Abstract
Disease resistance to the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) in the cultivated tomato, Lycopersicon esculentum, and the closely related L. pimpinellifolium is triggered by the physical interaction between plant disease resistance protein, Pto, and the pathogen avirulence protein, AvrPto. To investigate the extent to which variation in the Pto gene is responsible for naturally occurring variation in resistance to Pst, we determined the resistance phenotype of 51 accessions from seven species of Lycopersicon to isogenic strains of Pst differing in the presence of avrPto. One-third of the plants displayed resistance specifically when the pathogen expressed AvrPto, consistent with a gene-for-gene interaction. To test whether this resistance in these species was conferred specifically by the Pto gene, alleles of Pto were amplified and sequenced from 49 individuals and a subset (16) of these alleles was tested in planta using Agrobacterium-mediated transient assays. Eleven alleles conferred a hypersensitive resistance response (HR) in the presence of AvrPto, while 5 did not. Ten amino acid substitutions associated with the absence of AvrPto recognition and HR were identified, none of which had been identified in previous structure-function studies. Additionally, 3 alleles encoding putative pseudogenes of Pto were isolated from two species of Lycopersicon. Therefore, a large proportion, but not all, of the natural variation in the reaction to strains of Pst expressing AvrPto can be attributed to sequence variation in the Pto gene.
THE specificity of interactions between plants and pathogens is often determined by single genes in both host and parasite. The genetic basis for disease resistance in plants and virulence in pathogens was first determined in detail by Flor (1956). Studies of flax and flax rust showed that resistance occurs only when the dominant resistance allele (R-gene) in the plant is matched by a dominant allele for avirulence in the pathogen (Flor 1971; Islam and Mayo 1990; Islam and Shepherd 1991). This “gene-for-gene” model led to the hypothesis that pathogens express ligands that are detected by receptors in resistant plants (Pryor 1987). Specific recognition of a pathogen by these R-gene receptors initiates a signal transduction cascade leading to resistance (reviewed in Dangl and Jones 2001; Hammond-Kosack and Parker 2003; Innes 2004). In the absence of matching avirulence and resistance genes, a compatible interaction ensues and the pathogen successfully colonizes the host.
The highly specific gene-for-gene interaction between host and pathogen creates complex and dynamic selection pressures on both host and pathogen. Reciprocal selection on plant and pathogen populations results in the coevolution of genes determining specificity in both genomes. Negative frequency-dependent selection acting on loci determining resistance was recognized over 50 years ago as potentially important in maintaining polymorphism (Haldane 1949). Modeling of frequency-dependent selection in a two-allele system in host and parasite confirmed this potential (Barrett 1988; Seger 1988; Leonard 1994). However, elevated levels of polymorphism are not predicted in all scenarios of host-parasite coevolution. Under certain circumstances a novel R-gene may spread to fixation. Directional selection for resistance and virulence, rather than negative frequency-dependent selection, could result in an “arms race” between host and pathogen and the sequential evolution and fixation of R-genes (May and Anderson 1983; Frank 1992).
Extensive intraspecific variation in phenotypic disease resistance has been documented, often as the result of germplasm screens. However, little is known about the evolutionary dynamics of disease resistance in natural plant populations. Population genetic analyses of allelic variation in Arabidopsis (Rps2, Rpm1, and Rps5) and Lycopersicon spp. (Cf-9) revealed that pathogen recognition specificity corresponded to sequence variation at these loci (Caicedo et al. 1999; Stahl et al. 1999; van der Hoorn et al. 2001; Tian et al. 2002; Mauricio et al. 2003). Additional sequence-based analyses combined with functional tests are needed to explore the evolutionary consequences of interactions between hosts and pathogens.
