Abstract
Using quantitative trait locus (QTL) mapping, we studied the genetic basis of the difference in pigmentation between two sister species of Drosophila: Drosophila yakuba, which, like other members of the D. melanogaster subgroup, shows heavy black pigmentation on the abdomen of males and females, and D. santomea, an endemic to the African island of São Tomé, which has virtually no pigmentation. Here we mapped four QTL with large effects on this interspecific difference in pigmentation: two on the X chromosome and one each on the second and third chromosomes. The same four QTL were detected in male hybrids in the backcrosses to both D. santomea and D. yakuba and in the female D. yakuba backcross hybrids. All four QTL exhibited strong epistatic interactions in male backcross hybrids, but only one pair of QTL interacted in females from the backcross to D. yabuka. All QTL from each species affected pigmentation in the same direction, consistent with adaptive evolution driven by directional natural selection. The regions delimited by the QTL included many positional candidate loci in the pigmentation pathway, including genes affecting catecholamine biosynthesis, melanization of the cuticle, and many additional pleiotropic effects.
DURING the modern synthesis, the dominant view of the genetics of species differences was that of Ronald Fisher (1930), who believed that such differences were almost invariably due to the accumulation of many genes, each of small phenotypic effect. Tests of this proposition, however, were limited by the lack of genetic markers in most crossable but differentiated species, although some data suggested that species differences could occasionally be due to genes of large effect (Orr and Coyne 1992).
Recently, however, the advent of molecular techniques has improved our ability to study the genetics of species differences. Two innovations have been crucial. First, quantitative trait locus (QTL) mapping enables us to localize genes responsible for species differences by determining their association with molecular markers at known sites. Second, molecular techniques such as germline transformation enable us to determine directly whether a candidate gene affects a species difference. Orr (2001) describes these innovations and the results of recent genetical studies using them. Although most data derive from a small number of organisms (Orr's study describes only 13 analyses, 6 from Drosophila and 4 from the monkeyflower genus Mimulus), the results show that while differences in traits between species can be polygenic, genes of large effect are involved more frequently than previously suspected. Here we describe a genetic analysis of a striking character difference—the degree of abdominal pigmentation—between two sister species of Drosophila: Drosophila yakuba and D. santomea.
D. yakuba is widely distributed in open habitats across sub-Saharan Africa and the islands near the continent (including Madagascar). In contrast, D. santomea is endemic to the 860-km2 volcanic island of São Tomé, in the Gulf of Benin 255 km west of the coast of Gabon (Lachaise et al. 2000). On the mountain Pico de São Tomé, D. yakuba occurs at elevations below 1450 m, while D. santomea occupies the mist forests at elevations between 1153 and 2024 m. (D. yakuba is also widespread throughout lowland São Tomé, probably as a result of a secondary invasion after the initial common ancestor evolved into D. santomea.) Between ∼1100 and 1450 m in elevation, the ranges of the two species overlap, with the ratio of D. yakuba/D. santomea shifting from 2:1 to 1:20 as one moves upward through this zone. The species show substantial sexual isolation when tested in the laboratory (Lachaise et al. 2000), and, using morphological criteria, one finds a low frequency (∼1%) of hybrids in the zone of overlap.
Molecular evidence puts the divergence between D. yakuba and D. santomea at ∼400,000 years ago (Llopart et al. 2002a). In interspecific crosses, F1 male hybrids are sterile but female hybrids are fertile and thus can be crossed to either parental species (Lachaise et al. 2000; Cariou et al. 2001). This fertility permits genetic analysis using backcross individuals.
The diagnostic differences between the species include male genital morphology and sex-comb tooth number (Lachaise et al. 2000; Coyne et al. 2004), but the most striking difference involves abdominal pigmentation. Among the nine species in the D. melanogaster subgroup, eight of them, including D. yakuba, have similar patterns of dark pigmentation: males possess thin black stripes along the posterior portions of tergites 2, 3, and 4, while tergites 5–7 are completely black. Females of these species have stripes along the posterior portions of all tergites, and tergites 5–7 show substantial but not complete black pigmentation. In contrast, D. santomea males show virtually no pigmentation, and females show only very light striping on the posterior parts of tergites 2–5, with no pigmentation on other tergites. (Photographs of these differences are given in Lachaise et al. 2000, Figures 1 and 2, and in Llopart et al. 2002a, Figure 1). Given the dark pigmentation in all but one species in the subgroup, including the outgroup species D. orena and D. erecta, it is nearly certain that the absence of dark pigmentation in D. santomea is a novel derived trait.
Figure 1.
QTL affecting variation in pigmentation between D. yakuba and D. santomea. (A) F1 females [from D. yakuba (males) × D. santomea (females)] backcrossed to D. yakuba males. (B) F1 females [from D. yakuba (females) × D. santomea (males)] backcrossed to D. santomea males. Molecular makers are indicated as triangles on the x-axis. Plots are likelihood-ratio (LR) test statistics for pigmentation differences between males (teal) and females (magenta) as determined by composite interval mapping. Significance thresholds for each cross were determined by permutation and are ∼LR = 10 for each cross, denoted by the dashed horizontal line.
Figure 2.
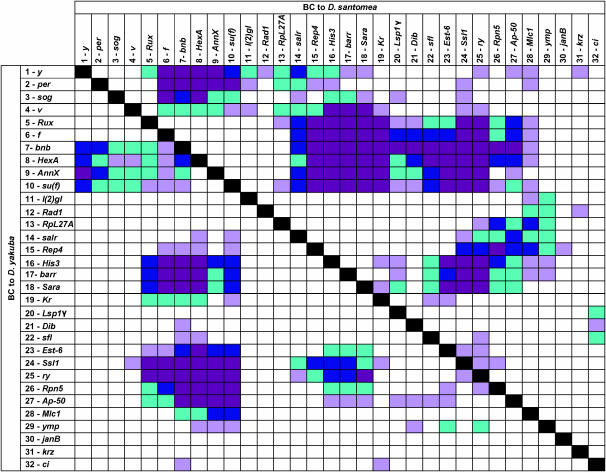
Pairwise epistasis between markers. The significance of all pairwise interactions between markers for males from the backcross to D. santomea is indicated above the diagonal, and for interactions for males from the backcross to D. yakuba it is shown below the diagonal. (Magenta box) P < 0.0001 (Bonferroni correction); (blue box) 0.0001 < P < 0.001; (teal box) 0.001 < P < 0.01; (light purple box) 0.01 < P < 0.05.
The adaptive significance of this pigmentation difference, if any, is unknown. Although these species show strong sexual isolation (Coyne et al. 2002), this does not diminish when flies are tested in the dark, suggesting that the pigmentation difference is not a cue for mate discrimination (Llopart et al. 2002a,b). Moreover, the relationship between pigmentation and temperature is opposite to that expected from other studies of Drosophila: individuals within a species or closely related species living under colder conditions are almost invariably darker (e.g., David et al. 1985; Gibert et al. 1998), yet D. santomea, which lives at higher altitudes than D. yakuba, is lighter.
In previous genetic analyses using three morphological markers and eight molecular markers, we determined that at least three genes were involved in the pigmentation difference between D. santomea and D. yakuba females and five genes between males. In each case, the genes resided on all three major chromosomes, with the X chromosome having a particularly strong effect in males (Llopart et al. 2002a).
In this study we extend and refine our previous analysis, using a more accurate method of measuring pigmentation as well as a QTL analysis employing 32 molecular markers, which enables us to map “pigmentation genes” more accurately. Our goals are to determine the number of genes involved in this morphological difference, their chromosomal locations, whether the same genetic regions affect the pigmentation difference in both males and females, and whether QTL from a given species tend to affect the character in the same direction, implying that the species difference evolved by natural selection (Orr 1998). Finally, the mapping of QTL to fairly restricted regions of the genome may eventually allow us to identify specific loci involved in this species difference.
MATERIALS AND METHODS
Drosophila strains:
All flies were maintained in 8-dram vials containing standard cornmeal-agar-Karo media on a 12 hr:12 hr light:dark cycle at 24°. One isofemale strain was used from each species. The D. yakuba Taï 18 strain was derived from a female collected by D. Lachaise in 1983 in the Taï rainforest on the border between Liberia and the Ivory Coast. The D. santomea STO.4 stock was derived from a female collected in March 1998 in the Obo Natural Reserve on São Tomé Island. These two strains were used (and further described) in our previous work on the genetics of pigmentation in these species (Llopart et al. 2002a). The strains are homosequential in chromosome banding pattern except for the right arm of the second chromosome: the D. yakuba Taï 18 strain is polymorphic for inversion 2Rn, which covers ∼40% of the right arm of chromosome 2 (Lemeunier and Ashburner 1976).
