Abstract
Certain configurations of maize Ac/Ds transposon termini can undergo alternative transposition reactions leading to chromosome breakage and various types of stable chromosome rearrangements. Here, we show that a particular allele of the maize p1 gene containing an intact Ac element and a nearby terminally deleted Ac element (fAc) can undergo sister-chromatid transposition (SCT) reactions that generate large flanking deletions. Among 35 deletions characterized, all begin at the Ac termini in the p1 gene and extend to various flanking sites proximal to p1. The deletions range in size from the smallest of 12,567 bp to the largest of >4.6 cM; >80% of the deletions removed the p2 gene, a paralog of p1 located ∼60 kb from p1 in the p1-vv allele and its derivatives. Sequencing of representative cases shows that the deletions have precise junctions between the transposon termini and the flanking genomic sequences. These results show that SCT events can efficiently generate interstitial deletions that are useful for in vivo dissection of local genome regions and for the rapid correlation of genetic and physical maps. Finally, we discuss evidence suggesting that deletions induced by alternative transposition reactions can occur at other genomic loci, indicating that this mechanism may have had a significant impact on genome evolution.
DELETIONS have long been recognized as very efficient tools for genetic mapping. One of the best examples of the use of deletions for genetic fine-structure analysis is Benzer's classic work on the phage T4 rII gene. In this study, ∼2400 rII mutants were first crossed with seven overlapping deletions that span the rII region. On the basis of their ability to generate functional recombinants, all the mutants were easily and unambiguously localized to one of the seven major deletion intervals. Further crosses of the mutants with smaller deletions in each of the major deletion intervals yielded more precise map data. By this approach, the relative order and position of the ∼2400 mutants were determined using 25,000 crosses (Benzer 1961, 1962), whereas >2 million crosses would have been required to obtain the same results using a two- or three-factor method. Similarly, deletions have been successfully used for the physical mapping of part of the Drosophila X chromosome (Snyder et al. 1985) and for localization of the lettuce dm3 mutation (Meyers et al. 1998).
In addition to genetic mapping, deletions are also useful for mutation screening in diploid organisms due to their pseudodominance phenotype. If a deletion heterozygote is used as starting material to perform mutagenesis, any nonlethal recessive mutation located within the deleted region can be detected in the M0 generation; otherwise recessive mutations can be detected only in the following M1 generation when they become homozygous (Klug and Cummings 1991).
The most widely used treatment to induce deletions is gamma irradiation (Anderson et al. 1996; Cecchini et al. 1998). However, high-energy irradiation can also induce other undesirable chromosome rearrangements and point mutations that can complicate the recovery and analysis of deletion mutants. In maize, the r-X1 allele can induce terminal deletions, but the viability of large terminal deletions is poor (Birchler and Levin 1991; Lin et al. 1997). For target genes that are not near telomeres, in most cases it will not be possible to recover a viable deletion large enough to include the gene.
Recently, the cre/lox site-specific recombination system was used to generate deletions of up to 3–4 cM in mice (Ramirez-Solis et al. 1995; Li et al. 1996; Wagner et al. 1997; Zeh et al. 1998). The cre/lox system has also been applied to plant species such as tobacco and Arabidopsis to generate deletions, inversions, and reciprocal translocations (Dale and Ow 1990; Bayley et al. 1992; Russell et al. 1992; Odell et al. 1994; Medberry et al. 1995; Osborne et al. 1995). Deletions have been generated in plants using cre/lox and the Ac/Ds transposable element system as follows: Plants were transformed with a construct containing two lox sites—one lox site within a Ds element and a second lox site within the transgene, but outside Ds. In the presence of Ac-encoded transposase, the Ds element in the construct can transpose to a new site in the genome. Subsequent expression of cre recombinase can induce recombination between the lox sites in the original transgene locus and the transposed Ds element. If the transposed Ds element is on the same chromosome as the original transgene insertion, cre-induced recombination of the lox sites will generate either a deletion or an inversion, depending upon the relative orientation of the lox sites. If Ds transposed to another chromosome, cre-induced recombination will produce a reciprocal translocation. Using this system, researchers have produced a number of deletions in tobacco and Arabidopsis (Medberry et al. 1995; S. Zhang et al. 2003).
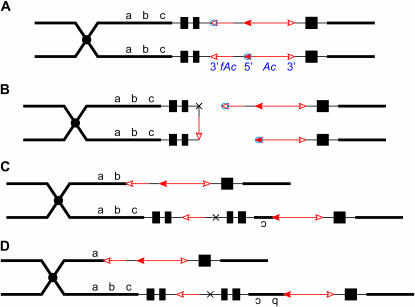
Previously we have shown that large deletions and inverted duplications could be generated in maize via transposition reactions involving Ac/Ds termini located on sister chromatids [sister-chromatid transposition (SCT), previously termed nonlinear transposition; Zhang and Peterson 1999]. The maize p1-vv9D9A allele carries an intact Ac element and a terminally deleted Ac element with their 5′ and 3′ termini in direct orientation. In the SCT model, Ac transposase can excise the 3′ fAc terminus and the 5′ Ac terminus on the two sister chromatids. The two chromatid ends at the site of Ac/fAc excision are ligated together as in a standard Ac transposition reaction, forming a covalent linkage between the sister chromatids (chromatid bridge). Reinsertion of the excised transposon ends into the chromatid bridge generates structurally altered sister chromatids containing a reciprocal deletion and inverted duplication (Zhang and Peterson 1999) (Figure 1; see also an animated version in supplemental material at http://www.genetics.org/supplemental/). In this article, we describe the isolation and molecular characterization of 35 interstitial deletions derived from the p1-vv9D9A allele. As predicted by the SCT model, all the deletions start at the Ac/fAc insertion site in p1-vv9D9A and end at various sites in the region proximal to the p1 locus. These results provide further support for the SCT mechanism and demonstrate the utility of SCT for the production of deletions in plants.
Figure 1.
Model for formation of deletions by sister-chromatid transposition. (For animated version, see supplemental material at http://www.genetics.org/supplemental/.) The diagram pertains to the structure of the p1-vv9D9A allele, which is the progenitor of the p1-ww deletion alleles described in the text. The two lines indicate sister chromatids joined at the centromere, which is indicated by a solid circle. The solid black boxes indicate the three exons of the p1 gene; the 5′-end of the p1 gene is closer to the centromere (Zhang and Peterson 1999). The red arrows indicate the Ac or fAc elements inserted into the second intron of the p1 gene, and the open and solid arrowheads indicate the 3′- and 5′-ends, respectively, of Ac/fAc. The short black line between Ac and fAc indicates a 112-bp rearranged p1 sequence (rP) present in the p1-vv9D9A allele (not to scale). (A) Following DNA replication, identical sister chromatids are joined at the centromere. Ac transposase (small circles) binds to the 5′ terminus of Ac in one chromatid and to the 3′ terminus of fAc in the sister chromatid. (B) Cuts are made at the Ac and fAc termini to excise the transposon ends. The two nontransposon ends join together at the site marked by the black X to form a chromatid bridge. (C) Reinsertion of the excised transposon ends into the chromatid bridge between b and c generates two reciprocal chromatids; one carries a deletion of c and the other carries an inverted duplication of c. (D) Same as for C, except that reinsertion between a and b generates one chromatid with a deletion of b c and one with an inverted duplication of b c. For simplicity, the model depicts fully replicated sister chromatids at the time of transposition. In reality, transposition may occur when the chromosomes are partially replicated.
