Abstract
Dispensable, supernumerary (B) chromosomes are found in diverse eukaryotic species. The origin and genetic consequences of B chromosomes have been the subjects of speculation for more than a century. Until now, there has been no molecular evidence that B chromosome DNA is transcribed and there is no unequivocal evidence as to their origin. B chromosomes are considered to be genetically inert although they appear to cause a variety of phenotypic effects. We report that members of one of two ribosomal RNA gene families that are confined to the B chromosomes of a plant, Crepis capillaris, are transcribed—thus providing the first molecular evidence of gene activity on B chromosomes. Sequence analysis of part of the A and B chromosome rRNA genes, together with comparisons with related species, indicates that the B chromosome rRNA genes originate from the A chromosome.
B chromosomes (B's) are among the oldest conundrums in genetics and evolution (Wilson 1907). These chromosomes are supernumerary to the basic (A chromosome) set and variation in their numbers produces major intraspecific genome size polymorphism. B chromosomes differ from A chromosomes in morphology and pairing behavior and are not required for normal growth and development (reviewed by Jones and Rees 1982; Jones 1995; Jones and Houben 2003). The origin of B chromosomes remains obscure but it is likely that they arose in different ways in different organisms. It has been argued that B's could be derived from A chromosomes in, e.g., Crepis capillaris (Jamilena et al. 1994), or from sex chromosomes in, e.g., Leiopelma hochstetteri (Sharbel et al. 1998). The spontaneous generation of B's following interspecific crosses has been reported in, e.g., hybrids of Coix aquaticus and C. gigantea (Sapre and Deshpande 1987). The de novo formation of B's is most likely a rare event, as analyses of different B chromosome variants within species suggest a close relationship among different variants (Houben et al. 1999).
Most supposed B-specific sequences have been found subsequently, at least in low copy numbers, on the A's of the host species or in a close relative and probably it would be more accurate to describe these as B-amplified sequences. One possible exception to this generality comes from a recent detailed study using amplified fragment length polymorphism analyses (AFLP) and fluorescence in situ hybridization (FISH) in the cyprinid fish Alburnus alburnus which has one of the largest supernumerary chromosomes in vertebrates. This B chromosome contains specific sequences with strong homology to a retrotransposon from Drosophila and medaka (Oryzias latipes). The sequence is highly abundant on the B chromosome but undetectable in the normal A chromosome complement or in the B chromosome of the closely related species, Rutilus rutilus. These results suggest that the supernumerary chromosome of A. alburnus is not derived from the normal chromosome complement (Ziegler et al. 2003). In contrast, Page et al. (2001), investigating sequences isolated from the maize B chromosome (Alfenito and Birchler 1993), found that homologous sequences were confined to the centromere of chromosome 4 in the A genome. This indicates that a diversity of sequences compose centromeric regions in maize and also suggests that the centromere of chromosome 4 is related to that of the B chromosome (Page et al. 2001).
B chromosomes appear to carry very few identified genes other than commonly occurring clusters of 18S, 5.8S, and 25S ribosomal RNA (rRNA) genes (here referred to as 45S rDNA as this is the size of the unprocessed RNA transcript). These genes have been recognized on B's by their capacity to organize extra nucleoli (Green 1990), by in situ hybridization, and, in a few instances, their activity has been inferred from positive silver nitrate staining (Jones 1995). It has been argued that active nucleolus organizing regions (NORs) are visualized by silver staining (e.g., Hubbell 1985) and more recently Sirri et al. (2000) identified subunits of RNA polymerase I and a transcription factor (UBF) as the silver-staining proteins that remain associated with NORs at the metaphase of mitosis. Until now there has been no direct molecular evidence of transcription of B chromosome DNA in any species.
In situ hybridization demonstrated the presence of ribosomal genes dispersed throughout the B chromosomes of Rattus rattus but despite the clear demonstration of silver-staining NORs on the A chromosomes, no signal is apparent on the B chromosomes. The authors suggest that the accessory chromosomes of this species originated from one of the smaller NOR-carrying chromosome pairs and that, in the course of evolution, repetitive sequences invaded this supernumerary element. They suggest that its ribosomal DNA content was dispersed throughout the chromosome where it was inactivated by heterochromatinization that precluded access by transcription factors (Stitou et al. 2000).
The B chromosomes of the Australian daisy Brachycome dichromosomatica C. R. Carter have a clear secondary constriction and a cluster of rRNA genes detectable by FISH that is often seen associated with a nucleolus at mitotic prophase (Donald et al. 1995). These loci do not silver stain and Donald et al. (1997) were unable to detect a transcription product using PCR, of reverse-transcribed total RNA from leaf tissue of plants containing B chromosomes, with primers specific to the internal transcribed spacer 2 (ITS2) of the B chromosome rRNA gene.
The B chromosomes of C. capillaris (L.) Wallr (2n = 6) were first described and investigated by Rutishauser (1960), who recognized two B types termed monocentric and dicentric. An accession derived from the original collection contains only one type of B, a small metacentric chromosome (Małuszyńska and Schweizer 1989), which is morphologically identical to the B chromosome present in natural populations in Britain (Parker et al. 1989). Jones et al. (1989) studied synaptonemal complexes in C. capillaris plants derived from two British populations and found that single B chromosomes showed predominantly foldback meiotic pairing resulting in a symmetrical univalent hairpin loop, which suggests that the B is an isochromosome. The B chromosomes paired freely and extensively within and among themselves but there was no indication of pairing with the A chromosomes (Jones et al. 1989). The C. capillaris B may have originated from the standard genome because any repeated DNA sequences that they contain are also present on the A chromosomes (Jamilena et al. 1994, 1995). Histochemical silver staining and in situ hybridization of mitotic chromosomes suggested the presence of active rDNA on both ends of the B chromosomes of C. capillaris, as well as the major functional NORs located on chromosome 3 (Małuszyńska and Schweizer 1989; Małuszyńska 1990).
In this article we report two C. capillaris B-chromosome-specific rRNA gene types that differ substantially in sequence from those located on the A chromosomes. We present the first evidence that some of these rRNA genes are transcribed in leaf and flower bud tissues. We also report the outcome of an extensive search for a possible donor species for this unique B chromosome rDNA.