Crosses:
Backcross (BC) hybrids were produced by crossing 4-day-old virgin D. yakuba Taï 18 females to virgin D. santomea STO.4 males, and then backcrossing virgin F1 females to males from both species. Genetically, female BC hybrids to D. yakuba are either homozygous D. yakuba or heterozygous D. yakuba/santomea and have mitochondrial DNA from D. yakuba. Similarly, female BC hybrids to D. santomea are either homozygous D. santomea or heterozygous D. yakuba/santomea and have mitochondrial DNA from D. yakuba. Male BC hybrids have the same autosomal and mitochondrial genotypes as females, but the X-linked loci are either pure D. santomea or D. yakuba, with the Y chromosome from the parental male used in the backcross. We scored pigmentation in ∼50 males and 50 females from each of the pure species and the reciprocal F1 hybrids and in between 73 and 544 males and females from each of the two backcrosses (Table 1). To improve the precision of QTL mapping, we selected backcross individuals with extreme and intermediate pigmentation phenotypes for subsequent pigmentation scoring and genotyping. For each of the first three BC genotypes listed in Table 1, we selected flies of each sex from a sample of ∼12,500 individuals (∼50 bottles, each containing ∼250 flies of each sex). One-third of the total individuals chosen were judged by eye to have very dark pigmentation, one-third to have very light pigmentation, and the remaining third were chosen randomly from individuals with intermediate phenotypes. (Equal numbers of all three classes were chosen from each bottle inspected until we had accumulated ∼500 flies of each sex in each backcross. Thus for each sex we selected ∼4% of total individuals inspected; this stringent selection facilitates the precision of mapping.) However, females from the backcross of F1 individuals to D. santomea showed little variation in pigmentation, and so all of these were chosen randomly.
TABLE 1.
Pigmentation scores of pure D. yakuba and D. santomea and of F1 hybrids from the reciprocal crosses and backcross individuals
| Genotype | Sex | Mean score (SE) | N |
|---|---|---|---|
| D. yakuba Taï 18 | M | 14.22 (0.30) | 51 |
| F | 9.85 (0.15) | 53 | |
| D. santomea STO.4 | M | 0.63 (0.05) | 50 |
| F | 1.02 (0.06) | 56 | |
| F1 (Y × S) | M | 11.92 (0.21) | 51 |
| F | 4.62 (0.18) | 50 | |
| F1 (S × Y) | M | 3.72 (0.21) | 51 |
| F | 4.29 (0.12) | 51 | |
| Backcross (F1 × Y) | M | 7.26 (0.18) | 544 |
| F | 7.42 (0.09) | 544 | |
| Backcross (F1 × S) | M | 4.50 (0.16) | 517 |
| F | 2.05 (0.13) | 73 |
Y, D. yakuba; S, D. santomea (Taï 18 and STO.4 strains were used in all crosses). In all crosses the genotype of the female parent is given first. All F1 females were produced by crossing D. yakuba females to D. santomea males.
Pigmentation scores:
All scoring of pigmentation was done on 4-day-old virgin flies. We scored only the three posterior tergites of each fly (segments 5, 6, and 7) by examining the fly under a dissecting microscope. A pigmentation score was assigned on the basis of both the percentage of the tergite that was pigmented and the degree of pigmentation. First, the proportion of the tergite covered by black pigment was estimated to the nearest 0.05 (5%). Then, the relative degree of pigmentation was measured within the pigmented area. Using color standards, we assessed the degree of pigmentation within the pigmented area using a five-point scale ranging from 1 (very light pigmentation, slightly darker than background color) to 5 (dark, shiny black), with intermediate numbers representing intermediate degrees of pigmentation. Unpigmented areas were given a score of 0. We limited ourselves to assigning only three shades of black to each tergite. The percentage of the area of each tergite covered by each shade of pigmentation was then multiplied by the intensity of pigmentation, and these areas were summed. This gives each tergite a minimum possible pigmentation score of 0 (no area pigmented) to 5 [tergite completely covered with very dark pigmentation (1.0 × 5)]. These areas were summed for all three tergites, yielding a minimum possible pigmentation score for a given fly of 0 and a maximum possible score of 15. As shown in Table 1, this procedure discriminates well between the pigmentations of these species.
Molecular markers:
We identified single nucleotide polymorphisms (SNPs) and insertion/deletion variants (INDELs) that discriminated between the D. yakuba Taï18 and the D. santomea STO.4 strains for 41 nuclear regions: y, per, sog, v, sn, Rux, f, bnb, Hex-A, AnnX, su(f), l(2)gl, Rad1, RpL27A, Gart, salr, vkg, Rep4, Adh, His3, barr, Sara, Hex-C, Ngp, Kr, Lsp1-γ, RpL14, dib, sfl, Sod, Est6, Ssl1, hb, Xdh (ry), Rpn5, AP-50, Mlc1, ymp, janB, krz, and ci. Newly reported sequences were deposited in GenBank under accession nos. DQ068949 (Gart_T18) and DQ068950 (Gart_sto4); otherwise see Llopart et al. (2005, this issue) for details. To determine nucleotide sites differentially fixed between D. yakuba Taï18 and D. santomea STO.4, we tested 20 individuals (10 per strain) for each region by direct sequencing of PCR products obtained from single-fly DNA extractions (Ashburner 1989), using D. melanogaster or D. yakuba primers. We purified PCR products using the Wizard Magnesil PCR clean-up system (Promega, Madison, WI) and sequenced them directly with an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, CA). We edited the sequences with the Sequencher 3.0 software (Gene Codes, Ann Arbor, MI) and aligned them using the ClustalX program (Thompson et al. 1997).
In total, we sequenced ∼17.5 kb in each of the 20 flies tested. We detected 263 nucleotide differences fixed between Taï18 and STO.4, that is, 70% of the total nucleotide variation. Among these fixed differences we selected 32 that affect a restriction endonuclease site to be used as markers in the genotyping procedure. Table 2 lists the 32 markers, their relative order within the D. yakuba chromosomes, and the conditions for genotyping. We inferred the relative order of markers within each chromosome in D. yakuba/D. santomea from the D. yakuba genome project (http://www.genome.wustl.edu/projects/yakuba/; version 040407).
TABLE 2.