MATERIALS AND METHODS
Mutation screening:
The maize p1 gene controls red phlobaphene pigmentation of husks and floral organs, including kernel pericarp and cob glumes. The p1-vv9D9A allele confers variegated pericarp and cob (Zhang and Peterson 1999), and the P1-wr allele confers colorless pericarp and red cob (Anderson 1924). The r-m3∷Ds allele contains a Ds element inserted in the r1 gene required for kernel aleurone pigmentation; Ac-induced excision of Ds from r-m3∷Ds results in purple aleurone sectors (Kermicle 1980). SCT reactions involving the p1-vv9D9A allele are predicted to result in deletions extending from the Ac/fAc insertions in p1 intron 2 toward the 5′-end of the p1 gene. Deletions that extend into and beyond exons 1 and 2 would remove the Myb-homologous DNA-binding domain and thus should abolish p1 function, leading to a p1-ww phenotype (colorless pericarp and cob). Therefore, we screened ears from plants of genotype p1-vv9D9A/P1-wr pollinated with P1-wr, r-m3∷Ds for multiple-kernel sectors of colorless pericarp or whole colorless-pericarp ears. From a total of 4000 ears produced on plants grown in two generations, we obtained 45 ears with large multiple-kernel colorless pericarp sectors, 54 ears with completely colorless pericarp, and 1 ear with a large twinned colorless pericarp sector, described in Zhang and Peterson (1999). From the colorless pericarp sectors, we selected purple-spotted kernels for progeny analysis as these were predicted to contain an Ac element linked with the desired deletion alleles. Plants grown from these kernels were self-pollinated to homozygose the new p1-ww alleles. In the following generation, plants were screened for the presence of colorless tassel glume margins to distinguish homozygous p1-ww plants from sibling plants heterozygous or homozygous for the P1-wr allele, as described previously (Athma and Peterson 1991). In addition, putative mutant plants were also screened for the occurrence of browning at the cut ends of silks (Levings and Stuber 1971), an indicator of the presence of maysin, a C-glycosyl flavone whose synthesis is coregulated by the p1 and p2 genes (Byrne et al. 1996; P. Zhang et al. 2003; Szalma et al. 2005). New mutant alleles with colorless pericarp and cob were designated p1-ww, followed by a numerical indicator of culture number, according to standard nomenclature. The alleles p1-ww1 and p1-ww2 described here were formerly named p1-ww-def1 (Zhang and Peterson 1999) and p1-del2 (P. Zhang et al. 2003).
Genomic DNA extractions and Southern blot hybridization:
Total genomic DNA was prepared using a modified CTAB extraction protocol (Saghai-Maroof et al. 1984). Agarose gel electrophoresis and Southern hybridizations were performed as described (Sambrook et al. 1989), except that hybridization buffers contained 250 mm NaHPO4, pH 7.2, 7% SDS, and wash buffers contained 20 mm NaHPO4, pH 7.2, 1% SDS. The RFLP probes csu814, npi286, csu392, and asg69 were provided by T. Muskett and M. McMullen, University of Missouri, Columbia, Missouri. Hybridization signals were quantified using ImageQuant 5.0.
PCR amplifications:
PCR amplifications were performed using the following oligonucleotide primers: p1-1, ATCCATCGCCCAACCCCAACC; p1-2, TGAACACTAAATACTCAATCGTGGCAT; p1-3, ACGCGCGACCAGCTGCTAACCGTG; p1-4, GAATTCCGCCCGAAGGTAGTTGATCC; p1-5, CTGGCGAGCTATCAAACAGGACA; Ac6, ATTTTACCGACCGTTACCGACC; Ac7, ATCTTCCACTCCTCGGCTTTAG; and p1-8, GACCGTGACCTGTCCGCTC. Reactions were heated at 94° for 3 min, cycled 35 times at 94° for 20 sec, 60° for 30 sec, and 72° for 1 min per 1-kb length of expected PCR product, and finally extended at 72° for 8 min.
Pulsed-field gel electrophoresis:
Characterization of high-molecular-weight maize genomic DNA was done following the protocols described by (Kaszas and Birchler 1996, 1998). Pulsed-field gel electrophoresis (PFGE) was conducted on a CHEF-DRII apparatus (Bio-Rad, Hercules, CA), and membranes were hybridized as described above.
RESULTS AND DISCUSSION
Identification of SCT-generated deletion mutants:
Upon sister-chromatid transposition involving the Ac and fAc elements in p1-vv9D9A, the excised Ac/fAc ends could, theoretically, reinsert anywhere in the maize genome. Insertions into sites distal to p1 on chromosome 1s would be predicted to generate acentric molecules and large, nontransmissable, terminal deficiencies. However, insertions into sites proximal to p1 would restore centromere linkage and generate sister chromatids containing reciprocal deletion and inverted duplication products as previously shown (Zhang and Peterson 1999) (Figure 1). If the Ac/fAc reinsertion site is in or upstream of exon 2 of p1, the p1 gene function would be destroyed due to loss of exon 2, which encodes a part of the R2R3-Myb DNA-binding domain (Grotewold et al. 1991). The resulting deletion mutants are expected to have a p1-ww phenotype (colorless pericarp and cob), which in large multiple-kernel sectors or whole ears is easily distinguishable from the variegated pericarp and cob phenotype of the p1-vv9D9A progenitor allele. However, not all p1-ww alleles derived from p1-vv9D9A are expected to contain SCT-generated deletions; at least three other types of structural changes in the p1-vv9D9A allele could also destroy p1 function. First, the inverted duplication alleles (p1-ww-id) generated via SCT as the reciprocal product of deletions also have disrupted the p1 gene and thus have a p1-ww phenotype (Zhang and Peterson 1999). Second, p1-ww alleles can arise from Ac transposition-induced recombination between two 5.2-kb direct repeat sequences that flank the p1 gene, leading to the loss of the entire p1 coding region (Athma and Peterson 1991; Xiao et al. 2000). Third, p1-ww alleles might arise due to intragenic transposition of Ac, if the insertion and associated 8-bp target site duplication (TSD) occurred at an essential sequence of the p1 gene (Athma et al. 1992; Moreno et al. 1992).
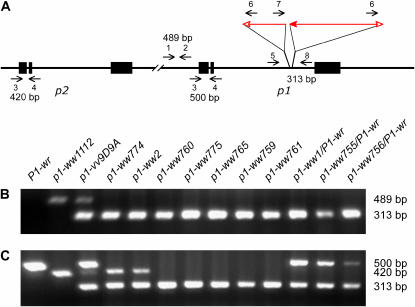
From 4000 ears produced by plants of genotype p1-vv9D9A/P1-wr, we selected 100 ears with completely colorless pericarp or with large multiple-kernel colorless pericarp sectors (see materials and methods). The SCT model (Figure 1) predicts that SCT-induced deletions should lose sequences upstream of the fAc insertion site in p1-vv9D9A, while retaining the p1 gene sequence downstream of the Ac sequence. In contrast, null alleles generated by recombination of the flanking repeats would lack the entire p1 gene and Ac element (Athma and Peterson 1991), while alleles carrying inverted duplications or new insertions of Ac would retain both upstream and downstream p1 sequences. To distinguish SCT-generated deletions from these other classes of mutations, a multiplex PCR assay was performed to test for the presence of sequences upstream and downstream of the fAc/Ac insertions in the p1 gene (Figure 2). Primers p1-1 + p1-2 were used to detect losses of the p1 5′ region, while primers Ac-6 + p1-8 were used to detect retention of the junction of Ac and the p1 3′ sequence. Moreover, the Ac-6 + p1-8 primer pair can serve as positive internal controls for this multiplex PCR assay. Among 100 p1-ww mutants screened in this assay, we identified 35 cases that exhibit a loss of the 5′ p1 gene sequences, but retain the 3′ p1 sequence as expected for SCT-generated deletions (Figure 2B).
Figure 2.
