Abstract
The compact genome of the tiger pufferfish, Takifugu rubripes (fugu), has been sequenced to the “draft” level and annotated to identify all the genes. However, the assembly of the draft genome sequence is highly fragmented due to the lack of a genetic or a physical map. To determine the long-range linkage relationship of the sequences, we have constructed the first genetic linkage map for fugu. The maps for the male and female spanning 697.1 and 1213.5 cM, respectively, were arranged into 22 linkage groups by markers heterozygous in both parents. The resulting map consists of 200 microsatellite loci physically linked to genome sequences spanning ∼39 Mb in total. Comparisons of the genome maps of fugu, other teleosts, and mammals suggest that syntenic relationship is more conserved in the teleost lineage than in the mammalian lineage. Map comparisons also show a pufferfish lineage-specific rearrangement of the genome resulting in colocalization of two Hox gene clusters in one linkage group. This map provides a foundation for development of a complete physical map, a basis for comparison of long-range linkage of genes with other vertebrates, and a resource for mapping loci responsible for phenotypic differences among Takifugu species.
THE tiger pufferfish, Takifugu rubripes (fugu), was proposed as a genomic model because of its compact genome size, which is about eight times smaller than that of the human genome (Brenner et al. 1993). Since fishes have a body plan and physiological systems similar to mammals, the compact genome of fugu can help to discover genes and gene regulatory regions in the human genome and serve as a reference to understand the evolution of vertebrate genomes and karyotype (Grutzner et al. 1999; Hedges and Kumar 2002). Three years ago, a “draft” sequence of fugu was generated purely by a whole-genome shotgun strategy (Aparicio et al. 2002). The draft sequence comprises of 12,381 scaffolds >2 kb and efforts are currently underway to obtain the complete sequence of the genome. Since fugu has 22 pairs of chromosomes (Miyaki et al. 1995), the ultimate objective is to merge the draft sequence scaffolds into 22 “superscaffolds” corresponding to the 22 chromosomes. However, the linkage relationship of the scaffolds is currently unknown due to the lack of a genetic linkage map or a genome-wide physical framework for this fish. This has restricted the use of the fugu genome sequence for global comparison with other vertebrate genomes to address questions related to genome architecture and evolution of vertebrate chromosomes. Thus, it is recognized that a genetic linkage map is essential for completing the genome sequence of fugu and using it in comparative genomic studies.
In addition to fugu, genomes of two other teleosts, the zebrafish (Danio rerio) and medaka (Oryzias latipes), are being sequenced. These two species are excellent genetic models. Fugu and medaka are grouped together under the common superorder Acanthopterygii and are phylogenetically closer to each other than to zebrafish, which are classified under the more basal superorder Ostariophysi (Nelson 1994). Molecular clock analyses of the mitochondrial sequences have suggested that the common ancestors of the pufferfish and zebrafish diverged ∼280 million years ago (Kumazawa et al. 1999). Although there are no estimates for the divergence time of the medaka and pufferfish lineages, since the fossil evidence shows that fishes of the order Tetraodontiformes have been around for ∼95 million years (Tyler and Sorbini 1996), it is assumed that the two lineages diverged >95 million years ago.
Excellent gene maps of zebrafish and medaka have been generated, using ∼1500 and ∼800 gene or EST sequences, respectively (Woods et al. 2000; Naruse et al. 2004). Recently, the draft genome sequence of another pufferfish, the spotted green pufferfish, Tetraodon nigroviridis, has been generated (Jaillon et al. 2004). A partial physical map of the Tetraodon genome has been constructed by anchoring 64.4% of the genome assembly to its 21 chromosomes (Jaillon et al. 2004). The Tetraodon lineage has been estimated to have diverged from the fugu lineage ∼18–30 million years ago (Crnogorac-Jurcevic et al. 1997).
Comparisons of gene maps of zebrafish, medaka, and Tetraodon with humans have helped to infer the number and content of the chromosomes of the last common ancestor of teleosts and mammals (Woods et al. 2000; Jaillon et al. 2004; Naruse et al. 2004). However, there has been no attempt to compare the extent of genome reorganization between the teleost species and the mammalian species. A genetic linkage map of fugu would be useful for such comparative studies among teleost lineages and between teleost and mammalian lineages and would help to understand the process of teleost genome evolution.
In addition to comparative genomics, genetic linkage maps are invaluable in forward genetic analysis for the identification of gene loci responsible for genetic traits. Pufferfishes belonging to the genus Takifugu are mainly distributed in coastal regions of East Asia and include >20 species (Matsuura 1990). Interestingly, hybrids produced by artificial fertilization of eight Takifugu interspecies crosses, including fugu and Takifugu niphobles, were found viable (Fujita 1967; Miyaki 1992). Fugu and T. niphobles show marked differences in body size, body color pattern, body shape, the number of meristic skeletons, temperature preference, parasite resistance, and behavior (Uno 1955; Ogawa 1991). Given the the ability to generate fertile crosses between Takifugu species, the availability of the draft genome sequence of fugu provides an unprecedented opportunity to understand the genetic basis of the evolution of natural species of these vertebrates. Availability of a genetic map of fugu would greatly facilitate such studies.
Here we report a genetic linkage map for fugu, the first map for a pufferfish. The map consists of genetic markers physically linked to the assembled genome sequences, allowing us to establish a long-range relationship of the scaffold sequences and to compare gene maps between fugu and other vertebrates. Comparison of parental maps showed sex-specific differences in recombination ratio. Comparison of gene maps revealed a tendency for highly conserved synteny among teleost genomes as well as a lineage-specific rearrangement in the pufferfish lineages. This genetic map will serve as a foundation for obtaining the complete sequence of the fugu genome, a basis for comparison of the genome map with those of other vertebrates, and an important tool for the future genome-wide analysis of phenotypic differences observed within and between Takifugu species.
MATERIALS AND METHODS
Mapping population:
Wild female and male fugu were caught in the Sea of Japan at Toyama Bay and in the Pacific Ocean off Shizuoka prefecture, respectively. A mapping population was obtained by in vitro crossing (Fujita 1967; Matsuyama et al. 1997). A total of 64 full-sib progeny from the single cross were used for developing a linkage map.
DNA extraction:
Muscle tissue (50–100 mg) or whole bodies of juveniles (∼100 mg each) were preserved in 400 μl of TNES-urea buffer (10 mm Tris-HCl, pH 7.5, 125 mm NaCl, 10 mm EDTA, 1% sodium dodecyl sulfate, 8 m urea) at room temperature for months. Genomic DNA was extracted from the preserved samples using standard phenol-chloroform technique with slight modifications (Asahida et al. 1996). Approximately 3–15 μg of purified DNA was obtained from each preserved sample.
