 (dark stipe2).
TABLE 3.
Complementation tests between dark-stipe mutant strains
| Strain | Uar801 F1#140 | R1428 F1#52 | Uad290 | Uad351 | H1-1280 F1#5 |
|---|---|---|---|---|---|
| Uar801F1#140 | DS | WT | WT | WT | |
| R1428F1#52 | WT | WT | WT | ||
| Uad290 | NT | DS | |||
| Uad351 | DS | ||||
| H1-1280F1#5 |
WT, wild type; DS, dark stipe; NT, not tested.
Cloning of the dst1 gene:
If the dst1 gene responsible for the mutation in REMI strain R1428 were tagged by the plasmid, it would be possible to perform a plasmid rescue for cloning the dst1 gene. However, F1 analysis of a cross between R1428 and the wild-type homokaryon, 5302, revealed that the dst1 gene in R1428 was not tagged: the dst1 mutation segregated independently from the hygromycin-resistance in the progeny (data not shown). Therefore, we chose to first map the dst1 gene to a chromosome and then screen a cosmid library from the chromosome for a clone that complements the mutation. We crossed another dst1 mutant strain, Uar801 (A12B12 dst1-1), with the wild-type homokaryon, 5302 (A2B2), and analyzed its F1 progeny. We found that the dst1 gene is linked to Hiroshima VII-1#58, an RFLP marker on chromosome IX (Arima et al. 1996), at a map distance of 25% (4/16). We also confirmed that the dark-stipe phenotype was caused by mutation in a single chromosomal gene: of 186 progeny analyzed, 86 exhibited the wild-type phenotype and 100 the dark-stipe phenotype (P for 1:1 = 0.50–0.30).
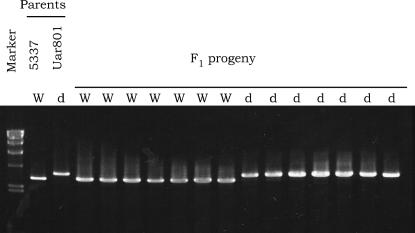
On the basis of the above result suggesting that the dst1 gene is located on chromosome IX, we constructed a cosmid library from chromosome IX and screened it for a clone that complements the dst1 mutation. We found that the dst1 mutation was rescued in 32% (112/346) of trp+ transformants by DNA from cosmid clone 1A1. As a negative control, we transformed strain T140 (A3B1 trp1-1,1-6 dst1-1) with plasmid pCc1003 carrying the wild-type trp1 gene: all 598 trp+ transformants isolated showed the dark-stipe phenotype. We scored 16 progeny from a cross between Uar801 and the wild-type homokaryon, 5337, for length polymorphism of a 3-kb fragment in clone 1A1, showing that the 1A1 region is closely linked to the dst1 locus (Figure 3). We then defined the dst1 region within a 6-kb PstI-XbaI fragment by digestion of clone 1A1 with various restriction enzymes, followed by examination of each digest for the dst1 activity.
Figure 3.
Cosegregation between length polymorphism of a 3-kb region in 1A1 and the dark-stipe phenotype in F1 progeny from the cross Uar801 × 5337. The progeny were scored for fruiting phenotypes. A selection of the dark-stipe mutant and wild-type progeny were then scored for length polymorphism of a 3-kb genomic region in 1A1 amplified using primers A4F3 and 3DR3-2 (see Figure 4A). W, wild type; d, dark stipe. Marker, the HindIII digest of λDNA.