Detection of deletions by PCR analysis. (A) Structure of the p1-vv9D9A haplotype, including p1 (right) and its paralog p2 (left). Symbols have the same meaning as in Figure 1. Short horizontal arrows indicate the orientations and approximate positions of the primers used in PCR analysis. (B) Screening for deletions of sequences 5′ of the p1 gene using primer pair p1-1 + p1-2, which gives a 489-bp product in p1-vv9D9A. The primer pair Ac-6 + p1-8 detects a 313-bp band from the junction of the 3′-end of Ac with the 3′ sequence of p1 intron 2. PCR was performed using genomic DNA from plants of the genotypes indicated above each lane. The lane marked P1-wr contains DNA from the W22 inbred. The P1-wr allele has been previously shown to contain a tandem array of p1 genes (Chopra et al. 1998), whereas no p2 gene was detected in 16 diverse maize inbred lines containing P1-wr (Szalma et al. 2005). The negative result in the P1-wr lane would suggest that P1-wr alleles also lack (or are polymorphic for) the sequence upstream of p1 in p1-vv. The p1-ww1112 allele contains a deletion of p1 (Athma and Peterson 1991) and retains the p2 gene (Zhang et al. 2000). (C) Screening for deletions of the 5′-end of the p2 gene using primer pair p1-3 + p1-4, which gives a 420-bp product from the p2 gene and a 500-bp product from the p1 gene. As in B, primer pair Ac-6 + p1-8 detects a 313-bp band derived from the junction of the 3′-end of Ac with the 3′ sequence of p1 intron 2.
Identification of deletions extending to the p2 gene and beyond:
The p1 gene is linked with a second, highly similar gene termed p2; the p1 and p2 genes were proposed to have been generated by a segmental duplication followed by retroelement insertions to separate the two paralogs (Zhang et al. 2000). If the p2 gene is located 5′ of p1, then some of the SCT-generated deletions would be expected to have deletions that include p2. Consistent with this hypothesis, a p1-ww allele derived from p1-vv9D9A (p1-ww2) was previously characterized and found to have a deletion extending from p1 to the p2 gene (P. Zhang et al. 2003). To determine the frequency at which SCT deletions remove the p2 gene, we used a second PCR assay with primers p1-3 + p1-4. This primer pair amplifies the 5′ region of both p1 and p2; due to sequence polymorphisms, the products derived from p2 and the p1 alleles used in this cross differ in size. (Primers Ac-6 + p1-8 again serve the same role as in the first PCR assay.) Among 35 p1-ww deletion alleles tested, 6 alleles retain the 420-bp p2 fragment whereas the other 29 p1-ww alleles lack this product (Figure 2C), suggesting that p2 was deleted in these 29 cases. Among the 29 alleles with deletions of p2, 7 alleles are homozygous lethal. These 7 cases were maintained as heterozygotes with the P1-wr allele, which produces a 500-bp product in the PCR assay (Figure 2C).
The region in the vicinity of p1 has been identified as a major QTL for the control of levels of silk maysin, a C-glycosyl flavone that deters feeding by corn earworm (Byrne et al. 1996; Lee et al. 1998; McMullen et al. 1998). Maysin accumulation is correlated with a phenotype termed silk browning, in which the cut ends of silks turn brown due to the oxidation of flavones (Byrne et al. 1996; Lee et al. 1998; McMullen et al. 1998; Guo et al. 2001; Rector et al. 2003). Both p1 and p2 genes are expressed in maize silk (Zhang et al. 2000), and both encode highly similar R2R3-Myb proteins with a similar potential to activate flavonoid biosynthesis in transgenic cell lines (P. Zhang et al. 2003). Previous studies have shown that a stock that contains both p1 and p2 genes has high maysin levels and strong silk browning. In contrast, a previously characterized deletion allele (p1-ww774), which has a deletion of p1 but retains p2, conditions light-browning silks and low, but significant, maysin levels (P. Zhang et al. 2003). To further test the role of the p1 and p2 genes in the control of silk maysin and silk browning, we examined the silk browning phenotype of 28 deletion lines that are homozygous viable (the remaining 7 deletions were not informative because they were maintained as heterozygotes with the P1-wr allele that specifies silk browning). Among the 22 homozygous-viable deletion lines that lack the 5′-ends of both the p1 and p2 genes, all had nonbrowning silks, whereas among the 6 deletion lines that lack the 5′-end of p1 but retain the 5′-end of p2, 5 exhibited light-browning silks, and 1 line (p1-ww2) exhibited nonbrowning silks. Interestingly, this latter line contains a deletion into the 3′-end of the p2 gene (see below). Although our data do not rule out the possibility of an additional factor involved in maysin biosynthesis located between p1 and p2, the simplest interpretation of our results is that the p2 gene is sufficient to confer weak maysin levels, while p1 and p2 together produce higher maysin levels and stronger silk browning. These results further support the hypothesis that the p1 and p2 genes are essential coregulators of maysin biosynthesis (Byrne et al. 1996; P. Zhang et al. 2003; Szalma et al. 2005).
Sequences of deletion endpoints contain precise junctions with Ac/fAc termini:
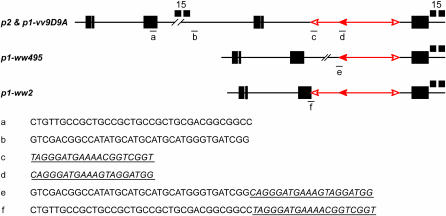
The SCT model predicts that deletion endpoints are determined by transposase-mediated insertion of Ac or fAc termini into flanking genomic sequences. If the SCT reaction is mechanistically similar to standard transposition, then the deletion endpoints should contain precise junctions of the Ac or fAc termini and the flanking genomic sequences with no loss of sequences at either the transposon termini or the genomic sequence. An 8-bp TSD is predicted to occur at the insertion site, with one copy present at the deletion endpoint and the second copy present in the inverted duplication structure formed as a reciprocal product of the SCT reaction (Figure 1). We previously demonstrated that an 8-bp TSD was present in both the deletion and the inverted duplication alleles generated from a single SCT event, indicating no loss of sequences at the genomic insertion site (Zhang and Peterson 1999). To further investigate the structures of deletion endpoints, we sequenced the junctions of the Ac/fAc termini with the genomic DNA in two additional cases. The first case (p1-ww495) was identified in the course of DNA gel-blot hybridizations with p1 locus probes that were performed on a subset of deletions to check the results of the PCR assays presented in Figure 2. These results (not shown) suggested that p1-ww495 had an endpoint upstream of p1 within a region that was previously cloned and sequenced (AF209212). PCR using primers from the Ac and the p1 genomic sequences flanking the estimated insertion site were used to amplify the Ac/p1 junction. Sequencing of the PCR product indicated that the deletion junction occurred exactly at the 5′-end of Ac at a genomic site 12,567 bp upstream of the 5′-end of Ac (Figure 3). The second case (p1-ww2) was previously shown by DNA gel-blot analysis to have an endpoint in the 3′ region of the p2 gene (P. Zhang et al. 2003). This result is consistent with PCR analysis showing that p1-ww2 lost the 5′ region of p1 but retained the 5′ portion of p2 (Figure 2). Further PCR and sequencing analysis indicated that the endpoint of p1-ww2 is in exon 3 of the p2 gene at a site 63 bp upstream of the p2 translation stop codon (Figure 3; 9588 in AF210616) (P. Zhang et al. 2003). In this case, the p1-ww2 deletion endpoint occurs exactly at the fAc 3′-end. These results demonstrate that, at least in these three cases, the deletion endpoints occurred precisely at the Ac or the fAc terminus as predicted by the SCT model. It is possible that imprecise junctions exist among the 32 other deletions derived from p1-vv9D9A; however, the three junctions sequenced to date support the hypothesis that the SCT-induced deletion endpoints occur precisely at the site of insertion of the Ac/fAc termini.
Figure 3.
Nucleotide sequences at endpoints of p1-ww495 and p1-ww2 deletion alleles. The top three lines show the structures of the indicated alleles and the locations of the sequences given below. Sequences a–d are from the progenitor allele p1-vv9D9A. Sequences e and f are from the derivative alleles p1-ww495 and p1-ww2, respectively. Sequences in italics and underlined represent Ac or fAc sequences. Note that the deletion endpoint in p1-ww495 is joined to the Ac 5′-end, while the deletion endpoint of p1-ww2 is joined to the 3′ fAc end. Small black boxes indicate the locations of sequences that hybridize with p1 genomic fragment 15. Other symbols have the same meaning as in Figure 1.