Microsatellite marker:
The CA repeat is the most common microsatellite interspersed in the genomes of vertebrates, including fugu (Aparicio et al. 2002). We first identified 160 CA-repeat loci by querying (CA)50 sequence against the fugu draft sequence data on the website at the Joint Genome Institute (http://www.jgi.gov/fugu/index.html). An additional 40 loci were then chosen by scanning the fugu scaffolds starting from the largest scaffold (no. 1). To increase the map density and compare the linkage relationships of orthologs among fugu, medaka, and zebrafish, we searched the CA-repeat loci physically linked to fugu orthologs of medaka and zebrafish genes that have been mapped to their respective maps (see Sequence comparison below) and identified 152 loci. The analysis was carried out on the August 2002 freeze data set for the fugu genome. Primers flanking the CA repeats were designed and used for PCR to find heterozygous markers in one or both parents. Construction of initial male and female genetic maps of fugu using 192 microsatellite markers and their consolidation identified 23 linkage groups. Comparison of this map with gene maps of medaka and Tetraodon indicated that two of the fugu linkage groups might be merged into one linkage group (LG2). We therefore increased the density of microsatellite markers on LG2 to obtain adequate markers for merging the two linkage groups. Details of primer sequences are shown in supplementary Table S1 at http://www.genetics.org/supplemental/ as well as on our website at http://park.itc.u-tokyo.ac.jp/suijitsu/. Primer sequences have also been deposited in DDBJ/EMBL/GenBank with accession nos. AB213693–AB214112.
Genotyping:
PCR was performed in 20-μl reaction volume of 100 mm Tris-HCl (pH 8.3) containing 0.5 m KCl, 2.5 mm dNTPs, 0.5 μm of each primer, Taq DNA polymerase (Takara, Tokyo, Japan), and 5 ng of genomic DNA. The following PCR protocol was used: initial denaturation at 94° for 1 min, 35 cycles of 94° for 30 sec, and 59° for 2 min, followed by a final extension period at 59° for 3 min. The PCR products were heat denatured with formamide loading buffer and fractionated on a denaturing 6% acrylamide gel (Long-Ranger, ABI, Columbia, MD) with 8 m urea. The gels were stained with SYBR gold and visualized with a fluorescent imaging system (FLA3000, FUJIFILM).
Genotypic classification:
The progeny data are subdivided into two independent data sets that contain the meiotic segregation data from each parent. Ancestry-unknown markers in the three-generation pedigree were analyzed within a phase-known model of inbred pedigree and converted to ancestry-known markers following the method of Sewell et al. (1999).
Map construction:
The map was constructed using MAPMAKER (Lander et al. 1987). Linkage groups were determined using a two-point analysis. Local order was established by multipoint analysis. Marker orders were modified after visual analysis of the distribution of marker genotypes in the reference mapping panel.
Sequence comparison:
Fugu orthologs of medaka, zebrafish, and human genes were identified by BLAST searching these sequences against the fugu draft sequence data with a cutoff E-value of e−10. For the search, we used 819 gene and EST sequences for medaka and their human orthologs and 792 gene and EST sequences for zebrafish and their human orthologs reported by Naruse et al. (2004) and Woods et al. (2000). In cases where the searches hit two or more scaffolds with a less-than-threefold difference in the E-value, we did not assign any orthology. In particular, to identify fugu orthologs to medaka and zebrafish genes, we chose 239 gene and EST sequences whose orthologs have been predicted between medaka and zebrafish (Naruse et al. 2000) and BLAST searched their sequences (a maximum E-value of e−10) against the fugu database. Fugu ortholog was predicted if both medaka and zebrafish sequences showed the highest similarity to the same fugu gene. In cases where the searches hit two or more scaffolds, fugu orthologs were identified on the basis of phylogenetic analysis. For the phylogenetic analysis, fugu protein sequences identified in the BLAST search were aligned with medaka, zebrafish, and human gene family members and phylogenetic analysis was carried out using the program Clustal-X (Thompson et al. 1997) and the neighbor-joining method (Galtier et al. 1996). Medaka and zebrafish mapping information and orthologous relationships among medaka, zebrafish, and human were obtained from Naruse et al. (2004) and Woods et al. (2000), who compiled the latest peer-reviewed medaka and zebrafish maps available. Medaka and zebrafish gene and EST sequences were obtained from DDBJ (http://getentry.ddbj.nig.ac.jp) or the Zebrafish Information Network (http://www.zfin.org) using accession numbers listed in Naruse et al. (2004) or Woods et al. (2000). Map position of human and mouse orthologs were determined using the NCBI human genome resources (http://www.ncbi.nlm.nih.gov/genome/guide/human/). Fugu genes on the same scaffolds are represented as a single locus in our analysis to prevent an overestimation of the extent of conserved synteny. To find putative orthologous segments of the fugu scaffolds in the “partial physical map” of Tetraodon, at least three DNA sequences of ∼10 kb were chosen from the 5′-end, middle, and 3′-end regions of the fugu scaffolds and searched for BLAT (Kent 2002) sequence similarity (a minimum score of 2000) against the Tetraodon draft sequence data (http://www.genoscope.cns.fr/). When the fugu scaffold size was <30 kb, the entire scaffold sequence was used for the search.
Nomenclature:
All loci identified are simple sequence length polymorphisms and are named in accordance with laboratory rules, e.g., f35, where “f” denotes both fugu and the fisheries laboratory at the University of Tokyo, and the number indicates the assay number. Ultimately, once the map is completed and physically anchored to each chromosome, each marker should receive a locus name following the system used for the dog, mouse, pig, and rat maps.
RESULTS
Markers:
To obtain informative markers for linkage analysis in the progeny from the cross between noninbred parents, we examined the segregation types of the CA-repeat locus in both parents and four progeny. PCR reactions successfully amplified 376 loci in a total of 389 primer pairs that were designed on the basis of the fugu draft genome sequence. The father and mother were heterozygous for 184 (48.9%) and 188 (50%) of these markers, respectively. Among them, 160 markers were heterozygous in both parents with ab × cd or ab × ac type of segregation and allowed the identification of homologous pairs of linkage groups of the male and female parents. In total, 212 markers (56.4%) were informative for segregation analyses in the mapping population. An additional 60 progeny were scored for these markers to construct the linkage map.
Linkage map:
The male map consists of 24 linkage groups with 169 microsatellite loci that are physically linked to multi-gene-sized DNA sequences assembled in the fugu draft genome database (Table 1). Among the informative markers in male meiosis, 91.8% of the markers showed detectable linkage to another marker. The map spans 697.1 cM and the average spacing between two markers is 4.1 cM. The sizes of linkage groups range from 0 to 75 cM (mean 29 cM). The number of markers per linkage group varied from 3 to 17, with an average of 7 markers per group.
TABLE 1.
