Abstract
Arabidopsis thaliana ecotypes differ in their susceptibility to Fusarium wilt diseases. Ecotype Taynuilt-0 (Ty-0) is susceptible to Fusarium oxysporum forma specialis (f.) matthioli whereas Columbia-0 (Col-0) is resistant. Segregation analysis of a cross between Ty-0 and Col-0 revealed six dominant RESISTANCE TO FUSARIUM OXYSPORUM (RFO) loci that significantly contribute to f. matthioli resistance in Col-0 relative to Ty-0. We refer to the locus with the strongest effect as RFO1. Ty-0 plants in which only the Col-0 allele of RFO1 (RFO1Col-0) was introduced were resistant to f. matthioli. Surprisingly, RFO1Col-0 also conferred resistance to f. raphani, demonstrating that RFO1-mediated resistance is not race specific. Expression of resistance by RFO2, RFO4, or RFO6 was dependent on RFO1Col-0. Map-based cloning of RFO1Col-0 showed that RFO1 is identical to the previously named Arabidopsis gene WAKL22 (WALL-ASSOCIATED KINASE-LIKE KINASE 22), which encodes a receptor-like kinase that does not contain an extracellular leucine-rich repeat domain. Consistent with these results, a Col-0 rfo1 loss-of-function mutant was more susceptible to f. matthioli, f. conglutinans, and f. raphani. Thus, RFO1 encodes a novel type of dominant disease-resistance protein that confers resistance to a broad spectrum of Fusarium races.
IN most examined cases, plant genes that confer dominant resistance to pathogens have been shown to encode proteins involved in race-specific pathogen recognition. For a variety of practical reasons, the best studied of these resistance (R) genes confer strong resistance in a gene-for-gene manner to pathogens that express a corresponding avirulence (avr) gene (Nimchuk et al. 2003).
R genes have been shown to encode two broad categories of leucine-rich-repeat (LRR) proteins that can be distinguished by protein domain structure and site of pathogen perception (Jones and Takemoto 2004). The nucleotide-binding site (NBS)-LRR-containing R proteins mediate recognition of an intracellular pathogen-derived signal. Thus far, NBS-LRR proteins have been shown to function in resistance signaling only in response to pathogen. The second category of R proteins is inserted in the plasma membrane and minimally consists of an extracellular LRR domain and a transmembrane (TM) domain (Jones and Takemoto 2004). Some of these transmembrane LRR proteins also have an intracellular protein kinase (PK) domain and belong to the larger class of receptor-like protein kinases (RLKs). The extracellular LRR domain of LRR-TM and LRR-TM-PK proteins is thought to function as the receptor for an extracellular pathogen-derived signal. The signal that is recognized by an R protein can assume a variety of forms, including a viral coat protein, a secreted effector protein, the enzymatic activity of an effector protein, or a fungal polyketide metabolite (Nimchuk et al. 2003; Bohnert et al. 2004). In addition to pathogen recognition, LRR-TM-PK proteins function in signal transduction in a variety of plant processes, including development, maintenance of meristem identity, and brassinolide hormone perception (Torii 2004). Moreover, at least two LRR-TM-PKs function in both pathogen resistance and a seemingly unrelated plant process. The wild-type allele of the classical visible marker erecta (er) is necessary for cell proliferation throughout the plant. ERECTA has also been shown to confer resistance to the bacterial wilt pathogen Ralstonia solanacearum (Godiard et al. 2003). In tomato, the LRR-TM-PK BRI1 gene is necessary for the perception of both the plant steroid hormone brassinolide and the peptide elicitor systemin, which functions in a systemic signaling pathway that confers resistance to herbivorous insects (Scheer and Ryan 2002).
In contrast to the strong resistant phenotypes mediated by single R genes, host resistance is often oligogenic, which is referred to as “horizontal” resistance, and detected as multiple quantitative trait loci (QTL). In Arabidopsis, horizontal resistance to pathogens with a variety of different lifestyles is observed (Wilson et al. 2001; Godiard et al. 2003; Bohman et al. 2004; Denby et al. 2004). Importantly, because the components of horizontal resistance are very poorly defined, the relationship between oligogenic and monogenic resistance remains unclear.
The fungus F. oxysporum causes a vascular disease that is commonly known as wilt, root rot, or yellows (Talboys 1972; Beckman and Roberts 1995). Most soil-borne F. oxysporum is innocuous and is frequently isolated as an endophyte within the cortex of asymptomatic roots (Gordon and Martyn 1997). Rare pathogenic isolates have a narrow host range but can be devastating to a monoculture crop (Armstrong and Armstrong 1975). Control measures applied to infested fields are often impracticable or imperfect. However, natural resistance to Fusarium wilt within a species or genus is common, and traditional plant breeding has been successful in controlling the disease (Sherbakoff 1949).
The host specificity of a particular F. oxysporum isolate is described by its forma specialis (f.) (Armstrong and Armstrong 1975). A pathogenic strain can be further defined by its race, which describes the differential range among varieties of the host species.
Both dominant monogenic and oligogenic resistance to F. oxysporum are observed in various crop species (Sherbakoff 1949; Beckman and Roberts 1995). Although the genetic basis is unknown in most cases, it is common for different host varieties to possess different levels of resistance to Fusarium vascular disease. Without regard for the genetic nature of resistance, when both disease symptoms and fungal infection have been examined carefully, as with cabbage and tomato, symptoms have been found to correlate with degree of vascular colonization, and resistance level is quantitatively related to the success in restricting vascular colonization (Smith and Walker 1930; Anderson and Walker 1935; Gao et al. 1995a,b).
Six Immunity (I) loci that provide resistance to F. oxysporum f. lycopersici have been identified in Lycopersicon genomes (Sela-Buurlage et al. 2001). I, I-2, and I-3 are utilized for resistance in cultivated tomatoes (Beckman and Roberts 1995). The relationship between f. lycopersici races and the resistance loci I and I-2 is complicated. The I locus of L. pimpinnelifolium confers strong resistance to race 1 and no resistance to race 2, whereas the syntenic I locus from L. pennellii displays partial resistance to race 1 as well as strong resistance to race 2. Similarly, the I-2 locus of L. pimpinnelifolium confers strong resistance to race 2, but the syntenic I-2 locus of L. pennellii gives only partial resistance to race 2.
The best-characterized locus, I-2, is a cluster of at least seven related NBS-LRR gene sequences (Simons et al. 1998). Three of these I-2 homologous-coding sequences have been shown to confer resistance to race 2. As a transgene in a susceptible tomato line, coding sequence for what is designated as I-2 gives complete resistance while I-2C-1 and I-2C-5 introduce partial resistance to race 2 (Ori et al. 1997; Sela-Buurlage et al. 2001).
I-3 resistance has been delimited to a 0.3 cM interval in the tomato genome and cosegregates with a cluster of genes from the S-receptor gene family (Hemming et al. 2004). Among the I-3-linked genes, LpSRLK-1 encodes a S-locus RLK homologous to those involved in self-recognition in the self-incompatibility system of Brassica species.
Here we describe six dominant RESISTANCE TO FUSARIUM OXYSPORUM (RFO) loci in the Arabidopsis Col-0 ecotype. Among the six loci, RFO1 is the largest contributor to resistance. We identify At1g79670/WAKL22 as the gene responsible for the resistance expressed by RFO1, one of a limited number of genes corresponding to QTL that have been cloned in Arabidopsis (Koornneef et al. 2004). We also show that resistance expression of three other RFO loci is completely dependent RFO1. Although RFO1 is defined by resistance to f. matthioli, RFO1 is not race specific and plays a role in conferring resistance to three distinct F. oxysporum formae speciales.
MATERIALS AND METHODS
Arabidopsis lines, growth conditions:
Seeds of Arabidopsis ecotypes, including Ty-0 (CS6878), and Salk insertion lines, 077975 and 140705, were provided by Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH). A list of ecotypes tested and their reaction to F. oxysporum can be found at http://ausubellab.mgh.harvard.edu/ecotypesxFo. Plants were grown in flats (10.75 × 21.5 × 2 in.) or pots (2 × 2.25 × 2 in.) in Metromix 200 (Scotts-Sierra, Marysville, OH), in Fafard 2 Mix (Conrad Farfard, Agawam, MA), or on 30-mm Jiffy-7 peat pellets (Jiffy Products, Shippagan, New Brunswick, Canada). Plants were kept at 22° in a greenhouse with supplemental fluorescent lighting to maintain a 12-hr day length. When seed was limiting or when transgenic seedlings were selected for kanamycin resistance, disinfected seeds were sown on plant nutrient (PN) medium with 0.8% Bacto agar alone (PNA) or with 0.5% sucrose (PNS). PN is 5 mm KNO3, 2.5 mm KH2PO4, 2 mm MgSO4, 2 mm Ca(NO3)2, 50 μm FeNa(EDTA), 70 μm H3BO3, 14 μm MnCl2, 0.5 μm CuSO4, 1 μm ZnSO4, 0.2 μm Na2MoO4, 10 μm NaCl, and 0.01 μm CoCl2. Seeds were disinfected in 10% household bleach for 15 min and subsequently washed three times with excess sterile water. Petri plates were sealed with paper tape (Medline Ind., Mundelein, IL). For kanamycin selection, seeds were sown on PNS supplemented with 20 μg ml−1 kanamycin A (Sigma-Aldrich, St. Louis).
Molecular biology:
Total Plant DNA was purified from rosette leaves using a protocol recommended for Qiagen DNA-affinity columns (Qiagen, Valencia, CA). Total Plant RNA was purified from rosette leaves using a standard LiCl precipitation protocol. The BAC genomic clone F20B17—refer to GenBank accession no. AC010793 for nucleotide sequence—was obtained from the ABRC. The DNA probe for Northern and Southern blot hybridization was a BglII fragment originating from BAC F20B17, covering nucleotides 40,439–41,458, and was labeled with [α-32P]dCTP by random-primer extension. Techniques for manipulation and detection of DNA and RNA were derived from standard molecular biology protocols (Ausubel et al. 1998). The At1g79670 genomic sequence was PCR amplified from Col-0 and Ty-0 DNA with the following primer pair: At1g79670-P6, 5′-TGTGATGGAACCTTAACCAACA-3′, and At1g79670-R, 5′-TGAGAGAATTTGTTATCACAGCAC-3′. The PCR product was sequenced by the DNA Core Facility (Department of Molecular Biology, Massachusetts General Hospital, Boston) as overlapping sequence runs using appropriate primers. Sequence flanking the left or right border of insertion 077975 was PCR amplified and sequenced with primer pairs AT1G79670-P5, 5′-GAGATTTAATGTGAACAACTCC-3′, and LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′, or SALK_077975-RP, 5′-CGTTGGTGAATAGTCAATTTCCTGA-3′, and RBb1, 5′-ACGTTGCGGTTCTGTCAGTTCC-3′, respectively. The left and right borders are located at nucleotides 41,827 and 41,915, respectively, of BAC F20B17. The difference between the T-DNA borders leaves a deletion in coding sequence of 31 amino acid residues. In 12 genotyped T3 plants from insertion line 077975, the Rfo phenotype and kanamycin-resistance phenotype were consistent in the subsequent T4 generation. The segregation of 90 or 89 kanamycin resistant and 31 or 32 kanamycin sensitive (P > 10% for 3 resistant:1 sensitive, using χ2 test), respectively, in the progeny of two line 077975 heterozygotes suggests that one insertion is present. Sequence flanking the left or right border of insertion 140705 was PCR amplified and sequenced with primer pairs SALK_140705-LP and LBb1 or SALK_140705-RP and RBb1, respectively. The genomic position for the right and left border of the 140705 insertion coincided.