The dst1 gene encodes a putative blue-light receptor:
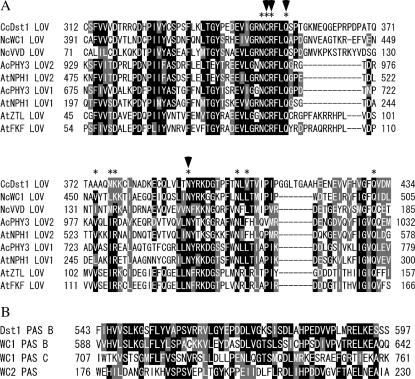
Genomic DNA sequencing, together with 5′- and 3′-RACE experiments, identified an ORF interrupted by a single intron of 68 nucleotides (nt), which encodes 1175 amino acids (Figure 4A). The 5′ splice site, GTACGT, agrees with the consensus sequence GTRNGT found for filamentous fungi and the 3′ splice site, CAG, with the consensus sequence YAG (Gurr et al. 1987). 5′-RACE experiments showed that the dst1 mRNA has a 113-nt 5′-untranslated region, while 3′-RACE experiments identified two kinds of transcripts with distinct poly(A) sites, 3′-untranslated regions of which are predicted to be 564 and 629 nt, respectively (Figure 4A). Database searches using the BLAST procedure (Altschul et al. 1990) revealed that the predicted Dst1 protein has a high similarity to WC-1 (Ballario et al. 1996), a blue-light receptor of N. crassa (Froehlich et al. 2002; He et al. 2002). Motif Scan analysis using the program Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/) showed that, like WC-1, Dst1 has the PAS A domain (LOV domain), the PAS B domain, a coiled-coil structure, and the C-terminal glutamine-rich region, but unlike WC-1, it lacks the PAS C domain, a zinc-finger DNA-binding motif, and the N-terminal glutamine-rich activation region (Figure 4B). It was noted that the LOV and PAS B domain of Dst1 are highly similar to those of WC-1, respectively (Figure 5). Although SMART did not identify the PAS C domain in Dst1, manual comparison revealed a low similarity (11.9% identify) between the PAS C region in WC1 and the corresponding region in Dst1.
Figure 5.
(A) Alignment of amino acid sequences of LOV domains from Dst1, WC-1 (Ballario et al. 1996), VVD (Heintzen et al. 2001), A. capillus-veneris PHY3 (Nozue et al. 1998), and Arabidopsis thaliana phototropins NPH1 (Huala et al. 1997), ZTL (Somers et al. 2000), and FKF1 (Nelson et al. 2000). The two LOV domains of PHY3 and NPH1 are both aligned. Asterisks mark the 11 FMN-interacting residues identified in the LOV2 domain of PHY3 (Crosson and Moffat 2001). Arrowheads mark the four residues that are essential for the light function of WC-1 (Cheng et al. 2003b). The similar and identical residues are shaded and solid, respectively, when >5 of 9 residues are similar or identical. The numbers on the left and right of each sequence indicate the numbers of the N-terminal and C-terminal residues in the sequence. (B) Alignment of amino acid sequences of PAS domains from Dst1, WC-1 (Ballario et al. 1996), and WC-2 (Linden and Macino 1997). The numbers are as in A.
dst1-1 mutant allele:
We performed PCR amplifications on the genomic DNA of the dst1-1 mutant using primers that were designed on the basis of the wild-type dst1 sequence and sequenced the PCR products directly. Comparison of the dst1-1 mutant allele with the wild-type dst1 gene of the parental CopD5-12 strain as well as with that of strain 5302, from which the cosmid library was constructed, revealed a one-nucleotide substitution (T to G), which yields a stop codon (TGA) at 3200–3202 bp downstream from the translational start site, truncating the C-terminal 109 amino acids.
Developmental regulation of dst1 transcription:
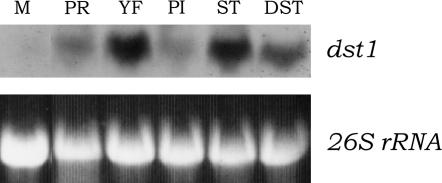
Total RNAs from various development phases of mycelia and tissues were subjected to Northern analysis for examination of dst1 transcription. As expected from cDNA analysis, we identified a transcript at ∼4.3 kb in all mycelia and tissues examined (Figure 6). The transcription of dst1 was higher in fruiting-body primordia than in vegetative mycelia (Figure 6, lanes 1 and 2). The transcription level was also higher in young fruiting bodies, and the level was higher in young stipes as compared with that in young pileus (Figure 6; lanes 3–5). The level of dst1 transcription was also high in the dark stipe (Figure 6, lane 6). The results that the primordia and dark stipes exhibited high levels of dst1 transcription are explainable, because both have to regulate their development in response to faint light (Kamada et al. 1978). However, we cannot give any explanation for a high level of dst1 transcription in young fruiting bodies at present, because young fruiting bodies in this developmental stage undergo stipe elongation and pileus development irrespective of light conditions (Kamada et al. 1978).
Figure 6.
Expression of the wild-type dst1 gene. For each lane, 15 μg of total RNAs from the following were electrophoresed in a 1.0% agarose formaldehyde gel: vegetative mycelia cultured for 3 days (M), fruiting-body primordia of ∼2 mm in height (PR), young fruiting bodies on the day when fruiting-body maturation occurs (YP), pilei from young fruiting bodies (PI), stipe from young fruiting bodies (ST), and dark stipes (DST). After the ribosomal RNA was visualized under UV, the gel was blotted and hybridized. The probe was the 2640-bp PCR product including almost the entire ORF of dst1 (see Figure 4A). After hybridization, the blot was exposed to X-ray film. Similar results were reproducibly obtained in three independent experiments.