Molecular polymorphisms discriminating D. yakuba and D. santomea
| Marker | Cytological location | Primer sequence (5′-3′) | Type | PCR TA (°) | Restriction endonuclease |
|---|---|---|---|---|---|
| y | 1A5 | CGCTGCGTGTTTGTTTATTT | S | 55 | AvaII |
| GCGAATGTTCAAAGAATAATTTC | |||||
| per | 3B1–2 | TTCCAGTTCTCCGAATCAGC | S | 55 | BbvI |
| CCTTAGGGCTGAGCCACTCT | |||||
| sog | 13E1 | GCTGGCGTACAACATTGAAA | S | 57 | XhoI |
| CTCGGTGGCCACATTCAC | |||||
| v | 9F11 | AGACTCCCTTCCTGCCTTTC | S | 55 | SspI |
| TGAGAGCTCCAGTTCCGACT | |||||
| rux | 5D2 | CATTTGCTCATCCGTTTCCT | S | 55 | HpyCH4IV |
| GTGCTTGTAGCGCGTTGTC | |||||
| f | 15F4–7 | CTCGCCGAATGGCAGCAT | S | 55 | HpaII |
| AATGTACGTCCGCCTGGAT | |||||
| bnb | 17D6 | TTCCTTCTCCTGCTCCTTGA | S | 55 | PvuII |
| CCGAGAAGAAGTCCATCGAG | |||||
| Hex-A | 8E10 | GGTACCCAGCTCTTCGATCA | S | 57 | HhaI |
| GGCAATGGCATCCTTTAGAA | |||||
| AnnX | 19C1 | AAACCAGAGAGCTGCCTTCA | S | 55 | Taq1α |
| ATTCTCCTTGCGACGTCTTG | |||||
| su(f) | 20E | TGGTGGGCAAAAGTCAAAAT | ID | 57 | NcoI |
| AAAATCTTAGCCGCCTGGAC | |||||
| l(2)gl | 21A5 | TGACGTCGCTGAAGTTCTTG | S | 54 | MseI |
| GATGGGCCAGCTTATATTGC | |||||
| Rad1 | 23A1 | ATGAATGTGCTGTCCGAGTG | S | 55 | HpyCH4IV |
| GTTCGTGGAACACCTTCGAT | |||||
| RpL27A | 24F3 | ATCAAGCGGAAGAAGACCAG | S | 55 | NcoI |
| GACCTTGCCGAAGTAACCAG | |||||
| salr | 32E4–F1 | AGCTGACTGATCCCAACCAG | S | 60 | ScrF1 |
| GATGATGCCGTTGGAGAACT | |||||
| Rep4 | 34B4 | TCACGGAGTACGAACACCAA | S | 57 | Taq1α |
| TACGGGTCAGTTCCTCCTTG | |||||
| His3 | 39D3–E1 | TTTCAGGACCACAAACCACA | ID | 57 | MfeI |
| CCGTTTGCCCCTTATAAACA | |||||
| barr | 38B1–2 | GCAGTGCAGGATGAAGATCA | S | 53 | HaeIII |
| TTGGAGTCCACCTCCAGAAC | |||||
| Sara | 57E6 | CGACCACAAACCCTGAATTT | S | 55 | HphI |
| CATGTTATCCGGCACCCATA | |||||
| Kr | 60F5 | ACCAGCCATGAGTGGAGATT | S | 56 | MlyI |
| CTACAGAGCTGGCTCCATCC | |||||
| Lsp1γ | 61A6 | CAAAACCCACCACAAGCAG | S | 52 | AluI |
| CCTTGTACTCCTTCTCGTACATGAT | |||||
| dib | 64A5 | AGTCCTTTTCTCCCCAGGAA | S | 52 | NruI |
| ATTGGGCCTGGCTGAGTT | |||||
| sfl | 65B3–4 | GGGTAATCCCTGTGACGATG | S | 52 | NsiI |
| TTCCGATGGAAAGAAGTCCA | |||||
| Est-6 | 69A1 | TCCTGCCTACGCTTTTGTCT | ID | 52 | MseI |
| AAAAGTAGTCGTCGCCATGC | |||||
| Ssl1 | 80B2 | GGTGCCCAGTAGTGGTGAGT | S | 52 | BsrI |
| GACGCACATTTTCGAGATCA | |||||
| ry | 87D9 | CGCTTTGAGCAAAAATCCA | S | 52 | SacI |
| GAAGAACAAGCTCACCACCA | |||||
| Rpn5 | 83C4 | TACCGAGGGCAAGATTTACG | S | 57 | MseI |
| TGCTGATCTTCTTGGCAATG | |||||
| AP-50 | 94A15–16 | AGTGCAAGTTCGGCATCAA | S | 57 | HaeIII |
| GAATGGCAGCGAAATGTCTT | |||||
| Mlc1 | 98A14–15 | TGCAAACAGAGTTCGTCCAG | S | 52 | Tsp5091 |
| AACGGGCATTATCAGCATGT | |||||
| ymp | 96E | CCTCGAGACCCGCAGTAGT | S | 53 | HaeIII |
| CACCTCGCACTTCTGATTGA | |||||
| janB | 99D3 | CATGGCTTCACGAAATACGG | S | 57 | SalI |
| CTTACCCTGGAGGTGCCATA | |||||
| krz | 100E3 | CGCATGTTGTCAAATAAAATCG | S | 55 | MseI |
| TTTTTGGGATAACCCATTATTCA | |||||
| ci | 102A1–3 | AGCCCTTGCAGTGAAGACTC | S | 50 | HpaI |
| TGGTAGGTCTGCTACGTCC |
Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976). The order of the markers in the first column reflects their relative positions in D. yakuba/D. santomea chromosomes inferred from the D. yakuba genome project (http://www.genome.wustl.edu/projects/yakuba/). Marker types: S, SNP; ID, insertion/deletion. The PCR protocol for all markers is 1 cycle at 94° for 2 min; 35 cycles at 94° for 30 sec, annealing temperature (TA) for 30 sec, and 72° for 30 sec; and 1 cycle at 72° for 4 min, where the TA is listed. PCR products were digested with a restriction endonuclease, run on a 3% agarose gel stained with ethidium bromide, imaged with the Bio-Rad Chemi Doc System PC RS-170 using Quantity One (version 4.2.1) software, and manually genotyped.
Marker genotypes:
All BC individuals from the pigmentation assays were stored at −80° in 0.5-ml Eppendorf tubes. Genomic DNA was extracted from each BC individual using the Puregene (Gentra Systems, Minneapolis) single-fly DNA extraction protocol. The sample consisted of 506 BC D. santomea males, 73 BC D. santomea females, 537 BC D. yakuba males, and 526 BC D. yakuba females. The genotypes of the 1642 BC hybrids were determined for all 32 markers (i.e., 52,544 genotypes).
The 32 molecular markers were designed using sequence data from the parental strains of D. santomea and D. yakuba. The aligned sequences were used to develop PCR primers, using Primer3 (Rozen and Skaletsky 2000) and restriction enzyme (RE) digestions. Genotyping was performed using restriction fragment length polymorphism analysis by PCR amplification from genomic DNA, using RedTaq DNA polymerase (Sigma, St. Louis) followed by RE digestion (see Table 2 for primers, RE, and conditions). All REs were purchased from New England Biolabs (Beverly, MA) and primers were purchased from MWG Biotech (High Point, NC). The digested PCR products were run on a 3% agarose gel stained with ethidium bromide, imaged with the Bio-Rad ChemiDoc System PC RS-170 using Quantity One (version 4.2.1) software, and manually genotyped by assigning a “0” (homozygous D. santomea), “1” (D. yakuba/D. santomea heterozygote), or “2” (homozygous D. yakuba) to each marker genotype. A recombination map based on the 1642 BC hybrids (Table 3) was constructed using the Haldane mapping function.
TABLE 3.
Molecular markers and map positions
|
D. santomea
|
D. yakuba
|
|||||
|---|---|---|---|---|---|---|
| Marker | Marker name | Cytological location | r | Genetic distance (cM) | r | Genetic distance (cM) |
| Chromosome X | ||||||
| 1 | y | 1A5 | 0.0432 | 0.0 | 0.0245 | 0.0 |
| 2 | per | 3B1–2 | 0.1572 | 4.5 | 0.1468 | 2.5 |
| 3 | sog | 13E1 | 0.1883 | 23.4 | 0.1364 | 19.9 |
| 4 | v | 9F11 | 0.3005 | 47.0 | 0.2690 | 35.8 |
| 5 | rux | 5D2 | 0.0570 | 93.0 | 0.0913 | 74.4 |
| 6 | f | 15F4–7 | 0.0777 | 99.0 | 0.1110 | 84.5 |
| 7 | bnb | 17D6 | 0.0363 | 107.5 | 0.0329 | 97.2 |
| 8 | Hex-A | 8E10 | 0.0760 | 111.2 | 0.0593 | 100.6 |
| 9 | AnnX | 19C1 | 0.0501 | 119.5 | 0.0254 | 106.9 |
| 10 | su(f) | 20E | 0.0000 | 124.7 | 0.0000 | 109.5 |
| Chromosome 2 | ||||||
| 11 | l(2)gl | 21A5 | 0.0639 | 0.0 | 0.0865 | 0.0 |
| 12 | Rad1 | 23A1 | 0.1054 | 6.8 | 0.0922 | 9.5 |
| 13 | RpL27A | 24F3 | 0.1364 | 18.7 | 0.1665 | 19.7 |
| 14 | salr | 32E4–F1 | 0.1002 | 34.6 | 0.1001 | 39.9 |
| 15 | Rep4 | 34B4 | 0.2073 | 45.8 | 0.2023 | 51.2 |
| 16 | His3 | 39D3–E1 | 0.0345 | 72.6 | 0.0132 | 77.1 |
| 17 | barr | 38B1–2 | 0.1140 | 76.1 | 0.1477 | 78.4 |
| 18 | Sara | 57E6 | 0.2694 | 89.1 | 0.2653 | 96.0 |
| 19 | Kr | 60F5 | 0.0000 | 127.8 | 0.0000 | 133.8 |
| Chromosome 3 | ||||||
| 20 | Lsp1γ | 61A6 | 0.2729 | 0.0 | 0.1966 | 0.0 |
| 21 | Dib | 64A5 | 0.2712 | 39.5 | 0.1345 | 25.0 |
| 22 | sfl | 65B3–4 | 0.2314 | 78.5 | 0.2700 | 40.6 |
| 23 | Est-6 | 69A1 | 0.3057 | 109.6 | 0.2493 | 79.5 |
| 24 | Ssl1 | 80B2 | 0.2211 | 156.9 | 0.1637 | 114.0 |
| 25 | ry | 87D9 | 0.2159 | 186.1 | 0.1797 | 133.8 |
| 26 | Rpn5 | 83C4 | 0.0967 | 214.3 | 0.1176 | 156.1 |
| 27 | AP-50 | 94A15–16 | 0.2297 | 225.1 | 0.1844 | 169.5 |
| 29 | Mlc1 | 98A14–15 | 0.1036 | 255.8 | 0.1072 | 192.5 |
| 30 | ymp | 96E | 0.2107 | 267.4 | 0.1110 | 204.6 |
| 28 | janB | 99D3 | 0.2055 | 294.8 | 0.1326 | 217.1 |
| 31 | krz | 100E3 | 0.0000 | 321.3 | 0.0000 | 232.5 |
| Chromosome 4 | ||||||
| 32 | ci | 102A1–3 | 0.0000 | NA | 0.0000 | NA |
r is the recombination rate between two adjacent markers. The genetic distance d was inferred from r using the Haldane map function, 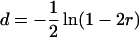 . Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976).
. Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976).
QTL mapping:
QTL affecting variation in pigmentation between D. yakuba and D. santomea were mapped in each BC population using composite interval mapping (CIM; Zeng 1994) and implemented using QTL Cartographer software (Basten et al. 1999). CIM tests whether an interval between two markers contains a QTL affecting the trait while simultaneously controlling for the effect of QTL located outside the interval using multiple regression on marker cofactors. Marker cofactors were chosen by forward selection-backward elimination stepwise regression. The likelihood-ratio (LR) test statistic is −2 ln(L0/L1), where L0/L1 is the ratio of the likelihood under the null hypothesis (i.e., there is no QTL in the test interval) to the alternative hypothesis (there is a QTL in the test interval). LR test statistics were computed every 2 cM with marker cofactors 10 cM or more from the test location. We used permutation analysis to determine appropriate significance thresholds that take into account the multiple tests performed and correlations among markers. We permuted trait and marker data 1000 times and recorded the maximum LR statistic across all intervals for each permutation. LR statistics calculated from the original data that exceed the 50th greatest LR statistic from the permuted data are significant at the experimentwise 5% level under the null hypothesis (Churchill and Doerge 1994; Doerge and Churchill 1996). The approximate boundaries of regions containing QTL were determined by taking 2 LOD intervals (9.22 LR) surrounding the point of greatest significance and interpolating the cytological location of the interval on the basis of the observed amount of recombination between flanking markers.
We estimated the effects of each QTL as the difference between the appropriate homozygous genotypes and heterozygous D. santomea/D. yakuba genotypes at the peak LR, scaled by the phenotypic standard deviation. The effects in females and autosomes of both sexes are thus estimates of a − d in the cross to D. yakuba and −a − d in the cross to D. santomea, where −a and a are, respectively, the genotypic values in D. santomea and D. yakuba, and d is the heterozygous effect (Falconer and Mackay 1996). The effects of X-linked QTL in males are estimates of 2a.
We evaluated pairwise epistatic interactions between all possible marker pairs by running ANOVA models to account for the main effects of all significant markers and of one pairwise interaction between markers (Dilda and Mackay 2002). The ANOVAs were performed with the PROC GLM procedure, using SAS (Cary, NC) 8.02 software. Interactions with P-values <0.0001 are significant on the basis of a Bonferroni correction for 496 tests per BC population. We estimated the effects of significant two-locus interactions from the least-squares means of the four marker locus classes as 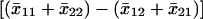 , where the first subscript is 1 if the marker has a homozygous genotype for either parental species and 2 if the marker has a heterozygous genotype, and the second subscript takes on the same values for the other marker in the interaction. Standard errors of the interaction effects were estimated as described by Dilda and Mackay (2002).
, where the first subscript is 1 if the marker has a homozygous genotype for either parental species and 2 if the marker has a heterozygous genotype, and the second subscript takes on the same values for the other marker in the interaction. Standard errors of the interaction effects were estimated as described by Dilda and Mackay (2002).
Candidate genes:
Cytological bands in D. melanogaster of the markers that define the interval under the QTL peak were obtained using FlyBase (Drysdale and Crosby 2005). Using the known cytological positions of these markers in D. melanogaster, we determined the corresponding positions in D. yakuba (Lemeunier and Ashburner 1976; Ashburner 1989). This defined an interval both in D. melanogaster and in D. yakuba that allowed us to search for candidate genes within that interval. We obtained a complete list of candidate genes involved in pigmentation in D. melanogaster from FlyBase (Drysdale and Crosby 2005). Candidate genes were identified on the basis of the markers that delimit each QTL and the presence of pigmentation genes between these two markers (Table 6).
TABLE 6.
Candidate genes
| Symbol | Gene name | Cytological location | Functiona |
|---|---|---|---|
| cin | cinnamon | 1A1 | Mo- molybdopterin cofactor biosynthesis |
| y | yellow | 1A5 | Cuticle pigmentation |
| a(1)HM26 | abnormal abdomen HM26 | 1B1–5C2 | Mutants affect abdominal tergite and sternite |
| mk | murky | 1B1–5C2 | Oogenesis |
| svr | silver | 1B5–7 | Cuticle biosynthesis |
| su(b) | suppressor of black | <1B8 | Interacts genetically with black |
| su(s) | suppressor of sable | 1B13 | Transcriptional repressor activity |
| dor | deep orange | 2B5 | Pteridine biosynthesis |
| A | Abnormal abdomen | 3A5 | Mutations affect the abdominal tergite |
| l(1)3B2 | lethal(1)3B2 | 3B2 | Mutations affect the tergite |
| omb | optomotor-blind | 4C3–4 | Patterning the pigment band |
| lac | lacquered | 4C6 | Mutations are body color defective |
| amb | amber | 4C6–D1 | Mutations are recessive body color defective |
| pt | platinum | 7F1 | Mutations are body color defective |
| t | tan | 8A1–B8 | β-Alanyl-dopamine hydrolase activity |
| s | sable | 11F1–12A1 | Encodes a product involved in pigmentation |
| e(y)1 | enhancer of yellow-1 | 16E1 | Interacts genetically with y, w, z, ct, and sc |
| e(y)3 | enahncer of yellow-3 | 18C–D | Interacts genetically with y, w, z, ct, and sc |
| mel | melanized | 19B3–C3 | Mutations are recessive body color defective |
| mal | maroon-like | 19D1 | Mo-molybdopterin cofactor biosynthesis |
| mel1 | melanized-like | 19E1+ | Mutations affect the abdominal tergite |
| vao | varied outspread | 19E7 | Mutations are body color defective |
| su(f) | suppressor of forked | 20E | Encodes a product with putative poly(A) binding |
| b | black | 34D5 | Glutamate decarboxylase activity |
| yellow-c | yellow-c | 35B8 | Cuticle pigmentation |
| Catsup | Catecholamines up | 37B11 | Regulation of catecholamine metabolism |
| Dox-A2 | Diphenol oxidase A2 | 37B12 | Endopeptidase activity |
| Ddc | Dopa decarboxylase | 37C1 | Dopamine biosynthesis; pigmentation patterning |
| amd | α-methyl dopa-resistant | 37C1 | Involved in cuticle biosynthesis |
| l(2)37Ca | lethal(2)37Ca | 37C5 | Interacts genetically with Ddc |
| tyr1 | tyrosine-1 | 38A6–C1 | Mutations are body color defective |
| pr | purple | 38B3 | Involved in pteridine biosynthesis |
| Bkd | Blackoid | 43E18–52D7 | Mutations are body color defective |
| dkb | dark bubbly | <49D7 | Mutations are body color defective |
| Cp1 | Cysteine proteinase-1 | 50C18–20 | Mutations affect the abdominal segment 1–5 |
| U | Upturned | 53A | Mutations are body color defective |
| Bc | Black cells | 54F6 | Involved in melanization defense response |
| Pu | Punch | 57C7–8 | Tetrahydrobiopterin biosynthesis |
| D | Dichaete | 70D3 | Mutations affect the abdominal segment 3–7 |
| db | dark body | 73C1–D2 | Mutations are body color defective |
| Crn | Crown | <77B3 | Mutations are dominant body color defective |
| kkv | krotzkopf verkehrt | 83A1 | Chitin synthase activity |
In cases where the function is unclear, mutant phenotypes are listed. All information, including cytological locations, was retrieved from the FlyBase website (http://www.flybase.org) (Drysdale and Crosby 2005).