Fusarium strains, growth conditions:
F. oxysporum crucifer isolates 726 (f. matthioli race 2), 777 (f. conglutinans race 1), and 815 (f. raphani) as well as F. oxysporum isolates MN-25 (f. lycopersici race 3), FORL-D69 (f. radicis-lycopersici), NRRL25609, and NRRL25367 (f. cubense) are from Corby Kistler (University of Minnesota, St. Paul) (Kistler et al. 1991). We maintained a frozen stock of each Fusarium isolate in 50% glycerol at −80°. Fusarium was thawed and grown on Czapek-Dox (CzD) plates (CzD broth with 1.5% Bacto agar, Becton Dickinson, Sparks, MD) at 22°–28°. Liquid cultures were inoculated from plates.
Soil infection assay:
Fusarium bud cells were cultured in 700 ml CzD broth in a 2-liter Erlenmeyer flask by shaking at 250 rpm at 22°–28° for 5 days. To harvest bud cells, the culture was filtered with Steri-Pad gauze pads (Johnson & Johnson Medical, Arlington, TX), settled by centrifugation at 8000 rpm for 15 min, washed with water, and finally resuspended in water. The soft bud-cell pellet was washed by resuspending bud cells in water and settling by centrifugation three consecutive times. Bud-cell culture density was measured using a hemocytometer, and bud cells were diluted with sterile water. Unless stated otherwise, the inoculum density was 1 × 106 bud cells ml−1. For soil inoculation, 5 ml of bud-cell suspension was applied ∼1 in. below the surface of the soil with a pipet. Peat pellets were inoculated by placing them in a 50-ml beaker with 15 ml of bud-cell suspension for 1 min. Infested plantings were incubated in a growth chamber (Percival Scientific, Perry, IA) with a 12-hr day, under a light density of 120 μE m−2 sec−1 and at 26° and 70% relative humidity. Disease was evaluated using the disease index (DI) described in Figure 1. Often, intermediate 0.5 scores were included in the DI to improve resolution but rounded to integers for presentation. To measure stunting, we determined the distance from the stem to the distal end of the midrib (leaf length). The three longest leaves of each rosette were included in each measurement.
Figure 1.
Disease symptoms of Fusarium wilt. The soil of 3-week-old Ty-0 plants growing in peat pellets was infested with f. matthioli bud cells. Plants are shown at 6 weeks. (A) Below each DI score, a representative plant exhibits the symptoms of that score: 0, the plant is dead; 1, older leaves are dead and younger leaves are severely stunted; 2, older leaves are chlorotic, yellow, or dead and younger leaves are stunted; 3, older leaves have vascular chlorosis and the rosette appears compact because leaves are stunted; 4, leaf petioles are stunted; and 5, plants are indistinguishable from mock-inoculated plants. (B) Each plant represents the disease symptoms that are observed after infestation with f. matthioli (bottom left), infestation with f. raphani (bottom right), or mock infestation with water (top center).
Plant genotyping:
PCR analysis of SSLP markers was performed on crude tissue preparations. One or both cotyledons (or same amount of leaf) were placed in a microcentrifuge tube, frozen on dry ice, and then ground briefly with a pestle. The tissue was thawed and resuspended in 20 μl 0.5 m NaOH, and the tube was heated in boiling water for 2 min. The homogenate was neutralized with 200 μl 0.2 m Tris base, pH 8.0, 1 mm EDTA. One microliter of crude DNA was used in a 20-μl PCR preformed as recommended by the Taq polymerase supplier (Roche Applied Science, Mannheim, Germany). All SSLP markers were size-separated in 3% MetaPhor agarose (Cambrex Bio Science, Rockland, ME). The set of SSLP markers, including F21M12, ciw12, nga280, nga111, ciw2, nga162, ciw11, ciw4, nga6, ciw5, ciw6, ciw7, nga1107, CTR1, ciw8, PHYC, and ciw9, are described in Lukowitz et al. (2000). The description of SSLP markers PLS7, C4H, BIO2, and nga172 is found in TAIR. We named new SSLP markers after the originating BACs. The name of new markers and oligonucleotide pairs for PCR amplification follow: F23A5, 5′-CTATGATAATATTAGTCAGTAGGG-3′ and 5′-CTTTAACAGTTATTGTAATCAGTC-3′; F3F9, 5′-AGTTCTGTATCTGCAAATTTCT-3′ and 5′-CCTGTTCCCTTTTAGCTTCTCC-3′; F9K20, 5′-CGTCAGCTTACGAGCTTCTCTT-3′ and 5′-GCTTCCGATTGGTCTGACTTGG-3′; F19K16, 5′-GAGTGGGGGATGCTTTGTGTTTTG-3′ and 5′-TTGCTTGATCATATTCTCTCTTTG-3′; F15O4, 5′-CTAATGACGATAATAATTGTTAC-3′ and 5′-TTCTAAGTTTCTTTGTGTTCAG-3′; MOA2, 5′-ATCCAGAAAATCATGTAATGCATGA-3′ and 5′-CTAATGTGATGTCAGTTGTCACTCA-3′; and MBK5, 5′-GAGCATTTCACAGAGACG-3′ and 5′-ATCACTGTTGTTTACCATTA-3′. SALK Insertion lines were genotyped by a PCR protocol recommended by The Salk Institute Genomic Analysis Laboratory (SIGnAL, http://signal.salk.edu/tdnaprimers.html). Flanking primers for genotyping either line 140705 or 077975 are SALK_140705-LP, 5′-AGCCGTCTCGTTGGAATTTGG-3′, and SALK_140705-RP, 5′-GTGTTACCGCCGCATCATCTC-3′, or AT1G79670-P5 and SALK_077975-RP. The T-DNA primer is LBb1.
RFO genetic linkage:
Kendall's rank correlation coefficient (τ) was calculated using Excel (Microsoft, Redman, WA) (Campbell 1996). τ was measured for each SSLP marker from the tabulated DI scores and SSLP marker genotypes of 239 plants derived from a backcross (BC) of an F1 Ty-0/Col-0 plant to Ty-0. For the RFO1Col-0 and RFO1Ty-0 subpopulations, τ was measured with data for either the 124 BC plants with genotype C/T or the 114 BC plants with genotype T/T, respectively, at SSLP marker F19K16. Excel functions were configured to compute τ,
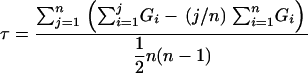 |
where n is the number of BC plants and GR is the genotype for BC plant at rank R. Arbitrarily, the values for genotypes T/T and C/T were set at +1 and −1, respectively. For the most susceptible BC plant, R = 1, and, for the most resistant BC plant, R = n. BC plants were sorted into a rank by increasing DI scores from 19, 15, and 12 dpi. For our analysis in Table 1, we favored a rank sorted first by DI score on 19 dpi; then, equivalent ranks from 19 dpi were sorted by DI score on 15 dpi; and finally, the remaining equivalent ranks from the preceding order were sorted by DI score on 12 dpi. Most BC plants had equivalent DI scores with other BC plants at all three time points. For example, the 239 BC plants were sorted into 65 levels with equivalent scores. The final rank of BC plants with equivalent DI scores was arbitrarily set by the initial order of the BC plants before sorting. Because the correlation coefficient value could be skewed by the initial order of BC plants, we recalculated the coefficient values at each SSLP marker 11 additional times with a different starting order of BC plants. The correlation coefficients in Table 1 are the mean value of the 12 trials. For all significant correlation coefficients, the 95% confidence limit was less than 5% of the mean value. We acquired genome-wide significance levels from a distribution of 240,000 correlation coefficients, or 10,000 simulated values at each of the 24 SSLP markers. A. Diener wrote a BASIC computer program that calculates correlation coefficient values from the BC genotype data after random permutation of plant ranking. The 500th and 100th highest values were taken as the correlation coefficient at the P = 0.05 and P = 0.01 threshold. The genetic linkage maps in Figure 4 were assembled by Map Manager QTXb17 software (Roswell Park Cancer Institute, Buffalo) using the Kosambi mapping function.
TABLE 1.
Correlation coefficient for association of SSLP marker genotype and disease index score
| Correlation coefficienta
|
|||||
|---|---|---|---|---|---|
| Chromosome | Marker | Allb | RFO1Col-0c | RFO1Ty-0d | Locus |
| 1 | F21M12 | 0.28 | 0.73 | −0.13 | RFO2 |
| 1 | ciw12 | 0.26 | 0.60 | −0.07 | |
| 1 | F15O4 | 0.18 | 0.38 | −0.05 | |
| 1 | nga280 | 0.21 | 0.18 | −0.02 | |
| 1 | nga111 | 0.49 | −0.03 | 0.12 | |
| 1 | F3F9 | 0.72 | — | — | RFO1 |
| 2 | ciw2 | 0.06 | −0.01 | 0.09 | |
| 2 | PLS7 | 0.19 | 0.16 | 0.17 | |
| 2 | C4H | 0.21 | 0.24 | 0.19 | |
| 2 | BIO2 | 0.08 | 0.19 | 0.00 | |
| 3 | nga172 | 0.19 | 0.25 | 0.35 | |
| 3 | nga162 | 0.30 | 0.30 | 0.58 | RFO3 |
| 3 | ciw11 | 0.13 | 0.10 | 0.13 | |
| 3 | ciw4 | 0.03 | 0.08 | −0.06 | |
| 3 | nga6 | −0.12 | −0.06 | −0.28 | |
| 4 | ciw5 | 0.09 | 0.32 | −0.11 | RFO4 |
| 4 | ciw6 | 0.09 | 0.17 | 0.00 | |
| 4 | ciw7 | 0.03 | −0.07 | −0.02 | |
| 4 | nga1107 | 0.05 | −0.16 | 0.18 | |
| 5 | CTR1 | 0.13 | 0.19 | 0.08 | |
| 5 | ciw8 | 0.24 | 0.14 | 0.23 | RFO5 |
| 5 | PHYC | 0.26 | 0.38 | 0.10 | |
| 5 | ciw9 | 0.16 | 0.35 | −0.05 | RFO6 |
| 5 | MBK5 | −0.06 | 0.05 | −0.20 | |
Significant (P ≤ 0.05) correlation coefficient values are in italics. For significance, threshold values from genome-wide evaluation are derived from permutation of the BC population data (see materials and methods).
Threshold values at P = 0.05 and P = 0.01 are, respectively, 0.22 and 0.26 for the whole BC population.
Threshold values at P = 0.05 and P = 0.01 are, respectively, 0.30 and 0.36 for the RFO1Col-0/RFO1Ty-0 BC subpopulation.
Threshold values at P = 0.05 and P = 0.01 are, respectively, 0.31 and 0.37 for the RFO1Ty-0/RFO1Ty-0 BC subpopulation.
Figure 4.
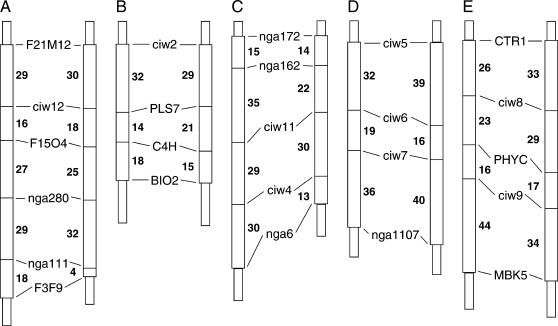
Genetic maps for (A) Arabidopsis chromosome 1, (B) chromosome 2, (C) chromosome 3, (D) chromosome 4, and (E) chromosome 5. Chromosomal genetic maps are represented by vertical bars and oriented with the start of AGI sequence at the top. The map positions of SSLP markers are located at horizontal lines that segment the chromosomal maps. The vertical length along each chromosome is proportional to genetic distance, and genetic distances between SSLP markers are given next to the segment separating the markers. The chromosomal map to the left was calculated from recombination frequencies between marker loci in our BC population. The chromosomal map to the right represents the established genetic map, which was previously derived from Col-0/Ler RI lines.