The interaction between species of Lycopersicon and the bacterial pathogen, Pseudomonas syringae pv. tomato (Pst), provides an excellent opportunity for evolutionary studies because both the pathogen ligand and resistance protein have been extensively characterized at the molecular level (reviewed in Sessa and Martin 2000; Bogdanove 2002; Wu et al. 2004). Since the physical interaction of these molecules elicits the resistance response (Scofield et al. 1996; Tang et al. 1996), they provide the possibility to study the origin and recognition specificity of these molecules and the functional consequences of amino acid variation on an important aspect of plant fitness. Furthermore, Lycopersicon has many useful attributes for population genetic studies. All nine species are diploids and there are extensive, well-documented collections from natural populations of all species in the group (http://tgrc.ucdavis.edu); also Lycopersicon esculentum, the cultivated tomato, is a model species for plant genetics and detailed genetic maps exist for several others (e.g., Tanksley et al. 1992; van Ooijen et al. 1994; Monforte and Tanksley 2000).
The Pto gene confers resistance to strains of Pst expressing avrPto and was introgressed into the cultivated species, L. esculentum, from the sister species L. pimpinellifolium (Pilowsky and Zutra 1982; Martin et al. 1993). Pto is a small gene; the open reading frame (ORF) consists of 963 nucleotides, has no introns, and encodes a functional serine-threonine kinase (Martin et al. 1993; Loh and Martin 1995). Specific residues within the Pto resistance protein that are required for ligand binding, autophosphorylation, and downstream signaling have been identified through site-directed mutagenesis and domain swaps between Pto and an allele of a related paralog, Fen, that encodes a protein lacking AvrPto recognition (Scofield et al. 1996; Tang et al. 1996; Frederick et al. 1998; Rathjen et al. 1999; Sessa et al. 2000; Wu et al. 2004). The current model for Pto activation involves Pto binding to AvrPto within the plant cell and then becoming activated, possibly by a change in protein conformation induced through ligand binding. The activated Pto protein then transduces the AvrPto signal and is dependent upon Prf to elicit the resistance response (Rathjen et al. 1999; Sessa and Martin 2000; Bogdanove 2002).
The Pto resistance gene belongs to a small multigene family of five to six members in Lycopersicon (Martin et al. 1993). The genomic region containing the Pto gene family has been sequenced from a susceptible L. esculentum cultivar and a resistant cultivar containing the introgressed Pto locus from L. pimpinellifolium (D. Lavelle and R. Michelmore, unpublished results; GenBank accession nos. AF220602 and AF220603). Orthologous relationships of the Pto gene family members between the resistant and susceptible cultivars were determined on the basis of positional information and sequence identity. The two haplotypes share five orthologous, clustered genes (Pth2, Pth3, Pth4, Pth5, and Fen). Paralogs of Pto range in nucleotide sequence identity to Pto from 79 to 91%. One of the paralogs, Fen, confers sensitivity to the insecticide fenthion (Martin et al. 1994). Functions have not been ascribed to the other paralogs, although most are expressed and several are capable of downstream signaling (Chang et al. 2002). Members of the Pto gene family have also been sequenced in another species of this genus, L. hirsutum (Riely and Martin 2001).
The pathogen, Pst, invades the intercellular space of its host and secretes proteins into the host cells via a type III secretion system (Jin et al. 2003). One of these secreted proteins is AvrPto. The specific recognition of AvrPto by the host Pto protein results in the induction of the hypersensitive response (HR), which is characterized by localized cell death (Ronald et al. 1992; Martin et al. 1993). A second avirulence gene, avrPtoB, whose product is also recognized by Pto, has been isolated from Pst (Kim et al. 2002). This gene has little overall sequence similarity to avrPto. However, some of the same residues that are required for interaction of AvrPto with Pto are present in AvrPtoB (Bernal et al. 2005). Avirulence conferred by avrPtoB is Pto dependent, indicating that Pto has dual recognition specificity for the AvrPto and AvrPtoB ligands (Kim et al. 2002).
In this article, we describe variation in disease resistance within and among seven species of Lycopersicon. We determined the nucleotide sequences of 52 alleles of Pto and tested 16 alleles for specific recognition of the AvrPto ligand and activation of HR in planta. This analysis allowed the functional characterization of 41 variant amino acid positions among these alleles. Additionally we identified three putative pseudogenes of Pto segregating within natural populations of species of Lycopersicon. The implications of the naturally occurring protein variation found at Pto and variation in host resistance are discussed in the context of host-parasite coevolution.