DISCUSSION
In this study, we characterized a photomorphogenetic mutant, dst1-1, of C. cinereus. The mutant produces dark stipes under light conditions that normally promote the development of fruiting-body primordia into mature fruiting bodies. Because the dark stipe is the figure that the primordium forms when left in the dark, it seems that the mutant is defective in perception of light. dst1, the gene responsible for the mutation, was found to encode a protein (Dst1) containing a LOV domain, which has strong similarities to those of the blue-light receptors in N. crassa, WC-1 and VVD, and of the blue-light receptors in plants, phototropins ZTL and FKF (Figure 5A). It has been demonstrated in the fern Adiantum capillus-veneris that the 11-amino-acid residues in the LOV2 domain of PHY3 interact with its chromophore, flavin mononucleotide (FMN) (Crosson and Moffat 2001). Also, it has been shown in N. crassa that 7 of the 11 residues are conserved in WC-1 and that amino acid replacement at any of 4 residues (429, 430, 432, and 471) from among the 7 residues completely eliminates blue-light responses in this fungus (Cheng et al. 2003b). In the LOV domain of Dst1, 6 of the 11 residues, including all of those corresponding to the 4 residues, are conserved (Figure 5A). These findings strongly suggest that Dst1 is a blue-light photoreceptor in C. cinereus.
All known LOV-containing photoreceptors except VVD are multi-domain proteins containing regulatory and/or protein-protein interaction domains in addition to the LOV domain. Phototropins contain a serine-threonine protein kinase domain (Huala et al. 1997). ZTL and FKF1 have an F-box and six kelch repeats (Nelson et al. 2000; Somers et al. 2000). In addition to the LOV domain (PAS A), WC-1 has the PAS B domain, the PAS C domain, a zinc-finger DNA-binding domain, and two glutamine-rich AD domains, one at the N terminus and the other at the C terminus (Cheng et al. 2002; He et al. 2003). Dst1 is similar to WC-1 in that it contains, in addition to the LOV domain, the PAS B domain, a coiled-coil structure, and a putative, glutamine-rich AD domain at the C terminus. However, Dst1 lacks three regions corresponding to the N-terminal AD domain, the PAS C domain, and the zinc-finger domain in WC-1. Very recently, a gene encoding a LOV-containing protein lacking a zinc-finger DNA-binding domain, designated BWC1, was identified in C. neoformans (Idnurm and Heitman 2005).
We found that the dst1-1 mutant allele carries a nonsense mutation truncating the whole of the putative, glutamine-rich AD at the C terminus. This finding demonstrates that the glutamine-rich AD is essential for normal function of Dst1. This in turn suggests that Dst1 works in direct or indirect interaction with a protein containing a DNA-binding domain, which is lacking in Dst1 as described above. This hypothesis is supported by the fact that Dst1 contains the PAS B domain, which potentially plays a role in protein-protein interaction (Taylor and Zhulin 1999), and a coiled-coil structure. In N. crassa, WC-1 interacts with WC-2, one of the central components in the blue-light signal transduction pathway of this fungus (Talora et al. 1999; Denault et al. 2001; Cheng et al. 2002, 2003a). By analogy to the WC-1-WC-2 interaction, we might be able to hypothesize that Dst1 collaborates with a presumptive WC-2 homolog in C. cinereus. However, it is not clear at present whether a region in Dst1 corresponding to PAS C, which has been demonstrated to play a significant role for interaction with WC-2 (Cheng et al. 2003a), is functional as a PAS domain. In addition, we failed to identify a WC-2 homolog in the draft sequence of the C. cinereus genome (http://www.broad.mit.edu/annotation/fungi/coprinus_cinereus). A presumptive, interacting partner of Dst1, therefore, seems to be much different in amino acid sequence from WC-2. A candidate for the interacting partner of Dst1 might be a protein encoded by another gene, dst2, a mutation of which causes the dark-stipe phenotype, as described above. Cloning of dst2 is now in progress.