MATERIALS AND METHODS
Plant material:
Achene samples of C. capillaris that originated from a C. capillaris population near Schaffhausen, Switzerland, in which B chromosomes were first identified (Rutishauser 1960; D. Schweizer, personal communication) were provided by J. Małuszyńska (Poland). Leaf tissue of living plants was collected directly from sites in the northeastern Harz region of Germany and documented by voucher specimens (Table 1). Herbarium material was selected from the collection of the Institute of Plant Genetics and Crop Plant Research Gatersleben (GAT), including those species naturally occurring in the surroundings of Schaffhausen, northernmost Switzerland (Hess et al. 1972). The classification follows Babcock (1947a,b), Sell (1976), and Jäger and Werner (2002).
TABLE 1.
Crepis specimens from the herbarium of the Institute of Plant Genetics and Crop Plant Research (GAT) of which dry leaf material has been analyzed and the voucher specimens of the living material from the northeastern Harz region investigated in 2004
| C. alpestris (Jacq.) Tausch | GAT 3686 Alpbach (Tirol): Gratspitz, leg. N. Reich 20.6.1934 (8.10.2004:11) |
| C. aspera L. | GAT 3667 Jerusalem: Mt. Scopus, leg. I. Amdurski 30.4.1931 (10.12.2004) |
| C. biennis L. | GAT 3636, 3637, 3638 northeastern Harz foreland: Gatersleben, railway station, leg. K. Pistrick (K.P.), A. Houben (A.H.) et J. Timmis (J.T.) 30.9.2004 (l, 30.9.2004:3, 4, 5) |
| GAT 3646, 3647, 3648 northeastern Harz foreland: Schadeleben, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:11, 12, 13) | |
| GAT 3654, 3655, 3656 northeastern Harz foreland: Warnstedt, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:19, 20, 21) | |
| GAT 3664 Harz: Kreuztal, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:29) | |
| C. capillaris (L.) Wallr. | GAT 3634, 3635 northeastern Harz foreland: Gatersleben, Gutspark, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:1, 2) |
| GAT 3639, 3640, 3641/3642 northeastern Harz foreland: Gatersleben, cemetery, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:6, 7, 8) | |
| GAT 3643, 3644/3645 northeastern Harz foreland: Concordia-lake, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:9, 10) | |
| GAT 3649, 3650 northeastern Harz foreland: Cochstedt, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:14, 15) | |
| GAT 3651, 3652, 3653 northeastern Harz foreland: Westerhausen, Königstein, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:16, 17, 18) | |
| GAT 3660 Harz: Neuwerk, Krockstein, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:25) | |
| C. dioscoridis L. | GAT 3674 Genbank Gatersleben: CRE 2 (D 2544/79) leg. H. Ohle 23.7.1979 (8.10.2004:20) |
| C. foetida L. subsp. foetida | GAT 3676 Könnern, leg. P. Schuster 23.7.1952 (8.10.2004:1) |
| C. foetida subsp. rhoeadifolia (M. Bieb.) Celak. | GAT 3688 Ungarn: Bács. Kysáč, leg. S. Kupčok 7.1909 (8.10.2004:15) |
| C. mollis (Jacq.) Asch. | GAT 3680 Altvater mountains: Grosser Kessel, leg. H. Laus 7.1930 (8.10.2004:6) |
| C. nicaeensis Balb. | GAT 3665 Aveyron: St. Paul les Fonto, leg. A. Bec 20.6.1910 (4.1.2005) |
| C. paludosa (L.) Moench | GAT 3658, 3659 Harz: Treseburg, leg. K.P., A.H. et J.T. 30.9.2004 (l, 30.9.2004:23, 24) |
| C. praemorsa (L.) Walther | GAT 3683 Wiener Wald: Mödling, leg. O. Schwarz 21.5.1944 (8.10.2004:8) |
| C. rubra L. | GAT 3691 Italien, leg. V. Engelhardt 15.5.1935 (8.10.2004:16) |
| C. setosa Haller f. | GAT 3692 Slowakei: Nová Baňa, leg. K. Hammer (K.H.), P. Hanelt (P.H.) et F. Kühn (F.K.) 1.8.1974 (8.10.2004:17) |
| C. taraxacifolia Thuill. | GAT 3694 Rheinland: Bingen, leg. P. Schuster 22.6.1938 (8.10.2004:19) |
| C. tectorum L. | GAT 3679 Spremberg: Slamener Heide, leg. R. Fritsch 8.7.1971 (8.10.2004:4) |
| Hieracium spec. | GAT 3685 Taunus: Eggenheim, leg. M. Schuster 8.1956 (8.10.2004:10) |
Dates (DD.MM.YYYY) when the material was collected and sample number are in parentheses.
Isolation of RNA and DNA:
Total cellular RNA and DNA of C. capillaris were prepared from young leaves and flower buds as previously described (Donald et al. 1997). DNA of herbarium specimens of Crepis species was isolated by using a DNeasy plant kit (QIAGEN, Chatsworth, CA).
Chromosome microdissection, PCR isolation, and cloning of rRNA gene sequences:
DNA was prepared from 15 microdissected B chromosomes according to Houben et al. (2001). PCR was used to amplify the ITS1-5.8S-ITS2 region (the two internal transcribed spacers and the 5.8S encoding region of the rRNA gene) from microisolated B chromosome DNA and from total genomic DNA of 0B C. capillaris plants using the primers ITS4 (5′-TCC TCC GCT TAT TGA TAT GC) and ITS5 (5′-GGA AGT AAA AGT CGT AAC AAG G) (White et al. 1990). PCR products were ligated into pGEM-T Easy (Promega, Madison, WI) and propagated in Escherichia coli strain DH5α. PCR was used to amplify the C. capillaris ITS1 region. Primer P1 (5′-GGA AGG ATC ATT GTC GAA CCC TGC) spanned the junction of 18S and ITS1 and primer P2 (5′-GGG ATT CTG CAA TTC ACA CCA G) was located in the central region of the 5.8S sequence (see Figure 2). The PCR conditions for primers P1 and P2 were 94°, 2 min (94°, 30 sec; 59°, 1 min; 72°, 5 min) for 30 cycles.
Figure 2.
Alignment of the consensus sequences of part of the A and B chromosome rRNA genes. The dark shading indicates the NsiI restriction site and the primer sequences and their corresponding labels. The light shading indicates the 5.8S rRNA encoding sequence. Asterisks (*) indicate conservation of the sequence in the three gene types.