MATERIALS AND METHODS
Plant materials:
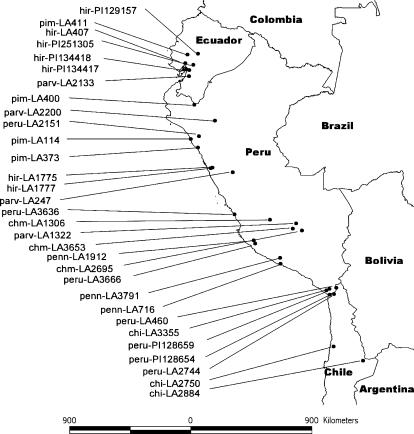
Populations of each species of Lycopersicon were sampled across their ranges (Figure 1). Plants were grown from seed collected from natural populations in Peru and Chile (Table 1). More information on these collections is available from the Tomato Genetics Resource Center (TGRC; http://tgrc.ucdavis.edu). Seed from additional populations was obtained from the U.S. Department of Agriculture, Agricultural Research Service Plant Genetic Resources Unit in Geneva, New York. For the outbreeding species (L. chilense, L. hirsutum, L. pennellii, and L. peruvianum), this study utilized the primary seed collected from native populations. For the inbreeding species (L. chmielewskii, L. parviflorum, and L. pimpinellifolium), selfed seed was used. Seeds were soaked in a 50% bleach solution for 30 min and incubated on germination paper (Anchor, St. Paul) at 22° with 24-hr fluorescent light. Seedlings were transferred to soil 2 weeks after germination and grown under greenhouse conditions at Davis, California. Cuttings to provide replicated materials were made when the plants were ∼3 months old.
Figure 1.
Map of western coast of South America indicating locations of populations used in this study. The populations are labeled by an abbreviated form of the species name and an accession number corresponding to the accession numbers in Table 1.
TABLE 1.
Origin and resistance phenotype of individuals in this study
| Population (individual identifier)a | Locality | Pst + empty vectorb | Pst + AvrPto | Pto allelec | Trans. assaye |
|---|---|---|---|---|---|
| L. pimpinellifolium | |||||
| LA114 | Pacasmayo, Peru | S | S | pim114ψ | |
| LA373 | Culebras, Peru | S | R | pimPto2 | |
| LA400 | Haciendo Buenos Aires, Peru | S | R | pimPto | HR |
| LA411 | Pichilingue, Ecuador | S | R | pimPto2 | |
| L. parviflorum | |||||
| LA247 | Chavinillo, Peru | S | S | NDd | |
| LA1322 | Limatambo, Peru | R | R | parv80 | HR |
| LA2200 | Choipiaco, Peru | R | R | parv94 | HR |
| LA2133 | Ona, Ecuador | R | R | ND | |
| L. chmielewskii | |||||
| LA1306 | Tambo, Peru | R | R | chm106 | HR |
| LA2695 | Chihuanpampa, Peru | R | R | chm115 | HR |
| LA3653 | Matara, Peru | S | S | chmPto | HR |
| L. hirsutum | |||||
| LA 407 (c1) | Mirador, Ecuador | S | R | hir540 | |
| LA 407 (c2) | Mirador, Ecuador | S | R | ||
| LA1775 (7216) | Rio Casma, Peru | R | R | ND | |
| LA1775 (7219) | Rio Casma, Peru | hir46 | No HR | ||
| LA1777 (7220) | Rio Casma, Peru | R | R | ND | |
| PI 129157 (c1) | Banos, Ecuador | R | R | hir132 | HR |
| PI 129157 (c2) | Banos, Ecuador | S | R | ||
| PI 134417 (c1) | Ferrocarril a la Costa, Ecuador | R | R | hir183 | HR |
| PI 134417 (c2) | Ferrocarril a la Costa, Ecuador | S | R | ||
| PI 134418 (c1) | Ferrocarril a la Costa, Ecuador | R | R | hir186 | HR |