Introduction of RFO1Col-0 into Ty-0 background:
RFO1Col-0 was isolated from other Col-0 alleles of RFO loci in BC plant 4E9. In 4E9, SSLP markers on all chromosomes except chromosome 1 were T/T. Markers F21M12 and ciw12 at one end of chromosome 1 and closest to RFO2 were T/T. The genotype of 4E9 was C/T for much of chromosome 1 extending from at least marker F15O4 to the telomeric marker F23A5. Ignoring chromosomal ends and possible double recombination between markers, 4E9 contains RFO1Col-0 in a largely Ty-0 background. Among the F2 progeny of BC plant 4E9, we selected a plant 1A3 in which the T/T genotype was extended to all markers on chromosome 1 except marker F3F9 at the end where RFO1 is located. Appearance and growth of uninfected 1A3 plants and the Ty-0 parent were indistinguishable.
RFO1 positional cloning:
Two plants 4C5 and 5D7 of 239 BC plants had recombination breakpoints both between F3F9 and F23A5 and on either side of RFO1, placing RFO1 between these markers. The genotype of 4C5 was T/T at F23A5 and C/T at F3F9, and the genotype of 5D7 was C/T at F23A5 and T/T at F3F9. At a new marker F19K16 between F3F9 and F23A5, 4C5 was T/T and 5D7 was C/T. In the self-cross progeny of either 5D7 or 4C5, resistance failed to segregate with markers linked to RFO1Col-0 and implied that 5D7 and 4C5 were both homozygous for RFO1Ty-0. This placed RFO1 between F3F9 and F19K16. We further restricted the RFO1 interval by screening 65 self-crossed progeny of BC plant 4E9 for 2 plants, 6 and 47, with recombination breakpoints between F3F9 and F19K16. The genotype of 6 was T/T at F3F9 and C/T at F19K16, and the genotype of 47 was C/T at F3F9 and T/T at F19K16. At a new marker F9K20 between F3F9 and F19K16, the genotype of both 6 and 47 was T/T. In the self-crossed progeny of 6 and 47, a lack of resistance implied that 6 was a RFO1Ty-0 homozygote, and the presence of F19K16-linked resistance implied that 47 was RFO1Col-0/RFO1Ty-0. Plant 47 placed RFO1 between markers F9K20 and F19K16.
Plant transformation:
Arabidopsis ecotype Ty-0 was transformed by Agrobacterium tumefaciens strain GV3101 using the inflourescence-dip protocol (Clough and Bent 1998). GV3101 was transformed with recombinant T-DNA plasmids by electroporation (Mattanovich et al. 1989). A restriction-digest fragment of BAC genomic clone F20B17 was subcloned into the polylinker of T-DNA binary vector pPZP212 (Hajdukiewicz et al. 1994). The subcloning was confirmed by concordance between the restriction-digest pattern of a subclone and the pattern that is predicted from the F20B17 sequence in GenBank accession no. AC010793. For plasmid constructs A, B, or C (see Figure 6A), an EcoRI-digest fragment of BAC F20B17, covering nucleotides 18,409– 28,996, 28,996–36,277, or 36,277–43,623, respectively, was subcloned into the EcoRI site of pPZP212. For construct D, an AgeI-digest fragment of BAC F20B17, covering nucleotides 41,603–48,054, was subcloned into the XmaI site of pPZP212. For construct E, a PstI-digest fragment of BAC F20B17, covering nucleotides 36,833–48,749, was subcloned into the PstI site of pPZP212. To make construct F, construct C was digested with XbaI and then religated, removing one end of genomic sequence in construct C. Construct F covers the nucleotide sequence 39,220–43,623 of F20B17. T1 seed of Agrobacterium-treated Ty-0 plants was selected, and green kanamycin-resistant transformants were transplanted to peat pellets and later assayed for resistance to f. matthioli. We derived a T2 homozygous 5B1 line from a representative T1 plant harboring construct C. Of 55 kanamycin-selected seeds, all expressed kanamycin resistance and displayed enhanced resistance to f. matthioli.
Figure 6.
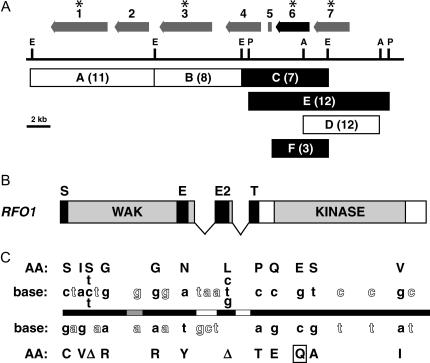
Suppression of Ty-0 susceptibility with Col-0 allele of RFO1. (A) Restriction map of BAC F20B17 region with RFO1 candidate genes. Vertical bars mark restriction sites: A is AgeI, E is EcoRI, and P is PstI. Above the restriction map, arrows delimit putative start and stop codons of TIGR-annotated genes and point in the sense direction. The annotated genes are labeled by number, where 1 is At1g79620, 2 is At1g79630, 3 is At1g79640, 4 is At1g79650, 5 is At1g79660, 6 is At1g79670/RFO1 (highlighted in black), and 7 is At1g79680. An asterisk is above each RFO1 candidate. Below the restriction map, a box delimits the genomic sequence that is contained in each transgenic construct: A, B, C, D, E, or F. Each box is solid or open to indicate whether the transgene conferred enhanced resistance or no more resistance than the Ty-0 parent, respectively. The number of tested T1 plants is in parentheses. (B) Encoded domains of RFO1 are depicted: S is a signal peptide, WAK is the extracellular wall-associated kinase domain, E is an EGF2 domain, E2 is an EGF-Ca domain, T is transmembrane domain, and KINASE is an RLK kinase domain (Verica et al. 2003). (C) The horizontal bar represents the RFO1 genomic sequence and is solid for coding sequence, shaded for an alternative-spliced sequence, and open for intronic sequence. The sequence from Ty-0 is in GenBank accession no. DQ023268. Corresponding nucleotide bases in either Col-0 (above the bar) or Ty-0 (below the bar) in the RFO1 genomic sequence and resulting differences in translation (AA, amino acids) are depicted along the RFO1 genomic sequence. Nucleotide bases in outline type are within codons that are synonymous in Col-0 and Ty-0. A Δ indicates the absence of three nucleotides in the Ty-0 sequence. A box encloses the nonconserved glutamine residue (Q) in Ty-0.
RESULTS
Ecotypes differ in susceptibility to Fusarium:
We adapted a root-dip inoculation method for F. oxysporum infection to the model plant Arabidopsis. Because Arabidopsis poorly tolerates transplanting, 2- to 3-week-old Arabidopsis plants were inoculated by delivering F. oxysporum to soil in which roots were growing. As expected for Fusarium wilt, infected susceptible plants began to display symptoms of the disease ∼9 days after soil infestation (Jimenez-Gasco et al. 2004).
We found that the appearance and severity of wilt disease was determined by both the pathogen forma specialis and the host ecotype. In particular, inoculation of Arabidopsis with pathogenic isolates from a related crucifer host, cabbage (f. conglutinans), radish (f. raphani), or stock (f. matthioli), produced disease symptoms. In contrast, isolates from unrelated hosts such as tomato (f. lycopersici race 3 and f. radicis-lycopersici) or banana (f. cubense) failed to induce visible disease symptoms on any of several tested Arabidopsis ecotypes, even when soil was infested with a very high inoculum of 5 × 106 bud cells ml−1 (data not shown). Because Arabidopsis was not susceptible to all pathogenic forms of F. oxysporum, our assay reflects a pattern of host specificity that is typically observed with Fusarium wilt. The range of observed symptoms allowed us to develop a DI, which is exemplified in Figure 1A, to provide a rough quantitative measure of disease severity.
Discrimination of pathogenic races by disease symptoms is another common feature of Fusarium wilt (Jimenez-Gasco et al. 2004). Indeed, as shown in Figure 1B, different symptoms in Arabidopsis were elicited by each of the three crucifer-specific formae speciales. Stunting of petioles and vascular chlorosis were prominent in infections with f. conglutinans (not shown) and matthioli. Browning of the stem and petioles was pronounced with f. raphani and matthioli infection. Distal yellowing on rosette leaves was characteristic of f. raphani infection.
As others have observed among varieties of a particular host species, we found that different levels of resistance to Fusarium wilt can be observed among Arabidopsis ecotypes (Beckman and Roberts 1995). Furthermore, the level of resistance expressed by an ecotype was specific for each forma specialis. Most ecotypes, including Col-0, exhibited strong resistance to f. matthioli, and only 7 of 83 tested ecotypes (8%) displayed any significant chlorosis with a high inoculum of 1 × 106 bud cells ml−1 (data not shown). Among the ecotypes tested, Ty-0 was the most susceptible to f. matthioli, displaying symptoms identical to those reported for its natural host (Mathiola incana) (compare Figure 1 to Figure 2 in Baker 1948). In contrast to infection with f. matthioli, inoculation with f. conglutinans or f. raphani produced pronounced symptoms in many ecotypes (data not shown). The susceptibility of Ty-0 to f. matthioli presumably reflects a race-specific susceptibility and not a general deficiency in disease resistance because Ty-0 exhibits resistance to f. raphani that was comparable to many other ecotypes, including Col-0 (in Figure 2, A and C, and data not shown).
Figure 2.
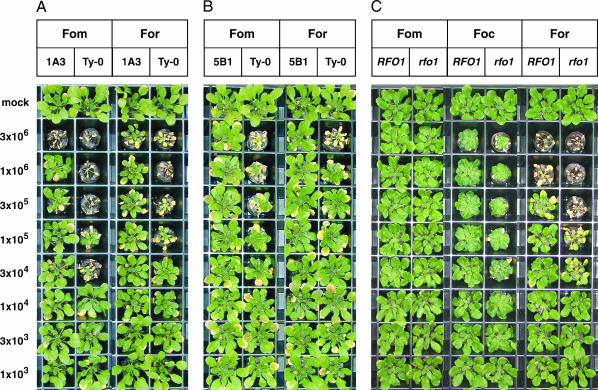
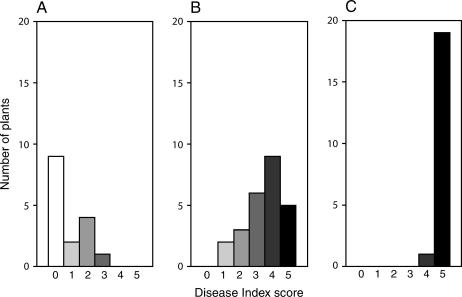
RFO1 resistance to crucifer races of Fusarium oxysporum. At 3 weeks of age, the peat under each plant was infested with dilutions of a Fusarium bud cell suspension or with water (mock). The bud cell densities of the inocula are on the left side. Six-week-old plants are shown. Each plant that is shown displays the median DI score of five replicates. Plants in each column are the same genotype and were infested with the same forma specialis. The forma specialis administered is indicated above the columns. Fom is F. oxysporum f. matthioli, Foc is F. o. f. conglutinans, and For is F. o. f. raphani. The plant lineage is also given above each column. (A) The 1A3 line is the Ty-0 background with the end of chromosome 1 introduced from the Col-0 ecotype, including RFO1. (B) The 5B1 line is the Ty-0 ecotype with a homozygous RFO1Col-0 transgene. (C) Homozygous rfo1 is compared to the wild-type (RFO1) segregant from the 079775 insertion line.