Number of markers, genetic length, and total scaffold size for each linkage group
| Male
|
Female
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| LG | No. of markers | Total scaffold size (bp) | No. of gene models | LG | No. of markers | Length (cM) | LG | No. of markers | Length (cM) |
| LG1 | 17 | 3,284,077 | 394 | LG1 | 16 | 64.8 | LG1-F1 | 3 | 12.6 |
| LG1-F2 | 11 | 75.2 | |||||||
| LG1-F3 | 2 | 21.2 | |||||||
| LG2 | 17 | 3,003,930 | 481 | LG2 | 16 | 75 | LG2 | 13 | 117.4 |
| LG3 | 12 | 2,521,477 | 231 | LG3 | 10 | 31.4 | LG3 | 11 | 96 |
| LG4 | 12 | 2,344,834 | 233 | LG4 | 11 | 38.8 | LG4 | 11 | 86.1 |
| LG5 | 14 | 1,844,110 | 279 | LG5-M1 | 4 | 3.2 | LG5-F1 | 4 | 17.4 |
| LG5-M2 | 6 | 11.1 | LG5-F2 | 8 | 65.8 | ||||
| LG6 | 7 | 1,543,293 | 190 | LG6 | 7 | 56.5 | LG6 | 7 | 76.2 |
| LG7 | 10 | 1,743,445 | 197 | LG7-M1 | 2 | 11.7 | LG7 | 10 | 69 |
| LG7-M2 | 6 | 22.5 | |||||||
| LG8 | 9 | 1,005,816 | 110 | LG8 | 7 | 34.6 | LG8 | 7 | 68.9 |
| LG9 | 8 | 1,526,000 | 146 | LG9 | 7 | 21.2 | LG9 | 8 | 63.4 |
| LG10 | 12 | 2,414,664 | 299 | LG10 | 9 | 49.8 | LG10-F1 | 7 | 38.5 |
| LG10-F2 | 4 | 19.3 | |||||||
| LG11 | 8 | 1,695,911 | 135 | LG11 | 7 | 35.3 | LG11-F1 | 2 | 23.7 |
| LG11-F2 | 5 | 24 | |||||||
| LG12 | 12 | 3,690,823 | 451 | LG12 | 10 | 20.5 | LG12 | 12 | 46.4 |
| LG13 | 5 | 1,403,550 | 65 | LG13 | 5 | 11 | LG13 | 4 | 43.1 |
| LG14 | 7 | 582,240 | 40 | LG14 | 5 | 12.6 | LG14 | 5 | 42.6 |
| LG15 | 8 | 2,018,465 | 186 | LG15 | 5 | 6.3 | LG15-F1 | 4 | 31.9 |
| LG15-F2 | 3 | 4.7 | |||||||
| LG16 | 5 | 645,346 | 76 | LG16 | 4 | 42 | LG16 | 3 | 34.9 |
| LG17 | 8 | 906,885 | 139 | LG17 | 7 | 43.3 | LG17-F1 | 4 | 20.9 |
| LG17-F2 | 3 | 7.9 | |||||||
| LG18 | 3 | 992,949 | 92 | LG18 | 2 | 0 | LG18 | 3 | 27 |
| LG19 | 5 | 1,286,233 | 136 | LG19 | 5 | 36.7 | LG19 | 3 | 23.2 |
| LG20 | 6 | 1,409,226 | 111 | LG20 | 6 | 22.1 | LG20 | 5 | 22.5 |
| LG21 | 6 | 2,031,407 | 254 | LG21 | 5 | 22.6 | LG21 | 3 | 22.4 |
| LG22 | 9 | 1,302,580 | 207 | LG22 | 7 | 24.1 | LG22 | 5 | 11.3 |
| Total | 200 | 39,197,261 | 4452 | 169 | 697.1 | 171 | 1213.5 | ||
The female map consists of 29 linkage groups with 171 microsatellite markers. Among the informative markers in female meiosis, 90.9% of the markers tested showed detectable linkage to another marker. The maps span 1213.5 cM and the average distance between two markers is 7.1 cM. The sizes of linkage groups range from 4.7 to 117.4 cM (mean 41.8 cM). The number of markers per linkage group varied from 2 to 13, with an average of 5.8 markers per group.
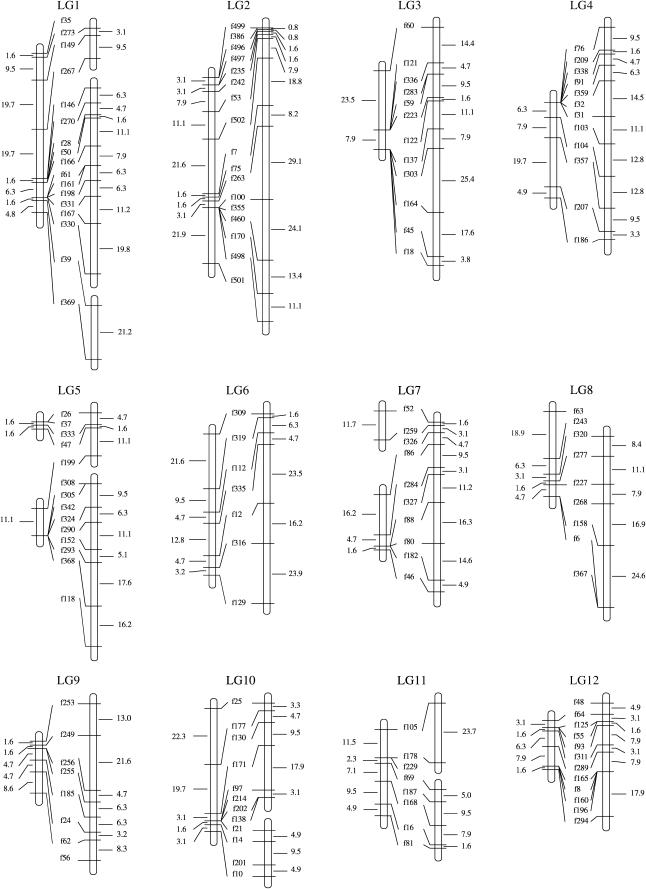
By using heterozygous markers in both parents, we identified homologous pairs of linkage groups of the male and female parents and arranged these maps in 22 linkage groups, designated LG1–LG22 (Figure 1 and Table 1). The resulting map consists of 200 markers physically linked to the assembled genome sequences ranging from 1,145,486 to 1447 bp. The total length of genomic sequences mapped is 39.2 Mb and contains 4452 gene loci (http://www.jgi.gov/fugu/index.html).
Figure 1.
Male and female genetic linkage maps of fugu. Allelic bridges are indicated by lines connecting male (left) and female (right) linkage maps. Genetic distances between adjacent markers are shown (Kosambi mapping function).