| PI 134418 (c2) | Ferrocarril a la Costa, Ecuador | R | R | ||
| PI 251305 (c1) | Sibambe, Ecuador | R | R | hir137 | HR |
| PI 251305 (c2) | Sibambe, Ecuador | R | R | ||
| L. pennellii | |||||
| LA716 | Atico, Peru | S | S | ND | |
| LA1912 | Cerro Locari, Peru | S | S | ND | |
| LA3791 | Caraveli, Peru | S | S | ND | |
| L. chilense | |||||
| LA460 (c1) | Palca, Peru | R | R | ||
| LA460 (c2) | Palca, Peru | R | R | ||
| LA2884 (7177) | Ayaviri, Chile | chi487 | No HR | ||
| LA2884 (7179) | Ayaviri, Chile | chi489 | HR | ||
| LA2884 (7180) | Ayaviri, Chile | chi493 | |||
| LA2884 (7183) | Ayaviri, Chile | chi497 | |||
| LA2884 (7184) | Ayaviri, Chile | chi502 | |||
| LA2884 (7185) | Ayaviri, Chile | S | R | chi580 | HR |
| LA2750 (1) | Mina La Despreciada, Chile | S | S | chi558 | No HR |
| LA2750 (2) | Mina La Despreciada, Chile | S | S | chi591 | |
| LA3355 (7186) | Cacique de Ara, Peru | R | R | chi151 chi551 | HR |
| LA3355 (7189) | Cacique de Ara, Peru | S | R | chi151 chi620 | HR |
| LA3355 (7190) | Cacique de Ara, Peru | R | R | chi566 chi565 | |
| L. peruvianum | |||||
| LA2744 (1) | Sobraya, Chile | S | S | peru554 | HR |
| LA2744 (2) | Sobraya, Chile | S | S | peru554 | HR |
| LA2744 (7232) | Sobraya, Chile | peru505ψ peru602ψ | |||
| LA2744 (7233) | Sobraya, Chile | peru603 | |||
| LA2744 (7234) | Sobraya, Chile | S | S | peru515 peru605 | |
| LA2744 (7235) | Sobraya, Chile | peru517 peru518 | HR | ||
| LA2744 (7236) | Sobraya, Chile | peru608 peru521 | |||
| LA3636 (7252) | Coayllo, Peru | S | R | peru592 | No HR |
| LA3636 (7258) | Coayllo, Peru | S | R | peru570 | No HR |
| LA3636 (7259) | Coayllo, Peru | S | R | peru594 | |
| LA3636 (7260) | Coayllo, Peru | S | R | peru567 | HR |
| PI126444 (1) | Rio Canta, Peru | S | S | peru245 peru246 | |
| PI126444 (2) | Rio Canta, Peru | S | R | ||
| PI128654 (1) | Azapa Valley, Chile | S | R | peru234 peru236 | |
| PI128654 (2) | Azapa Valley, Chile | S | S | ||
| PI128659 (1) | Tacna, Peru | S | S | peru238 peru239 | |
| PI128659 (2) | Tacna, Peru | S | R | peru243 | |
| LA2151 (1) | Morochupa, Peru | R | R | ||
| LA2151 (2) | Morochupa, Peru | R | R | ||
| LA3218 (1) | Arequipa, Peru | R | R | ||
| LA3666 (1) | Ica, Peru | R | S | ||
Identifier of specific individuals of these populations.
Plants resistant (R) or susceptible (S) when inoculated with isogenic strains of Pst expressing empty vector or avrPto based on pathogen growth.
Name of Pto allele isolated from this individual.
Pto allele not detected (ND) even though multiple independent PCR reactions were attempted. In all individuals, alleles of other paralogs were amplified and sequenced, indicating that the gene family does exist in these individuals.
Results of transient expression of these Pto alleles in N. benthamiana (HR, hypersensitive response).