Few susceptible F2 plants from a self-crossed hybrid:
The strong resistance of Col-0 and consistent susceptibility of Ty-0 to f. matthioli, which are depicted in Figure 3, A and B, let us investigate the genetic basis of ecotype differences in defense against Fusarium wilt. We crossed the Col-0 and Ty-0 parents and observed that 90% of tested F1 hybrid plants showed the same high level of resistance as their Col-0 parent, whereas 10% displayed mild disease symptoms as shown in Figure 3C. Because the Col-0 parent was always asymptomatic, we concluded that, while the resistance of Col-0 was dominant, the dominance was incomplete in the F1 hybrid background.
Figure 3.
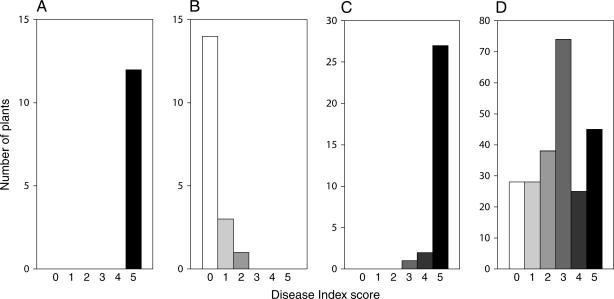
Disease symptoms in three generations. Histograms show the distributions of disease index scores for the parental ecotypes (A) Col-0 and (B) Ty-0, (C) the F1 Col-0/Ty-0 hybrid, and (D) the F1 backcrossed to Ty-0. Three-week-old plants were exposed to f. matthioli, and plants were scored at 6 weeks.
To reveal the pattern of inheritance for f. matthioli resistance, we generated a recombinant F2 population by self-crossing the F1 hybrid. The soil under 3-week-old F2 plants was infested with f. matthioli, and 3 weeks later the symptom severity of each rosette was scored using the disease index. The distribution of DI scores suggested that resistance was an oligogenic trait because the vast majority of F2 plants were highly resistant, and only a small fraction, just 8 of 128 tested plants, developed any symptoms (DI < 5). The simplest explanation for this susceptible:resistant ratio of 1:15 is the independent segregation of two dominant-resistance loci. However, because all 8 diseased F2 plants displayed milder symptoms than the susceptible Ty-0 parent, it is likely that more than two genetic loci are responsible for resistance in Col-0.
Phenotypic variety in a backcross population:
To increase the frequency of phenotypic variety in the mapping population, a second recombinant population was made by backcrossing the F1 Col-0/Ty-0 hybrid to the recessive Ty-0 parent. A backcross (BC) population is not typically used as a mapping population in Arabidopsis because the self-fertilizing F1 plant produces a F2 population without intervention. However, a recessive phenotype is more highly represented in a BC especially when multiple loci contribute to resistance. Furthermore, the possible genotypes in a BC population are simpler. In our case, a BC plant is either Ty-0/Ty-0 (T/T) or Col-0/Ty-0 (C/T) at any locus. This also means that only dominant disease-resistance genes from Col-0 can contribute to resistance in the BC population.
As anticipated and depicted in Figure 3D, the BC population expressed a greater diversity of disease phenotypes than the F2 population. In total, 239 BC plants were tested and scored. This experiment was conducted using an alternative infection protocol that gives essentially the same results as the soil inoculation protocol described above. BC plants were grown in Jiffy peat pellets and inoculated by bottom watering with 1 × 106 f. matthioli bud cells ml−1. As with the F2 population, the distribution of DI scores in the BC population suggested that more than one locus contributed to resistance. For simple monogenic inheritance, a segregation ratio of 1 susceptible:1 resistant would be expected. Rather, as shown in Figure 3D, the majority of plants displayed an intermediate susceptibility (2 ≤ DI ≤ 3.5) as compared to either the Ty-0 parent (DI ≤ 2) or the F1 hybrid (with one exception, DI > 3.5).
A genetic map from the BC population:
To carry out linkage analysis of the complex pattern of inheritance of resistance in the Ty-0 × Col-0 cross, we were compelled to evaluate the genotype of our BC population throughout the genome. In doing so, we were able to assemble a genetic linkage map of the whole genome. For this linkage analysis, we took advantage of a comprehensive set of simple sequence length polymorphism (SSLP) markers that were designed to distinguish between Col-0 and Landsberg erecta (Ler) chromosomes. Of these 22 markers, 17 were also found to distinguish between Col-0 and Ty-0. The gaps in genomic coverage were filled in with 7 additional SSLP markers: We obtained 4 SSLP markers from The Arabidopsis Information Resource database (TAIR, www.arabidopsis.org) and designed 3 new SSLP markers (see materials and methods).
A genetic map for each chromosome was assembled from the genotype of the 239 BC individuals at all 24 SSLP markers and is presented in Figure 4. The median genetic distance between SSLP markers was found to be 21 cM with the shortest and longest lengths equal to 14 and 44 cM, respectively. In Figure 4, this BC genetic map is aligned with the standard genetic map that is used by the Arabidopsis community as derived from Col-0/Ler recombinant inbred (RI) lines. The genetic distances across each chromosome were roughly comparable between the BC map and the standard RI map with one exception. The genetic distance of chromosome 3 on the BC map was 40% greater than the corresponding distance on the standard RI map. Coincidentally, a similar higher than expected recombination frequency on chromosome 3 was previously reported by another group also using a recombinant BC population, in this case, from a Col-0 × Ler cross (Copenhaver et al. 1998).
Distribution of DI scores in BC population:
We tried to gain accuracy in our disease assessment by scoring each of the 239 BC plants on 12, 15, and 19 days after infestation (dpi). Comparison of the three scores for each plant showed good correspondence: Later scores were approximately the same or higher than earlier scores. As shown in Figure 3D, at 19 dpi, the distribution of the BC population was roughly symmetric over the DI range and was centered on a median score of 3.0. Similar results were obtained at 12 and 15 days (not shown).
However, a large proportion of BC plants had extreme DI scores in Figure 3D. At each of the time points, 15% of plants had no obvious disease and almost a quarter had very mild symptoms (DI ≥ 4). By the last time point, 12% of plants were dead. At the upper end of the DI range, phenotypic discrimination was diminished. Because inheritance of all detected RFO loci was not necessary to attain the highest DI score, the apparent linkage to resistance loci in the most resistant individuals of the BC population was probably somewhat obscured.
Measuring correlation between genotype and resistance:
We wished to use the phenotypic data of the BC population to detect genetic linkage of SSLP markers to RFO loci. Because the DI measurement for resistance is a discontinuous rank with finite ends rather than a normal distribution, the association between genotype and resistance was evaluated with Kendall's rank correlation coefficient (see materials and methods). Although this test was more appropriate for our data, we note that the linkage results were essentially the same when we assumed a normal distribution for DI scores and computed significance with available QTL software (data not shown).
Kendall's test required that we order the BC population by rank: the most susceptible plant to most resistant plant. To better differentiate the rank of BC plants, we incorporated DI scores for all three time points and we made two alternative rankings that gave priority to either the early score at 12 dpi (early phenotype) or the late score at 19 dpi (late phenotype). For the late phenotype ranking, the BC plants were sorted in succession first by the DI score from 19 dpi, then, those plants with the same score at 19 dpi were sorted by the score from 15 dpi, and finally, those plants with the same score at both late time points were sorted by the score from 12 dpi. For the early phenotype, the input order of DI scores was reversed for sorting the population.
In assessing RFO linkage, we were testing the association of genotype and DI score at each of the SSLP markers. The genotype of each BC plant at any particular locus was either C/T or T/T. Because the F1 phenotype showed that resistance was dominant we expected only the C/T genotype to be correlated with resistance. In Table 1, the correlation coefficient enumerates the degree of association at the listed SSLP marker. An absolute association of marker genotype and resistance would yield a correlation coefficient of 1.00, and perfect lack of correlation would be zero. A positive correlation associates C/T with resistance, while a negative correlation associates T/T with resistance.
Genetic linkage of RFO loci:
Significant correlation (P ≤ 0.05) between Col-0 genotype and disease resistance was detected at SSLP markers that are linked to six loci and are distributed on four of the five Arabidopsis chromosomes. The six loci are tentatively named RESISTANCE TO FUSARIUM OXYSPORUM 1 (RFO1) through RFO6, although it should be pointed out that each locus might represent more than one gene. In Table 1, each RFO locus is assigned to the marker that exhibits the strongest linkage.
Using the early phenotype, resistance was significantly linked to three major resistance loci (RFO1, RFO2, and RFO3). The same three loci exhibited significant linkage to resistance using the late phenotype, although the correlation was stronger with the early phenotype. In contrast, the correlation coefficient at SSLP markers linked to the minor loci (RFO4, RFO5, and RFO6) was greater for the late phenotype than for the early. Overall, it appeared that the loci responsible for the earliest indication of resistance at 12 dpi were the same as the loci that determine resistance at later time points. However, because more loci were detected at later times, the analysis in Table 1 uses data derived from the late phenotype.
RFO1 resistance enhanced by RFO2, RFO4, and RFO6:
The largest correlation coefficient was at marker F3F9, which was designated RFO1. As we describe below, RFO1 conferred resistance when separated from the other five RFO loci. Nevertheless, the magnitude of the correlation at RFO1 depends on interactions with RFO2, RFO4, and RFO6. In particular, the large correlation coefficient of F21M12, which is linked to RFO2, was dependent on the genotype of RFO1. Of the 20% of BC plants with the strongest resistance, 94% had a genotype of C/T at both RFO1- and RFO2-linked markers.
The interaction of RFO1 with RFO2, RFO4, and RFO6 was evident when the BC population was divided into two subpopulations, one with RFO1Col-0, the Col-0 allele of RFO1, and the other with RFO1Ty-0. The correlation between marker genotype and DI rank and its significance were retested. If other loci interact with RFO1 to affect the resistance phenotype, we expect a difference in correlation between the two subpopulations. Indeed, as shown in Table 1, significant positive correlation in the RFO1Col-0 subpopulation was measured at F21M12, ciw5, and ciw9, markers linked to RFO2, RFO4, and RFO6, respectively. The contribution of RFO2Col-0, RFO4Col-0, or RFO6Col-0 to resistance in the RFO1Col-0 subpopulation is contrasted in Table 1 with the low and negative correlation at these loci in the RFO1Ty-0 subpopulation. If the contribution of RFO2, RFO4, or RFO6 were independent of RFO1 genotype, we would have expected to see a common trend in both RFO1 subpopulations. Moreover, the complete lack of correlation in the RFO1Ty-0 subpopulation shows that these three loci are completely dependent on RFO1Col-0.
RFO resistance independent of RFO1:
In contrast to RFO2, RFO4, and RFO6, the resistance provided by the remaining two resistance loci, RFO3 and RFO5, was independent of RFO1. The correlation between Col-0 genotype at the RFO3-linked marker nga162 and resistance was positive and significant within both RFO1 subpopulations. At RFO5-linked marker ciw8, both RFO1 subpopulations also gave positive correlations, but only the whole BC population gave significant correlation.
Because RFO1Col-0 plays a central role in resistance, the RFO1Col-0 subpopulation was composed of many more individuals with strong resistance than the RFO1Ty-0 subpopulation was. As mentioned above, apparent linkage to RFO loci was obscured among the most resistant plants. This hidden linkage would result in a reduced correlation coefficient. Therefore, for RFO loci that conferred resistance independent of RFO1, a stronger correlation in the RFO1Ty-0 subpopulation than in the RFO1Col-0 subpopulation was anticipated; indeed, this was observed for both RFO3 and RFO5.
