Abstract
Lamins are intermediate filaments that line the inner surface of the nuclear envelope, providing structural support and making contacts with chromatin. There are two types of lamins, A- and B-types, which differ in structure and expression. Drosophila possesses both lamin types, encoded by the LamC (A-type) and lamin Dm0 (B-type) genes. LamC is nested within an intron of the essential gene ttv. We demonstrate that null mutations in LamC are lethal, and expression of a wild-type LamC transgene rescues lethality of LamC but not ttv mutants. Mutations in the human A-type lamin gene lead to diseases called laminopathies. To determine if Drosophila might serve as a useful model to study lamin biology and disease mechanisms, we generated transgenic flies expressing mutant LamC proteins modeled after human disease-causing lamins. These transgenic animals display a nuclear lamin aggregation phenotype remarkably similar to that observed when human mutant A-type lamins are expressed in mammalian cells. LamC aggregates also cause disorganization of lamin Dm0, indicating interdependence of both lamin types for proper lamina assembly. Taken together, these data provide the first detailed genetic analysis of the LamC gene and support using Drosophila as a model to study the role of lamins in disease.
LAMINS belong to a family of structural proteins known as intermediate filaments (Stuurman et al. 1998). All intermediate filaments, except lamins, localize to the cytoplasm where they impart physical strength to a cell. In contrast, lamins localize to the inner surface of the nuclear envelope, providing structural support for the nucleus and making contacts with other nuclear components (Zastrow et al. 2004). Lamins possess an N-terminal head domain, a central α-helical rod domain, and a C-terminal globular domain (Stuurman et al. 1998). Dimerization occurs through interactions in the rod domain; dimers associate in a head-to-tail fashion to form protofilaments, which then align in antiparallel orthogonal arrays to form a meshwork called the lamina (Stuurman et al. 1998). The nuclear lamina possesses additional protein components, including the so-called “LEM” domain proteins, named after a conserved domain found in LAP2, Emerin, and MAN1 (Laguri et al. 2001). Components of the nuclear lamina make connections with histones (Goldberg et al. 1999), transcription factors (Ozaki et al. 1994), and nucleic acids (Rzepecki et al. 1998). These interactions are likely to play a role in spatially organizing the genome and regulating gene expression.
There are two types of lamins, A- and B-types, which differ in their protein structure and expression patterns (Hutchison 2002). In humans, B-type lamins are encoded by the genes LMNB1 and LMNB2. B-type lamins are ubiquitously expressed and possess a C-terminal CaaX box (C, cysteine; a, an aliphatic amino acid; X, any amino acid) that is isoprenylated and carboxy methylated, serving as a membrane anchor. Mammalian cells require at least one B-type lamin for viability (Harborth et al. 2001). In humans, A-type lamins are encoded by the LMNA gene. Alternative splicing generates messages encoding two isoforms: lamin A and lamin C. Expression of A-type lamins is limited to terminally differentiated somatic cells. Human lamin A possesses a CaaX box that is proteolytically removed to form the mature protein, whereas no CaaX box is encoded in the primary amino acid sequence for lamin C.
A-type lamins are not required for viability in mammalian cell culture (Harborth et al. 2001); however, in humans, mutations in LMNA are associated with a range of diseases known as laminopathies. Many of these diseases have tissue-restricted phenotypes, such as Emery-Dreifuss muscular dystrophy (EDMD) (Bonne et al. 2000) and familial dilated cardiomyopathy (Brodsky et al. 2000) that affect skeletal and cardiac muscle; Dunnigans's familial partial lipodystrophy (Shackleton et al. 2000) that affects adipose tissue; and Charcot-Marie Tooth syndrome type 2 that is a neuropathy (Chaouch et al. 2003). For a few laminopathies, the phenotype appears to be more systemic; these diseases include Hutchinson-Gilford progeria syndrome (HGPS) (Eriksson et al. 2003) and atypical Werner's syndrome (Chen et al. 2003), both diseases of premature aging. It is unclear how different mutant forms of A-type lamin cause these various disease phenotypes (Burke and Stewart 2002). With the possible exceptions of Dunnigan's familial partial lipodystrophy and HGPS, the amino acid substitutions responsible for a given laminopathy do not map to a specific domain of the lamin protein (Burke and Stewart 2002). Therefore, functional studies are required to determine how distinct molecular defects arise from particular amino acid substitutions in specific protein domains.
To address the molecular defects associated with expression of mutant A-type lamins in disease, genetically tractable model organisms are invaluable. Analyses of LMNA gene knock-out and mutant LMNA gene knock-in transgenic mice have provided important insights into disease progression. Knocking out LMNA or LMNB1 in mice causes lethality shortly after birth (Sullivan et al. 1999; Vergnes et al. 2004). Knock-in mice expressing mutated lamin A/C proteins (H222P and L530P) exhibit phenotypes similar to EDMD and HGPS (Mounkes et al. 2003; Arimura et al. 2005). Nevertheless, a comprehensive study of the numerous lamin mutations associated with these diseases would benefit from the rapid generational analysis that is possible in a well-characterized invertebrate model organism. Most invertebrates, however, do not possess the two distinct lamin types found in humans. An exception is Drosophila melanogaster, which possesses two genes encoding nuclear lamins. lamin Dm0 encodes a B-type lamin on the basis of its constitutive expression pattern, possession of a CaaX box, and in vitro assembly properties (Riemer et al. 1995; Klapper et al. 1997). Mutations in lamin Dm0 show defects in locomotion, tracheal development, and nuclear positioning in the oocyte and eye (Lenz-Bohme et al. 1997; Guillemin et al. 2001; Patterson et al. 2004). In contrast, Lamin C (LamC) encodes an A-type lamin, on the basis of its developmentally regulated pattern of gene expression and lack of a CaaX box (Riemer et al. 1995).
To understand the function of the A-type lamins in Drosophila, we undertook a formal genetic and molecular analysis of LamC. We demonstrate that LamC is an essential gene nested within another essential gene called tout-velu (ttv). LamC null mutants die at the prepupal stage, consistent with a critical role in differentiating tissues. To assess whether LamC functions similarly to its human counterpart, we have generated transgenic stocks expressing mutant forms of LamC modeled after mutations linked with human disease. These transgenic animals exhibit nuclear phenotypes that are remarkably similar to those observed in mammalian cell culture upon expression of mutant human A-type lamins. Collectively, these data provide a foundation for using an insect model to dissect the biology of nuclear lamins and their role in disease.
MATERIALS AND METHODS
Drosophila culture and imprecise excision:
Drosophila stocks were raised at room temperature on standard sucrose/cornmeal medium. All crosses were performed at room temperature unless otherwise stated. Crosses for complementation tests, rescue experiments, and transgene expression assays were carried out in vials. For heat-shock-induced expression, 45-min heat-shock treatments were administered daily throughout development by placing vials in a 37° water bath and then returning the cultures to room temperature for recovery.