Seven pairs of markers that are linked in the male map remain unlinked in the female map whereas two pairs of markers in the female map remain unlinked in the male map (Figure 1 and Table 1). This could be explained by differences in recombination ratio between sexes as seen in most of the linkage groups. For example, while f330 and f39 are linked in the male map of LG1, they are not linked in the female map. In the adjacent intervals flanked by f330 and f167, female recombination rates are much higher than those observed in males. We expect that the pairs of unlinked markers will be linked as described in the map for the other sex when marker density increases.
Differences in recombination rate between male and female:
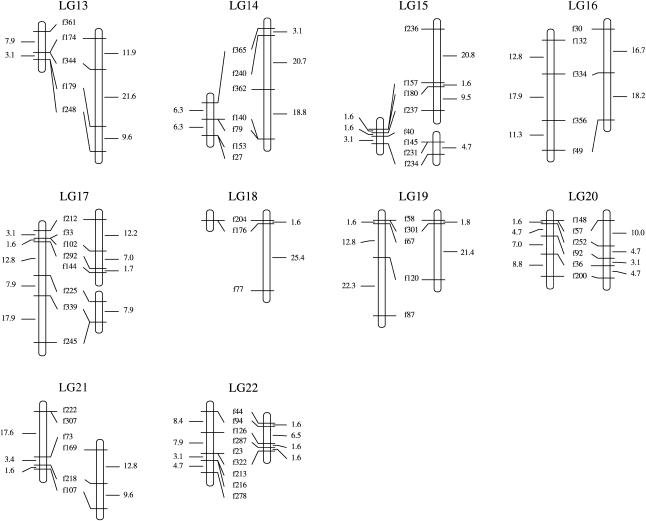
To estimate the relative rates of recombination between parents, we chose 104 pairs of adjacent markers heterozygous for both parents and compared sex-specific recombination rates. Figure 2 shows a comparison of male and female recombination ratio between the common intervals flanked by the paired markers from all the linkage groups. The number of intervals that shows a higher recombination ratio in females is larger than that in males. On the other hand, the number of intervals that shows expansion in the male map relative to the female map is small but not negligible. Summing up the length of the common interval for each linkage group on both the male and female maps gave a total length of 507.1 and 1100.8 cM, respectively. These results suggest that overall recombination is suppressed in male meiosis compared to female meiosis.
Figure 2.
Differences in recombination ratio between male and female. The common intervals flanked by adjacent markers from the 22 linkage groups are compared.
Comparison with T. nigroviridis:
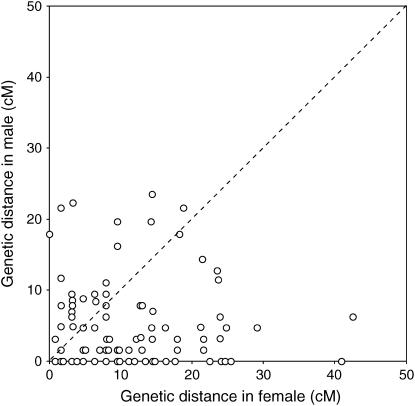
To compare the extent of conserved synteny between the two pufferfish genomes, we assigned 200 of the fugu scaffolds to putative orthologous segments of the Tetraodon genome. Among them, 152 segments have been anchored to Tetraodon chromosomes on its “partial physical map,” whereas 48 segments have not been anchored. This is consistent with the limited anchoring of the Tetraodon genome sequences to chromosomes (64.4%). In Figure 3, putative orthologous segments have been arrayed according to linkage groups of fugu and chromosomes of Tetraodon. The display shows that each of the fugu linkage groups shares putative orthologous segments with one Tetraodon chromosome except for LG4, LG8, LG10, and LG14. In the fugu linkage group LG10, six scaffolds (scaffold 4, 146, 429, 446, 1117, and 2289) were assigned to putative orthologous segments on Tetraodon chromosome 6, whereas two other scaffolds, scaffold 1628 and scaffold 572, showed similarity over large regions to Tetraodon chromosomes 18 and 17, respectively. (Figure 3 and supplementary Table S2 at http://www.genetics.org/supplemental/). This suggests that there were interchromosomal translocations of segments of the ancestral sequences of scaffolds 1628 and 572 in either the fugu or the Tetraodon lineage after the divergence of the two species. Similar rearrangements were also evident in the fugu linkage groups LG4, LG8, and LG14. Thus, of the 152 orthologous segments, only 6 segments indicate possible interchromosomal rearrangement events after the divergence of the two species of pufferfishes. This shows that most regions of the chromosomes have been conserved during the 18–30 million years of divergent evolution of the two lineages. Figure 3 also indicates that each of Tetraodon chromosomes 1, 3, 17, 18, and 20 shows conserved synteny with two to three linkage groups of fugu. The disruption of the one-to-one relationship of the Tetraodon chromosomes to fugu linkage groups may be due to interchromosomal translocations that include recent fusion of chromosomes in the Tetraodon lineage or fission in the fugu lineage after the divergence of the two pufferfish species. This could be related to the presence of an additional pair of chromosomes in fugu (n = 22) compared to Tetraodon (n = 21).
Figure 3.
Summary of the number of orthologous genome regions in fugu and T. nigroviridis. The linkage groups of fugu are arrayed as columns and the chromosomes of Tetraodon as rows. Genome sequences that have not been mapped in Tetraodon are displayed in the last row (UN, unknown). Numbers in boxes indicate the number of orthologous genome sequences. Details of mapped sequences are listed in supplementary Table S2 at http://www.genetics.org/supplemental/.