RFO1Col-0 in Ty-0 background:
We next examined the resistance mediated by RFO1Col-0 by itself in the susceptible Ty-0 background. Among our recombinant BC population, we selected plant 4E9 because we concluded, as explained in materials and methods, that 4E9 was heterozygous at the RFO1 locus, the Col-0 alleles of the other five RFO were absent, and the genome was homozygous Ty-0 outside of chromosome 1. Progeny of the 4E9 heterozygote RFO1Col-0/RFO1Ty-0 were infected with f. matthioli and scored for disease symptoms at 21 dpi. The genotype at the RFO1-linked marker F19K16 was ascertained for 61 of the 4E9 progeny, which were then grouped by genotype: C/C, C/T, or T/T. As depicted in Figure 5, RFO1 genotype clearly divided the population by phenotype, and resistance cosegregated with RFO1Col-0. Moreover, the resistance of homozygous RFO1Col-0 (in Figure 5C) appeared stronger than that of RFO1Col-0/RFO1Ty-0 heterozygote (in Figure 5B). Hence, the Col-0 and Ty-0 alleles of RFO1 appeared to be codominant. The RFO1Ty-0 homozygote appeared to be as susceptible as the Ty-0 ecotype (data not shown), which agreed with our conclusion that no other Col-0 alleles of RFO loci were present in plant 4E9.
Figure 5.
Cosegregation of the Col-0 allele of RFO1 with resistance. RFO1Col-0 is isolated from the other Col-0 RFO loci in the BC plant 4E9. The genotype of RFO1 can be followed using the SSLP marker F19K16, which is tightly linked to RFO1. Histograms show the distributions of disease index scores for plants with each possible RFO1 genotype: (A) Ty-0/Ty-0, (B) Col-0/Ty-0, and (C) Col-0/Col-0. Plants were infected with f. matthioli at 3 weeks of age and scored after 6 weeks.
The line 1A3, which was propagated from BC plant 4E9 and selected to be homozygous RFO1Col-0, bred true for stronger resistance to F. oxysporum than to the Ty-0 parent. The fact that RFO1-linked marker F19K16 and resistance cosegregated in the offspring of 4E9 confirmed that the resistance in 1A3 was linked to RFO1. We measured the improvement in resistance by infesting soil with a serial dilution of Fusarium bud cells. In preliminary experiments, we noted that disease severity was dependent on the density of inoculum. In Figure 2A, a lethal dose for f. matthioli-infected 1A3 plants was ∼3 × 106 bud cells ml−1, which was between 10- and 100-fold higher than the lethal dose for the Ty-0 ecotype. Surprisingly, the 1A3 line was also significantly more resistant to f. raphani. Comparable symptoms were induced by f. raphani in Figure 2A at a 10-fold higher inoculum in 1A3 as compared to the Ty-0 ecotype.
Positional cloning of RFO1:
We took advantage of the strong influence of RFO1 on resistance to identify the corresponding gene. Of the 239 plants in the BC population, 50 exhibited complete resistance (DI ≥ 4.5) at 19 dpi, and all 50 resistant BC plants were C/T at both telomere-proximal marker F3F9, which is shown in Figure 4A, and a new telomeric marker F23A5. At nga111, ∼20 cM from the end of chromosome 1, the genotype of just 40 of the 50 resistant individuals was C/T. Therefore, the strongest resistance in the BC population showed tight linkage to the end of chromosome 1. As described in materials and methods, RFO1 was further mapped to a four BAC/YAC interval between markers F9K20 and F19K16.
We surveyed The Institute for Genome Research (TIGR) Arabidopsis database (http://www.tigr.org/tdb/e2k1/ath1/) for candidate genes between markers F9K20 and F19K16. Among 108 putative gene loci within this interval, 6 were provisionally considered RFO1 candidates because each contained homology to a domain found in a known resistance gene, i.e., either an RLK or a LRR domain. We focused our attention on 4 of these candidate genes, which are located in a ∼30-kb region of AGI-sequenced BAC K21H1. The genomic region of these 4 candidate genes, including upstream and downstream intergenic sequence, was subcloned from a BAC library clone. Because the BAC libraries were constructed from Col-0 genomic DNA, we could introduce the Col-0 allele of each candidate gene into the Ty-0 background by transformation of Ty-0 with an appropriate subclone. For each of the five subclones, which we designate A–E in Figure 6A, we tested multiple T1 plants for enhanced resistance to f. matthioli. Constructs C and E, both of which included At1g79670 (gene 6; Figure 6A), consistently conferred resistance. In contrast, no significant resistance was seen in T1 plants without At1g79670, including construct D, which possessed a homologous gene, At1g79680.
In addition to At1g79670, both constructs C and E also contained an adjacent, short open reading frame At1g79660, as depicted in Figure 6A. To determine whether At1g79670 or At1g79660 or both encode RFO1, we made construct F (derived from construct C) that did not contain At1g79660. Three transgenic Ty-0 T1 plants with this shorter construct F also displayed resistance that was similar to resistance expressed by plants with constructs C and E.
From a representative T1 plant, harboring construct C, we derived a homozygous T2 plant line 5B1 (see materials and methods). In Figure 2B, we inoculated 21-day-old 5B1 and Ty-0 plants with dilutions of either f. matthioli or f. raphani or with water alone (mock). In comparison to Ty-0, 5B1 plants exhibited enhanced resistance to both formae speciales.
As shown in Figure 2, A and B, the transgenic Col-0 allele of At1g79670 in Ty-0 recapitulates the nonspecific resistance of RFO1Col-0 introduced to the Ty-0 background. As depicted in Figure 6B, At1g79670 has been annotated as WALL-ASSOCIATED KINASE-LIKE KINASE 22 (WAKL22) (Verica et al. 2003). Polymorphisms in At1g79670/WAKL22 distinguish RFO1Col-0 and RFO1Ty-0: We compared the sequence of the wild-type RFO1/WAKL22 alleles from Col-0 and Ty-0 as shown in Figure 6C. Both the 5′- and 3′-ends of RFO1 in the TAIR database correspond to a previously sequenced full-length cDNA clone (GenBank no. AY078961). The RFO1-coding sequence and the intervening introns in the Col-0 parent used in our laboratory were identical to the sequence generated by the Arabidopsis Genome Initiative (AGI) and the Ty-0 sequence is described in GenBank accession no. DQ023268. The Col-0 and Ty-0 RFO1 alleles were distinguished by 21 nucleotide polymorphisms, including two 3- bp deletions/insertions, and 10 of the polymorphisms accounted for amino acid residue substitutions in the Ty-0 allele as compared to the Col-0 allele. Almost all of the amino acid substitutions did not correspond to conserved amino acid residues among the 24 WAK/WAKL protein sequences that are annotated in the Col-0 genome. However, one polymorphism (nucleotide G1652 to C) does change a highly conserved glutamic acid, in the Col-0 RFO1 allele and most WAKs in Col-0, to a glutamine in Ty-0.
T-DNA insertions in the RFO1/At1g79670 locus:
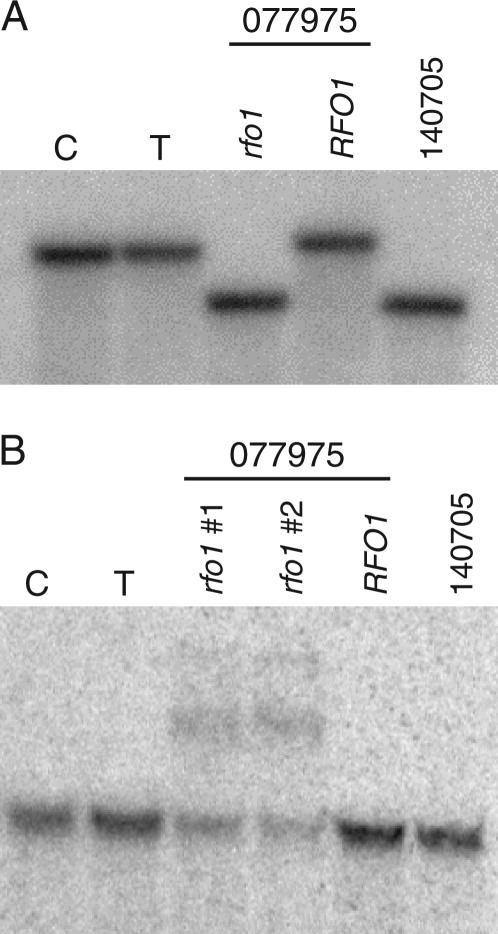
Thus far, we have defined RFO1Col-0 as a dominant allele in the Ty-0 background. However, because RFO1Col-0 and RFO1 Ty-0 appeared to be codominant in the 4E9 offspring (see above), it remained to be determined whether RFO1Col-0 was a loss-of-function allele of a susceptibility gene or a gain-of-function allele of a resistance gene. We therefore sought a Col-0 loss-of-function allele in the Salk T-DNA insertion collection. Salk insertion line 077975 contains a T-DNA that disrupts the first exon and deletes coding sequence for amino acid residues 24–54 of At1g79670 according to TAIR annotation (see materials and methods). We isolated T3 plants homozygous for the At1g79670 and for comparison a wild-type segregant from insertion line 077975. We also isolated a T3 plant from another insertion line 140705 with a T-DNA that is inserted 626 nucleotide basepairs upstream of the start codon of At1g79670 (see materials and methods). In Figure 7A, detection of a single distinct DNA restriction fragment length polymorphism (RFLP) at the At1g79670 locus in genomic DNA confirmed the isolation of a plant line homozygous for either T-DNA insertion. A wild-type sibling from line 077975 shared the same RFLP as either the Col-0 or the Ty-0 parent.
Figure 7.
Aberrant transcripts of At1g79670 in rfo1-1. (A) A Southern blot of EcoRI-digested genomic DNA was hybridized to a genomic BglII-fragment that is within the At1g79670 (RFO1) gene. From left to right, the sample lanes are C (Col-0), T (Ty-0), rfo1 (homozygote for a T-DNA insertion in salk line 077975), RFO1 (homozygote for the absence of a T-DNA insertion in salk line 077975), and 140705 (homozygote for a T-DNA insertion between At1g79670 and At1g79680). (B) A Northern blot of 5 μg total RNA per lane was hybridized to the probe that was used in the above Southern blot. Equal loading in all lanes was confirmed by ethidium bromide staining of the gel and blot (data not shown). The source and lane order of RNA samples are the same as above with the exception that we obtained RNA from two different lines (rfo1 1 and rfo1 2) of a screen for a homozygous T-DNA insertion within At1g79670 from the T3 pool of Salk line 077975.
Detection of aberrant WAKL22 mRNA:
A single message and the same expression level of RFO1/WAKL22 RNA was detected in Col-0 and Ty-0 ecotypes, as shown in Figure 7B. In addition, RFO1 expression in either the wild type or the insertion line 140705 homozygote was identical to the Col-0 parent. In contrast, three RFO1-hybridizing bands were detected in the RNA of the homozygotes isolated from insertion line 077975. The two larger messages were not apparent in either ecotype or the insertion line 140705 and may represent readthrough messages that include T-DNA insertion sequences.
The lowest-molecular-weight band was equivalent in size to the RFO1 RNA detected in Col-0 and Ty-0, but with less intensity. This residual signal was probably cross-hybridized with the RNA of a WAKL gene cluster that has significant homology to RFO1/WAKL22 (Verica et al. 2003). In support of this interpretation, we were unable to PCR amplify a ∼500-bp product across the T-DNA insertion site using genomic DNA of the insertion line 077975 homozygote. The absence of any product probably reflects inefficient extension across the T-DNA insertion. In contrast, we did obtain product from the DNA of Col-0, Ty-0, or the wild-type segregant of insertion line 077975 (data not shown).
rfo1 confers susceptibility to Fusarium:
We examined the phenotype of the insertion line 077975 homozygote (rfo1) after infection with f. matthioli. Because multiple RFO loci contributed to resistance in the Col-0 background, we anticipated that the loss of resistance in rfo1 might be modest. Indeed, although f. matthioli-infected rfo1 plants displayed enhanced susceptibility, both stunting and chlorosis, in comparison to infected wild-type Col-0, rfo1 remained significantly more resistant than the Ty-0 ecotype.