RESULTS
Pigmentation of pure species and F1 hybrids:
Table 1 gives the mean pigmentation scores, standard errors, and sample sizes for the pure species, the reciprocal F1 hybrids, and the backcross hybrids used for genotyping. The difference between the pure species is substantial: the mean pigmentation scores of D. yakuba males and females are 14.22 and 9.85, respectively, and for D. santomea males and females 0.63 and 1.02, respectively. As seen in our previous analysis (Llopart et al. 2002a), reciprocal F1 hybrid males show a large effect of the X chromosome on pigmentation: these males have pigmentation scores fairly close to those of males from the species of the maternal parent. The difference in pigmentation scores between the two classes of F1 males is highly significant (t = 27.1, 100 d.f., P < 0.001). The relative effect of the X chromosome in male pigmentation can be judged as the percentage of the total difference between males of the two species explained by the difference between the reciprocal F1 males; this effect is ∼60%. This effect is much larger than the relative size of this chromosome [constituting ∼21% of the haploid genome (Table 3)] and suggests that either the X chromosome carries a disproportionate number of genes affecting pigmentation or individual X-linked genes have disproportionately large effects. (The QTL analysis below shows that the second explanation is most likely to be correct.)
In contrast to males, the F1 females do not differ significantly in pigmentation scores (Table 1; t = 1.55, 99 d.f., P = 0.12). There is thus no evidence for a maternal or mitochondrial effect affecting pigmentation of these females, who are identical in nuclear genotype. The mean score of all F1 females (4.45) is slightly lighter than the average score of females for the two species (5.43), showing a small amount of dominance for the D. santomea phenotype.
QTL affecting variation in pigmentation in BC hybrids:
We mapped four QTL with large effects on pigmentation (Table 4, Figure 1). The same four QTL were detected in male hybrids in the backcrosses to both D. santomea and D. yakuba: two QTL were on the X chromosome (between markers 1 and 2 and markers 6 and 10), one QTL was on the second chromosome (between markers 15 and 18), and one QTL was on the third chromosome (between markers 23 and 26). The magnitude of the QTL effects ranged from 0.49 to 1.42 phenotypic standard deviations in the backcross to D. santomea and accounted for 67% of the total phenotypic variation. Similarly, the QTL effects ranged from 0.62 to 1.62 phenotypic standard deviation in the backcross to D. yakuba and accounted for 58% of the total phenotypic variation. In both backcrosses, the sum of the estimated QTL effects equaled or exceeded that expected from the difference between the parental genotypes. In the backcross to D. santomea, the expected difference in pigmentation is −11.29 [i.e., the difference in pigmentation between D. santomea males (0.63) and (Y × S) F1 males (11.92)], whereas the sum of the QTL effects was −11.64. In the backcross to D. yakuba, the expected difference in pigmentation is 10.5 [i.e., the difference in pigmentation between D. yakuba males (14.22) and (S × Y) F1 males (3.72)], whereas the sum of the QTL effects is 16.15. Thus, it is likely that we have detected all of the QTL affecting variation in pigmentation in this hybridization and that our selective genotyping protocol led to overestimation of effects (Lynch and Walsh 1998). It is also possible that estimates of main effects have been biased by epistatic interactions (see below).
TABLE 4.
QTL affecting variation in pigmentation between D. yakuba and D. santomea
| Backcross population | Sex | QTL | Peak LRa | LRa | Effect (SE)b | Effect/σpc | R2d |
|---|---|---|---|---|---|---|---|
| F1 females × D. yakuba males | F | 1A5–13E1 | 1A5 | 26.71 | 0.68 (0.18) | 0.31 | 0.0169 |
| 15F4–20E | 8E10 | 87.08 | 1.44 (0.56) | 0.66 | 0.0584 | ||
| 34B4–57E6 | 34B4 | 441.74 | 1.81 (0.76) | 0.83 | 0.4382 | ||
| 69A1–83C4 | 80B2 | 130.57 | 1.69 (0.69) | 0.78 | 0.1135 | ||
| M | 1A5–13E1 | 3B1-2 | 65.13 | 2.60 (0.65) | 0.62 | 0.0327 | |
| 15F4–20E | 17D6 | 525.94 | 6.75 (2.24) | 1.62 | 0.3186 | ||
| 34B4–57E6 | 34B4 | 193.19 | 3.29 (0.70) | 0.79 | 0.1060 | ||
| 69A1–83C4 | 80B2 | 216.23 | 3.51 (0.91) | 0.84 | 0.1227 | ||
| F1 females × D. santomea males | F | 15F4–20E | 19C1 | 53.485 | −1.39 (0.68) | −1.27 | 0.4280 |
| M | 1A5–13E1 | 1A5 | 67.88 | −2.16 (0.45) | −0.61 | 0.0409 | |
| 15F4–20E | 17D6 | 450.81 | −5.03 (1.84) | −1.42 | 0.3964 | ||
| 34B4–57E6 | 34B4 | 199.18 | −1.73 (0.61) | −0.49 | 0.1397 | ||
| 69A1–83C4 | 80B2 | 133.46 | −2.72 (0.76) | −0.77 | 0.0966 |
QTL regions are estimated from 2 LOD support intervals (P ≤ 0.05). The peak is the cytological location with the highest likelihood ratio (LR). Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976).
Effects were estimated from the least-squares means of the two marker locus classes as: 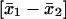 , where the subscript is 1 if the marker has a homozygous genotype and 2 if the marker has a heterozygous or hemizygous genotype. The standard error (SE) is listed in parentheses.
, where the subscript is 1 if the marker has a homozygous genotype and 2 if the marker has a heterozygous or hemizygous genotype. The standard error (SE) is listed in parentheses.
Effect divided by the phenotypic standard deviation. See footnote a for the calculation of the effect.
The proportion of variance explained by the QTL and estimated by 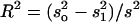 , where s2 is the variance of the trait,
, where s2 is the variance of the trait,  is the sample variance of the residuals, and
is the sample variance of the residuals, and 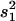 is the variance of the residuals (Basten et al. 1999).
is the variance of the residuals (Basten et al. 1999).
Four QTL in the same positions affected variation in pigmentation in the female D. yakuba BC hybrids. The magnitude of the QTL effects ranged from 0.31 to 0.83 phenotypic standard deviations and accounted for 63% of the total phenotypic variance. The sum of the estimated QTL effects slightly exceeded that expected from the difference between the parental genotypes in this backcross, again suggesting that we have detected all of the QTL affecting variation in pigmentation in this hybridization. The expected difference in pigmentation is 5.39 [i.e., the difference in pigmentation between D. yakuba females (9.85) and F1 females (4.46 on average)], whereas the sum of the QTL effects is 5.62. We observed only a single X chromosome QTL (between markers 6 and 10) in the D. santomea BC females, accounting for 43% of the total phenotypic variance. This could be attributable to a lack of power to detect QTL in this cross, since only 73 flies were assessed for genotype-phenotype associations, compared to >500 individuals in each of the other crosses. Indeed, this QTL accounted for only 40% of the expected difference in pigmentation [−3.44; i.e., the difference in pigmentation between D. santomea females (1.02) and F1 females (4.46 on average)].
The QTL effects were largely additive within loci. In the BC to D. yakuba, the effects of the two X chromosome QTL in females (a − d) were approximately half that of the effects in males (2a), consistent with d = 0. In addition, the effects of the chromosome 3 QTL in males from the backcrosses to D. yakuba and D. santomea were equal and opposite, as expected if d = 0. The second chromosome QTL had a larger effect in the BC to D. yakuba than to D. santomea, suggesting partial dominance of the D. santomea genotype. Since dominance of D. santomea QTL reduces the power to detect QTL in the backcross to D. santomea, this could also account for our failure to detect this QTL in females from this backcross.