To generate deletions of LamC, a P-element excision scheme (Adams and Sekelsky 2002) was performed as follows: females bearing the G00158 green fluorescent protein (GFP) exon-trap allele (Morin et al. 2001) over a CyO balancer chromosome (w/w; G00158/CyO; +/+) were mated to males carrying a second chromosome balanced over SM1 and a transposase source, Δ2-3, marked with Sb on the third chromosome (w/Y; +/SM1; Δ2-3Sb/TM6, Tb). Individual F1 male progeny that were heterozygous for the G00158 allele and the transposase source (w/Y; G00158/SM1; Δ2-3Sb/+) were outcrossed to a homozygous white stock with dominantly marked second chromosomes (y1w67c23/ y1w67c23; Sco/SM1). Resulting white mutant F2 male progeny carrying a marked second chromosome were individually backcrossed to flies of the maternal genotype (y1w67c23/y1w67c23; Sco/SM1); the resulting flies carrying potential deletions of LamC were balanced over SM1.
Lethal phase analysis:
To determine the lethal phase of a LamC deletion, the putative null excision allele, LamCEX296, and a deficiency for the region, Df(2R)trix, were used. LamCEX296/CyO-GFP males were crossed to Df(2R)trix/CyO-GFP virgin females. Resulting embryos were collected in cornmeal/yeast bottles overnight at room temperature and aged for 5 days. A total of 200–250 larvae were collected from the food and scored for GFP using a Leica MZ12 dissecting microscope equipped with a fluorescent light source (Kramer Scientific, Yonkers, NY). Approximately 60 GFP-negative and GFP-positive larvae were collected per assay. Larvae were transferred to 350-mm petri dishes containing Whatman filter paper moistened with water and pulvarized cornmeal/sucrose media. The number of individuals at each developmental stage was recorded daily. Larval stages were identified by the morphology of mouth hooks and/or anterior spiracles. As a control, LamCEX296/CyO-GFP and Df(2R)trix/CyO-GFP siblings were scored in parallel. As an external control, +/CyO-GFP males were crossed to Df(2R)trix/CyO-GFP virgin females, and the +/Df(2R)trix progeny were scored.
Transgene design:
A full-length LamC cDNA was amplified by PCR from 18- to 21-hr embryonic RNA [purchased from CLONTECH (Palo Alto, CA)]. The cDNA was cloned into pCR2.1-TOPO (Invitrogen, San Diego) and sequenced. A comparison of the LamC DNA sequence to that present in FlyBase (http://flybase.bio.indiana.edu/) revealed seven silent mutations; these could represent naturally occurring strain polymorphisms and/or errors generated during PCR. The full-length LamC cDNA was used as a template for in vitro mutagenesis (QuikChange, Invitrogen) to generate the R401K mutant transgene. For the N-terminal truncation, a 33-amino-acid deletion generated in the human A-type lamin (Spann et al. 1997) served as a model. Alignment of human lamin A/C and Drosophila LamC amino acid sequences identified amino acid position 48 in the Drosophila protein as the equivalent end point of the truncation. The following primers were designed for Pfu-Ultra (Stratagene, La Jolla, CA) amplification of the N-terminal deletion mutant: 5′-CAAACATGGAACTGCAGCATTTGAACGATC-3′, which encodes a consensus translation start sequence, and 5′-CTAGAAGAGCAGGGAGAAGAG-3′, which includes the last 6 amino acids and a termination codon. cDNAs encoding wild-type LamC, R401K, and the N-terminal truncation were cloned into the two P-element germline transformation vectors pUAST (GAL4 inducible) and pCaSpeR-hs/act (heat-shock inducible). Constructs were injected according to standard procedures. For each transgenic line, Southern analysis was performed to examine transgene integrity and copy number; Western analysis was performed to examine LamC expression levels (see below).
Molecular characterization of the excision alleles:
For Southern analysis of LamC alleles, genomic DNA was isolated from 100 adult flies (Bender et al. 1983). For each sample, 3 μg of DNA was digested for 3 hr by the appropriate restriction endonucleases (New England Biolabs, Beverly, MA). The DNA was ethanol precipitated, resuspended in dH2O, separated by agarose gel electrophoresis, and transferred to positively charged nylon membrane (Hybond N; Amersham, Arlington Heights, IL). Hybridization was carried out using a nonradioactive LamC cDNA digoxygenin (DIG)-labeled probe (DIG High Prime DNA labeling and detection kit II; Roche, Indianapolis). Anti-DIG-AP conjugate antibody was used to detect hybridization, followed by CSPD chemiluminescence reaction and exposure to X-ray film. All procedures were carried out according to manufacturer's guidelines.
To generate adults homozygous for a given excision allele, chromosomes carrying excisions were balanced over a CyO chromosome possessing a GFP reporter gene and crossed into a background containing an X-linked wild-type Lamin C transgene. Daily heat shocks of 45 min in a 37° water bath were administered throughout development, and DNA was extracted from individual homozygous (straight wing) adults (Gloor et al. 1993). PCR was performed to determine the integrity of genomic exons, using primers that would distinguish them from the sequences within the LamC transgene present in the background. The regions encompassing the deletion breakpoints in the excision stocks LamCEX187 and LamCEX296 were amplified by Pfu-Ultra (Stratagene), cloned into pCR2.1-TOPO (Invitrogen), and sequenced (University of Iowa DNA Core Facility).
Western analysis:
To determine the expression levels of LamC, proteins were extracted from third instar larvae or adults (Friedman et al. 1992) and separated by size on 10–12% polyacrylamide gels, transferred to nitrocellulose membrane, and incubated with anti-LamC LC28.26 anti-mouse IgG (Riemer et al. 1995) used at 1:5000–1:8000 dilution or anti-α-tubulin anti-mouse IgG1 [Sigma (St. Louis) no. T5168] used at 1:400,000 dilution. An HRP-conjugated anti-mouse IgG [Pierce (Rockford, IL) no. 31446] used at a 1:20,000 dilution served as a secondary antibody. Detection was carried out using the SuperSignal West Pico chemiluminescent substrate (Pierce no. 34080). Signal from the membranes was collected from an Epi Chemi II darkroom unit fitted with a CCD camera (UVP, San Gabriel, CA) and the resulting data were quantified using LabWorks Image Acquisition software (UVP) and/or Image J software (http://rsb.info.nih.gov/ij/). Three independent protein isolations were performed for each genotype. Means and standard deviations were calculated; the formula shown below was used to calculate the standard error of the variance between the expression level in a mutant relative to that in the normalized wild-type controls:
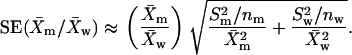 |
SE represents the standard error, 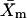 represents the mean of the LamC:tubulin ratio in the mutant,
represents the mean of the LamC:tubulin ratio in the mutant, 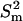 represents the square of the variance of the LamC:tubulin ratio in the mutant,
represents the square of the variance of the LamC:tubulin ratio in the mutant, 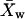 represents the mean of the LamC:tubulin ratio in the wild type, and
represents the mean of the LamC:tubulin ratio in the wild type, and 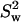 is the square of the variance of the LamC:tubulin ratio in the wild type. n is the number of independent experiments performed.
is the square of the variance of the LamC:tubulin ratio in the wild type. n is the number of independent experiments performed.