In both maize and Arabidopsis, transposition of simple Ac/Ds elements can generate sequence changes (commonly termed footprints), including small deletions at the site of transposon excision (Rinehart et al. 1997). Evidence indicates that these sequence changes are the result of cellular functions acting to repair the site of transposon excision (Yu et al. 2004). Deletions associated with excision of a simple transposon are usually relatively small (i.e., <50 bp), although a deletion >700 bp associated with excision of a single Ac element has been reported (Dooner et al. 1988). Some large deletions have been found in maize following excision of compound elements composed of Ac/Ds termini flanking genomic sequences (Ralston et al. 1989; Dowe et al. 1990). More recently, Page et al. (2004) identified several large deletions (≥100 kb) in Arabidopsis that were apparently generated during Ac-induced excision of a simple Ds element. The authors suggest that these large deletions were formed through a two-step process in which normal transposition of Ds is followed immediately by intrachromosomal excision of a hybrid Ds element. However, the deletion endpoints reported by Page et al. (2004) do not end precisely at the Ds termini, suggesting that the formation of these deletions probably involved other cellular functions in addition to the Ac transposase. Transposition of simple Ac/Ds elements is not known to induce large deletions in maize, and their occurrence in Arabidopsis may reflect a loss of normal transposition controls in the nonnative host.
Use of deletions to determine the physical distance between p1 and p2:
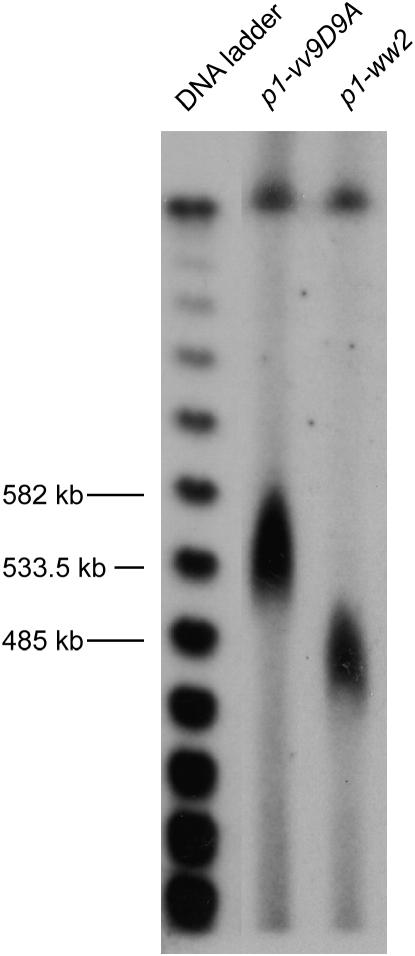
The fact that the p2 gene is lost in 29 of 35 SCT-generated deletion alleles suggests that p2 is tightly linked with p1. We used CHEF gel analysis to determine the physical distance between p1 and p2 by comparing the progenitor allele p1-vv9D9A with the deletion allele p1-ww2 in which the deletion endpoint lies within the p2 gene exon 3. Agarose blocks containing protoplasted cells from plants homozygous for p1-vv9D9A or p1-ww2 were digested with NotI and subjected to CHEF gel electrophoresis. The DNA fragments were transferred to membranes and hybridized with p1 fragment 15, which detects a sequence located both 5′ and 3′ of the p1 gene (Figure 3). Because the SCT-induced deletions retain the p1 3′ sequence, fragment 15 can be used to detect the NotI fragments containing this sequence in both alleles. The size difference of the signals from the two alleles is ∼70 kb (Figure 4). After accounting for the p1 5′ sequences and the p2 3′ sequences that are deleted in p1-del2, we estimate that the intergenic distance between the p1 and p2 genes is ∼60 kb.
Figure 4.
Determination of the physical distance between p1 and p2 by CHEF gel analysis. Cells from plants of the indicated genotypes were protoplasted, embedded in agarose, and digested with NotI endonuclease. DNAs were separated by CHEF gel electrophoresis, transferred to membrane, and hybridized with genomic probe fragment 15 from the p1 gene (Figure 3). Left lane contains λDNA concatemers as size standards.
We previously reported that the p1 gene is oriented with its 5′-end toward the centromere (Zhang and Peterson 1999). The p1-ww2 allele has a deletion of the 5′ portion of the p1 gene, the 3′ portion of the p2 gene, and the intervening sequences. Assuming that no other rearrangements occurred during the formation of p1-ww2, we can infer that the p2 gene has the same orientation as that of the p1 gene and that it is located between p1 and the centromere in the following arrangement: 3′-p1-5′, 3′-p2-5′, centromere. This conclusion is consistent with previous results showing that the p1 and p2 genes are derived from a segmental duplication, followed by retroelement insertions to separate the p1 and p2 genes (Zhang et al. 2000).
Interval mapping of the SCT-generated deletions:
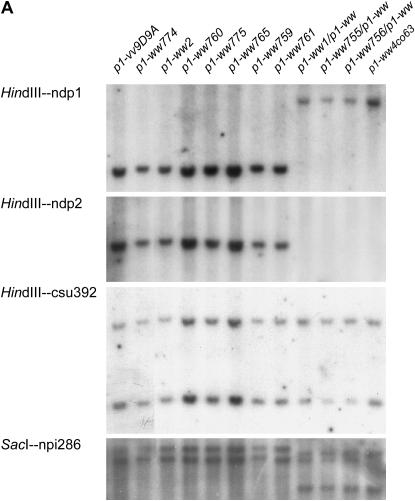
To determine the relative sizes of the other deletions, six p1-linked probes (ndp1, ndp2, csu814, npi286, csu392, asg69) were used for genomic DNA gel-blot analysis of 10 representative deletions (including p1-ww2). We previously described genomic fragments ndp1 (formerly p1.5B22) and ndp2 (formerly pJZPX): ndp1 was isolated from the endpoint of inverted duplication allele p1-ww12:27-3, which was derived from p1-vv9D9A. ndp2 was isolated from a second inverted duplication allele, p1-ww-id1, also derived from p1-vv9D9A. ndp1 and ndp2 were mapped at 3.5 and 4.6 cM proximal to p1, respectively (Zhang and Peterson 1999). For Southern analysis, genomic DNA was digested with HindIII and hybridized with the ndp1 and ndp2 probes. Several alleles are homozygous inviable (see below), and these were tested as heterozygotes with a p1-ww allele from inbred line 4Co63. For ndp1, three alleles (p1-ww1, p1-ww755, and p1-ww756) contain a band of the same size as that in the p1-ww [4Co63] parent, but they lack the band corresponding to the DNA fragment from the chromosome carrying the p1-vv9D9A allele. The same three alleles also lack a band hybridizing with ndp2. We conclude that these alleles are deleted for the loci represented by the ndp1 and ndp2 probes (Figure 5A).
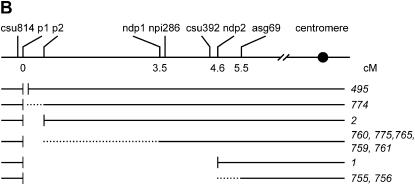
Figure 5.
Genomic DNA gel-blot analysis of the SCT-induced deletion alleles using probes linked to the p1 gene. (A) Genomic DNA of the genotypes indicated above each lane was digested with HindIII or SacI and hybridized with the indicated probes. See text for details. (B) Summary of endpoint mapping data of the SCT-generated p1-ww deletions. Schematic at top shows the positions of probe fragments (the solid boxes) and genetic distances in centimorgans, where known. Lines below show the extent of deletion found in the alleles indicated by the numbers to the right. Short vertical lines indicate deletion endpoints defined by cloned sequences. Dotted lines indicate the intervals into which those deletion(s) map. The relative order of ndp1 and npi286 cannot be determined on the basis of hybridization data reported here; the positions shown are based on prior recombination data showing a genetic distance from p1 of 3.5 and 3.6 cM for Ndp1 and npi286, respectively (http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=143431). See text for details.