Conservation of synteny with other teleosts:
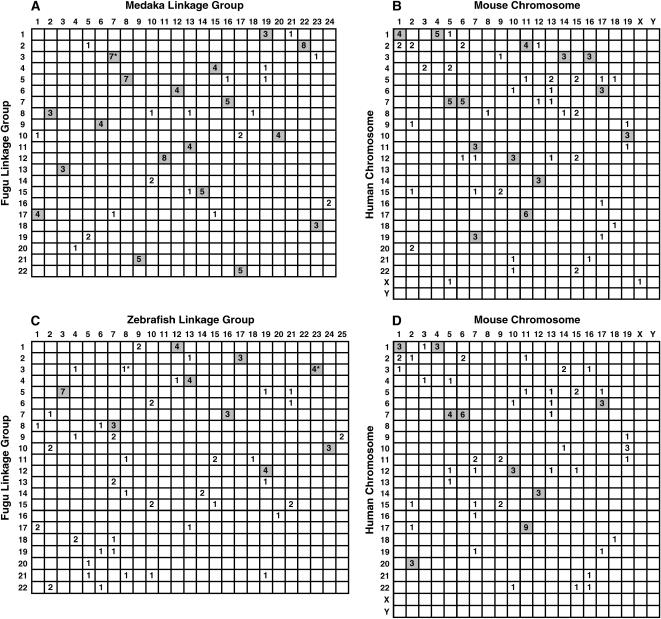
To compare the extent of conserved synteny between the fugu and medaka genomes, we identified fugu orthologs to medaka genes chosen from a medaka linkage map (Naruse et al. 2004) and found that 108 of them were present in the fugu map. In the array of orthologs assigned on the basis of the linkage groups for each species, we were able to identify the cluster of orthologous gene pairs that are located on conserved syntenic segments in fugu and medaka (Figure 4A). Among 108 putative orthologs, 94 orthologs fell in 22 conserved synteny segments containing two or more genes between the two species. Furthermore, 18 pairs of linkage groups shared conserved syntenic segments containing at least three orthologous gene pairs. These correlations imply that these pairs of linkage groups are derived from a common ancestral chromosome. Of the 101 genes from these 18 fugu linkage groups, 86 genes (85.1%) are assigned to orthologous linkage groups in medaka. These results suggest that the syntenic relationship of a significant fraction of the genome has been remarkably conserved in the fugu and medaka lineages. Using a similar strategy, we compared syntenic relationships between the fugu and zebrafish genomes by using 88 pairs of orthologous genes (Figure 4C). Among them, 61 orthologs fell in 22 conserved syntenic segments, each containing two or more orthologous genes. Indeed, 9 of the conserved syntenic segments contain three or more orthologous gene pairs between the linkage groups of the two fishes (Figure 4C).
Figure 4.
Conservation of synteny among fugu, medaka, zebrafish, human, and mouse. The putative orthologous genes from two species are arrayed according to linkage group or chromosome for each species. Numbers in boxes indicate the number of orthologous gene pairs. The boxes containing more than three orthologous gene pairs are shaded. (A) Fugu-medaka comparison; (B) human-mouse comparison based on genes orthologous to data set in A; (C) fugu-zebrafish comparison; (D) human-mouse comparison based on genes orthologous to data set in C. (*) Three of the orthologs in comparisons between A and B and two of the orthologs in comparisons between C and D, respectively, have been excluded as we failed to identify their mouse orthologs. Details of the orthologous genes are listed in supplementary Table S2 at http://www.genetics.org/supplemental/.
To compare the degree of conserved synteny between the teleost and mammalian genomes, we defined conserved syntenic segments using 105 quadruplets of orthologous genes from our data set of fugu, medaka, human, and mouse (Figure 4, A and B). This analysis revealed that whereas only 14 orthologous gene pairs did not fall in conserved syntenic segments between fugu and medaka (Figure 4A), 28 orthologous gene pairs between the human and mouse did not fall in conserved syntenic segments (Figure 4B). Since the evolutionary divergence period between fugu and medaka is similar or even longer than that of human and mouse, this result suggests that syntenic relationships are more conserved in the teleost lineage than in the mammalian lineage. We did similar analysis using 86 quadruplets of orthologous genes for fugu, zebrafish, human, and mouse and defined syntenic segments in the teleost and mammalian genomes. In this analysis, 26 and 32 orthologous genes for fugu and zebrafish, and the two mammals, respectively, did not fall in conserved syntenic segments (Figure 4D). Such a similar degree of conserved synteny between the fugu and the zebrafish and the human and mouse, despite the longer divergence period between the two fishes, further supports a lower frequency of interchromosomal rearrangements in the teleost lineage than in the mammalian lineage.
Hox clusters:
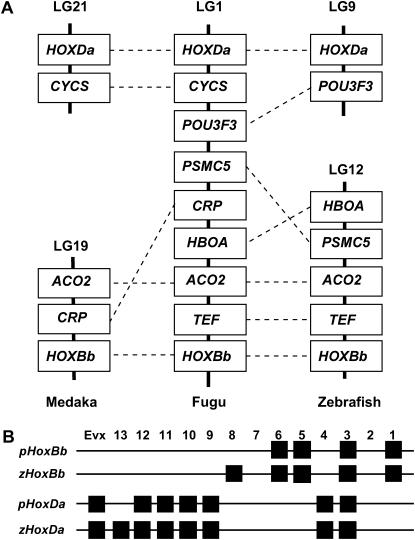
In vertebrates, Hox genes are present in tight clusters of up to 14 genes and these clusters have been often used as landmarks for comparison of gene maps (Amores et al. 1998). We mapped the Hox clusters of fugu to our genetic linkage map and compared the location of Hox clusters in the gene maps of fugu, Tetraodon, medaka, and zebrafish. To determine the position of Hox clusters in the fugu map, we first identified nine scaffolds that contained members of six Hox clusters (scaffold 47 for HoxAa, scaffold 330 and 5310 for HoxAb, scaffold 1439 and 706 for HoxBa, scaffold 1245 and 2182 for HoxBb, scaffold 93 for HoxCa, and scaffold 214 for HoxDa; also see supplementary Table S2 at http://www.genetics.org/supplemental/) by BLAST search and obtained genetic markers from these sequences. A polymorphic marker for the HoxDb cluster could not be obtained because we could identify only one scaffold that contained a member of the HoxDb cluster (scaffold 19,911 for HoxDb9) in the fugu database and this scaffold contained no CA-repeat locus. The six markers physically linked to the six Hox clusters (HoxAa, HoxAb, HoxBa, HoxBb, HoxCa, HoxDa) were then mapped to the linkage groups of fugu (Table 2). Unexpectedly, the six markers mapped to only five linkage groups, with two Hox clusters, HoxBb and HoxDa, being located in the same linkage group, LG1. To determine whether the genetic events that led to the two Hox clusters being located to the same linkage group in fugu involved any of the flanking genomic regions, we compared the syntenic relationship of the Hox clusters and their known genetically linked genes such as CYCS, POU3F3, PSMC5, CRP, HBOA, ACO2, and TEF among fugu, medaka, and zebrafish (Figure 5A). The comparison revealed that whereas the syntenic segments of both HoxBb and HoxDa together with their linked non-Hox genes were located in the same linkage group in fugu, the two syntenic clusters were assigned to different linkage groups in both medaka and zebrafish, suggesting that the two Hox clusters and their flanking regions were translocated into one linkage group in an ancestor of fugu after it diverged from the medaka lineage. We also identified the location of Hox clusters in the “partial physical map” of Tetraodon by BLAT searching with the fugu scaffold sequences containing Hox genes and found that both Tetraodon HoxBb and HoxDa clusters are localized on chromosome 2 (Table 2). This suggests that the translocation event occurred before the separation of the fugu and Tetraodon lineages.
TABLE 2.