Epistatic interactions:
We assessed all possible epistatic interactions between pairs of markers within each cross and sex (Table 5, Figures 2–4). We observed significant epistasis (after correcting for multiple tests) in males from both backcrosses between markers in regions encompassed by the QTL, but not between QTL regions and regions without main effects or between two regions with no main effects on pigmentation (Figure 2). The significant interactions were between the two X chromosome QTL, between the X chromosome QTL between markers 6 and 10 and the chromosome 2 QTL, between the X chromosome QTL between markers 6 and 10 and the chromosome 3 QTL, and between the chromosome 2 and chromosome 3 QTL (Figure 2, Table 5). The nature of these interactions is illustrated in Figure 3, where the effect of a Y-S substitution at the second locus is shown in the form of reaction norms, conditional on the genotype of the first locus (where Y denotes a D. yakuba allele and S denotes a D. santomea allele at the QTL). In the absence of epistasis, the effect of the substitution at the second locus would be independent of the genotype of the first, and the reaction norms would be parallel.
TABLE 5.
Epistatic effects of QTL affecting variation in pigmentation between D. yakuba and D. santomea
| Backcross population | Sex | Markers | Effect (SE)a | Effect/σpb | P-value |
|---|---|---|---|---|---|
| F1 females × D. yakuba males | M | 1A5 × 19C1 | 1.38 (0.30) | 0.33 | <0.0001 |
| 8E10 × 57E6 | −1.54 (0.32) | −0.37 | <0.0001 | ||
| 8E10 × 87D9 | −2.50 (0.33) | −0.60 | <0.0001 | ||
| 57E6 × 87D9 | −1.33 (0.17) | −0.32 | 0.0001 | ||
| F | 8E10 × 39D3–E1 | −1.04 (0.25) | −0.48 | <0.0001 | |
| F1 females × D. santomea males | M | 1A5 × 19C1 | 1.29 (0.30) | 0.36 | 0.0001 |
| 8E10 × 57E6 | 2.59 (0.58) | 0.73 | <0.0001 | ||
| 8E10 × 87D9 | 2.17 (0.47) | 0.61 | <0.0001 | ||
| 57E6 × 87D9 | 1.64 (0.36) | 0.46 | <0.0001 |
See text for details. The standard error (SE) is given in parentheses.
The QTL effect divided by the phenotypic standard deviation.
Figure 3.
Significant epistatic interactions between QTL, depicted as reaction norms. Values shown are the mean pigmentation scores (y-axis) for a particular genotype in the background of another genotype. (A) Backcrosses to D. santomea males. (B) Backcrosses to D. yakuba males. Epistatic interactions are shown for the significant markers common to both backcross populations, as shown in Figure 2 (M1 × M9, M8 × M18, M8 × M25, and M18 × M25). See Table 3 for marker definitions. Marker genotypes are indicated as D. santomea (A) or D. yakuba (B) homozygote (blue circle) and D. santomea/D. yakuba heterozygote (purple triangle).
Figure 4.
Mean pigmentation scores (y-axis) of the eight marker haplotypes derived from the four QTL with large effects on pigmentation. The letters denote the genotype of the QTL allele (S, D. santomea; Y, D. yakuba), and the order of the letters indicates the genotype for the QTL at the tip of the X chromosome, the QTL at the base of the X chromosome, the second chromosome QTL, and the third chromosome QTL, respectively. Haplotypes refer to the markers at the peak LR in the respective QTL analyses. See Table 3 for marker descriptions. Haplotypes for male backcross hybrids are hemizygous D. yakuba and/or D. santomea for the two X chromosome QTL. The X chromosome QTL in female backcross hybrids and all autosomal QTL are heterozygous or homozygous; the marker genotype of the nonrecurrent parent is indicated. (A) Males from the backcross to D. santomea. Haplotypes are for markers M1, M7, M15, and M24. (B) Males from the backcross to D. yakuba. Haplotypes are for markers M2, M7, M15, and M24. (C) Females from the backcross to D. yakuba. Haplotypes are for markers M1, M8, M15, and M24. The bar graphs are color coded (increasing ratio of black to yellow) to indicate increasing number of D. yakuba alleles and pigmentation scores.
In the backcross to D. santomea, we expect the hemizygous Y or heterozygous SY genotype at the second locus to be more pigmented than the hemizygous S or homozygous SS genotype at this locus. However, for all the interacting markers in this backcross, this is true only if the genotype at the first locus is Y (or SY). Either there is no difference between the genotypes at the second locus if the first is S (SS), or, for the case of the interaction between the second X chromosome QTL and the chromosome 2 QTL, the SS genotype at the chromosome 2 QTL is actually more pigmented than the SY genotype at this QTL when the X chromosome QTL is S (Figure 3). In other words, the effect of a Y-S substitution in an otherwise S background is smaller than the effect of an S-Y substitution at each QTL in the Y background. Equivalently, the effect of Y-S substitutions at two interacting loci in the homozygous S background is greater than additive, and the effect of S-Y substitutions at two interacting loci in the heterozygous SY background is less than additive. This is illustrated in Figure 4, where the sum of the effects of substituting single Y alleles at each QTL in the S background would yield a predicted pigmentation score of 4.09 for the YYYY haplotype, whereas the observed score is 9.43.
The epistatic interactions are more complicated in the backcross to D. yakuba. Here we expect the hemizygous Y or homozygous YY genotype at the second locus to be more pigmented than the hemizygous S or heterozygous SY genotype at this locus. In the interaction between the two X chromosome QTL (marker 1 × marker 9), this is true if marker 1 is Y. On the other hand, in the interactions with the chromosome 3 QTL (marker 8 × marker 25 and marker 18 × marker 25), this is true if marker 8 is S and marker 25 is SY (Figure 3). However, the interaction between the second X chromosome QTL and the chromosome 2 QTL (marker 8 × marker 18) is in the opposite direction to that expected: the SY genotype at marker 18 is actually more pigmented than the YY genotype at this marker, but only if marker 8 is Y. Overall, the effect of Y-S substitutions at two interacting loci in the heterozygous SY background is less than additive, and the effect of S-Y substitutions at two interacting loci in the homozygous YY background is greater than additive. Figure 4 shows that the sum of the effects of substituting single Y alleles at each QTL would yield a predicted pigmentation score of 15.15 for a YYYY haplotype, whereas the observed score is 13.43.
A single epistatic interaction was observed in females from the backcross to D. yakuba, between the second X chromosome QTL (markers 6–10) and the chromosome 2 QTL (data not shown). The direction of the epistatic effects between these QTL is the same as that in males from this hybridization. The effect of Y-S substitutions at the two interacting loci in the heterozygous SY background is less than additive, and the effect of S-Y substitutions at two interacting loci in the homozygous YY background is greater than additive. Figure 4 shows that the sum of the effects of substituting single Y alleles at each QTL would yield a predicted pigmentation score of 11.17 for a YYYY haplotype, whereas the observed score is 9.49.
DISCUSSION
We have mapped at least four QTL with large effects associated with the variation in pigmentation between D. yakuba and D. santomea. The QTL mapped to the same locations in both males and females in the backcrosses to D. yakuba and in males in the backcross to D. santomea: two QTL mapped to the X chromosome and one each to the second and third chromosomes. Thus, the loss of pigmentation in D. santomea involved evolutionary changes in several genes, which probably affected both sexes. (Although only a single QTL was detected in females in the backcross to D. santomea, it mapped to the same location as one of the X chromosome QTL, and the small sample size of this population and narrow range of pigmentation conspire to reduce the power to detect QTL with small effects.)
This study not only largely expands and refines the earlier results of Llopart et al. (2002a), in which pigmentation differences were assessed in backcross hybrids using eight molecular markers, but also provides the first accurate chromosomal locations of genetic factors associated with these differences. In Llopart et al. (2002a), the QTL of largest effect was also associated with AnnX at the base of the X chromosome, with a second QTL with smaller effect at the tip of the X chromosome associated with y. The locations of the autosomal QTL are also concordant between the two studies. The QTL with the smallest effect detected by Llopart et al. (2002a) was associated with the marker at bric-à-brac 1 (bab1) at the tip of 3L, which is only marginally significant in the backcross to D. yakuba females in this study (LR = 14 between the Lsp1γ and dib markers). One possible explanation for this small discrepancy is that the bab1 marker is in linkage disequilibrium with the chromosome 3 QTL mapped in this study. This, however, is not likely because the map distance between the major QTL detected on chromosome 3 and the bab1 region is >100 cM. It is possible that the discrepancy could be due to the fact that the methods used to score abdominal pigmentation in both studies, although correlated, are different.