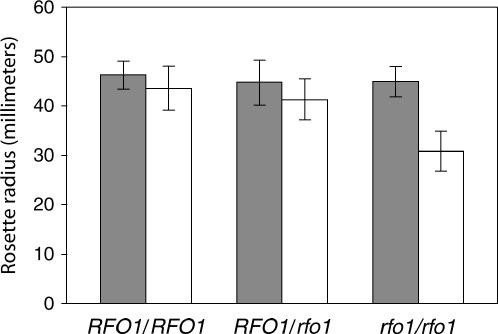
We correlated RFO1 genotype and disease symptoms to both ensure that susceptibility in the rfo1 was linked to the T-DNA insertion and determine how the rfo1 allele behaved. PCR genotyping of the original insertion line 077975 had detected the T-DNA insert and the intact RFO1 gene as codominant markers. The T4 offspring of a T3 RFO1/rfo1 heterozygote gave 22 RFO1/RFO1:40 RFO1/rfo1:30 rfo1/rfo1 (1:2:1 segregation satisfies χ2 test, P > 0.10). No disease symptoms were ever observed among 48 progeny that were mock infected. Three weeks after 44 progeny were infected with 3 × 106 f. matthioli bud cells ml−1, we measured the rosette radius of both infected and mock-infected plants. Because symptoms were modest, the rosette radius quantified the degree of stunting in Figure 8. Mock-infected wild-type (46 ± 3 mm SD) and rfo1 plants (45 ± 3 mm) were indistinguishable. However, while the rosette radius of wild-type plants (43 ± 4 mm) was slightly affected by infection, rfo1/rfo1 rosettes (31 ± 4 mm) were appreciably stunted. Importantly, the RFO1/rfo1 heterozygote (mock infected, 45 ± 5 mm, and f. matthioli infected, 41 ± 4 mm) was indistinguishable from wild type, showing that RFO1 is a dominant-resistance gene. After a fourth week, some vascular chlorosis was observed on 11 of 13 infected rfo1/rfo1, 1 of 20 RFO1/rfo1, and none of 11 RFO1/RFO1 plants. Thus, the disease symptoms and T-DNA insertion appeared to cosegregate in rfo1 plants.
Figure 8.
Susceptibility of rfo1 to f. matthioli. The peat under plants was either mock inoculated with water (solid bars) or infested with 3 × 106 f. matthioli bud cells ml−1 (open bars) when the plants were 3 weeks old. At 7 weeks, the rosette radius, the average length of the three longest rosette leaves, was measured for offspring of an RFO1/rfo1 plant. Data from each of the three possible genotypes were then combined. There were 11, 11, 19, 21, 17, and 13 plants, respectively, for RFO1/RFO1 (mock), RFO1/RFO1 (infected), RFO1/rfo1 (mock), RFO1/rfo1 (infected), rfo1/rfo1 (mock), and rfo1/rfo1 (infected). Error bars represent SD.
RFO1 mediated resistance to all three crucifer-specific races in Figure 2C. Col-0 plants, which exhibit an intermediate resistance to both f. conglutinans and f. raphani, were compromised by the T-DNA insertion to a similar extent. On the basis of the difference in disease symptoms from wild type, a ∼10-fold lower inoculum produced comparable symptoms in the rfo1 mutant.
Resistance depends on both SA-dependent and SA-independent pathways:
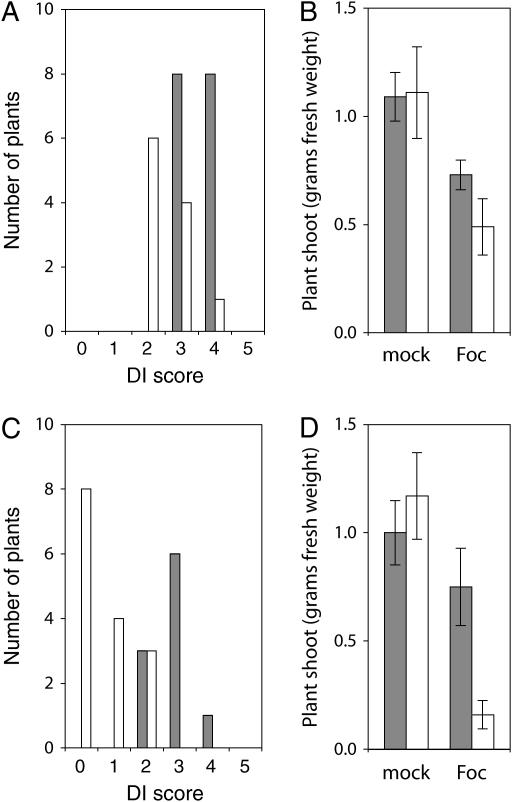
Similar to other infectious diseases that can be controlled by R genes, we found that Fusarium wilt disease was enhanced in mutants with defects in salicylic acid (SA) accumulation. For example, SA fails to accumulate in sid2 in response to virulent or avirulent Pseudomonas syringae or Erysiphe orontii infection, and sid2 has an enhanced disease susceptibility to these same pathogens (Nawrath and Metraux 1999). SID2/ISOCHORISMATE SYNTHASE 1 (ICS1) is thought to catalyze the initial step in SA biosynthesis (Wildermuth et al. 2001). Because sid2 and other mutants that we examined were in the Col-0 background, we infected mutants with f. conglutinans, to which Col-0 has an intermediate resistance. If mutants were infected with f. matthioli instead, a subtle difference in resistance might be missed. When sid2 and wild type were grown side by side in a common soil and infested with f. conglutinans, we observed a modest but significantly enhanced disease susceptibility (eds) in sid2. The median DI score in Figure 9A for wild type and sid2 were 3.5 and 2, respectively. The toll of disease on sid2 could also be seen in a significantly smaller shoot mass of sid2 plants as compared to wild type in Figure 9B. In the control experiment, shoot mass of wild type and sid2 were indistinguishable when soil was mock inoculated with water. A similar modest eds phenotype of sid2 was reproduced in two independent experiments (data not shown).
Figure 9.
Susceptibility of sid2 and pad4 to F. oxysporum f. conglutinans. Plants were grown in soil flats with wild type and mutant rows alternating. Flats were thinned and either mock inoculated with water or infested with 1 × 106 f. conglutinans bud cell ml−1 at 3 weeks after sowing. The distributions of disease index scores of 6-week-old plants are shown as solid bars for wild type and as open bars for either sid2 (A) or pad4 (C). The difference between disease symptoms of wild type and either sid2 or pad4 was determined to be significant (P < 0.05) using the Mann-Whitney U-test. At 6 weeks, the shoot mass was measured for each plant, and the mean fresh weight in grams is shown as solid bars for wild type and as open bars for either sid2 (B) or pad4 (D). Error bars represent the 95% confidence interval for the mean.
A more severe wilt disease was seen with pad4, a mutant with defects in both SA-dependent and SA-independent responses to pathogen (Zhou et al. 1998). When pad4 and wild type were grown in infested common soil, pad4 displayed a significantly more severe eds phenotype relative to wild type. In Figure 9C, while most wild-type plants had an intermediate DI score of 3, most pad4 plants were dead (a median DI score of 0). The severity of the eds phenotype was underscored by dramatic loss of shoot mass for diseased pad4 plants in Figure 9D.
In Table 2, we found that additional Arabidopsis mutants with previously defined connections to the SA-dependent response pathway exhibited an eds phenotype when infected with f. conglutinans (Glazebrook et al. 1996; Volko et al. 1998). However, no significant eds phenotype could be attributed to any of four npr1 alleles (npr1-1, npr1-2, npr1-3, or npr1-4), which is known to mediate response to SA (Dong 2004). The eds phenotype of pad4 was consistently stronger than that observed in sid2 or other mutants that we tested (data not shown).
TABLE 2.
Susceptibility of mutants to F. oxysporum f. conglutinans
| Genotype | Relative disease susceptibilitya |
|---|---|
| Wild type (Col-0) | + |
| eds3 | ++ |
| eds4 | ++ |
| eds5 | ++ |
| eds10 | ++ |
| sid2 | ++ |
| pad4 | +++ |
| npr1 | + |
| 35S∷NahG | +++ |
The relative disease susceptibility is the approximate dilution of Fusarium bud cells at which symptoms are equivalent to wild type. Wild type has an intermediate resistance (+). Mutants or transgenic line require a 3- to 10-fold lower inoculum (++), or a 10- to 100-fold lower inoculum (+++).
DISCUSSION
RFO1 is not a canonical R gene:
Our genetic data show that a difference in RFO1 alleles in wild Arabidopsis ecotypes accounts for a substantial difference in resistance to Fusarium wilt and that the phenotype of an rfo1 mutant is enhanced susceptibility to Fusarium infection. For these reasons, it is probably appropriate to classify RFO1 as an R gene rather than as a gene involved in conferring basal resistance to pathogens. However, as discussed below, how RFO1 should be categorized among R genes and whether RFO1 is a component of a canonical R-gene pathway remain unresolved. Our finding that RFO1 encodes a wall-associated receptor-like kinase is a first step in elucidating the role of RFO1 in conferring broad resistance as well as the oligogenic resistance conferred by RFO1 together with other RFO loci.
The identity of RFO1/WAKL22 is somewhat unexpected for an R protein. By far, most R proteins belong to the NBS-LRR class (Jones and Takemoto 2004). However, despite a lack of an extracellular LRR, WAKL22 does share a similar RLK structure with the previously characterized rice R proteins Xa21 and Xa26 (Song et al. 1995; Sun et al. 2004).
Additionally, resistance conferred by RFO1 alone is quantitative rather than qualitative. Typically, the hypersensitive response elicited by the interaction of an avirulent pathogen and a canonical R protein on a resistant host plant is qualitatively distinct from the disease symptoms elicited by a virulent pathogen on a susceptible host (Nimchuk et al. 2003). As shown in Figure 2C, however, resistance to F. oxysporum is quantitatively affected in plants carrying RFO1Col-0 compared to plants with only the RFO1Ty-0 or rfo1 allele. A 10-fold higher F. oxysporum dose elicited the same symptoms on plants carrying RFO1Col-0 as were elicited in RFO1Ty-0 or rfo1 plants at a lower dose.
Alhough RFO1Col-0 alone confers substantial resistance in Ty-0, stronger resistance is evident in combination with other RFOCol-0 loci, primarily with RFO2, but also with RFO4 and RFO6. On the other hand, no significant resistance is conferred by RFO2, RFO4, or RFO6 in the absence of RFO1Col-0. In most cases, NBS-LRR R genes provide resistance that is independent of the other R genes and behave as simple monogenic traits, whereas RFO1 and the three other RFO loci are interdependent.
An apparent lack of race specificity is another attribute that distinguishes RFO1 from most R genes. RFO1 confers resistance to all three crucifer-specific formae speciales of F. oxysporum. In contrast, R-mediated resistance is usually specific for a discrete subset of a bacterial or fungal species (Hammond-Kosack and Parker 2003). Also, race-specific resistance is a common host feature in Fusarium wilt diseases and the source of the only two previously identified R genes for resistance to F. oxysporum (Ori et al. 1997; Simons et al. 1998; Joobeur et al. 2004). However, at least two Arabidopsis R genes RPW8 and RPM1 are exceptional. RPW8, which also encodes a unique R protein structure, confers resistance to multiple species of powdery mildew (Xiao et al. 2001). The RPM1 protein has a typical NBS-LRR structure but recognizes two different P. syringae type III effectors, avrB and avrRpm1 (Bisgrove et al. 1994).