Primers were designed to be specific for ITS-B1 (5′-ACA CCA AAA CAA ACC GTT AGG ATG) (see Figure 2 and the text for explanation) and ITS-A (5′-A-CC CTA GCA GCT CAC AAC ACA CGG C) (see Figure 2 and the text for explanation) sequences when used in conjunction with P1 above (see Figure 2). The PCR conditions for these primers were 94°, 2 min (94°, 30 sec; 61°, 1 min; 72°, 5 min) for 30 cycles.
The ITS2 region was PCR amplified using the primers XF58S (5′-CTT CTA GAG CCT GGG CGT CAC G) and ER25S (5′-CGG AAT TCT GAC CTG GGG TCG C) for heterologous comparisons within the genus Crepis. The PCR conditions were 94°, 2 min (94°, 30 sec; 59°, 1 min; 72°, 5 min) for 30 cycles. The primer positions are shown in Figure 2.
Sequencing and sequence data analysis:
Sequence analysis of the clones was performed by the dideoxynucleotide-dye termination method. A neighbor-joining tree was calculated with maximum-likelihood distances (corresponding to the HKY85 + G + I model of sequence evolution) in PAUP*4.0 (Swofford 2002). Bootstrap values were calculated from 200 resamples using identical settings as in the initial neighbor-joining analysis.
RT-PCR:
Total RNA isolated from leaves and young flower buds was DNased with electrophoretically pure DNase (Ambion, Austin, TX) and 60 ng of RNA was reverse transcribed with random primers. Primers P1 and A and primers P1 and B1 were used to amplify the reverse-transcribed RNA as described above.
FISH:
The plasmid VER17 (Yakura and Tanifuji 1983) containing part of the 18S, the 5.8S, most of the 25S, and the internal transcribed spacers of the Vicia faba rRNA gene was used as an rDNA-specific probe. An Arabidopsis-type telomere probe was synthesized using PCR according to Ijdo et al. (1991). In situ hybridization probes were labeled by PCR with biotin-16-dUTP or digeoxigenin-11-dUTP. Young seedlings were pretreated in iced water for 16 hr, fixed in ethanol-glacial acetic acid (3:1, v/v) for 4 days at 4°, and stored at 4° in 70% ethanol. Preparation of mitotic chromosomes and FISH were performed as described by Leach et al. (1995) and Houben et al. (2001). Hybridization sites of digeoxigenin-labeled or biotin-labeled probes were detected using sheep antidigeoxigenin-rhodamine-rhodamine anti-sheep antibody or the streptavidin-Alexa 488 system, respectively. Epifluorescence signals were recorded with a cooled CCD camera and image manipulations were performed using Adobe Photoshop software.
RESULTS
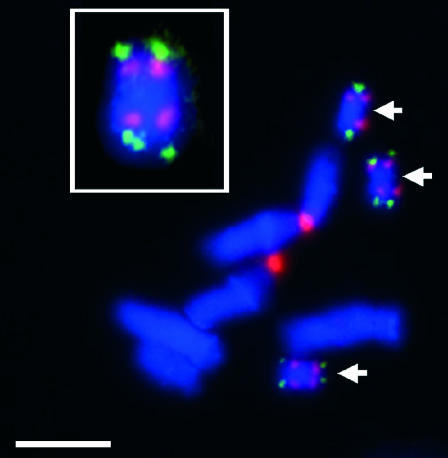
Fluorescence in situ hybridization confirmed clusters of 45S rRNA genes on both ends of the B chromosomes as well on chromosome 3 of C. capillaris (Figure 1). The B chromosome signal strength was considerably lower than that on chromosome 3, suggesting that the B's contribute a minority of additional rRNA genes to the cell. The rDNA signals were immediately proximal to telomere signals on the B chromosomes, which were consistently brighter than those seen on the A's, indicating a higher number of (AGGGTTT)n repeats on B's (Figure 1).
Figure 1.
The location of rDNA and telomeres on the A and B chromosomes of C. capillaris. Telomeres are labeled in green and 45S rRNA gene clusters in red. The chromosomal DNA is stained with DAPI. Bar, 5 μm. The inset shows an enlarged B chromosome.
The ITS1, 5.8S, and ITS2 rDNA regions differ between A and B chromosomes:
One type of rDNA sequence, derived from the A chromosomes, was recovered from plants without B chromosomes, while two additional types of sequences were recovered from microcloned B chromosome DNA. The nucleotide sequences of the ITS1, 5.8S, and ITS2 regions from 11 cloned inserts derived by PCR from DNA of microdissected B chromosomes of C. capillaris were determined and compared with four comparable A chromosome sequences isolated from plants that did not contain B chromosomes. A single type of rDNA sequence was found on the A chromosomes and two different ones were identified on the B chromosomes. The A chromosome sequences (called ITS-A) contained 643 bp, of which 265 bp were in ITS1, 152 encoded the 5.8S rRNA, and 226 bp were in ITS2. The corresponding consensus B chromosome sequences were 1 bp shorter, and one cloned insert was 676 bp long. Alignment revealed considerable sequence polymorphism between the ITS-A rDNA and the two distinct B sequences. Nine of the B chromosome clones were of one type, named ITS-B1, and the other two were a second type that was designated ITS-B2. Consensus sequences were determined for the three distinct types of sequences (Figure 2).