Assays for resistance to P. syringae pv. tomato:
A total of 51 individuals from seven species of Lycopersicon were tested with isogenic strains of Pst (strain T1) expressing a vector-borne copy of AvrPto or containing the empty vector to determine if the plants were resistant to this pathogen and whether the resistance was AvrPto dependent. Separate inoculations with isogenic strains of Pst-T1 containing pDSK519:AvrPto or pDSK519 (empty vector) were conducted for each plant genotype. Bacteria were grown overnight in King's B media [20 g/liter of Difco (Detroit) protease peptone no. 3, 1.5 g/liter of K2HPO4, 1.6 g/liter of MgSO4 × 7H20, and 10 ml/liter of glycerol] containing 50 μg/ml rifampicin and 20 μg/ml of kanamycin at 28° with shaking. Cells were washed in 10 mm MgCl2 and resuspended in 10 mm MgCl2 to an OD600 of 1.00. The concentrated Pst solution was then diluted in 10 mm MgCl2 to 1 × 105 colony-forming units (CFU)/ml. Two or more individuals from each population of the self-incompatible species, excluding L. pennellii, were tested, while only a single individual per population of each self-compatible species was tested. Previous studies had demonstrated that populations of the self-compatible species tended to be monomorphic at multiple loci and the greatest diversity could be detected by sampling between populations of self-compatible Lycopersicon species (Miller and Tanksley 1990). Inoculations with each bacterial genotype were replicated three times on each host. In results presented here, all independent replicates showed the same level of resistance to the corresponding bacterial strain. As described above, multiple replicates of host material were made by propagating cuttings of each plant. Cuttings were made before the plants were inoculated with Pst. The near-isogenic L. esculentum genotypes, cv. Rio Grande 76R that contains Pto, and cv. Rio Grande 76S that does not contain Pto (Seminis Seeds, Woodland, CA), were used as controls. Vacuum infiltration was used to separately inoculate plants of each genotype with each pathogen strain. Following inoculation, plants were placed in a growth chamber at 80% humidity, 25°, and 16 hr of light. Three and 7 days after inoculation, plants were scored for visible symptoms of the disease (i.e., water-soaked lesions, black specks surrounded by chlorotic halos). Three leaf discs per plant (28 mm2 in area) were also collected at 3 and 7 days after inoculations to estimate the number of bacteria within these plants. These leaf discs were ground in 1 ml of 10% glycerol solution and serial dilutions were plated on to King's B medium (see above) to estimate the number of colony-forming units. A plant was classified as resistant if at 3 and 7 days after inoculation the ground leaf discs showed the same or fewer CFUs as the Rio Grande 76R resistant control plant that had been inoculated with Pst expressing AvrPto during the same experiment. This corresponded to significantly <105 CFUs at 3 days after inoculation. A plant was classified as susceptible if at 3 and 7 days after inoculation it showed the same or more CFUs as compared to the respective susceptible controls (i.e., Rio Grande 76R inoculated with Pst expressing empty vector and Rio Grande 76S inoculated with either strain of Pst). This corresponded to >107 CFUs at 3 days after inoculation.
Analyses of genomic DNA:
DNA was isolated using a CTAB method (Doyle and Doyle 1987) from 2 g of leaf tissue collected from each plant. The DNA was resuspended in 300–1000 μl TE, depending on yield. Alleles of Pto from each species were PCR amplified using Pfu polymerase (Stratagene, La Jolla, CA). Reaction times and annealing temperatures varied depending on the primer used (supplementary Table S1 at http://www.genetics.org/supplemental/). However, the standard protocol was 94° for 5 min, 25× (94° for 30 sec, 50–60° for 30 sec, 72° for 90 sec), followed by 72° for 10 min. Products were gel purified using QIAGEN (Valencia, CA) Gene Clean or Prep-A-Gene (Bio-Rad, Hercules, CA) kits. These products were cloned into the pCR-Blunt, pCR2.1-TOPO, or pDONR vectors (Invitrogen, Carlsbad, CA). Multiple (∼10+) clones were sequenced from 49 individuals representing seven species of Lycopersicon and one species of Solanum. Sequencing was performed using an ABI 377 automated DNA sequencer. A total of 552 clones yielded 163 unique sequences of the Pto gene family. Nearly one-third of these unique sequences (51) encoded Pto alleles, while two-thirds (112) encoded paralogs of Pto including Fen, Pth2, Pth3, Pth4, and Pth5. Sequences were aligned by ClustalX (Thompson et al. 1997) and these alignments were refined visually. Phylogenetic analyses, including the Kishino-Hasegawa test, were completed using PAUP (Swofford 1999).