Linkage groups containing Hox clusters in zebrafish, medaka, T. nigroviridis, and fugu
| Hox cluster | Zebrafish | Medaka | Tetraodon | Fugu |
|---|---|---|---|---|
| HoxAa | LG19 | LG11 | Chromosome 21 | LG12 |
| HoxAb | LG16 | LG16 | Chromosome 8 | LG7 |
| HoxBa | LG3 | LG8 | Unmappeda | LG5 |
| HoxBb | LG12 | LG19 | Chromosome 2 | LG1 |
| HoxCa | LG23 | LG7 | Chromosome 9 | LG3 |
| HoxCb | LG11 | |||
| HoxDa | LG9 | LG21 | Chromosome 2 | LG1 |
| HoxDb | LG15 | Chromosome 17b | Unmapped |
Data on zebrafish and medaka are from Wood et al. (2000) and Naruse et al. (2004), respectively.
Tetraodon HoxBa genes were identified by BLAT search using fugu HoxBa sequence as query. Their chromosomal locations were not assigned in the Tetraodon map.
HoxDb genes of fugu and Spheroides nephalus were characterized by Amores et al. (2004). Their putative orthologs in Tetraodon have been labeled as HoxCb in Jaillon et al. (2004).
Figure 5.
(A) Comparative linkage map of HoxBb and HoxDa clusters and their flanking regions in fugu, medaka, and zebrafish. Genes are named according to the nomenclature for the orthologous genes of human or zebrafish. The zebrafish and medaka genes for which there is no mapping information are not shown in the linkage map. Orthologous genes are linked by dashed lines. (B) Genomic organization of HoxBb and HoxDa clusters in pufferfishes (fugu, Tetraodon, and Spheroides) (p) and zebrafish (z). Hox paralog group is shown along the top.
DISCUSSION
Genetic map and genetics:
We have constructed the male and female genetic maps for fugu and arranged them in 22 linkage groups in which 94.3% of the markers are linked to at least one other marker. While much effort has been concentrated on using the fugu as a “genome model,” less attention has been paid to the potential of this fish as a “genetic model.” One of the purposes of this study is to construct a tool for genome-wide genetic analysis of fugu. In our male and female maps, >94% of the markers are linked to other markers, suggesting a high probability that the markers may be linked to genetic traits. Such a map would be useful for the initial low-resolution genetic mapping of traits. Although we cannot expect that all markers that we used will be informative in other crosses—e.g., interspecific crossing of fugu and closely related species—the high level of polymorphic information content of the CA repeats in wild individuals will allow us rapid development of additional markers that are in close proximity to the noninformative markers by searching the genome sequence database.
The large collection of microsatellite markers will be also useful for aquaculture purposes, because fugu is becoming one of the most economically important fish for aquaculture (Statistics and Information Department 2001). These markers will be essential for the identification of loci responsible for commercially desirable traits in different strains of this fish. Molecular identification of such loci combined with the technology for marker-assisted selective breeding would be useful for establishing commercially better strains of fugu. The collection of markers is also critical for characterizing genetic background of wild and cultured fugu to maintain heterozygosity of stock and for tracking parentage.
Sex-specific difference in recombination ratio:
Among mammals, human, dog, pig, and mouse show reduced chromosomal recombination frequency in males, while cattle do not show such sex-specific differences (Dib et al. 1996; Dietrich et al. 1996; Marklund et al. 1996; Neff et al. 1999). Among teleost fishes, an overall lower recombination ratio has been identified in males of trout and zebrafish (Sakamoto et al. 2000; Singer et al. 2002). The recombination around the centromere region is preferentially lower in male meiosis than in female meiosis in these fishes. In addition, detailed studies in zebrafish have revealed that the recombination in the region near the telomere is preferentially lower in female meiosis than in male meiosis (Singer et al. 2002). Our analysis of the fugu genetic map shows that males have an overall reduction in recombination ratio relative to females. While a large number of the common intervals between the male map and female map show expansion in the female map, some intervals also show expansion in the male map. This tendency is similar to that observed in trout and zebrafish (Sakamoto et al. 2000; Singer et al. 2002). Since the male-specific reduction in the recombination ratio seems to exist in diverse species of teleosts such as trout, zebrafish, and fugu, it is likely that this feature is shared by all teleosts.
This finding has practical significance in facilitating efficient experimental design in the genome-wide linkage analysis for species differences among Takifugu species (Singer et al. 2002). A decreased rate of recombination in males is an advantage for mapping genetic traits in initial low-resolution analyses, especially when analyzing quantitative trait loci (Glazier et al. 2002). For example, if the phenotype shows complete or partial dominance in F1 hybrids between two Takifugu species, it is adequate to use a mapping population produced from a cross between male hybrid and a female wild species for primary mapping. On the other hand, it would be necessary to use a higher frequency of recombination in females for fine mapping of these loci.
Comparison of genomes:
Comparisons of the genetic map of fugu with that of other teleosts revealed that, as expected, the synteny between the two pufferfishes, fugu and Tetraodon, is highly conserved. However, we also found highly conserved synteny between fugu and medaka, which shared a common ancestor >95 million years ago. Our comparisons of the teleost maps and human and mouse maps showed that syntenic relationships are more conserved in the teleost fish lineage than in the mammalian lineage. For example, even though the divergence period between fugu and zebrafish is about threefold longer than that between the human and mouse, a similar degree of conserved synteny was found between the two fish lineages and between the two mammalian lineages. By comparing the complete physical map of human and the “partial physical map” of Tetraodon, Jaillon et al. (2004) reconstructed the karyotype of the last common ancestor of the teleost fish and mammals. Their model suggests a lower frequency of major interchromosomal exchanges in the teleost lineage than in the human lineage. Naruse et al. (2004) also inferred the ancestral karyotype using the medaka, zebrafish, and human maps. Their result suggested that karyotype evolution in teleosts occurred mainly by inversion and not by fusion or fission of chromosomes that is frequent in mammalian lineages. Our results independently demonstrate the lower frequency of interchromosomal exchanges in the teleost lineage and provide further support to the models proposed by Jaillon et al. (2004) and Naruse et al. (2004). We also showed that the extent of conserved synteny between fugu and zebrafish is less than that between fugu and medaka. This result is consistent with the phylogenetic positions of the three fish species, i.e., zebrafish being basal to both fugu and medaka. However, this can also have other explanations. The larger genome size of zebrafish compared to fugu and medaka (Venkatesh et al. 2000) implies that the genome of this teleost contains a significant proportion of transposable elements. Accumulation of transposable elements might have favored a higher frequency of interchromosomal rearrangements in the zebrafish (Grutzner et al. 1999).