Although RFO1 appears to be unusual, it is possible that the common attributes that are associated with the best-studied R genes, such as a simple inheritance pattern, race specificity, qualitative resistance, and a strong hypersensitive response, are a consequence of research methodology. That is, because R genes with these attributes are simpler to clone and better subjects for study, the attributes have become canonical.
On the basis of our present understanding of plant-microbe interactions, we envisage two scenarios to explain the role of RFO1 in resistance. RFO1 may be a component of a canonical NBS-LRR pathway, or RFO1 may function in a resistance pathway similar to the Nod factor-signaling pathway that mediates legume symbiosis with nitrogen-fixing bacteria.
Role for RFO1 in the guard hypothesis:
According to the recently formulated guard hypothesis, the role of some or all canonical NBS-LRR R proteins is to guard a plant target protein, or guardee, from the virulence activity of a pathogen-encoded effector (Dangl and Jones 2001). For example, the resistance conferred by either PBS1 or Pto kinase requires the surveillance of a guardian NBS-LRR protein, RPS5 or Prf, respectively (Swiderski and Innes 2001; Pedley and Martin 2003). Similarly, RFO1 could be a guardee of a corresponding NBS-LRR-resistance protein (perhaps RFO2). Another key feature of the guard hypothesis is that the guardee typically plays a role in basal resistance that is undermined by a virulence effector. Consistent with RFO1 playing a role in basal resistance, we observed that an rfo1 mutant is also more susceptible to both f. conglutinans and f. raphani. Indeed, the loss of resistance to f. conglutinans infection in rfo1 is more modest than the loss in a pad4 mutant but more severe than the loss of resistance in a sid2 mutant (A. C. Diener, unpublished results). PAD4 and SID2 have been previously shown to be key players in Arabidopsis basal defense pathways (Zhou et al. 1998; Nawrath and Metraux 1999).
In the BC population, RFO1 and RFO2 together account for much of the complete resistance to f. matthioli in the Col-0 ecotype. However, because Col-0 is susceptible to f. conglutinans and f. raphani, the strong resistance provided by RFO1Col-0 RFO2Col-0 appears to be race specific, consistent with the possibility that RFO2 is either a guard for RFO1 or yet another guardee that requires RFO1 activity for triggering resistance. As predicted for a guard, we failed to detect resistance by RFO2 on its own in the BC RFO1Ty-0 subpopulation. That is, without its corresponding guardee target, RFO2 is incapable of triggering a strong defense response.
RFO1/RFO2 as the receptor for a pathogen-specific signal:
In plants that form nitrogen-fixing nodules in association with rhizobia, perception of the symbiont-secreted Nod factor requires a combination of two related LysM-type RLKs (Riely et al. 2004). In the case of Lotus japonicus, the RLKs NFR1 and NFR5 are both required for full response to Nod factor (Radutoiu et al. 2003). As an alternative to RFO1 being a guardee as discussed in the previous section, genetic interdependence between RFO1Col-0 and RFO2Col-0 for strong resistance to f. matthioli could be explained if RFO1 and RFO2 were coreceptors for a Fusarium-encoded pathogen-associated molecule, similar to the roles of NFR1 and NFR5 in the Lotus response to Nod factor. Coincidentally, linkage analysis carried out with the BC population roughly places the RFO2 locus in the same region of chromosome 1 where 17 of the remaining 26 WAK/WAKL genes in Arabidopsis are clustered. Thus, if RFO2 is also a WAK/WAKL RLK, it is conceivable that a general Fusarium signal might be perceived by RFO1 alone and that a f. matthioli-specific signal might be perceived by the combination of RFO1 and RFO2, resulting in a stronger resistance phenotype.
Published work supports the possibility that WAKs function as receptors. Arabidopsis WAK1 binds the small glycine-rich cell wall protein AtGRP-3 in vitro and exposure of Arabidopsis protoplasts to exogenous AtGRP-3 promotes the association of both WAK1 and AtGRP-3 into a multimeric complex as well as the phosphorylation of OXYGEN-EVOLVING ENHANCER PROTEIN 2 (OEE2) (Park et al. 2001; Yang et al. 2003). Interestingly, OEE2 also appears to be phosphorylated after infection with avirulent P. syringae.
The structure of RFO1/WAKL22:
RFO1 has been classified as WAKL22 on the basis of the backbone structure of its extracellular domain that is unique to the WAK/WAKL family (Verica and He 2002). The WAK/WAKL family has been further divided into four subgroups. WAKL22 is a member of subgroup II with a previously characterized gene cluster that includes WAKL1 through WAKL6 (Verica et al. 2003). The genomic sequences adjacent to WAKL22 and the WAKL1/WAKL6 cluster, which are on opposite arms of chromosome 1, suggest that they originate from an ancestral duplication. Curiously, although WAKL10 is adjacent to WAKL22, WAKL22 is more closely related to the WAKL1/WAKL6 cluster than to WAKL10. Among the six WAKL proteins encoded by the WAKL1/WALK6 cluster, a high diversifying selection is evident in the unique WAK/WAKL domain and especially in five so-called variable subdomains. A similar diversifying selection is often found in comparisons of the LRR domain of closely related R genes and suggests rapid coevolution with a putative ligand (Michelmore and Meyers 1998).
As the name implies, WAK/WAKL proteins appear to be tightly bound to the plant cell wall. Antibodies that have been raised against either WAK1 or WAKL6 have been shown to recognize an epitope that can be removed from intact cell walls only with harsh chemical treatment or, in the case of anti-WAK1, after pectinase digestion (Wagner and Kohorn 2001; Verica et al. 2003). The anti-WAK1 epitope can also be readily retrieved from plasma membrane preparations from protoplasts (He et al. 1996). Intriguingly, pectin epitopes remain tightly bound to WAKs, even after dissociation from the cell wall (Wagner and Kohorn 2001). Fluorescent dye-labeled anti-WAK1 or anti-WAKL6 was also shown to specifically stain the outer surface of plants cells (Lally et al. 2001; Verica et al. 2003).
Assuming that RFO1Col-0 encodes a functional protein product, the nucleotide polymorphisms that distinguish the Ty-0 and Col-0 alleles suggest that RFO1Ty-0 also expresses a functional product. Only one polymorphism in the kinase domain of RFO1Ty-0 corresponds to a conserved amino acid residue among Arabidopsis WAKs. If this polymorphism were responsible for the resistance phenotype of the RFO1Col-0 allele, a dominant negative kinase activity for RFO1Ty-0 might explain why the RFO1Col-0/RFO1Ty-0 heterozygote expressed less resistance than the RFO1Col-0 homozygote. In contrast, the RFO1Col-0/rfo1 heterozygote exhibited resistance similar to the RFO1Col-0/RFO1Col-0 homozygote.
Potential role for WAK/WAKL family as cell wall guardian:
Roles in both development and stress-response signaling have been attributed to WAK/WAKL family members. An inducible reduction in WAK protein accumulation has been shown to reduce plant cell expansion and result in stunting of roots and shoots (Lally et al. 2001; Wagner and Kohorn 2001). On the other hand, ectopic overexpression of WAK1 has been shown to increase tolerance of roots to toxic aluminum concentrations (Sivaguru et al. 2003). Also, root elongation in loss-of-function mutations of either WAKL4 or WAKL14 is reported to be more or less tolerant to certain metal ion stresses (Jackson et al. 2004). Intriguingly, the pattern of increased tolerance or sensitivity to different cation stresses is inconsistent between wakl4 and wakl14.
As for biotic stress, WAK activity has been implicated in pathogen and SA response pathways. The expression of many WAK/WAKL genes is elevated by pathogen infection, wounding, and/or SA (He et al. 1998; Verica et al. 2003). Moreover, dominant positive WAK1 activity promotes resistance to otherwise toxic levels of exogenous SA, whereas dominant negative WAK1 activity enhances the sensitivity to exogenous SA (He et al. 1998). Because increased sensitivity to SA has also been observed in npr1 mutant, which is defective in responses to SA, WAK was proposed to positively regulate NPR1 activity. It seems unlikely, however, that the rfo1 phenotype is entirely due to sensitivity to accumulated SA. A sid2 mutant, which does not synthesize pathogen-induced SA, exhibited a more modest increase in susceptibility to F. oxysporum than did rfo1 (data not shown). In addition, we were unable to detect a significant enhanced disease susceptibility phenotype in Fusarium-infected npr1 plants.
One way that WAKs/WAKLs may be involved in both abiotic and biotic stress responses is through their tight association with the cell wall and specifically pectin. This may impart, for example, a mechanical sensing of cations binding to the anionic cell wall carbohydrate. Possibly, the loosening of the cell wall structure during either normal growth or glycolytic-digestion by a pathogen might be indirectly perceived via the cell wall-anchored WAK/WAKLs. Indeed, the WAK/WAKL family has been proposed to be the most attractive candidate for providing communication between the plant cell wall and the cytoskeleton (Kohorn 2000; Baluska et al. 2003).
We have yet to discern an obvious role for RFO1 in normal development. Growth of wild- type and homozygous Col-0 rfo1 mutants, including the root system, are indistinguishable on agar plates and in soil (A. C. Diener, personal observation). However, the existence of 26 other WAK/WAKL presents the possibility that functional redundancy and signal complexity might mask the full extent of RFO1 function.
Oligogenic resistance to F. oxysporum f. matthioli:
We identified six RFO loci that together account for much of the resistance of the Col-0 background to f. matthioli. At each RFO locus, the recessive Ty-0 allele contributes to the susceptibility of the Ty-0 ecotype. However, because we measured resistance as the difference between the Col-0 and Ty-0 alleles, and either of the ecotype alleles at any locus is not necessarily a null allele, we cannot infer from our data which RFO locus makes the greatest contribution to resistance.
In the context of our BC population, RFO1 is the most critical contributor to resistance. Of the quarter most resistant BC plants, 97% inherited the RFO1Col-0 allele, and, of the quarter most susceptible BC plants, 87% were homozygous RFO1Ty-0. In addition, the resistance that was conferred by RFO2, RFO4, or RFO6 appeared solely in the presence of RFO1Col-0. Despite the importance of RFO1, the rfo1 mutant still retained considerable resistance to f. matthioli. Modest disease symptoms on rfo1 were obtained by using a high inoculum and scoring plants at later times.
From analysis of the BC population, we could detect only dominant Col-0-resistance alleles. In the subsequent F1 generation derived from an individual plant in the BC population, we showed that homozygous RFO1Col-0 plants were even more resistant than the RFO1Col-0/RFO1Ty-0 heterozygote. The Col-0 and Ty-0 alleles of RFO3 and RFO5 may also be codominant. Alternatively, there may be additional RFO loci in the Col-0 background that are recessive and therefore not detected in the BC population. This may explain the strong residual resistance of the rfo1 mutant.
Arabidopsis is a good model host for Fusarium oxysporum:
In the field, the three formae speciales that we used in our experiments are consistently isolated from distinct crucifer hosts. However, during the 1950s Armstrong and Armstrong (1952) discovered that each forma specialis could produce disease symptoms in an overlapping set of crucifers in a laboratory setting. This observation led Armstrong and Armstrong (1966) to revise the nomenclature and make the three formae speciales into races of one f. conglutinans. Subsequently, the accepted nomenclature has reverted to the original designations in recognition of the fact that the broad, overlapping host range for any of the forma specialis is not observed in the field (Bosland and Williams 1987).
Arabidopsis can be a specific host for crucifer isolates of F. oxysporum. Although Arabidopsis may not be a natural host for the crucifer races of F. oxysporum, Arabidopsis ecotypes express specific resistance to these races. Similar to the partial breakdown of host specificity observed in the laboratory by Armstrong and Armstrong, Arabidopsis still appears to be completely resistant to pathogenic isolates from more unrelated hosts, tomato and banana. These features of the Arabidopsis-Fusarium infection model make it an excellent system for studying the molecular basis of Fusarium-host interactions.