Probe csu814 was mapped to the same position as that of p1, and npi286, csu392, and asg69 were mapped 3.6, 4.7, and 5.6cM proximal to p1, respectively (http://www.maizegdb.org/cgi-bin/displaymaprecord.cgi?id=143431). Genomic DNA was digested with HindIII or SacI, and the four RFLP markers were used as probes for Southern analysis. Probe csu814 produced a complex Southern pattern (not shown), but there was no evidence that this sequence was deleted in any of the alleles (not shown). This suggests that csu814 is probably distal to p1. Probe csu392 hybridized with two genomic HindIII restriction fragments, which are nonpolymorphic between the deletion stocks tested here and the 4Co63 inbred line (Figure 5A). The signal intensities of the two bands were measured using ImageQuant 5.0 (see supplemental Figure S1 at http://www.genetics.org/supplemental/). In Figure 5A, in lanes 1–8 (parental p1-vv9D9A and derivative alleles) and in lane 12 (inbred 4Co63), the signal for the upper band is slightly less than, or approximately equal to, that of the lower band, whereas in lanes 9–11 (deletion alleles p1-ww1, p1-ww755, and p1-ww756, each heterozygous with p1-ww [4Co63]) the signal of the upper band is approximately twofold greater than the signal from the lower band. These results suggest that the genomic HindIII fragment corresponding to the lower band is missing in these three alleles; the band of lower signal intensity in these lanes represents the corresponding HindIII fragment from the 4Co63 genotype. On the basis of this altered signal intensity, we infer that csu392 is probably deleted from the chromosomes carrying the deletion alleles p1-ww1, p1-ww755, and p1-ww756. According to the map data, csu392 is 0.1 cM proximal to ndp2; the fact that p1-ww1 appears to be deleted for csu392 suggests that csu392 is actually distal to ndp2, because ndp2 was derived from sequences adjacent to the endpoint of p1-ww1 (Zhang and Peterson 1999). Probe npi286 hybridizes with multiple bands; one of these bands is specifically missing in p1-ww1, p1-ww755, and p1-ww756 (Figure 5A). For asg69, no deletion was detected in any of the alleles (data not shown here).
A summary of the mapping data based on these Southern hybridizations and other information is presented in Figure 5B. Probe csu814 was not deleted in any alleles and hence is probably distal to p1. The p1-ww495 allele has the small (12,567 bp) deletion (described above), which ends just upstream of p1. The endpoint of p1-ww774 is placed between p1 and p2, and the p1-ww2 deletion ends within the p2 gene. Five deletion alleles (p1-ww759, p1-ww760, p1-ww761, p1-ww765, and p1-ww775) have endpoints in the interval between p2 and ndp1. The p1-ww1 allele has its endpoint adjacent to ndp2, and p1-ww755 and p1-ww756 have their endpoints between ndp2 and asg69. Finally, probes npi286 and csu392 are located in the interval between ndp1 and ndp2.
It is important to note that the genetic distances in the map presented in Figure 5B are based on previous genetic recombination data and are likely only approximate. However, the relative order of the markers indicated by the deletion mapping presented here should be robust, assuming that the deletions are simple and unidirectional.
Location of the p1-linked genes zygotic lethal 1 and defective kernel 1:
It was interesting to determine whether any of the deletions disrupted known genes in the vicinity of p1. The zygotic lethal 1 (zl1) mutation was mapped 1.5 cM proximal to p1 (Emerson 1939). The zl1 mutation does not affect viability of the male or female gametophyte, but it is homozygous lethal in the zygote. (The original zl1 mutant stock has apparently been lost, and no other zygotic lethal mutations have been described to date in maize.) Interestingly, among the 35 p1-ww alleles studied here, 7 confer a zygotic lethal phenotype. One of these is the previously characterized p1-ww1 allele. The p1-ww1 allele transmits normally through both the pollen and the ovule; however, no homozygous p1-ww1 plants were recovered from >120 progeny plants derived from the self-pollination of a p1-ww1/P1-wr plant. There is a negligible probability (1.01 × 10−15) that homozygous p1-ww1 plants were not recovered by chance from a planting of this size. Additionally, self-pollinated p1-ww1/P1-wr ears show some empty spaces and irregular kernel rows, which are typical signs of 25% semisterility. For the remaining 6 p1-ww alleles, no p1-ww homozygous plants were identified among 20 or more progeny plants derived from self-pollination of plants carrying each p1-ww mutation heterozygous with P1-wr. The probability that p1-ww homozygotes were not detected by chance among 20 progeny of each self-pollinated p1-ww/P1-wr heterozygote is 0.3%. We conclude that the homozygous lethality of these 7 p1-ww alleles is probably due to loss of the zl1 locus. Because we obtained 22 p1-ww alleles that removed p2 and yet are viable as homozygotes, the zl1 locus is placed on the centromeric side of p2. The smallest characterized deletion that has a zygotic lethal phenotype is p1-ww1, whose endpoint is at the site of probe ndp2. Thus, the zl1 locus must lie in the interval between p2 and ndp2, with the gene order of p1, p2, zl1, centromere.
A second gene known to be in the vicinity of p1 is defective kernel 1 (dek1), which was tentatively mapped at 0.8 cM proximal to p1 (Dooner 1980). The dek1 gene encodes a 2159-aa protein belonging to the calpain superfamily and is essential for kernel aleurone development (Becraft and Asuncion-Crabb 2000; Becraft et al. 2002; Lid et al. 2002; Wang et al. 2003). Some of the p1-ww deletions described here extend >4.6 cM proximal to p1; if dek1 were 0.8 cM proximal to p1, then it should be deleted in some of these cases. However, no dek1 kernels were obtained by self-pollination of any of the p1-ww alleles obtained in this study. To test whether the zygotic lethal phenotype of the seven largest deletions is the null phenotype of dek1, we crossed three large deletions, which conferred the zygotic lethal phenotype (p1-ww1, p1-ww755, and p1-ww756) to dek1/Dek1 heterozygous plants; again, no dek1 kernels were found. We conclude that the dek1 locus is probably distal to p1. This prediction is consistent with more recent mapping data indicating that dek1 is located 0.3 cM distal to p1 (http://www.maizegdb.org/cgi-bin/displayposrecord.cgi?id=258944).
Substrate preferences for Ac transposition:
Genetic studies have concluded that, in maize, transposition of simple Ac or Ds elements does not give rise to large deletions or other rearrangements at appreciable frequencies (Fedoroff et al. 1983; Fedoroff 1989; Kunze and Weil 2002); however, deletions, duplications, and chromosome breakage are readily produced through transposition reactions involving complex Ac/Ds elements. For example, the maize doubleDs element, which contains one Ds element inserted into a second identical Ds in opposite orientation, induces chromosome breakage at a high frequency (Courage et al. 1984; Doring et al. 1984, 1989). Molecular analyses have shown that Ds-induced breakage is associated with the formation of chromatid bridges by transposition reactions involving Ds termini located on sister chromatids (English et al. 1993; Weil and Wessler 1993). The deletions generated by SCT occur when the transposon termini reinsert into the chromosome from which they were excised (Figure 1).
What determines the competence of individual Ac/Ds termini to participate in transposition reactions? In maize, Ac/Ds transposes during or shortly after DNA replication, but only one of the Ac/Ds elements in the two sister chromatids is competent for transposition (chromatid selectivity) (Greenblatt and Brink 1962; Greenblatt 1984; Chen et al. 1987, 1992; Fedoroff 1989). Several lines of evidence show that the methylation status of Ac plays an important role in chromatid selectivity. Data from in vitro binding assays show that the Ac transposase binds to hemi-, holo-, and unmethylated Ac sequences with distinctly different affinities: strong binding occurs at hemi-methylated sites in which a particular strand is methylated, whereas sequences in which the opposite strand is methylated exhibit little binding (Kunze and Starlinger 1989). In addition, studies of Ds excision from extrachromosomal DNA introduced into petunia cells show that a Ds element hemi-methylated on one DNA strand has a 6.3-fold higher transposition frequency than an element methylated on the complementary strand (Ros and Kunze 2001). These and other data have led to a model for the control of transposition competence by differential binding of Ac transposase depending on methylation state (Wang et al. 1996). Similarly, the transposase of the prokaryotic IS10 element binds to hemi- and holo-methylated IS10 ends with different affinities, thus determining which IS10 copy is transposition competent after DNA replication (Roberts et al. 1985).