The ITS-A consensus sequence was 94.4% similar to ITS-B1, differing by 20 nucleotides in ITS1 and 18 in ITS2, and 95.6% similar to ITS-B2, differing by 13 nucleotides in ITS1 and 12 in ITS2. The 5.8S regions were identical in ITS-A and ITS-B1 and differed in this region from ITS-B2 by three nucleotides. The two B chromosome consensus sequences showed only 91.3% sequence identity. In addition to the three differences in 5.8S, ITS-B1 and ITS-B2 sequences differed by 26 nucleotides in ITS1 and 27 nucleotides in ITS2. An NsiI restriction site was unique to ITS1 of ITS-B1. We analyzed the mutational profile of these clones assuming the B chromosome rDNA is derived from that on the A chromosome. Among the 12 possible kinds of point mutations, C→T and G→T substitutions were the most prevalent types in A/B1 comparisons, accounting for almost half of all the mutations. They are significantly (P < 0.001) more frequent than any other types of substitution, including the reverse transitions T→C and T→G. C→T transitions (and G→A transitions on the opposite strand) are the hallmark of spontaneous deamination of 5-methylcytosine (5mC), which produces a G-T mismatch at the deaminated site. The mismatch can be restored to the G-C pair in one direction, but creates an A-T pair in the other direction (Holliday and Grigg 1993; Finnegan et al. 1998), resulting in T→C transitions (and A→G on the opposite strand). This observation is consistent with the known high level of C methylation in plant nuclear DNA (Ayliffe et al. 1998) and with assuming no purifying selection on the sequence of ITS2 as may be expected if the ITS-B1-type genes do not contribute to the ribosome population. The origin of the G→T substitutions is not clear and G→A is not overrepresented. Nonetheless, these distortions suggest that some nucleotides in the rDNA of the B chromosome are modified and have been subject to hypermutation—assuming that they are derived from the A chromosome copies. Comparison of the ITS-A and ITS-B2 sequences indicates that the mutations are more randomly distributed among the 12 possible types.
Specific PCR amplification of B chromosome rDNA:
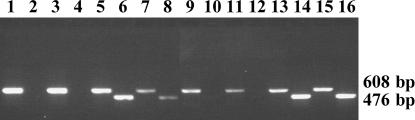
Primer P1 was used with primer B1 or primer A to determine specificity for their target sequences (Figure 3). Primers P1 and B1 amplified the expected product of 476 bp only from templates containing B chromosome DNA from either positive control clones of microdissected B rDNA (Figure 3, lanes 14 and 16) or total cellular DNA isolated from plants with two B chromosomes (lanes 6 and 8). This primer pair was unable to amplify any product from cloned A chromosome rDNA (lanes 2 and 4) or from total DNA isolated from plants lacking B chromosomes (lanes 10 and 12). It was concluded that primer B1 was specific for the ITS-B1-type ribosomal repeats that contribute the majority of rDNA that is present on the B chromosome. In contrast, the primer pair P1 and A amplified a product of 608 bp from all the plant DNA samples (odd-numbered lanes), including all the control plasmid DNAs (lanes 9, 11, 13, and 15), indicating that the 3′ mismatch incorporated into primer A was insufficient, at the stringency of annealing used here, to differentiate the two types of sequence. The reliability and universality of the specific amplification of B chromosome rDNA was confirmed when the P1 and B1 primers were applied to four independently isolated samples of DNA from 0B plants (i.e., plants containing no B chromosomes) and four plants containing two B chromosomes (2B plants) as well as positive and negative controls of cloned B chromosome and A chromosome rDNA sequences. In all cases complete specificity of the B1 primer for B chromosome rDNA was confirmed (data not shown).
Figure 3.
Gel electrophoresis of PCR-amplified DNA testing the specificity of the A and B1 primers. PCR products were amplified using P1 and A as primers from 0B total plant DNA (lanes 1 and 3), 2B total plant DNA (lanes 5 and 7), clones derived from 0B total plant DNA (lanes 9 and 11), and clones derived from microdissected B chromosome DNA (lanes 13 and 15) and using P1 and B1 as primers from 0B total plant DNA (lanes 2 and 4), 2B total plant DNA (lanes 6 and 8), clones derived from 0B total plant DNA (lanes 10 and12), and clones derived from microdissected B chromosome DNA (lanes 14 and 16). Fragment sizes are indicated in base pairs.
The B chromosome rDNA is transcriptionally active in leaves and flower buds:
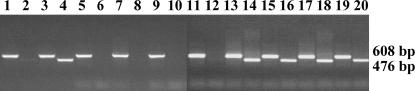
The availability of specific primers made possible a sensitive assay for transcription of the B-chromosome-located rRNA genes by probing total reverse-transcribed cellular RNA for the 45S precursor rRNA, which is the initial product of plant rRNA genes by RNA polymerase I (Kavanagh and Timmis 1988). Total RNA was isolated from leaves and young flower buds of individual 0B (Figure 4, lanes 5–12) and 2B (lanes 13–20) plants. The samples were DNased with electrophoretically pure DNase and 60 ng of RNA was reverse transcribed with random primers. Primers P1 and A (Figure 4, odd-numbered lanes) and primers P1 and B1 (Figure 4, even-numbered lanes) were used to amplify the reverse-transcribed RNA. In each sample derived from RNA of B-chromosome-containing plants, a product was amplified by primers P1 and B1 that was not present in any sample derived from the RNA of plants lacking B chromosomes. This result indicates that the B chromosome rDNA is transcribed because the 45S precursor is detectable in the total RNA of leaf and flower bud tissue.
Figure 4.
Gel electrophoresis of PCR-amplified products testing for transcription of rRNA genes from A and B chromosome DNA. PCR products were amplified using P1 and A as primers from 0B total plant DNA (lane 1), 2B total plant DNA (lane 3), 0B total flower bud RNA (lanes 5 and 9), 0B total leaf RNA (lanes 7 and 11), 2B total flower bud RNA (lanes 13 and 17), and 2B total leaf RNA (lanes 15 and 19) and using P1 and B1 as primers from 0B total plant DNA (lane 2), 0B total flower bud RNA (lanes 6 and 10), 0B total leaf RNA (lanes 8 and 12), 2B total flower bud RNA (lanes 14 and 18), and 2B total leaf RNA (lanes 16 and 20). Fragment sizes are indicated in base pairs.
Control experiments with and without DNase treatment, and with and without an initial reverse transcription step, confirmed that the PCR products in Figure 4 (lanes 14, 16, 18, and 20) are unequivocally dependent upon reverse transcription of RNA molecules originating from rRNA genes on B chromosomes (results not shown).