Nuclear staining and quantitation:
For nuclear morphology studies, third instar larvae were raised at room temperature in vials, administered a heat shock (45 min, 37°), and allowed to recover for ∼2 hr. Salivary glands ranging in age from early second instar to late third instar larval were dissected in phosphate-buffered saline solution (PBS). Additional tissues released upon salivary gland dissection were also stained; these included epithelial tissues, imaginal discs, brain, and gut. The total dissection time for an experiment was not >1.5 hr. For salivary glands, three to five pairs per genotype were placed in welled slides for fixation in 2% paraformaldehyde for 15–20 min, followed by 3 × 5-min washes in PBS2+ (130 mm NaCl, 7 mm Na2HPO4, 3 mm NaH2PO4, 10 mm EGTA, 0.1% Triton-X). The glands were blocked in PBS2+ + 0.1% BSA for 60 min and then incubated with 1:500 dilution of primary antibody (LC28.26 for LamC and ADL84.12 or ADL67.10 for lamin Dm0, University of Iowa Hybridoma Core Facility) in PBS2+ + 0.1% BSA. Incubation was carried out overnight at 4° in rotating 1.5-ml microfuge tubes. Following incubation with primary antibody, the salivary glands were washed 3 × 5 min in PBS2+, followed by blocking in PBS2+ + 0.1% BSA for 60 min, and then incubated for 1 hr in the dark with a 1:1000 dilution of Rhodamine-conjugated goat anti-mouse IgG + IgM secondary antibody (Jackson ImmunoResearch Labs, West Grove, PA, no. 115-025-068). The glands were then washed for 3 × 10 min in PBS2+ and treated with DAPI at a concentration of 250 ng/μl for ∼30 sec. The glands were washed for 3 × 10 min in PBS2+ and mounted in Vectashield H-1000 (Vector Laboratories, Burlingame, CA). The slides were placed in a dark box and left at 4° overnight. The nuclei were examined using a Leica DMLB compound microscope with fluorescent capabilities, and the images were collected and processed with a Spot RT-Slider CCD camera (Diagnostic Instruments) and Spot Advanced software. To determine the percentage of nuclei showing a lamin localization defect, nuclei within the first third of a gland (where visibility and antibody adsorption are best, due to reduced thickness of the tissue) were counted. The percentage of nuclei showing abnormal localization was calculated for at least three independent preparations per genotype.
RESULTS
Mutations in Lamin C are lethal:
To determine the function of A-type lamins in Drosophila, a genetic analysis of the Drosophila LamC gene was carried out. The LamC gene maps to cytological position 51B1 on the right arm of the second chromosome, spans ∼5 kb of genomic territory, and is nested within the fifth intron of the essential gene ttv, which encodes a protein involved in heparin sulfate proteoglycan biosynthesis (Bellaiche et al. 1998). Three lethal P-element transposon insertions, EP(2)2199, UM-8373, and G00158, all located within the first intron of LamC (Figure 1A), are available through stock centers and academic collections [EP(2)2199, http://expbio.bio.u-szeged.hu/fly/; UM-8373, http://www.drosdel.org.uk/; G00158, http://flytrap.med.yale.edu/). Due to the nested arrangement, it is unclear whether the lethality results from disruption of LamC, ttv, or both. To determine whether LamC and ttv encode genetically separable functions, we generated additional mutant LamC alleles and carried out complementation tests.
Figure 1.
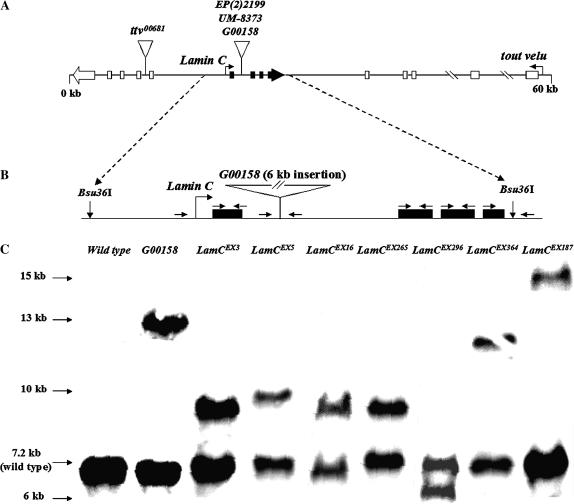
(A) Diagram of the LamC and ttv genomic region. LamC (solid exons) is nested within the fifth intron of the essential gene tout velu (open exons). EP(2)2199, UM-8373, and G00158 are lethal P-element insertions in Lamin C. ttv00681 is a lethal insertion into the sixth intron of tout velu. (B) Diagram of LamC with the location of the P-element insert in stock G00158 indicated. G00158 is mutant for LamC and ttv. The diagram shows the position of the primers (arrows) used for PCR and sequencing (Table 3). (C) Southern analysis of LamC alleles. Bsu36I cleavage sites flank the LamC genomic region and generate a 7.2-kb fragment containing the wild-type LamC gene. The membrane was hybridized with full-length LamC cDNA.
Initial complementation tests were performed with Df(2R)trix (a stock carrying a deletion of ∼60 genes including LamC and ttv), the three LamC P-element insertions, and the well-characterized ttv00681 null allele (The et al. 1999). Complementation between lesions in different, noninteracting genes results in trans-heterozygote adult viability and fertility. As anticipated, Df(2R)trix failed to complement all three LamC P-element insertion alleles and ttv00681 (Table 1). The P-element insertion stocks UM-8373 and G00158 also failed to complement EP(2)2199 and ttv00681, suggesting that UM-8373 and G00158 are doubly mutant for LamC and ttv. EP(2)2199, however, does complement ttv00681, indicating that the P-element insert in this stock specifically disrupts LamC function.
TABLE 1.
Complementation analysis of Lamin C alleles
| Df(2R)trix | ttv00681 | EP(2)2199 | UM8373 | G00158 | LamCEX3 | LamCEX5 | LamCEX16 | LamCEX187 | LamCEX265 | LamCEX296 | LamCEX364 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Df(2R)trix | − | − | − | − | − | − | − | − | − | − | − | |
| ttv00681 | + | − | − | + | + | + | + | + | + | + | ||
| EP(2)2199 | − | − | − | − | − | − | − | − | − | |||
| UM8373 | − | − | − | − | − | − | − | − | ||||
| G00158 | − | − | − | − | − | − | − | |||||
| LamCEX3 | − | − | − | − | − | − | ||||||
| LamCEX5 | − | − | − | − | − | |||||||
| LamCEX16 | − | − | − | − | ||||||||
| LamCEX187 | − | − | − | |||||||||
| LamCEX265 | − | − | ||||||||||
| LamCEX296 | − | |||||||||||
| LamCEX364 |
All alleles and the Df(2R)trix are balanced over the CyO chromosome.