Hox clusters:
In vertebrates Hox genes occur in tight clusters of up to 14 paralogous genes. They play an important role in anterior/posterior patterning in development, including skeletal complexity of animals. The expression domains of the genes in a cluster are colinear with the position of the genes in the cluster. In all tetrapods studied so far, four clusters of Hox genes (HoxA, HoxB, HoxC, and HoxD) have been identified. The clusters are located on different chromosomes (Scott 1992; Ladjali-Mohammedi et al. 2001). These four clusters are presumed to have arisen by two rounds of whole-genome or segmental duplication during the evolution of early vertebrates (Larhammar et al. 2002). The distribution of four Hox clusters on four chromosomes is often sighted as evidence for whole-genome (or whole-chromosome) duplications. Although extensive interchromosomal rearrangements are suggested in the evolution of amniote chromosomes (Bourque et al. 2004), all amniotes so far examined have retained the presumptive ancient organization of Hox clusters and chromosomes; that is, a single Hox cluster is located on each chromosome. In contrast to tetrapods, teleosts such as zebrafish, medaka, and pufferfishes (fugu, Spheroides, and Tetraodon) contain up to seven Hox clusters (Amores et al. 1998, 2004; Naruse et al. 2000). The additional Hox clusters in teleosts have been proposed to be the result of a whole-genome duplication (Amores et al. 1998) that occurred early during the evolution of the ray-finned fish (Christoffels et al. 2004). Presumably, one of the duplicate Hox clusters was lost following the whole-genome duplication. In the medaka and zebrafish, in which Hox clusters have been mapped to the genetic linkage groups, the seven clusters are located in different linkage groups. The genomic position of the Hox clusters in the pufferfish was not known until now. In our study, we unexpectedly found that two Hox clusters, HoxBb and HoxDa, along with their flanking genes are located in the same linkage group in fugu as well as in Tetraodon (Figure 5A, Table 2). Since these two Hox clusters are located in different linkage groups in the zebrafish and medaka, it seems likely that the two clusters were colocalized to the same chromosome through interchromosomal rearrangements, most likely through fusion of two chromosomes. Each of the pufferfish HoxBb and HoxDa clusters lacks one of the genes present in the zebrafish HoxB8b and HoxD13a (see Figure 5B). Although a detailed gene content of the medaka Hox clusters has not been analyzed, the overall organization of Hox clusters in medaka has been reported to be more similar to that in pufferfishes than in zebrafish (Amores et al. 2004) (see Table 2). For example, while two HoxD clusters, HoxDa and HoxDb, are present in medaka and pufferfishes, zebrafish have a single HoxDa cluster but two HoxC clusters (HoxCa and HoxCb). Thus, it is assumed that the contents of the HoxBb and HoxDa clusters in medaka are more similar to those in pufferfishes than to those in zebrafish. Pufferfish are the only known vertebrate group in which two Hox clusters are located on the same chromosome. It is not known if this has any influence on the expression pattern of the genes on these two clusters. A comparative study of the expression patterns of genes in these clusters in medaka and pufferfish should shed light on this.
Takifugu species as model systems for studies of the genetic basis of phenotypic diversity:
Teleost fishes such as cichlid and stickleback have recently been proposed as model vertebrate species for identifying the genetic variation that is responsible for phenotypic differences among related species. Viable crosses between closely related species of these fishes can be generated under laboratory conditions, and forward genetic analysis can be conducted (Cresko et al. 2004; Kocher 2004). Although many loci have been mapped onto genetic linkage maps of these teleost species, further progress in molecular identification of the locus has been hindered by lack of whole-genome sequences of these fishes. We propose that the fishes belonging to the genus Takifugu are also a good model system to study the genetic basis of phenotypic evolution in nature. A major drawback in using Takifugu fishes as a genetic model system was thought to be the difficulty of maintaining and breeding this marine species. This difficulty has, however, been largely overcome by developing suitable technology for controlled breeding under laboratory conditions (Miyaki et al. 1996; Matsuyama et al. 1997; Takaoka et al. 1998). The embryos of fugu have also been demonstrated to be amenable for molecular genetic investigations (Suzuki et al. 2002; Amores et al. 2004). More importantly, Takifugu can produce fertile progeny when crossed with closely related species (Miyaki 1992) and a draft sequence of the whole-genome of fugu has been generated. Thus, Takifugu species can be very useful tools to study the genetic basis of phenotypic variation between closely related species and to understand the process of speciation in vertebrates. The genetic map of fugu constructed by us and its further enrichment with additional genetic markers should greatly facilitate these types of studies.
Acknowledgments
We thank Kiyoshiro Kumasaka and Kazumitsu Honda (Nisshin Marinetech) for obtaining fish embryos; Kiyoshi Naruse, Osame Tabeta, and Yusuke Yamanoue for helpful discussion; and Kiyoshi Furukawa for technical advice. Hidekazu Suzuki and Naoki Mizuno provided technical support. This work was partially supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan. B.V. is supported by Singapore's Agency for Science, Technology and Research.
References
- Amores, A., A. Force, Y. L. Yan, L. Joly, C. Amemiya et al., 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711–1714. [DOI] [PubMed] [Google Scholar]
- Amores, A., T. Suzuki, Y. L. Yan, J. Pomeroy, A. Singer et al., 2004. Developmental roles of pufferfish Hox clusters and genome evolution in ray-fin fish. Genome Res. 14: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, S., J. Chapman, E. Stupka, N. Putnam, J. M. Chia et al., 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. [DOI] [PubMed] [Google Scholar]
- Asahida, T., T. Kobayashi, K. Saitoh and I. Nakayama, 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish. Sci. 62: 727–730. [Google Scholar]
- Bourque, G., P. A. Pevzner and G. Tesler, 2004. Reconstructing the genomic architecture of ancestral mammals: lessons from human, mouse, and rat genomes. Genome Res. 14: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S., G. Elgar, R. Sandford, A. Macrae, B. Venkatesh et al., 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366: 265–268. [DOI] [PubMed] [Google Scholar]
- Christoffels, A., E. G. Koh, J. M. Chia, S. Brenner, S. Aparicio et al., 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21: 1146–1151. [DOI] [PubMed] [Google Scholar]
- Cresko, W. A., A. Amores, C. Wilson, J. Murphy, M. Currey et al., 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA 101: 6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crnogorac-Jurcevic, T., J. R. Brown, H. Lehrach and L. C. Schalkwyk, 1997. Tetraodon fluviatilis, a new puffer fish model for genome studies. Genomics 41: 177–184. [DOI] [PubMed] [Google Scholar]
- Dib, C., S. Faure, C. Fizames, D. Samson, N. Drouot et al., 1996. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380: 152–154. [DOI] [PubMed] [Google Scholar]
- Dietrich, W. F., J. Miller, R. Steen, M. A. Merchant, D. Damron-Boles et al., 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149–152. [DOI] [PubMed] [Google Scholar]
- Fujita, S., 1967. Artificial interspecific and intergeneric hybridizations among the Tetraodontid puffers (preliminary report). Jpn. J. Michurin Biol. 3: 5–11. [Google Scholar]
- Galtier, N., M. Gouy and C. Gautier, 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12: 543–548. [DOI] [PubMed] [Google Scholar]
- Glazier, A. M., J. H. Nadeau and T. J. Aitman, 2002. Finding genes that underlie complex traits. Science 298: 2345–2349. [DOI] [PubMed] [Google Scholar]
- Grutzner, F., G. Lutjens, C. Rovira, D. W. Barnes, H. H. Ropers et al., 1999. Classical and molecular cytogenetics of the pufferfish Tetraodon nigroviridis. Chromosome Res. 7: 655–662. [DOI] [PubMed] [Google Scholar]
- Hedges, S. B., and S. Kumar, 2002. Genomics: vertebrate genomes compared. Science 297: 1283–1285. [DOI] [PubMed] [Google Scholar]
- Jaillon, O., J. M. Aury, F. Brunet, J. L. Petit, N. Stange-Thomann et al., 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher, T. D., 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nat. Rev. Genet. 5: 288–298. [DOI] [PubMed] [Google Scholar]
- Kumazawa, Y., M. Yamaguchi and M. Nishida, 1999. Mitochondrial molecular clocks and the origin of euteleostean biodiversity: familial radiation of perciforms may have predated the Cretaceous/Tertiary boundary, pp. 35–52 in The Biology of Biodiversity, edited by M. Kato. Springer-Verlag, Berlin/Heidelberg, Germany/New York.