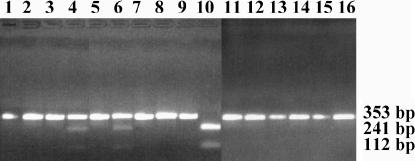
Having established that the B chromosome rRNA genes are transcribed, we determined the level of transcript relative to the number of ITS-B1 sequences present in the genomes. This was achieved by PCR amplification using primers P1 and P2 to amplify 353 bp spanning ITS1 and part of the 5.8S gene, followed by digestion of the products with NsiI for which there is a recognition site in ITS-B1 that is lacking in ITS-A. Figure 5 shows the PCR products before (odd-numbered lanes) and after (even-numbered lanes) complete NsiI digestion. The primers were applied to genomic DNA templates of 0B (Figure 6, lanes 1 and 2) and two independent 2B (lanes 3–6) plants. The product of the 0B plant is not digested by NsiI whereas a proportion of those of the two 2B plants is cleaved into the predicted fragments of 241 and 112 bp. Quantitative analysis of the Gel Doc (Bio-Rad, Hercules, CA) data file of Figure 5 (lanes 4 and 6) with the software Quantity One (Bio-Rad) indicated that 20.8 and 22.1% of the total product derived from the B-chromosome-containing genomes was digested by NsiI, consistent with each B chromosome contributing ∼10% of ITS-B1-type rRNA genes to the composite genome, assuming that the primers amplify the A and B chromosome sequences with equal efficiency. As controls to ensure that NsiI digestion was complete and specific, DNA from an ITS-A clone (lane 7) and an ITS-B1 clone (lane 9) was amplified and digested. As predicted, the products from the former were undigested by NsiI (lane 8) and those from the latter were completely digested into the expected fragments (lane 10). Similar experiments were performed with three of the DNased and reverse-transcribed RNA samples used in Figure 4 and the results are shown in Figure 5 (lanes 11–16). In no case was any NsiI digestion of the products observable. We conclude that, while there is demonstrable transcription of the B chromosome rDNA, the transcripts are present in the final 45S precursor pool at undetectable levels that are therefore disproportionately low compared with the number of genes present on the supernumerary B (∼10% of the total gene number per cell). Therefore, in the cellular RNA of leaves and flower buds of a 2B plant, nearly all the pre-rRNA molecules are derived from the A chromosome NOR.
Figure 5.
Gel electrophoresis of PCR-amplified products to quantify the level of transcription from the B chromosome. All PCR products were amplified using P1 and P2 with the reverse transcriptase step included for samples in lanes 11–16 and all RNA samples digested with DNase. All samples are in pairs not digested (odd numbers) and digested with NsiI (even numbers). Total plant DNA from 0B (lanes 1 and 2), 2B (lanes 3–6), clone 10 DNA from 0B (lanes 7 and 8), clone 3 from microdissected B chromosome DNA (lanes 9 and 10), total flower bud RNA from 2B plant (lanes 11 and 12), and total leaf RNA from 2B plant (lanes 13–16). Fragment sizes are indicated in base pairs.
Figure 6.
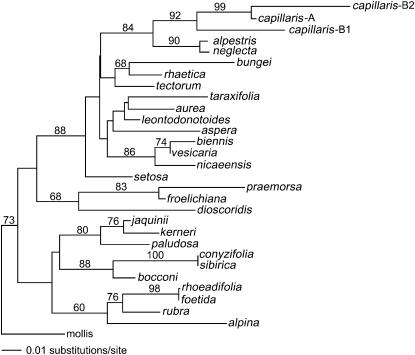
The phylogenetic relationships between the ITS2 sequences of 29 species within the genus Crepis and the corresponding sequences on the B chromosome of C. capillaris. The tree was prepared using neighbor-joining analysis. Numbers depict bootstrap values (>50%) derived from 200 bootstrap resamples.
A search for a donor of the B chromosome rDNA:
The observation that the B1 and B2 ITS regions were very significantly different from those of the A chromosome NOR suggested a search of species in central Europe that may have hybridized with C. capillaris and donated these rDNA types. The assumption was that, as in Hordeum interspecies (Kasha and Kao 1970) and some Poaceae (Laurie and Bennett 1989) or other intergeneric crosses, the chromosomes of one parent may be eliminated during embryogenesis. In most systems this leads to the formation of haploids containing only the chromosomes of one parent, but the process of elimination takes some time (Thomas and Pickering 1983; Pickering 1985) and some chromatin may survive provided that it becomes mitotically stable. If this event is followed by spontaneous chromosome doubling or backcrossing to one of the parents, then diploids may result in the formation of supernumerary chromosomes that may be genetically suppressed (Sapre and Deshpande 1987). We prepared DNA from herbarium specimens of 10 species and fresh tissue from 3 additional species collected in the northeastern Harz region in Germany, from which we amplified ITS2, which was sequenced and compared with ITS2 sequences of 18 additional Crepis species, available from the EMBL databases. In particular, we included those species occurring in the vicinity of Schaffhausen, in northernmost Switzerland (Hess et al. 1972), where the B-chromosome-positive C. capillaris population was first identified (Rutishauser 1960). We focused particularly on C. dioscoridis, C. tectorum, C. nicaeensis, C. leontodontoides, and C. aspera because chromosome fragments in artificially induced species hybrids have been described in crosses with C. capillaris (Navashin 1927; Avery 1930; Hollingshead 1930; Grob 1966; Doerschug and Miksche 1976).
Neighbor-joining phylogenetic analyses (Swofford 2002) of the sequence data (Figure 6) did not identify a species that contained an ITS2 sequence that was more similar to either of the B chromosome sequences than that of the C. capillaris A chromosomes. This reinforces the mutational analysis and suggests strongly that the B chromosome rDNA was derived from the A chromosome followed by divergence over time into major (ITS-B1) and minor (ITS-B2) gene clusters. A parsimony analysis of the ITS sequences resulted in identical taxon groups as found in the neighbor-joining tree (data not shown).
DISCUSSION
These experiments provide the first molecular demonstration of the transcription of B chromosome DNA. These chromosomes, which have the capacity to accumulate through generations or during mitosis, have generally been considered to be genetically, although not functionally, inert. There is ample evidence that they have a great variety of exo- and endo-phenotypic effects (Jones and Rees 1982; Puertas 2002).
Here we have shown that B chromosomes are not genetically inert but it appears unlikely, in the case of these rDNA clusters, that they make a pro rata contribution to the functional ribosome population. We cannot distinguish whether all the ITS-B1-type genes are transcribed at a low level or whether a few are transcribed and others are inactive. Of course, this same uncertainty applies for the A chromosome NOR, and it appears that most plants have a large excess of rRNA genes above the number that is capable of producing the amount of RNA required by somatic cells (Kavanagh and Timmis 1986).