To generate additional mutant alleles of LamC, imprecise excision (Adams and Sekelsky 2002) was performed using the P-element insertion stock G00158. The P element in this stock contains coding sequences for GFP flanked by splice donor and acceptor sites (“exon trap”) and a white+ reporter gene (Morin et al. 2001). Insertion of this P element within the intron of LamC generates a LamC fusion protein containing GFP sequences within the rod domain; the LamC-GFP fusion protein exhibits abnormal nuclear localization (Morin et al. 2001). Flies from the G00158 stock were crossed to flies expressing transposase. Mobilization of the P element was scored by the presence of a white eye phenotype and/or loss of GFP fluorescence. From a total of 236 independent P-element excision events, 24 generated a genetic lesion that was homozygous lethal. Of these 24, a complementation test showed that 10 were lethal for LamC and not for ttv. These lethal excision alleles failed to complement Df(2R)trix and the LamC P-element insertions; however, they all complemented ttv00681 (Table 1). Therefore, 10 novel mutant LamC alleles were generated.
Wild-type Lamin C transgenes rescue lethality of LamC mutants:
To confirm that LamC and ttv encode unrelated essential functions, we performed experiments to rescue the lethality of mutant LamC alleles by supplying a source of wild-type LamC protein. To this end, we generated transgenic stocks expressing wild-type LamC under control of either a heat-shock hsp70 promoter or a GAL4/UAS-driven promoter (Duffy 2002). Rescue data for heat-shock-induced expression of wild-type LamC are shown in Table 2. Viable, fertile adults were obtained for the trans-heterozygous combination of LamC P-element alleles UM-8373 and EP(2)2199 under heat-shock conditions (applied 45 min daily, with recovery at room temperature). Rescue of this combination of mutant alleles required heat-shock treatment; in addition, no rescue was observed when trans-heterozygous combinations included alleles that were lethal for ttv (UM-8373 and ttv00681). Only partial rescue was observed (27% of the expected class), possibly due to inappropriate levels of LamC produced by the daily heat-shock treatment. Rescue using GAL4/UAS-driven LamC gave similar results (data not shown). Thus, the lethality associated with mutations in LamC can be rescued by transgenic wild-type LamC expression, confirming that LamC is an essential gene.
TABLE 2.
Rescue of Lamin C lethality: Hsp70-Lamin C/Y; m1/CyO × w/w; m2/CyO
| No heat shock
|
Heat shock (37°/45 min)
|
|||
|---|---|---|---|---|
| F1 genotype (m1/m2) | Proportion of expected class | Total progeny scored | Proportion of expected class | Total progeny scored |
| UM-8373a/EP(2)2199 | 0 | 171 | 25/94b (27% rescue) | 399 |
| UM-8373a/ttv00681 | 0 | 288 | 0 | 369 |
UM-8373 is doubly mutant for Lamin C and tout velu.
The expected rescued class (trans-heterozgous females for two different mutant Lamin C alleles) represents one-sixth of the total progeny. Note that no trans-heterozygous males could be rescued (as the transgene was X-linked), which would represent another one-sixth of the total progeny. Balanced progeny represent two-thirds of the total, and this proportion was used to estimate the theoretical total number of progeny scored assuming complete viability for both sexes. Therefore, the expected number of rescued progeny is 94 (564/6). As anticipated, 0/94 males survived and 25/94 females survived, reflecting a 27% rescue.
Molecular characterization of LamC lethal excision alleles:
The molecular structure of the LamC P-element excision alleles was first determined by performing Southern analysis. Genomic DNA from flies heterozygous for a LamC excision allele and a CyO balancer chromosome (wild type for LamC) was digested with the restriction enzyme Bsu36I, which cleaves immediately upstream and downstream of the LamC coding region, but not within the P element present in stock G00158 (Figure 1B). A 7.2-kb fragment representing the wild-type LamC genomic region from the CyO balancer chromosome is present in all heterozygotes (Figure 1C). A second fragment, varying in size among the stocks, represents the LamC genomic region of the mutant chromosome. Six of the seven alleles analyzed exhibited a larger-sized fragment than that produced from the wild-type endogenous LamC gene, indicating these excision stocks have retained a portion of the original P-element insertion. LamCEX296 appears to have lost a considerable portion of the LamC genomic region, making it the best candidate for a LamC deletion mutant.
To more precisely determine the nature of the genetic lesions resulting from imprecise P-element excision, a high-resolution molecular characterization of the LamC excision stocks was performed using PCR and sequence analysis (Table 3). To obtain genomic DNA from adults homozygous for each excision allele, a wild-type LamC heat-shock-inducible transgene was used to rescue individuals homozygous for a particular excision allele. Seven novel LamC excision alleles were rescued by the wild-type LamC transgene, indicating that no second site lethal mutations exist in these stocks. Therefore, the only lethal mutations on these excision chromosomes are in LamC. PCR analysis revealed that six of the seven excision alleles retained a partial P-element insertion, while five of seven possessed intact LamC coding regions. Sequence analysis confirmed that two alleles, LamCEX296 and LamCEX187, possess deletions within the first exon of LamC, making them the best candidates for protein nulls.
TABLE 3.
Molecular structure of Lamin C excision alleles
| Mutant name | Coding region | No. of insertions | Insertion size (kb) | GFPa | white+/−b |
|---|---|---|---|---|---|
| G00158 (parent stock) | Intact | 1 | 6 | + | + |
| LamCEX3 | Intact | 1 | 1.5 | — | — |
| LamCEX5 | Intact | 1 | 1.8 | — | — |
| LamCEX16 | Intact | 1 | 1.5 | — | — |
| LamCEX187 | 357-bp deletion in first exon | 2 | ND | + | — |
| LamCEX265 | Intact | 1 | 1.4 | — | — |
| LamCEX296 | 560-bp deletion in first exon | 0 | NA | — | — |
| LamCEX364 | Intact | 1 | ND | + | — |
ND, not done; size exceeded standard PCR. NA, not applicable.
Determined by PCR.
Determined by eye color phenotype indicating presence of white reporter (+, red; −, white).
Mutations in LamC show reductions in LamC protein levels:
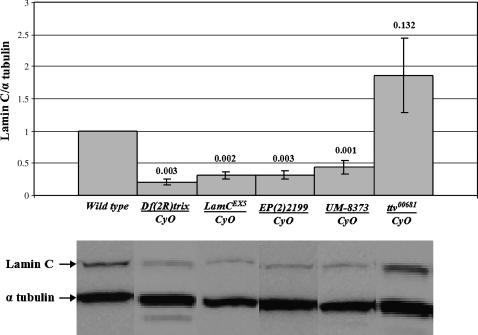
To determine the LamC protein levels in the various mutant stocks, Western analysis was performed using antibodies that specifically recognize LamC (Riemer et al. 1995). Initially, larvae heterozygous for the LamC alleles used in rescue were assayed for LamC protein expression levels. All three LamC P-element insertion stocks, the LamC excision allele, and the deficiency, showed at least a 50% reduction in LamC levels compared to the control stock. In contrast, ttv00681 exhibited wild-type levels of expression for LamC (Figure 2). Thus, at least one endogenous wild-type copy of LamC is required for viability in Drosophila.