Acknowledgments
We thank Jake Begun and Julia Dewdney for comments on and editing of the manuscript. This work was supported by National Institutes of Health grant GM48707 and National Science Foundation grant 02-SC-NSF-1017. A.D. is supported by National Research Service Award fellowship 5-F32-AI10634A-02 from the National Institutes of Health/National Institute of Allergies and Infectious Disease.
References
- Anderson, M. E., and J. C. Walker, 1935. Histological studies of Wisconsin hollander and Wisconsin ballhead cabbage in relation to resistance to yellows. J. Ag. Res. 50: 823–836. [Google Scholar]
- Armstrong, G. M., and J. K. Armstrong, 1952. Physiologic races of the Fusaria causing wilts of the Cruciferae. Phytopathology 42: 255–257. [Google Scholar]
- Armstrong, G. M., and J. K. Armstrong, 1966. Races of Fusarium oxysporum f. conglutinans; race 4, new race; and a new host for race 1, Lychnis chalcedonica. Phytopathology 56: 525–530. [Google Scholar]
- Armstrong, G. M., and J. K. Armstrong, 1975. Reflections on the wilt Fusaria. Annu. Rev. Phytopathol. 13: 95–103. [Google Scholar]
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. (Editors), 1998. Current Protocols in Molecular Biology. John Wiley & Sons, New York.
- Baker, K. F., 1948. Fusarium wilt of garden stock (Mathiola incana). Phytopathology 38: 399–403. [Google Scholar]
- Baluska, F., J. Samaj, P. Wojtaszek, D. Volkmann and D. Menzel, 2003. Cytoskeleton-plasma membrane-cell wall continuum in plants. Emerging links revisited. Plant Physiol. 133: 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman, C. H., and E. M. Roberts, 1995. On the nature and genetic basis for resistance and tolerance to fungal wilt diseases of plants. Adv. Bot. Res. 21: 35–77. [Google Scholar]
- Bisgrove, S. R., M. T. Simonich, N. M. Smith, A. Sattler and R. W. Innes, 1994. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohman, S., J. Staal, B. P. Thomma, M. Wang and C. Dixelius, 2004. Characterisation of an Arabidopsis-Leptosphaeria maculans pathosystem: resistance partially requires camalexin biosynthesis and is independent of salicylic acid, ethylene and jasmonic acid signalling. Plant J. 37: 9–20. [DOI] [PubMed] [Google Scholar]
- Bohnert, H. U., I. Fudal, W. Dioh, D. Tharreau, J. L. Notteghem et al., 2004. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16: 2499–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosland, P. W., and P. H. Williams, 1987. An evaluation of fusarium-oxysporum from crucifers based on pathogenicity isozyme polymorphism vegetative compatibility and geographic origin. Can. J. Bot. 65: 2067–2073. [Google Scholar]
- Campbell, R. C., 1996. Statistics for Biologists. Cambridge University Press, Cambridge, UK.
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., W. E. Browne and D. Preuss, 1998. Assaying genome-wide recombination and centromere functions with Arabidopsis tetrads. Proc. Natl. Acad. Sci. USA 95: 247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Denby, K. J., P. Kumar and D. J. Kliebenstein, 2004. Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 38: 473–486. [DOI] [PubMed] [Google Scholar]
- Dong, X., 2004. NPR1, all things considered. Curr. Opin. Plant Biol. 7: 547–552. [DOI] [PubMed] [Google Scholar]
- Gao, H., C. H. Beckman and W. C. Mueller, 1995. a The nature of tolerance to Fusarium oxysporum f. sp. lycopersici in polygenically field-resistant marglobe tomato plants. Phys. Mol. Plant Pathol. 46: 401–412. [Google Scholar]
- Gao, H., C. H. Beckman and W. C. Mueller, 1995. b The rate of vascular colonization as a measure of the genotypic interaction between various cultivars of tomato and various formae or races of Fusarium oxysporum. Phys. Mol. Plant Pathol. 46: 29–43. [Google Scholar]
- Glazebrook, J., E. E. Rogers and F. M. Ausubel, 1996. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143: 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godiard, L., L. Sauviac, K. U. Torii, O. Grenon, B. Mangin et al., 2003. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36: 353–365. [DOI] [PubMed] [Google Scholar]
- Gordon, T. R., and R. D. Martyn, 1997. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 35: 111–128. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Z. Svab and P. Maliga, 1994. The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K. E., and J. E. Parker, 2003. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14: 177–193. [DOI] [PubMed] [Google Scholar]
- He, Z. H., M. Fujiki and B. D. Kohorn, 1996. A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 271: 19789–19793. [DOI] [PubMed] [Google Scholar]
- He, Z. H., D. He and B. D. Kohorn, 1998. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 14: 55–63. [DOI] [PubMed] [Google Scholar]
- Hemming, M. N., S. Basuki, D. J. McGrath, B. J. Carroll and D. A. Jones, 2004. Fine mapping of the tomato I-3 gene for fusarium wilt resistance and elimination of a co-segregating resistance gene analogue as a candidate for I-3. Theor. Appl. Genet. 109: 409–418. [DOI] [PubMed] [Google Scholar]
- Jackson, A., X. Hou, H. Tong, J. A. Verica, L. Chae et al., 2004. Analyses of knockout mutants for the cell wall associated receptor like kinase genes reveal their important roles in Arabidopsis heavy metal responses. International Conference on Arabidopsis Research, Berlin, Poster abstract T04–086.
- Jimenez-Gasco, M. M., J. A. Navas-Cortes and R. M. Jimenez-Diaz, 2004. The Fusarium oxysporum f. sp. ciceris/Cicer arietinum pathosystem: a case study of the evolution of plant-pathogenic fungi into races and pathotypes. Int. Microbiol. 7: 95–104. [PubMed] [Google Scholar]
- Jones, D. A., and D. Takemoto, 2004. Plant innate immunity—direct and indirect recognition of general and specific pathogen-associated molecules. Curr. Opin. Immunol. 16: 48–62. [DOI] [PubMed] [Google Scholar]
- Joobeur, T., J. J. King, S. J. Nolin, C. E. Thomas and R. A. Dean, 2004. The Fusarium wilt resistance locus Fom-2 of melon contains a single resistance gene with complex features. Plant J. 39: 283–297. [DOI] [PubMed] [Google Scholar]
- Kistler, H. C., E. A. Momol and U. Benny, 1991. Repetitive genomic sequences for determining relatedness among strains of Fusarium-Oxysporum. Phytopathology 81: 331–336. [Google Scholar]
- Kohorn, B. D., 2000. Plasma membrane-cell wall contacts. Plant Physiol. 124: 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., C. Alonso-Blanco and D. Vreugdenhil, 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 141–172. [DOI] [PubMed] [Google Scholar]
- Lally, D., P. Ingmire, H. Y. Tong and Z. H. He, 2001. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell 13: 1317–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., C. S. Gillmor and W. R. Scheible, 2000. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattanovich, D., F. Ruker, A. C. Machado, M. Laimer, F. Regner et al., 1989. Efficient transformation of Agrobacterium spp. by electroporation. Nucleic Acids Res. 17: 6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R. W., and B. C. Meyers, 1998. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8: 1113–1130. [DOI] [PubMed] [Google Scholar]
- Nawrath, C., and J. P. Metraux, 1999. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk, Z., T. Eulgem, B. F. Holt, 3rd and J. L. Dangl, 2003. Recognition and response in the plant immune system. Annu. Rev. Genet. 37: 579–609. [DOI] [PubMed] [Google Scholar]
- Ori, N., Y. Eshed, I. Paran, G. Presting, D. Aviv et al., 1997. The I2C family from the wilt disease resistance locus I2 belongs to the nucleotide binding, leucine-rich repeat superfamily of plant resistance genes. Plant Cell 9: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, A. R., S. K. Cho, U. J. Yun, M. Y. Jin, S. H. Lee et al., 2001. Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 276: 26688–26693. [DOI] [PubMed] [Google Scholar]
- Pedley, K. F., and G. B. Martin, 2003. Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu. Rev. Phytopathol. 41: 215–243. [DOI] [PubMed] [Google Scholar]
- Radutoiu, S., L. H. Madsen, E. B. Madsen, H. H. Felle, Y. Umehara et al., 2003. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Riely, B. K., J. M. Ane, R. V. Penmetsa and D. R. Cook, 2004. Genetic and genomic analysis in model legumes bring Nod-factor signaling to center stage. Curr. Opin. Plant Biol. 7: 408–413. [DOI] [PubMed] [Google Scholar]
- Scheer, J. M., and C. A. Ryan, Jr., 2002. The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99: 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela-Buurlage, M. B., O. Budai-Hadrian, Q. Pan, L. Carmel-Goren, R. Vunsch et al., 2001. Genome-wide dissection of Fusarium resistance in tomato reveals multiple complex loci. Mol. Genet. Genomics 265: 1104–1111. [DOI] [PubMed] [Google Scholar]
- Sherbakoff, C. D., 1949. Breeding for resistance to Fusarium and Verticillium wilts. Bot. Rev. 15: 377–422. [Google Scholar]
- Simons, G., J. Groenendijk, J. Wijbrandi, M. Reijans, J. Groenen et al., 1998. Dissection of the fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 10: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru, M., B. Ezaki, Z. H. He, H. Tong, H. Osawa et al., 2003. Aluminum-induced gene expression and protein localization of a cell wall-associated receptor kinase in Arabidopsis. Plant Physiol. 132: 2256–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R., and J. C. Walker, 1930. A cytological study of cabbage plants in strains susceptible or resistant to yellows. J.Agric. Res. 41: 17–35. [Google Scholar]
- Song, W. Y., G. L. Wang, L. L. Chen, H. S. Kim, L. Y. Pi et al., 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sun, X., Y. Cao, Z. Yang, C. Xu, X. Li et al., 2004. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37: 517–527. [DOI] [PubMed] [Google Scholar]
- Swiderski, M. R., and R. W. Innes, 2001. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26: 101–112. [DOI] [PubMed] [Google Scholar]
- Talboys, P. W., 1972. Resistance to vascular wilt fungi. Proc.R. Soc. Lond. Ser B 18: 319–332. [Google Scholar]
- Torii, K. U., 2004. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. Int. Rev. Cytol. 234: 1–46. [DOI] [PubMed] [Google Scholar]
- Verica, J. A., L. Chae, H. Tong, P. Ingmire and Z. H. He, 2003. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis. Plant Physiol. 133: 1732–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verica, J. A., and Z. H. He, 2002. The cell wall-associated kinase (WAK) and WAK-like kinase gene family. Plant Physiol. 129: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volko, S. M., T. Boller and F. M. Ausubel, 1998. Isolation of new Arabidopsis mutants with enhanced disease susceptibility to Pseudomonas syringae by direct screening. Genetics 149: 537–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, T. A., and B. D. Kohorn, 2001. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13: 303–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth, M. C., J. Dewdney, G. Wu and F. M. Ausubel, 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Wilson, I. W., C. L. Schiff, D. E. Hughes and S. C. Somerville, 2001. Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession kashmir-1. Genetics 158: 1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S., S. Ellwood, O. Calis, E. Patrick, T. Li et al., 2001. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120. [DOI] [PubMed] [Google Scholar]
- Yang, E. J., Y. A. Oh, E. S. Lee, A. R. Park, S. K. Cho et al., 2003. Oxygen-evolving enhancer protein 2 is phosphorylated by glycine-rich protein 3/wall-associated kinase 1 in Arabidopsis. Biochem. Biophys. Res. Commun. 305: 862–868. [DOI] [PubMed] [Google Scholar]
- Zhou, N., T. L. Tootle, F. Tsui, D. F. Klessig and J. Glazebrook, 1998. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]