Genomic Southern blots were conducted to determine the complexity and level of haplotype variation within and between species of Lycopersicon. Genomic DNA of 25 individuals was digested with DraI, which digests each of the paralogs in L. pimpinellifolium and L. esculentum into individual fragments. The digested DNA was fractionated electrophoretically and transferred to a Hybond N+ membrane (Amersham Biosciences, San Francisco). The Pto ORF was used as the DNA template for probe synthesis using PCR. The blot was hybridized with the 32P-labeled probe at 65° and washed at 0.5× SSPE, 0.1% SDS. The membrane was exposed to film at −80°.
Functional in planta analysis of Pto alleles from multiple Lycopersicon species:
The function of 16 Pto alleles from wild Lycopersicon species was tested using Agrobacterium-mediated transient expression in a transgenic line of Nicotiana benthamiana that had been previously engineered to contain the avrPto gene under a dexamethasone (DEX)-inducible promoter (Chang et al. 2002). These 16 Pto alleles were selected to represent the sequence diversity found among the different species of Lycopersicon. The Pto genes that had been cloned in pCR2.1-TOPO were excised using XbaI and EcoRI, ligated into the T-DNA vector pCB302-3 (Xiang et al. 1999), and transformed into strain GV2260 of Agrobacterium tumefaciens by the freeze-thaw method. A 2-ml Luria broth (LB) culture was grown for 2 days at 28°. A 7.5-ml culture was inoculated with 0.2 ml of the 2-day-old culture and this culture was grown overnight in LB with 100 μm acetosyringone at 28°. The bacteria were washed in infiltration buffer (10 mm MES pH 5.6, 10 mm MgCl2, 150 μm acetosyringone) and diluted to an OD600 of 1.00. Leaves of 7-week-old transgenic N. benthamiana plants carrying the avrPto gene under the control of a DEX-inducible promoter (Chang et al. 2002) were pressure infiltrated with A. tumefaciens carrying alleles of Pto from the wild Lycopersicon species. Up to eight alleles were tested per leaf. Two days after infiltration, expression of avrPto was induced by swabbing the leaves with 30 μm DEX. One leaf per plant was induced with DEX and one was not. Leaves of nontransgenic control plants were also treated with DEX. Leaves were scored for macroscopic HR 24 hr after induction with DEX. Transient assays of each allele were replicated on at least two avrPto-expressing plants, one plant transformed with the empty vector and one nontransgenic N. benthamiana plant.
Site-directed mutagenesis and yeast two-hybrid analysis:
To investigate the functional effects of particular amino acid substitutions, additional functional analyses involving a subset of Pto alleles were carried out. Specifically site-directed mutagenesis was used to replace the alanine residue at position 313 in the protein sequence of Pto from L. pimpinellifolium and the L205D-Pto (constitutively active mutant) by a glutamic acid to investigate how a negatively charged amino acid at this position affected Pto function. Site-directed mutagenesis was carried out by recombinant PCR using a mixture of Klentaq (DNA Polymerase Technology, St. Louis) and Pfu polymerases. Oligonucleotides that introduced the desired mutations were used. Products were cloned in pDONR207 using the Gateway cloning system (Invitrogen). The pCB302-3 (Xiang et al. 1999) and pAS2.1 vectors were modified to be compatible with the Gateway system as described by the manufacturer. Clones from which the expected sequence was confirmed were transferred to the pCB302-3 and pAS2.1 vectors.
Yeast two-hybrid analysis of two alleles from L. hirsutum and one allele from L. pimpinellifolium was conducted using the MATCHMAKER GAL4 system following the protocols of the manufacturer (CLONTECH, Palo Alto, CA). These sequences were cloned in pAS2.1 and were cotransformed with avrPto in pACT2 (Scofield et al. 1996) into yeast strain Y190. Cotransformed yeast cells were selected by growth on selective media and replicated on filters (Whatman no. 1) prior to assaying for activation of β-galactosidase using the substrate 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-gal). The β-galactosidase assays were performed as described by CLONTECH.