Figure 2.
Western analysis of protein from LamC mutants used in rescue experiments. Western analysis of heterozygous adults carrying one mutant allele for LamC over the CyO balancer chromosome is shown. Protein extracts from a wild-type stock (y1w67c23) and heterozygous ttv00681 adults are also shown. LamC was detected with the antibody LC28.26 (Riemer et al. 1995). Quantitation and P-values (Student's t-test) for LamC levels from three independent samples per genotype are indicated on the histogram, and representative Western blots are depicted below. Values are normalized using the levels of α-tubulin and expressed as a ratio to the levels of LamC in the wild type, set at 1.0 (see materials and methods).
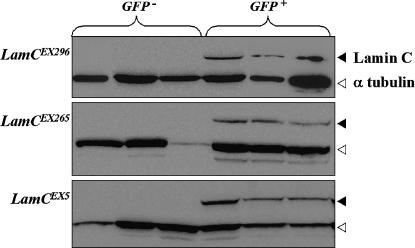
Western analysis was performed on selected excision stocks to determine which mutants were likely to be protein nulls for LamC. Stocks containing the excision alleles LamCEX296, LamCEX265, and LamCEX5 and the deletion Df(2R)trix were placed over the CyO chromosome possessing a GFP reporter gene and mated to one another. Individuals homozygous for an excision allele (LamCEX/LamCEX) or hemizygous for an excision allele (LamCEX/Df) were selected by loss of GFP fluorescence; heterozygotes (LamCEX/Cy0-GFP) were selected on the basis of the presence of GFP fluorescence. LamC protein was not detectable in the LamCEX296, LamCEX265, and LamCEX5 homozygotes and hemizygotes in this assay (Figure 3), suggesting they are all protein nulls.
Figure 3.
Three LamC excision alleles appear to be protein nulls. Larval protein extracts were isolated from LamC excision alleles or the Df(2R)trix over the CyO-GFP chromosome. GFP− samples represent LamCEX/LamCEX or LamCEX/Df(2R)trix); GFP+ samples represent LamCEX/CyO-GFP. Experimental triplicates are shown.
LamC lethal phase is prepupal:
The identification of the lethal phase for LamC mutants would determine when LamC function is essential for development. LamC is not maternally supplied (Riemer et al. 1995); therefore, null alleles were used in this assay. LamCEX296 is the best candidate for a protein null given that it possesses a deletion in the first exon (Table 3) and produces no detectable LamC protein on the basis of Western analysis (Figure 3). Therefore, the lethal phase was determined for individuals trans-heterozygous for LamCEX296 and Df(2R)trix. These mutants were placed over the CyO-GFP chromosome and mated to each other. The number of resulting progeny lacking GFP fluorescence [LamCEX296/Df(2R)trix] was scored for developmental stage for 14 days (for control genotypes, see materials and methods). The LamCEX296/Df(2R)trix individuals die predominantly as late third instar larvae or white prepupae. The lethal phase analysis also showed that development was delayed by ∼2 days for all stages in the hemizygote (data not shown). Therefore, loss of LamC produces a developmental delay and ultimately causes death at the prepupal stage, a time in development in which significant apoptosis and tissue differentiation occur (Ashburner 1989).
Expression of mutant LamC transgenes causes lethality and nuclear defects:
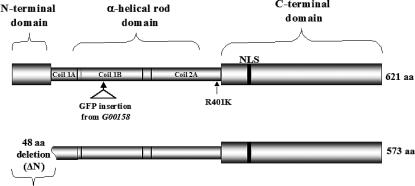
In mammals, expression of specific A-type lamin mutants causes nuclear phenotypes that include lamin aggregations and nuclear envelope blebbing (Ostlund et al. 2001). To examine functional similarities between human and Drosophila lamins, we generated transgenic animals expressing mutations in LamC that correspond to disease-causing mutant forms of A-type lamin in humans. A Drosophila LamC heat-shock-inducible transgene encoding an R401K amino acid substitution within the rod domain was generated (Figure 4). This substitution is homologous to the R386K missense mutation in human lamin A/C that leads to EDMD (Bonne et al. 2000). In addition, structural studies suggest that this residue plays a role in higher-order lamin assembly (Strelkov et al. 2004).
Figure 4.
Diagram of the mutant forms of LamC with principal domains labeled. NLS represents a putative nuclear localization signal. The top of the diagram shows the location of the P-element-encoded GFP insertion (present in stock G00158) and of the R401K amino acid substitution. The bottom of the diagram shows the extent of the N-terminal deletion; the first 48 amino acids are deleted, which removes the entire head domain and 8 amino acids from the first part of the rod domain.
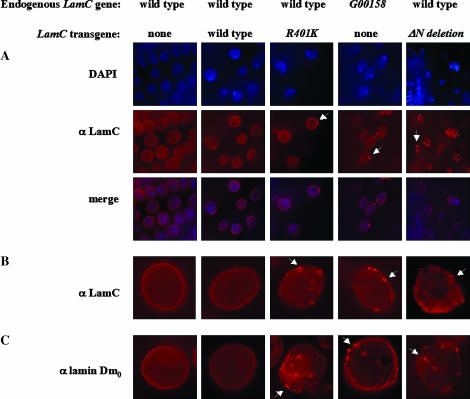
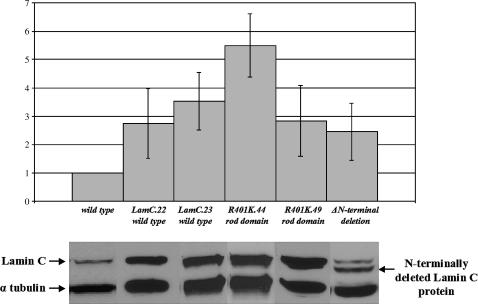
To examine the nuclear localization of the R401K mutant in Drosophila, a mixture of second instar larval tissues that included salivary glands, epithial tissue, imaginal discs, and gut was stained with antibodies against LamC. In all tissue types examined LamC aggregates appeared (Figure 5A and data not shown). We subsequently focused our cytological studies on third instar larval salivary glands due to their large size and ease of manipulation. Prominent nuclear rim staining was observed with LamC antibodies in control stocks (nontransgenic and transgenic stocks expressing wild-type LamC) with or without heat shock (Figure 5B). In contrast, daily heat-shock-induced expression of the R401K mutant resulted in a reduction in LamC at the nuclear periphery and a distinct LamC aggregation phenotype resembling O-rings (Figure 5B). The O-ring structures occurred in 50–100% of the nuclei examined, depending on the transgenic stock (see materials and methods). Interestingly, flies exhibiting this LamC localization defect are viable and have no obvious defects. Western analysis showed that the R401K protein was expressed at levels similar to that of the wild-type LamC transgenic control, indicating that the nuclear phenotype is a consequence of the specific mutation, and not merely resulting from high levels of transgene expression (Figure 6). A similar O-ring phenotype, but with reduced penetrance (∼40–50% of the nuclei scored), is evident in the G00158 heterozygote containing a GFP insertion within the rod domain (Figure 5B). Thus, an insertion or an amino acid substitution within the rod domain gives rise to lamin aggregation defects in Drosophila that are similar to those reported for rod domain mutations in human A-type lamins (Ostlund et al. 2001).