The results presented here raise the interesting possibility that the genetic basis of pigmentation differences between D. yakuba and D. santomea is fairly simple. We infer that we have detected all major QTL accounting for variation in pigmentation in these backcross hybrids (with the exception of females in the backcross to D. santomea), since the sum of the QTL effects equals or exceeds that expected from the difference between parental strain means. Further high-resolution mapping is required to determine whether single genes or multiple closely linked loci are responsible for the large QTL effects. Nevertheless, all QTL from the same species affected pigmentation in the same direction, suggesting that the species difference might have arisen by natural selection (Orr 1998). We were not able to formally test this hypothesis, since a minimum of six QTL are required to reject the null hypothesis (Orr 1998). However, sequencing of the relevant loci may show, by the ratio of coding vs. noncoding substitutions, whether selection was involved in their divergence.
Only two other studies have investigated the genetic basis of pigmentation differences between closely related species of Drosophila. Hollocher et al. (2000b) studied two Caribbean species in the D. cardini group having extremely different pigmentation patterns: D. arawakana (a light-colored sexually dimorphic species) and D. nigrodunni (the darkest sexually monomorphic species of the D. dunni subgroup). Using quantitative measures of abdominal pigmentation in F1 hybrids and backcross flies, the authors conclude that, at least for the posterior segment of the abdomen (Hollocher et al. 2000a, Figure 2, “area 3”), which is roughly equivalent to the area scored in our analysis, there are paternal and maternal effects, with no particular effect of the X chromosome. The second study mapped QTL for the difference in pigmentation between D. americana and D. novamexicana (Wittkopp et al. 2003), using 23 molecular markers. Five genes (y, e, Ddc, omb, and bab) previously implicated in the development and evolution of abdominal pigmentation were used as markers. The authors indicate that this species difference is polygenic with no significant effect of the X chromosome but with significant effects of three of the five autosomes. There is little genetic commonality between the results reported by these two studies and our results. Of course, unless there are a very limited number of genes that could be potentially responsible for differences in pigmentation, one does not expect the genetic architecture to be shared among distantly related species.
Our observations of epistatic interactions between QTL with main effects on pigmentation are consistent with genes corresponding to the QTL that are in the same pathway(s). In the absence of high-resolution mapping, however, we can only speculate about what candidate genes might correspond to the QTL. An obvious candidate for the QTL at the tip of the X chromosome is yellow (y) itself, and complementation tests to D. santomea using a y mutation in D. yakuba are consistent with a very small contribution of mutations at the y locus in the pigmentation difference between these species (Llopart et al. 2002a). In addition, two enhancers of y are located in the region embraced by the QTL at the base of the X chromosome; these could contribute to the interactions between the two X chromosome QTL.
Several candidate genes affecting body pigmentation have been identified by mutagenesis in D. melanogaster (Drysdale and Crosby 2005) and colocalize to the regions containing QTL affecting pigmentation differences between D. yakuba and D. santomea (Table 6). Catecholamines are required for proper melanization and sclerotization of the Drosophila cuticle (Wright 1987; Walter et al. 1996). The Ddc gene cluster on chromosome 2 [genetically defined by Df(2L)TW130; 37B9–C1,2;D1–2] contains at least 18 functionally related genes involved in the catecholamine pathway, including Catsup, Ddc, Dox-A2, amd, and l(2)37Ca (Stathakis et al. 1995). Mutations in 11 of the loci in this complex (including Ddc and amd) produce melanotic pseudotumors, indicating abnormal catecholamine metabolism (Wright 1996), and mutations in 14 of the loci affect the formation, sclerotization, or melanization of the cuticle (Wright 1996). The Ddc cluster colocalizes with the QTL on chromosome 2. Pu encodes GTP cyclohydrolase, the rate-limiting step in the synthesis of tetrahydobiopterin, the cofactor required for the phosporylation of tyrosine hydroxylase, which is in turn the rate-limiting step in the synthesis of dopamine (Stathakis et al. 1999). Pu also colocalizes with the QTL on chromosome 2. The silver (svr) gene, which encodes proteins that are members of the carboxypeptidase family (Settle et al. 1995), colocalizes with the QTL at the tip of the X chromosome. Mutations in svr affect pigmentation, wing shape, and catecholamine pools (Wright 1987). tan (t) is an excellent candidate gene corresponding to the QTL at the base of the X chromosome. t is probably the structural gene for β-alanyldopamine hydrolase activity; t mutants have reduced dopamine levels (Wright 1987).
Additional candidate genes in the pigmentation pathway include optomotor-blind (omb), black (b), Cysteine proteinase-1 (Cp1), and Black cells (Bc) (Wright 1987; Wittkopp et al. 2002, 2003). The developmental gene, omb, colocalizes with the QTL at the tip of the X chromosome. omb encodes a T-box transcription factor that is necessary for patterning the pigment band in each adult abdominal segment of D. melanogaster (Kopp and Duncan 1997, 2002). A recent study by Brisson et al. (2004) examined patterns of nucleotide variation at the omb locus in D. polymorpha, a species highly polymorphic for abdominal pigmentation. Two classes of haplotypes that appear to be under balancing selection were associated with variation in abdominal pigmentation in this species.
b, Cp1, and Bc all colocalize with the chromosome 2 QTL. b encodes a product involved in β-alanine biosynthesis; b mutants are heavily pigmented. Cp1 encodes a product with cathepsin L activity; deletion studies of Cp1 have shown complete female sterility and reduced pigmentation in abdominal segments 1–5 (Gray et al. 1998). Bc encodes a tyrosinase, which catalyzes the de novo synthesis of melanin from tyrosine (Wittkopp et al. 2003).
Conspicuously absent from the list of strong potential candidate genes are bab1 and bab2, two closely linked genes at the tip of chromosome 3L that are thought to be repress male-specific abdominal pigmentation in females (Kopp et al. 2000) and contribute significantly to variation of abdominal pigmentation in females of D. melanogaster (Kopp et al. 2003). The expression of bab is correlated with pigmentation across a diverse range of Drosophila species, such that species in which neither sex is pigmented exhibit similar expression of Bab in males and females, but species in which abdominal tergites of males are more pigmented than those of females have female-specific Bab expression (Kopp et al. 2000). Thus, it was possible a priori that overexpression of Bab in D. santomea could have resulted in loss of pigmentation in both sexes. This is not the case, however, since none of the QTL map in the vicinity of bab. Further, Bab2 protein is expressed in a dimorphic melanogaster-like pattern in D. santomea (Gompel and Carroll 2003), which is inconsistent with mutations at bab affecting the difference in pigmentation between D. santomea and D. yakuba.
While it is plausible that the loss of pigmentation in D. santomea was driven by natural selection, it is also possible that selection acted on pleiotropic effects of genes affecting pigmentation and not pigmentation itself. All of the candidate genes listed in Table 3 have highly pleiotropic effects on traits related to fitness, including reproduction and immune response. For example, Ddc catalyzes the final step in the biosynthesis of the neurotransmitters dopamine and serotonin. Dopamine is required in Drosophila for normal development (Neckameyer 1996); ovarian maturation, fecundity and sexual receptivity in females (Neckameyer 1996, 1998a); learning (Tempel et al. 1984; Neckameyer 1998b); locomotion (Pendleton et al. 2002); and aggressive behavior (Baier et al. 2002). Serotonin also regulates or modulates a variety of behaviors in many animal species, including aggression, feeding, learning, locomotion, sleep, and mood (Blenau and Baumann 2001). Further speculation about the nature of the pleiotropic effects (and sex-specific epistatic effects) of genes affecting variation in pigmentation between these species must await the positional cloning of these genes.
Acknowledgments
We thank Bethuel Mgumba and Eric Grossman for technical help as well as Amanda J. Moehring and Ted J. Morgan for helpful discussions. This work was funded by National Institutes of Health research grants to J.A.C. (GM 58260) and T.F.C.M. (GM45344 and GM 58260).
References
- Ashburner, M., 1989. Drosophila: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Baier, A., B. Wittek and B. Brembs, 2002. Drosophila as a new model organism for the neurobiology of aggression? J. Exp. Biol. 205: 1233–1240. [DOI] [PubMed] [Google Scholar]
- Basten, C. J., B. S. Weir and Z-B. Zeng, 1999. QTL Cartographer, Version 1.13. Department of Statistics, North Carolina State University, Raleigh, NC.