RESULTS
Variation in resistance within and between species:
Levels of disease resistance to two pathogen isolates differed between individuals of both self-compatible and self-incompatible species (Table 1). Resistance varied not only within species, but also within populations of the following species: L. hirsutum, L. chilense, and L. peruvianum. One-third of the plants tested were resistant specifically when the pathogen was expressing the AvrPto ligand and susceptible otherwise, consistent with the conservation of AvrPto recognition conferred by the Pto gene in these different species. One-quarter of the plants were susceptible whether or not the pathogen was expressing the avrPto gene.
Approximately 37% of the accessions were resistant to Pst regardless of the expression of AvrPto. These plants may recognize other avirulence proteins expressed by the T1 strain of Pst. Resistance to Pst independent of avrPto expression by the pathogen reflects the evolution of recognition involving other determinants of avirulence. An unexpected phenotype was observed in individuals from the LA3636 population of L. peruvianum. These plants were susceptible specifically to Pst with avrPto, but not to Pst expressing empty vector. One explanation for this unexpected phenotype is that these individuals may be capable of recognizing an as yet uncharacterized avirulence factor from Pst strain T1; however, this recognition may be specifically nullified by the action of AvrPto.
Functional analyses of Pto alleles:
Alleles from both susceptible and resistant individuals were expressed transiently in plants known to have an intact downstream Pto signaling pathway to determine whether these alleles encoded proteins capable of recognizing AvrPto and initiating downstream signaling. These alleles were derived from a total of 22 individuals from the different Lycopersicon species, except L. pennellii from which no Pto alleles could be amplified. Alleles from 11 individuals conferred an AvrPto-dependent HR (Table 1; Figure 2). These alleles conferring AvrPto recognition in the transient assay were designated A+ alleles. The majority of the A+ alleles (6/11) originated from individuals that were resistant to Pst, regardless of the expression of AvrPto by the pathogen. Since these individuals are also resistant when AvrPto is not expressed by the pathogen, they must be capable of detecting multiple avirulence determinants. Three of the A+ alleles originated from individuals that exhibited an AvrPto-dependent resistance when inoculated with Pst. Two A+ alleles (peru554 and chmPto) were isolated from plants that were susceptible to Pst whether or not AvrPto was expressed by the pathogen. The lack of resistance in these plants may be due to downstream genes being nonfunctional or the lack of expression of these Pto alleles.
Figure 2.
Variable amino acids in the alleles transiently expressed in the susceptible N. benthamiana line containing avrPto. The numbers at the top indicate the corresponding codon position in the predicted amino acid sequence of the Pto allele (LA400) from L. pimpinellifolium. Positions invariant among these 16 alleles are removed. A dot indicates an amino acid residue matching the LA400 sequence, while a dash indicates a gap. (A) The first 11 alleles conferred an avrPto-dependent HR (hypersensitive response). These alleles were designated A+ alleles. (B) The last five alleles did not confer an HR. These alleles were designated A− alleles. Amino acid substitutions limited to the alleles that did not confer an HR are underlined. The reaction of the genotypes from which each allele was derived when inoculated with Pst expressing avrPto is indicated to the right of the allele sequence (Res, resistant; Sus, susceptible; and n.t., not tested).
Five alleles did not confer an HR when transiently expressed in the presence of AvrPto. These alleles were designated the A− alleles. One of these A− alleles was isolated from a plant that was susceptible to both strains of Pst, consistent with this plant lacking a version of Pto that could recognize the AvrPto ligand. Two additional A− alleles were isolated from plants that showed AvrPto recognition in the inoculation assays with the pathogen; i.e., these plants possess A− alleles although they would have been expected to possess A+ alleles. Both of these plants were from the LA3636 population of L. peruvianum. Given the high levels of polymorphism observed in this species, it is possible that these individuals were heterozygous at Pto and only one Pto sequence was amplified from these individuals. Thus, these individuals could have possessed a second allele conferring an A+ phenotype. Alternatively, the product of another gene may recognize AvrPto in these plants. The remaining two A− alleles were isolated from plants that were not tested for resistance to Pst.