In the development of the fruiting-body primordium, light plays two roles. One is the inhibition of etiolation of the primordium and the other is the triggering of development of the primordium into the mature fruiting body (Kamada et al. 1978). The dst1-1 mutant is defective in the inhibition of etiolation, producing the dark stipe (Figure 2). Because the mutant Dst1 is predicted to lack the C-terminal AD, retaining other regions including LOV and PAS B domains and the coiled-coil structure, as described above, it is clear that AD is a prerequisite for the inhibition of etiolation. The dst1-1 mutant regularly developed mature fruiting bodies at the top of dark stipes after a prolonged cultivation. It may be that the mutant Dst1 without AD still functions in the photocontrol of fruiting-body maturation. Alternatively, there may be another photoreceptor in C. cinereus. A future challenge would be analysis of a null dst1 mutant as well as of dst1 mutants in which each of the domains was disrupted.
Acknowledgments
We thank Miriam E. Zolan for the REMI mutant strains, R1428 and H1-1280. This work was supported in part by a grant-in-aid from the Japanese Society for the Promotion of Science (K.T.).
Sequence data from this article have been deposited with the DDJB/EMBL/GenBank Data Libraries under accession no. AB195817.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Ambra, R., B. Grimaldi, S. Zamboni, P. Filetici, G. Macino et al., 2004. Photomorphogenesis in the hypogeous fungus Tuber borchii: isolation and characterization of Tbwc-1, the homologue of the blue-light photoreceptor of Neurospora crassa. Fungal Genet. Biol. 41: 688–697. [DOI] [PubMed] [Google Scholar]
- Arima, T., T. Okida and T. Morinaga, 1996. Behavior of chromosomes after meiosis in Coprinus cinereus. Mycoscience 37: 111–115. [Google Scholar]
- Ballario, P., P. Vittorioso, A. Magrelli, C. Talora, A. Cabibbo et al., 1996. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 15: 1650–1657. [PMC free article] [PubMed] [Google Scholar]
- Binninger, D. M., C. Skrzynia, P. J. Pukkila and L. A. Casselton, 1987. DNA-mediated transformation of the basidiomycete Coprinus cinereus. EMBO J. 6: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Flores, S., M. Rios-Momberg, M. Bibbins, P. Ponce-Noyola and A. Herrela-Estrella, 2004. BLR-1 and BLR-2, key regulatory elements of photoconidiation and mycelial growth in Trichoderma atroviride. Microbiology 150: 3561–3569. [DOI] [PubMed] [Google Scholar]
- Cheng, P., Y. Yang, K. H. Gardner and Y. Liu, 2002. PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady-state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol. 22: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, P., Y. Yang, L. Wang, Q. He and Y. Liu, 2003. a WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing, and transcription repression of wc-2. J. Biol. Chem. 278: 3801–3808. [DOI] [PubMed] [Google Scholar]
- Cheng, P., Q. He, Y. Yang, L. Wang and Y. Liu, 2003. b Functional conservation of light, oxygen, or voltage domains in light sensing. Proc. Natl. Acad. Sci. USA 100: 5938–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J. M., and W. R. Briggs, 2001. Blue light sensing in higher plants. J. Biol. Chem. 276: 11457–11460. [DOI] [PubMed] [Google Scholar]
- Crosson, S., and K. Moffat, 2001. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. USA 98: 2995–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings, W. J., M. Celerin, J. Crodian, L. K. Brunick and M. E. Zolan, 1999. Insertional mutagenesis in Coprinus cinereus: use of a dominant selectable marker to generate tagged, sporulation-defective mutants. Curr. Genet. 36: 371–382. [DOI] [PubMed] [Google Scholar]
- Denault, D. L., J. J. Loros and J. C. Dunlap, 2001. WC-2 mediates WC-1–FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J. 20: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P. F., 2002. Signs of the time: environmental input to the circadian clock. J. Exp. Bot. 53: 1535–1550. [DOI] [PubMed] [Google Scholar]
- Durand, R., and M. Furuya, 1985. Action spectra for stimulatory and inhibitory effects of UV and blue light on fruit-body formation in Coprinus congregatus. Plant Cell Physiol. 26: 1175–1183. [Google Scholar]
- Durand, R., and R. Jaques, 1982. Action spectra for fruiting of the mushroom Coprinus congregatus. Arch. Microbiol. 132: 131–134. [Google Scholar]
- Froehlich, A. C., Y. Liu, J. J. Loros and J. C. Dunlap, 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819. [DOI] [PubMed] [Google Scholar]
- Gurr, S. J., S. E. Unkles and J. R. Kinghorn, 1987. The structure and organization of nuclear genes of filamentous fungi, pp. 93–139 in Gene Structure in Eukaryotic Microbes, edited by J. R. Kinghorn. IRL Press, London.