Figure 5.
Nuclear defects associated with expression of mutant forms of LamC. (A) Tissues (salivary gland and epithelial) from second instar larvae stained with antibodies to LamC (red). Nuclei are indicated by DAPI staining (blue). (B) Salivary gland nuclei from third instar larvae stained with antibodies to LamC (red). (C) Salivary gland nuclei from third instar larvae stained with antibodies to lamin Dm0 (red). Arrows indicate representative examples of nuclear defects (LamC O-ring aggregates and lam Dm0 protrusions). Images in A were photographed using a 10× ocular and a 100× oil objective on a Leica DMLB microscope equipped with a Spot RT-Slider CCD camera (Diagnostic Instruments). B and C were photographed the same way, except using a 40× oil objective rather than the 100× objective.
Figure 6.
Western analysis of LamC levels in transgenic stocks used in rescue experiments and nuclear morphology assays. Protein extracts from a wild-type stock (y1w67c23) were used for normalization. Quantitation was carried out as described in Figure 3 and materials and methods.
Mutations within the N-terminal globular domain of human lamin A/C have been associated with EDMD, familial dilated cardiomyopathy, and Charcot-Marie-Tooth type 2 disease (Walter et al. 2005). We generated an N-terminal deletion of the Drosophila LamC protein that removes the first 48 amino acids, including 8 amino acids of the rod domain (Figure 4). This mutant was modeled after an N-terminally deleted version of human lamin A/C that has been functionally tested (Spann et al. 1997). Daily heat-shock-induced expression of the N-terminal LamC truncation was lethal; individuals died at the prepupal stage. A similarly lethal phenotype results when the same N-terminally deleted protein was overexpressed by ubiquitous GAL4 induction (data not shown). Prior to death, at the third instar larval stage, O-ring LamC aggregates were apparent in ∼50–70% of the salivary gland nuclei examined (Figure 5B). Western analysis showed that the N-terminal truncated protein is expressed at levels similar to that of the wild-type transgenic LamC and R401K mutant (Figure 6), suggesting that the phenotype is due to the specific mutant and not simply due to overexpression of lamin protein. Thus, expression of an A-type lamin lacking the N-terminal head domain is toxic to Drosophila.
A- and B-type lamins interact during in vitro assembly (Georgatos et al. 1988). Therefore, we tested whether expression of mutant A-type lamin in a wild-type LamC genetic background disrupts B-type lamin organization in vivo. Third instar larval salivary gland nuclei from individuals expressing the R401K and N-terminal truncated form of LamC were stained with antibodies specific for lamin Dm0, the Drosophila B-type lamin (Riemer et al. 1995). In nontransgenic and transgenic larvae expressing wild-type LamC, nuclear envelope staining characteristic of wild-type lamin Dm0 was apparent (Figure 5C). In transgenic larvae expressing mutant forms of LamC, lamin Dm0 exhibited aggregate structures that appear to bleb from the nuclear lamina (Figure 5C). A similar lamin Dm0 aggregation defect can be observed in the G00158 stock (Figure 5C). Thus, disruption of LamC organization affects other components of the nuclear lamina.
DISCUSSION
To our knowledge, Drosophila is the only well-characterized invertebrate model organism that appears to have both A- and B-type lamins. lamin Dm0 encodes the B-type lamin and has been the subject of several genetic and molecular studies (Riemer et al. 1995; Lenz-Bohme et al. 1997; Guillemin et al. 2001; Patterson et al. 2004). LamC encodes the A-type lamin (Riemer et al. 1995; Stuurman et al. 1999) and is nested within an intron of the essential gene ttv (Bellaiche et al. 1998; The et al. 1999), which complicates the genetic analysis of both genes. In this report, we provide data that functionally separate LamC and ttv. First, a previously identified lethal P-element insert in LamC, EP(2)2199, complements ttv00681, a confirmed null mutant allele of ttv. Second, a wild-type LamC transgene rescues trans-heterozygous combinations of LamC mutants, but not combinations of mutant alleles of ttv. Third, the lethal phases of LamC and ttv are distinct. Lethality due to loss of LamC occurs at the prepupal stage, whereas lethality due to the loss of ttv can occur during early embryogenesis, when the maternal contribution is removed (The et al. 1999).
Prepupal lethality of LamC is consistent with the timing of expression for this gene, as transcript levels peak during larval development (Riemer et al. 1995). This lethal phase is also consistent with a role for LamC in apoptosis and tissue differentiation. During the prepupal stage, larval tissues begin to histolyze via apoptotic pathways (Jiang et al. 1997) while precursors of adult structures (the imaginal discs) proliferate and initiate differentiation programs (Ashburner 1989; Sempere et al. 2002). Lamins are targets for caspases (Lazebnik et al. 1995), suggesting a role in apoptosis. In addition, A-type lamins may play important roles in differentiating tissues, accounting for the tissue specificity of laminopathic diseases in humans (Burke and Stewart 2002). Thus LamC is an essential gene that may play a conserved role in apoptosis and tissue differentiation.
Having determined that the A-type lamin in Drosophila encodes an essential function, we undertook a transgenic approach in Drosophila to study the effects of mutant A-type lamins in vivo. Mutations in human A-type lamins lead to inherited diseases, and the effects of mutant lamins on nuclear architecture and envelope integrity in mammalian cells have been investigated. In patient tissues, nuclear aberrations correlate with disease, but are also found at low frequency in control cells (Vigouroux et al. 2001). In mammalian cell culture, overexpression assays of mutant human A-type lamins produce conflicting results. Some experiments show nuclear aggregation and abnormal nuclear shapes, while other studies using the same mutant lamin show no nuclear phenotypes (Broers et al. 1999; Ostlund et al. 2001; Raharjo et al. 2001; Holt et al. 2003). The discrepancies are possibly due to the cell type used, expression levels of the mutant lamin, and/or tagging of the expressed protein.
For our transgenic analysis, we selected a missense mutation in the rod domain of the Drosophila LamC protein, since mutations throughout this domain in the human protein account for almost 40% of laminopathies (Strelkov et al. 2004). Specifically, we chose to mutate the residue R401K, which in humans corresponds to R386K that causes EDMD. This residue is conserved in both vertebrates and invertebrates and belongs to a subdomain within the rod that has been studied by crystallography (Strelkov et al. 2004). We also designed an N-terminal deletion mutant since this domain is essential for the head-to-tail assembly of lamin dimers into protofilaments (Stuurman et al. 1996; Sasse et al. 1998) and has recently been implicated in a neurogenic variant of EDMD (Walter et al. 2005).