- Blenau, W., and A. Baumann, 2001. Molecular and pharmacological properties of insect bioamine receptors: lessons from Drosophila melanogaster and Apis mellifera. Arch. Insect Biochem. Phys. 48: 13–38. [DOI] [PubMed] [Google Scholar]
- Brisson, J. A., A. R. Templeton and I. Duncan, 2004. Population genetics of the developmental gene optomotor-blind (omb) in Drosophila polymorpha: evidence for a role in abdominal pigmentation variation. Genetics 168: 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariou, M. L., J. F. Silvain, V. Daubin, J. L. DaLage and D. Lachaise, 2001. Divergence between Drosophila santomea and allopatric or sympatric populations of D. yakuba using paralogous amylase genes and migration scenarios along the Cameroon volcanic line. Mol. Ecol. 10: 649–660. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., S. Y. Kim, A. S. Chang, D. Lachaise and S. Elwyn, 2002. Sexual isolation between two species with overlapping ranges: Drosophila santomea and Drosophila yakuba. Evolution 56: 2424–2434. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., S. Elwyn, S. Y. Kim and A. Llopart, 2004. Genetic studies of two sister species in the Drosophila melanogaster subgroup, D. yakuba and D. santomea. Genet. Res. 84: 11–26. [DOI] [PubMed] [Google Scholar]
- David, J. R., P. Capy, V. Payant and S. Tsakas, 1985. Thoracic trident pigmentation in Drosophila melanogaster: differentiation of geographical populations. Genet. Sel. Evol. 17: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilda, C. L., and T. F. C. Mackay, 2002. The genetic architecture of Drosophila sensory bristle number. Genetics 162: 1655–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerge, R. W., and G. A. Churchill, 1996. Permutation tests for multiple loci affecting a quantitative character. Genetics 142: 285–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale, R. A., M. A. Crosby and The FlyBase Consortium, 2005. FlyBase: genes and gene models. Nucleic Acids Res. 33: D390–D395 (http://flybase.org/). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics, Ed. 4. Longman, London. [DOI] [PMC free article] [PubMed]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection: A Complete Variorum Edition. Oxford University Press, Oxford.
- Gray, Y. H. M., J. A. Sved, C. R. Preston and W. R. Engels, 1998. Structure and associated mutational effects of the cysteine proteinase (CP1) gene of Drosophila melanogaster. Insect Mol. Biol. 7: 291–293. [DOI] [PubMed] [Google Scholar]
- Gibert, P., B. Moreteau, J. C. Moreteau, R. Parkash and J. R. David, 1998. Light body pigmentation in Indian Drosophila melanogaster: a likely adaptation to a hot and arid climate. J. Genet. 77: 13–20. [Google Scholar]
- Gompel, N., and S. B. Carroll, 2003. Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424: 931–935. [DOI] [PubMed] [Google Scholar]
- Hollocher, H., J. L. Hatcher and E. G. Dyreson, 2000. a Evolution of abdominal pigmentation differences across species in the Drosophila dunni subgroup. Evolution 54: 2046–2056. [DOI] [PubMed] [Google Scholar]
- Hollocher, H., J. L. Hatcher and E. G. Dyreson, 2000. b Genetic and developmental analysis of abdominal pigmentation differences across species in the Drosophila dunni subgroup. Evolution 54: 2057–2071. [DOI] [PubMed] [Google Scholar]
- Kopp, A., and I. Duncan, 1997. Control of cell fate and polarity in the adult abdominal segments of Drosophila by optomotor-blind. Development 124: 3715–3726. [DOI] [PubMed] [Google Scholar]
- Kopp, A., and I. Duncan, 2002. Anteroposterior patterning in adult abdominal segments of Drosophila. Dev. Biol. 242: 15–30. [DOI] [PubMed] [Google Scholar]
- Kopp, A., I. Duncan, D. Godt and S. B. Carroll, 2000. Genetic control and evolution of sexually dimorphic characters. Nature 408: 553–559. [DOI] [PubMed] [Google Scholar]
- Kopp, A., R. M. Graze, S. Xu, S. B. Carroll and S. V. Nuzhdin, 2003. Quantitative trait loci responsible for variation in sexually dimorphic traits in Drosophila melanogaster. Genetics 163: 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise, D., M. Harry, M. Solignac, F. Lemeunier, V. Benassi et al., 2000. Evolutionary novelties in islands: Drosophila santomea, a new melanogaster sister species from Sao Tome. Proc. R. Soc. Lond. Ser. B 267: 1487–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeunier, F., and M. Ashburner, 1976. Relationship within the melanogaster subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc. R. Soc. Lond. Ser. B 193: 275–294. [DOI] [PubMed] [Google Scholar]
- Llopart, A., S. Elwyn, D. Lachaise and J. A. Coyne, 2002. a Genetics of a difference in pigmentation between Drosophila yakuba and D. santomea. Evolution 56: 2262–2277. [DOI] [PubMed] [Google Scholar]
- Llopart, A., S. Elwyn and J. A. Coyne, 2002. b Pigmentation and mate choice in Drosophila. Nature 419: 360. [DOI] [PubMed] [Google Scholar]
- Llopart, A., D. Lachaise and J. A. Coyne, 2005. Multilocus analysis of introgression between two sympatric sister species of Drosophila: Drosophila yakuba and D. santomea. Genetics 171: 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Neckameyer, W., 1996. Multiple roles for dopamine in Drosophila development. Dev. Biol. 176: 209–219. [DOI] [PubMed] [Google Scholar]
- Neckameyer, W., 1998. a Dopamine modulates female sexual receptivity in Drosophila melanogaster. J. Neurogenet. 12: 101–114. [DOI] [PubMed] [Google Scholar]
- Neckameyer, W., 1998. b Dopamine and mushroom bodies in Drosophila: experience-dependent and -independent aspects of sexual behavior. Learn. Mem. 5: 157–165. [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 1998. Testing natural selection vs. genetic drift in phenotypic evolution using quantitative trait locus data. Genetics 149: 2099–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., 2001. The genetics of species differences. Trends Ecol. Evol. 16: 343–350. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and J. A. Coyne, 1992. The genetics of adaptation: a reassessment. Am. Nat. 140: 725–742. [DOI] [PubMed] [Google Scholar]
- Pendleton, R. G., A. Rasheed, T. Sardina, T. Tully and R. Hillman, 2002. Effects of tyrosine hydroxylase mutants on locomotor activity in Drosophila: a study in functional genomics. Behav. Genet. 32: 89–94. [DOI] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Krawetz and S. Misener. Humana Press, Totowa, NJ (http://fokker.wi.mit.edu/primer3/). [DOI] [PubMed]
- Settle, Jr., S. H., M. M. Green and K. C. Burtis, 1995. The silver gene of Drosophila melanogaster encodes multiple carboxypeptidases similar to mammalian prohormone-processing enzymes. Proc. Natl. Acad. Sci. USA 92: 9470–9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D. G., E. S. Pentz, M. E. Freeman, J. Kullman, G. R. Hankins et al., 1995. The genetic and molecular organization of the Dopa decarboxylase gene cluster of Drosophila melanogaster. Genetics 141: 629–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D. G., D. Y. Burton, W. E. McIvor, S. Krishnakumar, T. R. F. Wright et al., 1999. The Catecholamines up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tryrosine hydroxlyase activity. Genetics 153: 361–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel, B. L., M. S. Livingstone and W. G. Quinn, 1984. Mutations in the dopa decarboxylase gene affect learning in Drosophila. Proc. Natl. Acad. Sci. USA 81: 3577–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M. F., L. L. Zeineh, B. C. Black, W. E. McIvor, T. R. F. Wright et al., 1996. Catecholamine metabolism and in vitro induction of premature cuticle melanization in wild type and pigmentation mutants of Drosophila melanogaster. Arch. Insect Biochem. Physiol. 31: 219–233. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., J. R. True and S. B. Carroll, 2002. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development 129: 1849–1858. [DOI] [PubMed] [Google Scholar]
- Wittkopp, P. J., S. B. Carroll and A. Kopp, 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19: 495–504. [DOI] [PubMed] [Google Scholar]
- Wright, T. R. F., 1987. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 24: 127–222. [PubMed] [Google Scholar]
- Wright, T. R. F., 1996. The Wilhelmine E. Key 1992 invitational lecture. Phenotypic analysis of the Dopa decarboxylase gene cluster mutants in Drosophila melanogaster. J. Hered. 87: 175–190. [DOI] [PubMed] [Google Scholar]
- Zeng, Z-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]