- He, Q., P. Cheng, Y. Yang, L. Wang, K. H. Gardner et al., 2002. White collar-1, a DNA binding tanscription factor and a light sensor. Science 297: 840–843. [DOI] [PubMed] [Google Scholar]
- Heintzen, C., J. J. Loros and J. C. Dunlap, 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464. [DOI] [PubMed] [Google Scholar]
- Huala, E., P. W. Oeller, E. Liscum, I. S. Han, E. Larsen et al., 1997. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278: 2120–2123. [DOI] [PubMed] [Google Scholar]
- Idnurm, A., and J. Heitman, 2005. Light controls growth and development via a conserved pathway in the fungal kingdom. PloS Biol. 3: e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada, K., Y. Morimoto, T. Arima, Y. Murata and T. Kamada, 2001. The clp1 gene of the mushroom Coprinus cinereus is essential for A-regulated sexual development. Genetics 157: 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada, T., R. Kurita and T. Takemaru, 1978. Effects of light on basidiocarp maturation in Coprinus macrorhizus. Plant Cell Physiol. 19: 263–275. [Google Scholar]
- Kües, U., 2000. Life history and development processes in the basidiomycete Coprinus cinereus. Microbiol. Mol. Biol. Rev. 64: 316–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., 2000. Plant blue-light receptors. Trends Plant Sci. 5: 337–342. [DOI] [PubMed] [Google Scholar]
- Linden, H., and G. Macino, 1997. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 16: 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B. C., 2000. The control of meiosis progression in the fungus Coprinus cinereus by light/dark cycles. Fungal Genet. Biol. 31: 33–41. [DOI] [PubMed] [Google Scholar]
- Muraguchi, H., and T. Kamada, 1998. The ich1 gene of the mushroom Coprinus cinereus is essential for pileus formation in fruiting. Development 125: 3133–3141. [DOI] [PubMed] [Google Scholar]
- Muraguchi, H., T. Takemaru and T. Kamada, 1999. Isolation and characterization of developmental variants in fruiting using a homokaryotic fruiting strain of Coprinus cinereus. Mycoscience 40: 227–233. [Google Scholar]
- Nelson, D. C., J. Lasswell, L. E. Rogg, M. A. Cohen and B. Bartel, 2000. FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101: 331–340. [DOI] [PubMed] [Google Scholar]
- Nozue, K., T. Kanegae, T. Imaizumi, S. Fukuda, H. Okamoto et al., 1998. A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc. Natl. Acad. Sci. USA 95: 15826–15830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila, P. J., and L. A. Casselton, 1991. Molecular genetics of the agaric Coprinus cinereus, pp. 126–150 in More Gene Manipulations in Fungi, edited by J. W. Bennett and L. L. Lasure. Academic Press, San Diego.
- Rao, P. S., and D. J. Niederpruem, 1969. Carbohydrate metabolism during morphogenesis of Coprinus lagopus (sensu Buller). J. Bacteriol. 100: 1222–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schwerdtfeger, C., and H. Linden, 2003. VIVID is a flavoprotein and serves as a fungal blue light photoreceptor for photoadaptation. EMBO J. 22: 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrzynia, C., D. M. Binninger, J. A. Alspaugh, II, and P. J. Pukkila, 1989. Molecular characterization of TRP1, a gene coding for tryptophan synthetase in the basidiomycete Coprinus cinereus. Gene 81: 73–82. [DOI] [PubMed] [Google Scholar]
- Somers, D. E., T. F. Schultz, M. Milnamow and S. A. Kay, 2000. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329. [DOI] [PubMed] [Google Scholar]
- Talora, C., L. Franchi, H. Linden, P. Ballario and G. Macino, 1999. Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J. 18: 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, B. L., and I. B. Zhulin, 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsusué, Y. M., 1969. Experimental control of fruit-body formation in Coprinus macrorhizus. Dev. Growth Differ. 11: 164–178. [DOI] [PubMed] [Google Scholar]
- Zolan, M. E., and P. J. Pukkila, 1986. Inheritance of DNA methylation in Coprinus cinereus. Mol. Cell. Biol. 6: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolan, M. E., J. R. Crittenden, N. K. Heyler and L. C. Seitz, 1992. Efficient isolation and mapping of rad genes of the fungus Coprinus cinereus using chromosome-specific libraries. Nucleic Acid Res. 20: 3993–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]