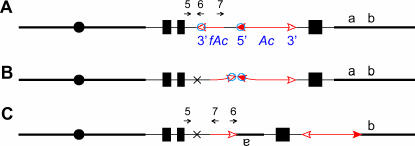
The methylation model for control of Ac transposition makes certain specific predictions regarding the transposition competence of the Ac and fAc termini in p1-vv9D9A. Immediately following replication of the p1-vv9D9A allele, the methylated DNA strand of the fAc element in one sister chromatid should be the same as that of the 3′-end of the Ac element in the other sister chromatid. Thus, the methylation hypothesis would predict that functional transposition complexes could involve (1) the 5′- and 3′-ends of the Ac element on one chromatid (standard transposition) or (2) the 3′-end of fAc and the 5′-end of Ac on different sister chromatids (sister-chromatid transposition). The former will result in Ac excision and generate a P1-rr allele with a fAc insertion, and the latter will generate deletions and corresponding inverted duplications. Indeed, both events are obtained, but the latter occurs at a lower frequency in the p1-vv9D9A allele (Zhang and Peterson 1999), whereas the methylation model predicts that transposition involving the 3′ fAc end and the 5′ Ac end on the same chromatid should not occur. If it did occur, then excision followed by reinsertion into the same chromosome would be predicted to generate inversions that include the Ac element and the genomic sequence to the reinsertion site (Figure 6). These inversions would disrupt p1 function and could be detected in our screen for new p1-ww alleles. To determine whether such inversions might exist among our collection of 100 p1-ww alleles derived from p1-vv9D9A, we tested 10 p1-ww alleles among the 65 cases that did not show deletion of p1 or p2 in the initial PCR screen. DNA from these 10 p1-ww alleles was used in PCR with primer pairs p1-5/Ac-6 and p1-5/Ac-7. As shown in Figure 6, excision of the 3′ fAc and 5′ Ac termini from the same chromatid would result in a fusion of the 5′-end of p1 intron 2 with the sequence adjacent to the 5′ breakpoint of the fAc element. If such a fusion occurred, the PCR product generated by p1-5/Ac-6 should disappear, and a new product from primers p1-5/Ac-7 should be formed. No such cases were identified among the 10 p1-ww alleles tested. These results suggest that when the fAc 3′-end and the Ac 5′-end are in direct orientation as in the p1-vv9D9A allele, the termini on sister chromatids are preferred transposition substrates. In contrast, we showed recently that when a fAc 3′-end and an Ac 5′-end are in reversed orientation (i.e., pointing toward each other), then transposition involving termini on the same chromatid can occur, generating inversions and other products (Zhang and Peterson 2004). Taken together, these results support the model of Wang et al. (1996) in which assembly of a functional transposition complex requires the interaction of 5′ and 3′ transposon termini whose individual competence is determined by strand-specific methylation patterns.
Figure 6.
Hypothetical transposition involving the fAc 3′-end and the Ac 5′-end on the same chromatid in p1-vv9D9A (symbols have the same meaning as in Figure 1). This type of transposition reaction would result in reorientation of the sequences hybridizing to oligonucleotide primers 6 (Ac-6) and 7 (Ac-7) (compare A and C) and thus could be detected by PCR. No such products were detected among 10 p1-ww alleles tested. See text for details. (A) Ac transposase binds to a fAc 3′-end and an Ac 5′-end in the same chromatid. (B) Cuts are made at the Ac and fAc termini; sequences at which the Ac and fAc termini were formerly inserted are joined together at the site marked by the X. (C) The excised transposon ends reinsert at a site between a and b. The DNA between fAc and the insertion site is inverted.
Evidence from McClintock for Ds-induced deletions on chromosome 9s:
Previous research indicates that the chromosome breaking (state I) Dissociation element originally identified by McClintock (1953) was a doubleDs element, which contains two copies of a simple Ds element, with one Ds inserted into the other in reversed orientation (Doring et al. 1984; English et al. 1993, 1995; Weil and Wessler 1993; Martinez-Ferez and Dooner 1997). Because doubleDs has two pairs of directly oriented 5′ and 3′ Ds termini pointing out from the element, hypothetically it could undergo SCT reactions to generate deletions on either side of the insertion, i.e., in both the proximal and distal directions. Hence it was interesting to determine whether any evidence of deletions was previously reported by McClintock. In one experiment, McClintock isolated a number of mutant alleles derived from a chromosome 9s containing a state I (chromosome-breaking) Ds element and dominant alleles of the colorless1 (c1), shrunken1 (sh1), and bronze1 (bz1) genes. The c1 gene specifies purple aleurone pigmentation, and the functional C1 allele is recessive to the dominant inhibitor allele C1-I. The sh1 and bz1 genes affect endosperm starch and aleurone color, respectively. The genes are linked in the order c1-(4 cM)-sh1-(2 cM)-bz1, and the Ds element was inserted in the c1-sh1 interval, very close to sh1 (McClintock 1953). SCT involving Ds could generate three classes of deletion mutants: proximal deletions that would remove sh1 (C1-I Ds Δsh1 Bz1) or both sh1 and bz1 (C1-I Ds Δsh1 Δbz1), and distal deletions that remove C1-I (Δc1 Ds Sh1 Bz1). Three stocks containing the chromosome of constitution C1-I Ds Sh1 Bz1 were crossed with a C1 sh1 bz1 stock, and progeny kernels were screened for the appearance of new mutants. Among an unspecified number of progeny kernels screened, McClintock reported finding 37 C1-I Ds sh1 Bz1, 12 C1-I Ds sh1 bz1, and 20 c1 Ds Sh1 Bz1 cases. Several lines of evidence suggest that many of these mutants may have been SCT-induced deletions:
Some C1-I Ds sh1 Bz1 mutants exhibited a pronounced decrease in crossover frequency between sh1 and bz1; in one case, no crossovers between sh1 and bz1 were detected among 3156 tested gametes. This result is consistent with deletions that extend into the sh1-bz1 interval. In contrast, the crossover frequency between c1 and sh1 was only slightly reduced. The observed small decrease in crossover frequency between c1 and sh1 could be expected as a consequence of the deletion of the short interval between Ds and sh1; the SCT-induced deletion should start from the Ds element and extend proximally to the sh1 locus, and it was known that the Ds element is tightly linked to sh1.
Among 12 C1-I Ds sh1 bz1 mutants, 6 showed decreased male and/or female transmission frequency, and 1 mutant was completely male and female sterile; similar transmission defects are a common feature of large deletions.
For all 20 cases showing losses of C1-I, no homozygous plants survived to maturity; all failed to germinate or died as seedlings. These results are consistent with deletion of essential genes, such as the dek12 gene, which is located in the interval between c1 and sh1 (McClintock 1953; Neuffer et al. 1997). McClintock (1953) reported that 10 of the 20 cases that showed losses of C1-I had lost the chromosome arm distal to the Ds element. For the remaining cases, however, there were no cytologically visible structural alterations in the short arm of chromosome 9.
The three lines of evidence described above are suggestive of the occurrence of deletions, but do not indicate how such deletions may have been generated. A possible clue to the mechanism is provided by McClintock (1953), who determined that Ds was still present on each of the mutant chromosomes she tested. This result is exactly what would be predicted for SCT of doubleDs: A chromosome-breaking Ds structure should be retained at the deletion junction, whereas deletions derived by standard transposition followed by excision of a macrotransposon as proposed by Page et al. (2004) would often not contain Ds in association with the deletion-bearing chromosome.