Togby (1943) described the evolution of C. fuliginosa Sibth. & Sm. (C. neglecta subsp. fuliginosa (Sibth. & Sm.) Vierh. (2n = 6) from C. neglecta L. (2n = 8) as a result of unequal interchange and loss, subsequently, of a small dispensable centric fragment. If this centric fragment is retained rather than lost, it could evolve into a B chromosome. Babcock (1942) presented an extensive analysis of the systematics, cytogenetics, and evolution in Crepis species and argued that species with lower chromosome numbers appeared to be derived from species with higher numbers. C. fuliginosa and C. capillaris are both species with the lowest diploid number found in the genus: 2n = 6.
If a similar kind of chromosome rearrangement was responsible for the evolution of the B chromosome in C. capillaris, it is still necessary to provide an explanation for the source, location, and activity of the rRNA genes found on the B chromosomes. First, it possible that the NOR-bearing chromosome was involved in the interchange and that the remaining centric fragment carried rRNA genes. This chromosome would have to undergo a misdivision of the centromere to generate the isochromosome now found in C. capillaris and such centromeric misdivisions are not uncommon in B chromosomes (see Jones and Rees 1982 for numerous examples). Second, most, if not all, Crepis species have distal rRNA gene clusters (see Jones 1995 for list) so that the position of this locus is not unusual. Finally, as the rRNA genes probably derive from the A genome, nucleolar dominance as seen, if only cytologically, in Crepis hybrids (Navashin 1934; Doerschug and Miksche 1976) would not be predicted in the nascent B chromosome. Sequence divergence may occur due to the relaxation of the constraints of concerted evolution due to lack of pairing of the B chromosome with the A's. However, there are a number of examples of either concerted evolution of different arrays of rRNA genes (Joly et al. 2004; Kovarik et al. 2004) or maintenance of different arrays (Rauscher et al. 2004) in various polyploid species. Thus either maintenance of sequence identity or sequence divergence is equally likely even in the absence of detectable pairing.
Jamilena et al. (1994) endeavored to determine the origin of the B chromosome of C. capillaris using in situ hybridization with a range of probes. Genomic in situ hybridization with DNA from plants with and without B's as probe indicated that the B chromosome had many DNA sequences in common with the A chromosomes, but showed no region rich in B-specific sequences. Additional DNA probes derived from the NOR chromosome (chromosome 3) were also used to test the possible origin of the B from this chromosome and only the 18S + 25S rDNA and the telomeric sequences were found to hybridize to the B's. Jamilena et al. (1994, p. 703) concluded that “the entire B of C. capillaris, although possibly having originated from the standard genome, did not derive directly from the NOR chromosome.”
If the B was derived from a centric fragment lacking rRNA genes, it is possible that the latter relocated as a result of a transposition event (Schubert and Wobus 1985). B chromosomes carry many transposable elements and numerous reports of variation in NOR location within individuals (Hall and Parker 1995) suggest that rRNA genes are mobile.
Babcock (1942) also reports that many variations from the typical diploid number are found in Crepis, including both trisomics and polyploids in C. capillaris and C. tectorum. Furthermore, Babcock and Navashin (1930) described a trisomic plant in C. tectorum in which the extra member was an atypical satellited chromosome. It is possible that this chromosome was in fact a B chromosome, although not recognized as such. The cohabitation of diploids and tetraploids of the same or closely related species provides an ideal environment for a scenario similar to that described by Sapre and Deshpande (1987) in Coix L. where a B chromosome was generated spontaneously as a result of the crossing of two allopatric species, C. gigantica (2n = 20) and C. aquatica (2n = 10), followed by natural backcrossing of the hybrid yielding a C. aquatica (2n = 10+1B) where the B was derived as a relic from C. gigantica. Crossing of diploid and tetraploid C. capillaris plants or crosses with C. tectorum (with which it forms hybrids; Babcock 1942) plants followed by natural backcrossing to the diploid could readily lead to the formation of a B chromosome. However, the phylogenetic analysis of the ITS2 regions from 28 other Crepis species, including C. nicaeensis (Grob 1966), C. leontodontoides (Avery 1930), C. aspera (Navashin 1927), C. tectorum, and C. dioscoridis (Doerschug and Miksche 1976) with which C. capillaris forms hybrids, did not indicate that any of them was a more likely donor of the B chromosome rRNA genes than was the A genome of C. capillaris.
As suggested above, a possible explanation for the extensive sequence variation between the ITS1 and ITS2 regions of the A and B chromosomes is that lack of meiotic pairing of the A's and B's and the stability of B's as univalents releases them from concerted evolution with the A chromosome rDNA (Liao 1999). This is further supported by the lack of identification of a donor species for the very different rRNA gene types that are present on the B chromosome, suggesting that the B genes have evolved from those on the A chromosome under different selection pressures and/or homogenizing mechanisms.
The presence of a transcript from the B chromosome rRNA genes indicates that these genes at least are not completely suppressed. However, transcripts are rare when compared with the number of rRNA genes on the B, suggesting that transcription is suppressed or the transcripts are quickly degraded compared with those arising from the A chromosomes. The lack of any detectable association of the B's with a nucleolus may be attributable to the low level of transcription or, consistent with intermittent silver staining, the fact that transcription is confined to a few cells. The search for a possible donor of the B chromosome did not identify any candidate species. Rather, the phylogenetic analysis shows that the B's are more closely related to the C. capillaris A's than to those of any other species tested and this is consistent with mutational analyses of the A and B rDNA.
Acknowledgments
We are grateful to Margit Hantschmann and Katrin Kumke for excellent technical assistance and to Frank Blattner for his valuable advice on the phylogenic analysis. A.H. was supported by a grant of the Land Saxony-Anhalt. J.N.T. thanks the Australian-German Joint Research Co-operation Scheme for financial assistance.
Sequence data from this article have been deposited with EMBL/GenBank Data libraries under the following accession numbers: C. capillaris ITS-B2 (AJ876602), C. capillaris ITS-A (AJ876603), C. capillaris ITS-B1 (AJ876604), C. alpestris (AJ876605), C. tectorum (AJ876606), C. nicaeensis (AJ876607), C. taraxacifolia (AJ876608), C. aspera (AJ876609), C. rhoeadifolia (AJ876610), C. foetida (AJ876611), C. rubra (AJ876612), C. praemorsa (AJ876613), C. paludosa (AJ876614), C. dioscoridis (AJ876615), C. setosa (AJ876616), and C. mollis (AJ876617).