Heat-shock-induced expression of Drosophila LamC proteins with mutations that disrupt the rod domain and/or the N-terminal head domain results in nuclear defects including (1) reduced intensity of LamC antibody staining at the nuclear periphery, (2) aggregation of LamC into O-ring structures, and (3) aggregation of lamin Dm0 (Figure 5). These defects are specific for the expression of mutant protein, as they are not evident when a wild-type LamC is expressed at similar levels (Figure 6). The O-ring aggregates are not specific for salivary gland nuclei or nuclei from tissues undergoing histolysis prior to pupation as they were observed in epithelial tissue, imaginal discs, brain, and gut from second and third instar larvae (Figure 5 and data not shown). The LamC O-rings bear a striking resemblance to lamin aggregations observed in mammalian cell culture upon overexpressing human lamin rod domain mutants (Ostlund et al. 2001; Raharjo et al. 2001; Holt et al. 2003). Lamin aggregration phenotypes, including O-ring structures, are thought to reflect defects in higher-order lamin assembly, which requires an intact rod and head domain (Stuurman et al. 1996; Sasse et al. 1998). A recent report describing the X-ray crystallographic structure of a portion of the human A-type lamin rod domain has provided evidence that higher-order assembly involves electrostatic interaction between charged residues in the N- and C-terminal portions of the rod domain (Strelkov et al. 2004). Interestingly, these interactions include arginine 386 (human lamin A/C), a residue that is mutated in patients with EDMD (Bonne et al. 2000) and is homologous to the R401K substitution in Drosophila LamC analyzed in our study.
In Drosophila, expression of the R401K LamC mutant produces nuclear lamin aggregates, but no overt phenotype in the adult. Expression of the N-terminal truncation produces a similar lamin aggregation defect; however, it also causes prepupal lethality. This lethal phase is similar to that of the LamC null mutants. The N-terminal truncation end point was selected on the basis of studies in mammalian systems (Spann et al. 1997); however, it removes eight amino acids of the rod domain. Therefore, the nuclear aggregation phenotype may be due to defects in the rod domain, while the lethality might result from the loss of the globular head domain.
Overexpression of our N-terminal LamC truncation, but not our wild-type LamC or the 401K mutant, caused lethality in Drosophila. Previously, a larval (48- to 120-hr) stage-specific lethal phenotype was reported for overexpression of a wild-type LamC transgene (Stuurman et al. 1999). One possible explanation for this discrepancy is that we did not achieve levels of expression high enough with our wild-type transgenes to cause lethality. Our transgenic lines express wild-type LamC approximately twofold over endogenous LamC levels (Figure 6), which is sufficient for rescue of lethality (Table 2). Another explanation is that the LamC transgene employed in the Stuurman study contained a single-base deletion in the 3′ end (nucleotide position G1781), causing a frameshift that adds 59 unrelated amino acids to the C terminus and shortens the overall length of the protein by 9 amino acids. Although this altered protein appears to localize normally (Stuurman et al. 1999), it does not possess assembly properties similar to that of wild-type LamC (Klapper et al. 1997 and associated erratum), which might contribute to the reported stage-specific lethality (Stuurman et al. 1999).
In summary, we have demonstrated that the Drosophila A-type lamin gene, LamC, encodes an essential function that is required during the prepupal stages of development. An essential developmental role for A-type lamins in differentiating tissues is thought to contribute to the tissue restriction of disease phenotypes manifested in human laminopathies. Expression of mutant Drosophila LamC protein causes nuclear phenotypes similar to those observed in human cell culture and tissue biopsies, in addition to demonstrating an essential function for the N-terminal head domain. Our results strongly suggest that the biological functions of A-type lamins are conserved between humans and Drosophila, thereby establishing Drosophila as a model to dissect the role A-type lamins play in development, nuclear architecture, and gene expression with relevance to human disease etiology.
Acknowledgments
We thank Janine Martin for assistance with nuclear cytology, Paul Fisher for the LamC and lamin Dm0 antibodies, and Bridget Zimmerman of the University of Iowa Biostatistics Consulting Center for assistance with statistical analysis. This research was supported by a Muscular Dystrophy Association grant (MDA3605) to L.L.W. and P.K.G. S.R.S. is supported by an American Heart Association postdoctoral fellowship (0520106Z) and R.I. is supported by a National Institutes of Health Research Supplement for Underrepresented Minorities (GM06513).
References
- Adams, M. D., and J. J. Sekelsky, 2002. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 3: 189–198. [DOI] [PubMed] [Google Scholar]
- Arimura, T., A. Helbling-Leclerc, C. Massart, S. Varnous, F. Niel et al., 2005. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum. Mol. Genet. 14: 155–169. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bellaiche, Y., I. The and N. Perrimon, 1998. Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394: 85–88. [DOI] [PubMed] [Google Scholar]
- Bender, W., P. Spierer and D. S. Hogness, 1983. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J. Mol. Biol. 168: 17–33. [DOI] [PubMed] [Google Scholar]
- Bonne, G., E. Mercuri, A. Muchir, A. Urtizberea, H. M. Becane et al., 2000. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 48: 170–180. [PubMed] [Google Scholar]
- Brodsky, G. L., F. Muntoni, S. Miocic, G. Sinagra, C. Sewry et al., 2000. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation 101: 473–476. [DOI] [PubMed] [Google Scholar]
- Broers, J. L., B. M. Machiels, G. J. van Eys, H. J. Kuijpers, E. M. Manders et al., 1999. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 112: 3463–3475. [DOI] [PubMed] [Google Scholar]
- Burke, B., and C. L. Stewart, 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3: 575–585. [DOI] [PubMed] [Google Scholar]
- Chaouch, M., Y. Allal, A. De Sandre-Giovannoli, J. M. Vallat, A. Amer-el-Khedoud et al., 2003. The phenotypic manifestations of autosomal recessive axonal Charcot-Marie-Tooth due to a mutation in Lamin A/C gene. Neuromuscul. Disord. 13: 60–67. [DOI] [PubMed] [Google Scholar]
- Chen, L., L. Lee, B. A. Kudlow, H. G. Dos Santos, O. Sletvold et al., 2003. LMNA mutations in atypical Werner's syndrome. Lancet 362: 440–445. [DOI] [PubMed] [Google Scholar]