Generality and significance of SCT-induced deletions:
The model for SCT is mechanistically very similar to that of standard cut-and-paste transposition, but the products are very different: standard transposition results in movement of the transposon to a new site in the genome, while the SCT reaction generates deletions, duplications, and, potentially, other rearrangements (Zhang and Peterson 1999). These products are generated due to the altered topology of the transposon termini: in both the Ac/fAc and doubleDs events discussed here, at least one pair of Ac/Ds 3′ and 5′ termini are in direct orientation relative to each other. We have recently shown that another type of unconventional transposition reaction can occur when Ac termini are oriented toward each other (reversed-ends transposition; Zhang and Peterson 2004). Reversed-ends transposition can generate deletions, inversions, and, potentially, other rearrangements.
The evidence presented above indicates that SCT has generated extensive and overlapping deletions at the maize p1 locus on chromosome 1s and possibly also in the vicinity of the sh1 locus on chromosome 9s. SCT-induced deletions have also been reported in transgenic tobacco (English et al. 1995). Thus, the SCT reaction can probably occur at any genomic location containing Ac/Ds termini in the appropriate orientation. Whether the SCT reaction can also occur with other members of the hAT transposon family remains to be determined.
The detection of deletions and chromosome breakage in the above studies was facilitated by the proximity of genes controlling nonessential, visible phenotypes. It should be possible to reproduce the SCT reaction using transgenes containing Ac/Ds termini with appropriate marker genes. This approach would enable the isolation of a deletion series, similar to that described here for p1, at any genomic location containing the transgene construct. The ability to generate numerous overlapping deletions in specific regions of plant genomes could be highly advantageous for genetic and physical mapping and for functional genomics research. Finally, the ability of SCT to generate large deletions may have provided a significant counterbalance to the tendency toward genome enlargement over evolutionary time (Bennetzen and Kellogg 1997).
Acknowledgments
We thank T. Muskett and M. McMullen, University of Missouri, for probes and mapping data cited in this article, and K. Cone for the dek1/Dek1 stock. We thank W. Sheridan and J. Clark for helpful discussion, E. Kaszas for advice on pulsed-field gel analysis, and T. Olson for performing pulsed-field gel experiments and Southern blotting. This research was supported by National Science Foundation grant 0110170 to T.P. This journal paper of the Iowa Agriculture and Home Economics Experiment Station (Ames, IA) was supported by Hatch Act and State of Iowa funds.
References
- Anderson, E. G., 1924. Pericarp studies in maize II. The allelomorphism of a series of factors for pericarp color. Genetics 9: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P. A., P. A. Okubara, R. Arroyo-Garcia, B. C. Meyers and R. W. Michelmore, 1996. Molecular analysis of irradiation-induced and spontaneous deletion mutants at a disease resistance locus in Lactuca sativa. Mol. Gen. Genet. 251: 316–325. [DOI] [PubMed] [Google Scholar]
- Athma, P., and T. Peterson, 1991. Ac induces homologous recombination at the maize P locus. Genetics 128: 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma, P., E. Grotewold and T. Peterson, 1992. Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131: 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley, C. C., M. Morgan, E. C. Dale and D. W. Ow, 1992. Exchange of gene activity in transgenic plants catalyzed by the Cre-lox site-specific recombination system. Plant Mol. Biol. 18: 353–361. [DOI] [PubMed] [Google Scholar]
- Becraft, P. W., and Y. Asuncion-Crabb, 2000. Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127: 4039–4048. [DOI] [PubMed] [Google Scholar]
- Becraft, P. W., K. Li, N. Dey and Y. Asuncion-Crabb, 2002. The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129: 5217–5225. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J. L., and E. A. Kellogg, 1997. Do plants have a one-way ticket to genomic obesity? Plant Cell 9: 1509–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer, S., 1961. On the topography of the genetic fine structure. Proc. Natl. Acad. Sci. USA 47: 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benzer, S., 1962. The fine structure of the gene. Sci. Am. 206: 70–87. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., and D. M. Levin, 1991. Directed synthesis of a segmental chromosomal transposition: an approach to the study of chromosomes lethal to the gametophyte generation of maize. Genetics 127: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, P. F., M. D. McMullen, M. E. Snook, T. A. Musket, J. M. Theuri et al., 1996. Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc. Natl. Acad. Sci. USA 93: 8820–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, E., B. J. Mulligan, S. N. Covey and J. J. Milner, 1998. Characterization of gamma irradiation-induced deletion mutations at a selectable locus in Arabidopsis. Mutat. Res. 401: 199–206. [DOI] [PubMed] [Google Scholar]
- Chen, J., I. M. Greenblatt and S. L. Dellaporta, 1987. Transposition of Ac from the P locus of maize into unreplicated chromosomal sites. Genetics 117: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., I. M. Greenblatt and S. L. Dellaporta, 1992. Molecular analysis of Ac transposition and DNA replication. Genetics 130: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., P. Athma and T. Peterson, 1998. A maize Myb-homolog is encoded by a stable multicopy gene complex. Mol. Gen. Genet. 260: 372–380. [DOI] [PubMed] [Google Scholar]
- Courage, U., H. P. Doring, W. B. Frommer, R. Kunze, A. Laird et al., 1984. Transposable elements Ac and Ds at the shrunken, waxy, and alcohol dehydrogenase 1 loci in Zea mays L. Cold Spring Harbor Symp. Quant. Biol. 49: 329–338. [DOI] [PubMed] [Google Scholar]
- Dale, E. C., and D. W. Ow, 1990. Intra- and intermolecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene 91: 79–85. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., 1980. gay, a new mutant in chromosome 1. Maize Newsl. 54: 79–80. [Google Scholar]
- Dooner, H. K., J. English and E. J. Ralston, 1988. The frequency of transposition of the maize element Activator is not affected by an adjacent deletion. Mol. Gen. Genet. 211: 485–491. [DOI] [PubMed] [Google Scholar]
- Doring, H. P., E. Tillmann and P. Starlinger, 1984. DNA sequence of the maize transposable element Dissociation. Nature 307: 127–130. [DOI] [PubMed] [Google Scholar]
- Doring, H. P., B. Nelsen-Salz, R. Garber and E. Tillmann, 1989. Double Ds elements are involved in specific chromosome breakage. Mol. Gen. Genet. 219: 299–305. [DOI] [PubMed] [Google Scholar]
- Dowe, M. F., Jr., G. W. Roman and A. S. Klein, 1990. Excision and transposition of two Ds transposons from the bronze mutable 4 derivative 6856 allele of Zea mays L. Mol. Gen. Genet. 221: 475–485. [DOI] [PubMed] [Google Scholar]
- Emerson, R. A., 1939. A zygotic lethal in chromosome 1 of maize and its linkage with neighboring genes. Genetics 24: 368–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J., K. Harrison and J. D. Jones, 1993. A genetic analysis of DNA sequence requirements for Dissociation state I activity in tobacco. Plant Cell 5: 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J. J., K. Harrison and J. Jones, 1995. Aberrant transpositions of maize double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell 7: 1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N. V., 1989. Maize transposable elements, pp. 375–411 in Mobile DNA, edited by M. Howe and D. Berg. American Society for Microbiology, Washington, DC.