References
- Alfenito, M. R., and J. A. Birchler, 1993. Molecular characterization of a maize B chromosome centric sequence. Genetics 135: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery, P., 1930. Cytological studies of five interspecific hybrids of Crepis leontodontoides. Univ. Calif. Publ. Calif. Publ. Ag. Sci. 6: 135–167. [Google Scholar]
- Ayliffe, M. A., N. S. Scott and J. N. Timmis, 1998. Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol. Biol. Evol. 15: 738–745. [DOI] [PubMed] [Google Scholar]
- Babcock, E. B., 1942. Systematics, cytogenetics and evolution in Crepis. Bot. Rev. 8: 139–190. [Google Scholar]
- Babcock, E. B., 1947. a The genus Crepis. Part 1: the taxonomy, phylogeny, distribution and evolution of Crepis. Univ. Calif. Publ. Bot. 21: 1–198. [Google Scholar]
- Babcock, E. B., 1947. b The genus Crepis. Part 2: systematic treatment. Univ. Calif. Publ. Bot. 22: 199–1030. [Google Scholar]
- Babcock, E. B., and M. Navashin, 1930. The genus Crepis. Bib. Genet. 6: 1–90. [Google Scholar]
- Doerschug, E. B., and J. P. Miksche, 1976. DNA variation and ribosomal-DNA constancy in two Crepis species and the interspecific hybrid exhibiting nucleolar-organiser suppression. Heredity 37: 441–450. [Google Scholar]
- Donald, T. M., C. R. Leach, A. Clough and J. N. Timmis, 1995. Ribosomal RNA genes and the B chromosome of Brachycome dichromosomatica. Heredity 74: 556–561. [DOI] [PubMed] [Google Scholar]
- Donald, T. M., A. Houben, C. R. Leach and J. N. Timmis, 1997. Ribosomal RNA genes specific to the B chromosomes in Brachycome dichromosomatica are not transcribed in leaf tissue. Genome 40: 674–681. [DOI] [PubMed] [Google Scholar]
- Finnegan, E. J., R. K. Genger, W. J. Peacock and E. S. Dennis, 1998. DNA methylation in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 223–247. [DOI] [PubMed] [Google Scholar]
- Green, D. M., 1990. Muller's rachet and the evolution of supernumerary chromosomes. Genome 33: 818–824. [Google Scholar]
- Grob, R., 1966. Zytotaxonomische Untersuchen an Crepis capillaris und Crepis nicaneensis und ihren F1-Bastarden. Ber. Schweiz. Bot. Ges. 76: 306–351. [Google Scholar]
- Hall, K. J., and J. S. Parker, 1995. Stable chromosome fission associated with rDNA mobility. Chromosome Res. 3: 417–422. [DOI] [PubMed] [Google Scholar]
- Hess, H. E., E. Landolt and R. Hirzel, 1972. Flora der Schweiz. Bd. 3. Birkhäuser Basel, Stuttgart, Germany.
- Holliday, R., and G. W. Grigg, 1993. DNA methylation and mutation. Mutat. Res. 285: 61–67. [DOI] [PubMed] [Google Scholar]
- Hollingshead, L., 1930. Cytological investigations of hybrids and hybrid derivatives of Crepis capillaris and Crepis tectorum. Univ. Calif. Publ. Calif. Publ. Ag. Sci. 6: 55–94. [Google Scholar]
- Houben, A., N. Thompson, R. Ahne, C. R. Leach, D. Verlin et al., 1999. A monophyletic origin of the B chromosomes of Brachycome dichromosomatica (Asteraceae). Plant Syst. Evol. 219: 127–135. [Google Scholar]
- Houben, A., B. L. Field and V. Saunders, 2001. Micro-dissection and chromosome painting of plant B chromosomes. Methods Cell Sci. 23: 115–124. [PubMed] [Google Scholar]
- Hubbell, H. R., 1985. Silver staining as an indicator of active ribosomal genes. Stain Technol. 60: 258–294. [DOI] [PubMed] [Google Scholar]
- Ijdo, J. W., R. A. Wells, A. Baldini and S. T. Reeders, 1991. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 19: 4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger, E. J., and K. Werner (Editors), 2002. Rothmaler. Exkursionsflora von Deutschland. Bd.4. Gefässpflanzen: Kritischer Band. Spektrum Heidelberg, Berlin.
- Jamilena, M., C. Ruiz Rejón and M. Ruiz Rejón, 1994. A molecular analysis of the origin of the Crepis capillaris B chromosome. J. Cell Sci. 107: 703–708. [DOI] [PubMed] [Google Scholar]
- Jamilena, M., M. Garridoramos, C. Ruiz Rejón, M. Ruiz Rejón and J. S. Parker, 1995. Characterisation of repeated sequences from micro-dissected B chromosomes of Crepis capillaris. Chromosoma 104: 103–120. [DOI] [PubMed] [Google Scholar]
- Joly, S., J. T. Rauscher, S. L. Sherman-Broyles, A. H. D. Brown and J. J. Doyle, 2004. Evolutionary dynamics and preferential expression of homeologous 18S–5.8S–26S nuclear ribosomal genes in natural and artificial Glycine allopolyploids. Mol. Biol. Evol. 21: 1409–1421. [DOI] [PubMed] [Google Scholar]
- Jones, G. H., J. A. F. Whitehorn and S. M. Albini, 1989. Ultrastructure of meiotic pairing in B chromosomes of Crepis capillaris. I. One-B and two-B pollen mother cells. Genome 32: 611–621. [Google Scholar]
- Jones, N., and A. Houben, 2003. B chromosomes in plants: Escapees from the A chromosome genome? Trends Plant Sci. 8: 417–423. [DOI] [PubMed] [Google Scholar]
- Jones, R. N., 1995. Tansley review no. 85: B chromosomes in plants. New Phytol. 131: 411–423. [DOI] [PubMed] [Google Scholar]
- Jones, R. N., and H. Rees, 1982. B Chromosomes. Academic Press, London/New York.