- Duffy, J. B., 2002. GAL4 system in Drosophila: a fly geneticist's Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- Eriksson, M., W. T. Brown, L. B. Gordon, M. W. Glynn, J. Singer et al., 2003. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, T. B., J. B. Burnett, S. Lootens, R. Steinman and L. L. Wallrath, 1992. The urate oxidase gene of Drosophila pseudoobscura and Drosophila melanogaster: evolutionary changes of sequence and regulation. J. Mol. Evol. 34: 62–77. [DOI] [PubMed] [Google Scholar]
- Georgatos, S. D., C. Stournaras and G. Blobel, 1988. Heterotypic and homotypic associations between the nuclear lamins: site-specificity and control by phosphorylation. Proc. Natl. Acad. Sci. USA 85: 4325–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G. B., C. R. Preston, D. M. Johnson-Schlitz, N. A. Nassif, R. W. Phillis et al., 1993. Type I repressors of P element mobility. Genetics 135: 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg, M., A. Harel, M. Brandeis, T. Rechsteiner, T. J. Richmond et al., 1999. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc. Natl. Acad. Sci. USA 96: 2852–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin, K., T. Williams and M. A. Krasnow, 2001. A nuclear lamin is required for cytoplasmic organization and egg polarity in Drosophila. Nat. Cell. Biol. 3: 848–851. [DOI] [PubMed] [Google Scholar]
- Harborth, J., S. M. Elbashir, K. Bechert, T. Tuschl and K. Weber, 2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114: 4557–4565. [DOI] [PubMed] [Google Scholar]
- Holt, I., C. Ostlund, C. L. Stewart, N. Man, H. J. Worman et al., 2003. Effect of pathogenic mis-sense mutations in lamin A on its interaction with emerin in vivo. J. Cell Sci. 116: 3027–3035. [DOI] [PubMed] [Google Scholar]
- Hutchison, C. J., 2002. Lamins: Building blocks or regulators of gene expression? Nat. Rev. Mol. Cell Biol. 3: 848–858. [DOI] [PubMed] [Google Scholar]
- Jiang, C., E. H. Baehrecke and C. S. Thummel, 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683. [DOI] [PubMed] [Google Scholar]
- Klapper, M., K. Exner, A. Kempf, C. Gehrig, N. Stuurman et al., 1997. Assembly of A- and B-type lamins studied in vivo with the baculovirus system. J. Cell Sci. 110: 2519–2532 [erratum: J. Cell Sci. 111: following 1766 (1998)]. [DOI] [PubMed] [Google Scholar]
- Laguri, C., B. Gilquin, N. Wolff, R. Romi-Lebrun, K. Courchay et al., 2001. Structural characterization of the LEM motif common to three human inner nuclear membrane proteins. Structure 9: 503–511. [DOI] [PubMed] [Google Scholar]
- Lazebnik, Y. A., A. Takahashi, R. D. Moir, R. D. Goldman, G. G. Poirier et al., 1995. Studies of the lamin proteinase reveal multiple parallel biochemical pathways during apoptotic execution. Proc. Natl. Acad. Sci. USA 92: 9042–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme, B., J. Wismar, S. Fuchs, R. Reifegerste, E. Buchner et al., 1997. Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes, and accumulation of annulate lamellae. J. Cell Biol. 137: 1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, X., R. Daneman, M. Zavortink and W. Chia, 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounkes, L. C., S. Kozlov, L. Hernandez, T. Sullivan and C. L. Stewart, 2003. A progeroid syndrome in mice is caused by defects in A-type lamins. Nature 423: 298–301. [DOI] [PubMed] [Google Scholar]
- Ostlund, C., G. Bonne, K. Schwartz and H. J. Worman, 2001. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 114: 4435–4445. [DOI] [PubMed] [Google Scholar]
- Ozaki, T., M. Saijo, K. Murakami, H. Enomoto, Y. Taya et al., 1994. Complex formation between lamin A and the retinoblastoma gene product: identification of the domain on lamin A required for its interaction. Oncogene 9: 2649–2653. [PubMed] [Google Scholar]
- Patterson, K., A. B. Molofsky, C. Robinson, S. Acosta, C. Cater et al., 2004. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol. Biol. Cell 15: 600–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raharjo, W. H., P. Enarson, T. Sullivan, C. L. Stewart and B. Burke, 2001. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 114: 4447–4457. [DOI] [PubMed] [Google Scholar]
- Riemer, D., N. Stuurman, M. Berrios, C. Hunter, P. A. Fisher et al., 1995. Expression of Drosophila lamin C is developmentally regulated: analogies with vertebrate A-type lamins. J. Cell Sci. 108: 3189–3198. [DOI] [PubMed] [Google Scholar]
- Rzepecki, R., S. S. Bogachev, E. Kokoza, N. Stuurman and P. A. Fisher, 1998. In vivo association of lamins with nucleic acids in Drosophila melanogaster. J. Cell Sci. 111: 121–129. [DOI] [PubMed] [Google Scholar]
- Sasse, B., U. Aebi and N. Stuurman, 1998. A tailless Drosophila lamin Dm0 fragment reveals lateral associations of dimers. J. Struct. Biol. 123: 56–66. [DOI] [PubMed] [Google Scholar]
- Sempere, L. F., E. B. Dubrovsky, V. A. Dubrovskaya, E. M. Berger and V. Ambros, 2002. The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev. Biol. 244: 170–179. [DOI] [PubMed] [Google Scholar]
- Shackleton, S., D. J. Lloyd, S. N. Jackson, R. Evans, M. F. Niermeijer et al., 2000. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 24: 153–156. [DOI] [PubMed] [Google Scholar]
- Spann, T. P., R. D. Moir, A. E. Goldman, R. Stick and R. D. Goldman, 1997. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 136: 1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov, S. V., J. Schumacher, P. Burkhard, U. Aebi and H. Herrmann, 2004. Crystal structure of the human lamin A coil 2B dimer: implications for the head-to-tail association of nuclear lamins. J. Mol. Biol. 343: 1067–1080. [DOI] [PubMed] [Google Scholar]
- Stuurman, N., B. Sasse and P. A. Fisher, 1996. Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J. Struct. Biol. 117: 1–15. [DOI] [PubMed] [Google Scholar]
- Stuurman, N., S. Heins and U. Aebi, 1998. Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122: 42–66. [DOI] [PubMed] [Google Scholar]
- Stuurman, N., J. P. Delbecque, P. Callaerts and U. Aebi, 1999. Ectopic overexpression of Drosophila lamin C is stage-specific lethal. Exp. Cell Res. 248: 350–357. [DOI] [PubMed] [Google Scholar]
- Sullivan, T., D. Escalante-Alcalde, H. Bhatt, M. Anver, N. Bhat et al., 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The, I., Y. Bellaiche and N. Perrimon, 1999. Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Mol. Cell 4: 633–639. [DOI] [PubMed] [Google Scholar]
- Vergnes, L., M. Peterfy, M. O. Bergo, S. G. Young and K. Reue, 2004. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. USA 101: 10428–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigouroux, C., M. Auclair, E. Dubosclard, M. Pouchelet, J. Capeau et al., 2001. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 114: 4459–4468. [DOI] [PubMed] [Google Scholar]
- Walter, M. C., T. N. Witt, B. S. Weigel, P. Reilich, P. Richard et al., 2005. Deletion of the LMNA initiator codon leading to a neurogenic variant of autosomal dominant Emery-Dreifuss muscular dystrophy. Neuromuscul. Disord. 15: 40–44. [DOI] [PubMed] [Google Scholar]
- Zastrow, M. S., S. Vlcek and K. L. Wilson, 2004. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117: 979–987. [DOI] [PubMed] [Google Scholar]