- Ladjali-Mohammedi, K., A. Grapin-Botton, M. A. Bonnin and N. M. Le Douarin, 2001. Distribution of HOX genes in the chicken genome reveals a new segment of conservation between human and chicken. Cytogenet. Cell Genet. 92: 157–161. [DOI] [PubMed] [Google Scholar]
- Lander, E. S., P. Green, J. Abrahamson, A. Barlow, M. J. Daly et al., 1987. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1: 174–181. [DOI] [PubMed] [Google Scholar]
- Larhammar, D., L. G. Lundin and F. Hallbook, 2002. The human Hox-bearing chromosome regions did arise by block or chromosome (or even genome) duplications. Genome Res. 12: 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marklund, L., M. Johansson Moller, B. Hoyheim, W. Davies, M. Fredholm et al., 1996. A comprehensive linkage map of the pig based on a wild pig-Large White intercross. Anim. Genet. 27: 255–269. [DOI] [PubMed] [Google Scholar]
- Matsuura, K., 1990. The pufferfish genus Fugu Abe, 1952, a junior subjective synonym of Takifugu Abe, 1949. Bull. Nat. Sci. Mus Tokyo Ser. A 16: 15–20. [Google Scholar]
- Matsuyama, M., H. Chuda, Y. Ikeda, H. Tanaka and S. Matsuura, 1997. Induction of ovarian maturation and ovulation in cultured tigger puffer Takifugu rubripes by differential hormonal treatments. Suisanzousyoku 45: 67–73. [Google Scholar]
- Miyaki, K., 1992. Biological study of hybrid pufferfishes of the genus Takifugu, Tetraodontidae. Ph.D. Thesis, Nagasaki University, Nagasaki, Japan.
- Miyaki, K., O. Tabeta and H. Kayano, 1995. Karyotypes in six species of pufferfishes genus Takifugu (Tetraodontidae, Tetraodontiformes). Fish. Sci. 61: 594–598. [Google Scholar]
- Miyaki, K., K. Yoshikoshi and O. Tabeta, 1996. Transmission electron microscopic observations on spermatozoa in seven species of puffers genus Takifugu (Tetraodontidae, Tetraodontiformes). Fish. Sci. 62: 1996. [Google Scholar]
- Naruse, K., S. Fukamachi, H. Mitani, M. Kondo, T. Matsuoka et al., 2000. A detailed linkage map of medaka, Oryzias latipes: comparative genomics and genome evolution. Genetics 154: 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse, K., M. Tanaka, K. Mita, A. Shima, J. Postlethwait et al., 2004. A medaka gene map: the trace of ancestral vertebrate proto-chromosomes revealed by comparative gene mapping. Genome Res. 14: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M. W., K. W. Broman, C. S. Mellersh, K. Ray, G. M. Acland et al., 1999. A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics 151: 803–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J. S., 1994. Fishes of the World. John Wiley, New York.
- Ogawa, 1991. Redescription of Heterobothrium tetrodonis (Goto, 1984) (Monogenea: Diclidophoridae) and other related new species from puffers of genus Takifugu (Teleost: tetraodontidae). Jpn. J. Parasitol. 40: 388–396. [Google Scholar]
- Sakamoto, T., R. G. Danzmann, K. Gharbi, P. Howard, A. Ozaki et al., 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, M. P., 1992. Vertebrate homeobox gene nomenclature. Cell 71: 551–553. [DOI] [PubMed] [Google Scholar]
- Sewell, M. M., B. K. Sherman and D. B. Neale, 1999. A consensus map for loblolly pine (Pinus taeda L.). I. Construction and integration of individual linkage maps from two outbred three-generation pedigrees. Genetics 151: 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, A., H. Perlman, Y. Yan, C. Walker, G. Corley-Smith et al., 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160: 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistics and Information Department, 2001. Annual Statistics on Fisheries and Aquaculture Production 2001. Association of Agricultural and Forestry Statistics, Tokyo.
- Suzuki, T., T. Kurokawa, H. Hashimoto and M. Sugiyama, 2002. cDNA sequence and tissue expression of Fugu rubripes prion protein-like: a candidate for the teleost orthologue of tetrapod PrPs. Biochem. Biophys. Res. Commun. 294: 912–917. [DOI] [PubMed] [Google Scholar]
- Takaoka, O., S. Furuta, M. Gouda, T. Ido, Y. Mukai et al., 1998. Induced spawning of cultured tigger puffer in winter and fall. Bull. Fish. Lab. Kinki Univ. 6: 167–170. [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler, J. C., and L. Sorbini, 1996. New superfamily and three new families of tetraodontiform fishes from the Upper Cretaceous: the earliest and most morphologically primitive plectognaths. Smithsonian Contrib. Paleobiol. 82: 1–59. [Google Scholar]
- Uno, Y., 1955. Spawning habit and early development of a puffer, Fugu (Torafugu) niphobles (Jordan et Snyder). J. Tokyo Univ. Fish. 41: 169–183. [Google Scholar]
- Venkatesh, B., P. Gilligan and S. Brenner, 2000. Fugu: a compact vertebrate reference genome. FEBS Lett. 476: 3–7. [DOI] [PubMed] [Google Scholar]
- Woods, I. G., P. D. Kelly, F. Chu, P. Ngo-Hazelett, Y. L. Yan et al., 2000. A comparative map of the zebrafish genome. Genome Res. 10: 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]