- Kasha, K. J., and K. N. Kao, 1970. High frequency haploid production in barley (Hordeum vulgare L.). Nature 225: 874–876. [DOI] [PubMed] [Google Scholar]
- Kavanagh, T. A., and J. N. Timmis, 1986. Heterogeneity in cucumber ribosomal DNA. Theor. Appl. Genet. 72: 337–345. [DOI] [PubMed] [Google Scholar]
- Kavanagh, T. A., and J. N. Timmis, 1988. Structure of melon rDNA and nucleotide sequence of the 17–25S spacer region. Theor. Appl. Genet. 75: 673–680. [DOI] [PubMed] [Google Scholar]
- Kovarik, A., R. Matyasek, K. Y. Lim, K. Skalická, B. Koukalová et al., 2004. Concerted evolution of 18–5.8–26S rDNA repeats in Nicotiana allotetraploids. Biol. J. Linn. Soc. 82: 615–625. [Google Scholar]
- Laurie, D. A., and M. D. Bennett, 1989. The timing of chromosome elimination in hexaploid wheat × maize crosses. Genome 32: 953–961. [Google Scholar]
- Leach, C. R., T. M. Donald, T. K. Franks, S. S. Spiniello, C. F. Hanrahan et al., 1995. Organisation and origin of a B chromosome centromeric sequence from Brachycome dichromosomatica. Chromosoma 103: 708–714. [DOI] [PubMed] [Google Scholar]
- Liao, D., 1999. Concerted evolution: molecular mechanism and biological implications. Am. J. Hum. Genet. 64: 24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Małuszyńska, J., 1990. B chromosomes of Crepis capillaris (L.) Wallr. in vivo and in vitro. Ph.D. Thesis, Uniwersytet Sląski, Katowice, Poland.
- Małuszyńska, J., and D. Schweizer, 1989. Ribosomal RNA genes in B chromosomes of Crepis capillaris detected by non-radioactive in situ hybridization. Heredity 62: 59–66. [DOI] [PubMed] [Google Scholar]
- Navashin, M., 1927. Über die Veränderung von Zhal und der Chromosomen infolge der Hybridisierung. Zeitschr. Zellforschung Mikroskopische Anatomie 19: 195–233. [Google Scholar]
- Navashin, M., 1934. Chromosomal alterations caused by hybridisation and their bearing upon certain general genetic problems. Cytologia 5: 169–203. [Google Scholar]
- Page, B. T., M. K. Wanous and J. A. Birchler, 2001. Characterization of a maize chromosome 4 centromeric sequence: evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159: 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J. S., G. H. Jones, L. A. Edgar and C. L. Whitehouse, 1989. The population cytogenetics of Crepis capillaris. II. The stability and inheritance of B chromosomes. Heredity 63: 19–27. [Google Scholar]
- Pickering, R. A., 1985. Partial control of chromosome elimination by temperature in immature embryos of Hordeum vulgare L. × H. bulbosum. Euphytica 14: 869–874. [Google Scholar]
- Puertas, M. J., 2002. Nature and evolution of B chromosomes in plants: a non-coding but information-rich part of plant genomes. Cytogenet. Genome Res. 198: 198–205. [DOI] [PubMed] [Google Scholar]
- Rauscher, J. T., J. J. Doyle and A. H. D. Brown, 2004. Multiple origins and rDNA internal transcribed spacer homeologue evolution in the Glycine tomentella (Leguminosae) allopolyploid complex. Genetics 166: 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser, A., 1960. Fragmentchromosomen bei Crepis capillaris. Beih. Z. Schweiz. Forstvereins 30: 93–106. [Google Scholar]
- Sapre, A. B., and D. Deshpande, 1987. Origin of B chromosomes in Coix L. through spontaneous interspecific hybridisation. J. Hered. 78: 191–196. [Google Scholar]
- Schubert, I., and U. Wobus, 1985. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 92: 143–148. [Google Scholar]
- Sell, P. D., 1976. Crepis, pp. 344–357 in Flora Europaea, Vol. 4, edited by T. G. Tutin, V. H. Heywood, N. A. Burges, D. M. Moore, D. H. Valentine et al. Cambridge University Press, Cambridge, UK/London/New York.
- Sharbel, T. F., D. M. Green and A. Houben, 1998. B-chromosome origin in the endemic New Zealand frog Leiopelma hochstetteri through sex chromosome devolution. Genome 41: 14–22. [DOI] [PubMed] [Google Scholar]
- Sirri, V., P. Roussel and D. Hernandez-Verdun, 2000. The AgNOR proteins: qualitative and quantitative changes during the cell cycle. Micron 31: 121–126. [DOI] [PubMed] [Google Scholar]
- Stitou, S., R. Jimenez, R. D. de la Guardia and M. Burgos, 2000. Sex-chromosome pairing through heterochromatin in the African rodent Lemniscomys barbarus (Rodentia, Muridae). A synaptonemal complex study. Chromosome Res. 8: 277–283. [DOI] [PubMed] [Google Scholar]
- Swofford, D., 2002. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA.
- Thomas, H. M., and R. A. Pickering, 1983. Chromosome elimination in Hordeum vulgare × Hordeum bulbosum hybrids. 2. Chromosome behaviour in secondary hybrids. Theor. Appl. Genet. 66: 141–146. [DOI] [PubMed] [Google Scholar]
- Togby, H.A., 1943. A cytological study of Crepis fuliginosa, C. neglecta and their F1 hybrids and its bearing on a mechanism of phytogenetic reduction in chromosome number. J. Genet. 45: 67–111. [Google Scholar]
- White, T. J., T. Bruns, S. Lee and J. Taylor, 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, pp. 315–322 in PCR Protocols: A Guide to Methods and Applications, edited by M. A. Innis, D. H. Gelfand, J. J. Sninsky and T. J. White. Academic Press, San Diego.
- Wilson, E. B., 1907. The supernumerary chromosomes of Hemiptera. Science 26: 870. [Google Scholar]
- Yakura, K., and S. Tanifuji, 1983. Molecular cloning and restriction analysis of Eco RI fragments from Vicia faba rDNA. Plant Cell Physiol. 24: 1327–1330. [Google Scholar]
- Ziegler, C. G., D. K. Lamatsch, C. Steinlein, W. Engel, M. Schartl et al., 2003. The giant B chromosome of the cyprinid fish Alburnus alburnus harbours a retrotransposon-derived repetitive DNA sequence. Chromosome Res. 11: 23–35. [DOI] [PubMed] [Google Scholar]