- Fedoroff, N., S. Wessler and M. Shure, 1983. Isolation of the transposable maize controlling elements Ac and Ds. Cell 35: 235–242. [DOI] [PubMed] [Google Scholar]
- Greenblatt, I. M., 1984. A chromosome replication pattern deduced from pericarp phenotypes resulting from movements of the transposable element, Modulator, in maize. Genetics 108: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt, I. M., and R. A. Brink, 1962. Twin mutations in medium variegated pericarp maize. Genetics 47: 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., P. Athma and T. Peterson, 1991. Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of myb-like transcription factors. Proc. Natl. Acad. Sci. USA 88: 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, B. Z., Z. J. Zhang, R. G. Li, N. W. Widstrom, M. E. Snook et al., 2001. Restriction fragment length polymorphism markers associated with silk maysin, antibiosis to corn earworm (Lepidoptera: Noctuidae) larvae, in a dent and sweet corn cross. J. Econ. Entomol. 94: 564–571. [DOI] [PubMed] [Google Scholar]
- Kaszas, E., and J. A. Birchler, 1996. Misdivision analysis of centromere structure in maize. EMBO J. 15: 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and J. A. Birchler, 1998. Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150: 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J. L., 1980. Probing the component structure of a maize gene with transposable elements. Science 208: 1457–1459. [DOI] [PubMed] [Google Scholar]
- Klug, W. S., and M. R. Cummings, 1991. Variations in chromosome number and arrangement, pp. 187–215 in Concepts of Genetics, edited by M. R. Cummings. Macmillan, New York.
- Kunze, R., and P. Starlinger, 1989. The putative transposase of transposable element Ac from Zea mays L. interacts with subterminal sequences of Ac. EMBO J. 8: 3177–3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, R., and C. F. Weil, 2002. The hAT and CACTA superfamilies of plant transposons, pp. 565–610 in Mobile DNA II, edited by N. L. Craig, R. Craigie, M. Gellert and A. M. Lambowitz. American Society for Microbiology, Washington, DC.
- Lee, E. A., P. F. Byrne, M. D. McMullen, M. E. Snook, B. R. Wiseman et al., 1998. Genetic mechanisms underlying apimaysin and maysin synthesis and corn earworm antibiosis in maize (Zea mays L.). Genetics 149: 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings, C. S., III, and C. W. Stuber, 1971. A maize gene controlling silk browning in response to wounding. Genetics 69: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. W., G. Stark, J. Gotz, T. Rulicke, M. Gschwind et al., 1996. Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc. Natl. Acad. Sci. USA 93: 6158–6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid, S. E., D. Gruis, R. Jung, J. A. Lorentzen, E. Ananiev et al., 2002. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99: 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, B. Y., S. F. Peng, Y. J. Chen, H. S. Chen and C. F. Kao, 1997. Physical mapping of RFLP markers on four chromosome arms in maize using terminal deficiencies. Mol. Gen. Genet. 256: 509–516. [DOI] [PubMed] [Google Scholar]
- Martinez-Ferez, I. M., and H. K. Dooner, 1997. Sesqui-Ds, the chromosome-breaking insertion at bz-m1, links double Ds to the original Ds element. Mol. Gen. Genet. 255: 580–586. [DOI] [PubMed] [Google Scholar]
- McClintock, B., 1953. Mutation in maize. Carnegie Inst. Washington Yearb. 53: 227–237. [Google Scholar]
- McMullen, M. D., P. F. Byrne, M. E. Snook, B. R. Wiseman, E. A. Lee et al., 1998. Quantitative trait loci and metabolic pathways. Proc. Natl. Acad. Sci. USA 95: 1996–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medberry, S. L., E. Dale, M. Qin and D. W. Ow, 1995. Intra-chromosomal rearrangements generated by Cre-lox site-specific recombination. Nucleic Acids Res. 23: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B. C., D. B. Chin, K. A. Shen, S. Sivaramakrishnan, D. O. Lavelle et al., 1998. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10: 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, M. A., J. Chen, I. Greenblatt and S. L. Dellaporta, 1992. Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics 131: 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M. G., E. H. Coe and S. R. Wessler, 1997. Dek mutants, p. 310 in Mutants of Maize, edited by S. R. Wessler. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Odell, J. T., J. L. Hoopes and W. Vermerris, 1994. Seed-specific gene activation mediated by the Cre/lox site-specific recombination system. Plant Physiol. 106: 447–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne, B. I., U. Wirtz and B. Baker, 1995. A system for insertional mutagenesis and chromosomal rearrangement using the Ds transposon and Cre-lox. Plant J. 7: 687–701. [DOI] [PubMed] [Google Scholar]
- Page, D. R., C. Kohler, J. A. Da Costa-Nunes, C. Baroux, J. M. Moore et al., 2004. Intrachromosomal excision of a hybrid Ds element induces large genomic deletions in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, E., J. English and H. K. Dooner, 1989. Chromosome-breaking structure in maize involving a fractured Ac element. Proc. Natl. Acad. Sci. USA 86: 9451–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis, R., P. Liu and A. Bradley, 1995. Chromosome engineering in mice. Nature 378: 720–724. [DOI] [PubMed] [Google Scholar]
- Rector, B. G., G. Liang and Y. Guo, 2003. Effect of maysin on wild-type, deltamethrin-resistant, and Bt-resistant Helicoverpa armigera (Lepidoptera: Noctuidae). J. Econ. Entomol. 96: 909–913. [DOI] [PubMed] [Google Scholar]
- Rinehart, T. A., C. Dean and C. F. Weil, 1997. Comparative analysis of non-random DNA repair following Ac transposon excision in maize and Arabidopsis. Plant J. 12: 1419–1427. [DOI] [PubMed] [Google Scholar]
- Roberts, D., B. C. Hoopes, W. R. McClure and N. Kleckner, 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43: 117–130. [DOI] [PubMed] [Google Scholar]
- Ros, F., and R. Kunze, 2001. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics 157: 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, S. H., J. L. Hoopes and J. T. Odell, 1992. Directed excision of a transgene from the plant genome. Mol. Gen. Genet. 234: 49–59. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof, M. A., K. M. Soliman, R. A. Jorgensen and R. W. Allard, 1984. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81: 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Snyder, L. A., D. Freifelder and D. L. Hartl, 1985. General Genetics, pp. 205–208. Jones & Bartlett, Boston.
- Szalma, S. J., E. S. Buckler, IV, M. E. Snook and M. D. McMullen, 2005. Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor. Appl. Genet. 110: 1324–1333. [DOI] [PubMed] [Google Scholar]
- Wagner, K. U., R. J. Wall, L. St-Onge, P. Gruss, A. Wynshaw-Boris et al., 1997. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25: 4323–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., J. K. Barry, Z. Min, G. Tordsen, A. G. Rao et al., 2003. The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J. Biol. Chem. 278: 34467–34474. [DOI] [PubMed] [Google Scholar]
- Wang, L., M. Heinlein and R. Kunze, 1996. Methylation pattern of Activator transposase binding sites in maize endosperm. Plant Cell 8: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil, C. F., and S. R. Wessler, 1993. Molecular evidence that chromosome breakage by Ds elements is caused by aberrant transposition. Plant Cell 5: 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. L., X. Li and T. Peterson, 2000. Ac insertion site affects the frequency of transposon-induced homologous recombination at the maize p1 locus. Genetics 156: 2007–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J., K. Marshall, M. Yamaguchi, J. E. Haber and C. F. Weil, 2004. Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol. Cell. Biol. 24: 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh, K., M. Andahazy, S. O'Gorman and H. Baribault, 1998. Selection of primary cell cultures with cre recombinase induced somatic mutations from transgenic mice. Nucleic Acids Res. 26: 4301–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and T. Peterson, 1999. Genome rearrangements by nonlinear transposons in maize. Genetics 153: 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., and T. Peterson, 2004. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167: 1929–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., S. Chopra and T. Peterson, 2000. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12: 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Y. Wang, J. Zhang, M. Snook and T. Peterson, 2003. A maize QTL for silk maysin levels contains duplicated myb-homologus genes which jointly regulate flavone biosynthesis. Plant Mol. Biol. 52: 1–15. [DOI] [PubMed] [Google Scholar]
- Zhang, S., S. Raina, H. Li, J. Li, E. Dec et al., 2003. Resources for targeted insertional and deletional mutagenesis in Arabidopsis. Plant Mol. Biol. 53: 133–150. [DOI] [PubMed] [Google Scholar]