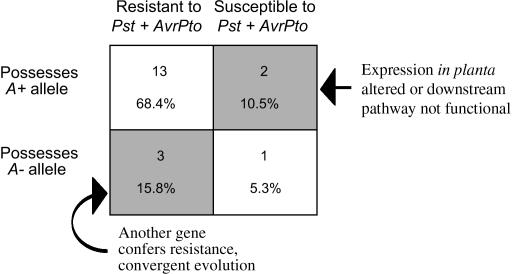
In summary, for nearly three-quarters (74%) of the plants for which both assays were conducted, a 1:1 correspondence was observed between recognition of Pst expressing avrPto and activation of HR in the presence of AvrPto in the transformation assay (Figure 3). In general, plants that were resistant to Pst expressing AvrPto were also likely to possess A+ alleles and vice versa. The exceptional cases, in which the outcomes of the two assays differed from each other (i.e., the plant was resistant but its Pto allele was A− or the plant was susceptible but its allele was A+), may reflect equally interesting biological phenomena, such as convergent evolution for resistance and/or the role of additional loci that are functionally relevant for resistance (Figure 3, shaded cells).
Figure 3.
Summary of results from plants tested for resistance to Pst expressing AvrPto and from which the alleles tested in the transient assay were derived. The top number is the number of observations per category and the bottom number is the percentage of observations per category. Open blocks show a 1:1 correspondence between the resistance assay and transformation assay. Shaded blocks indicate those observations that suggest that additional loci are involved in disease resistance.
Origin of A+ and A− alleles:
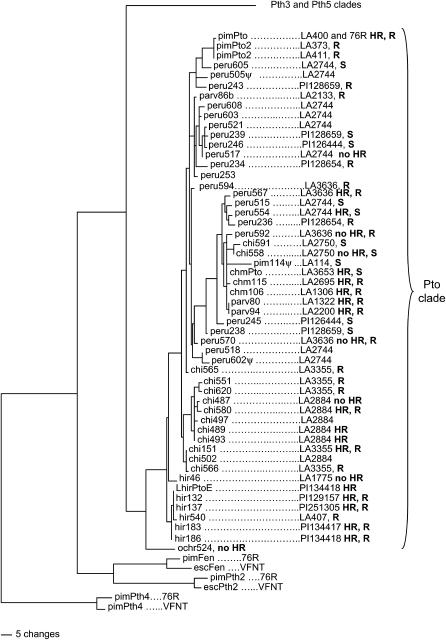
The nucleotide sequences of an additional set of 36 alleles were determined from these seven species of Lycopersicon. A phylogenetic analysis of the nucleotide sequences of all Pto alleles suggested that the A− alleles were independently derived, because the A− sequences are distributed across the tree rather than clustered together (Figure 4). To test for the independent origin of the A− alleles, maximum parsimony trees of the A+ and A− alleles were generated. A tree in which the A− alleles were constrained to be monophyletic was compared to the tree in which the A+ and A− alleles were not constrained. The constrained tree was significantly longer (i.e., a worse fit to the data) on the basis of the Kishino-Hasegawa test, rejecting the hypothesis that A− alleles have a single origin (P < 0.0001; Kishino and Hasegawa 1989). Unsurprisingly, no single amino acid change or group of changes united the A− alleles and distinguished them from the A+ alleles (Figure 2).
Figure 4.
One of 100 equally most parsimonious trees of the nucleotide sequences of alleles of Pto. The tree was rooted using the Pth2, Pth4, and Fen genes. Alleles are named by a short abbreviation of the species name followed by an arbitrary number. The source of the allele according to population is indicated further to the right by the LA and PI accession numbers. The responses of alleles tested in the transient assay are indicated by HR (hypersensitive response) or no HR. The resistance responses to Pst expressing AvrPto of the plants from which each allele was derived are indicated (R, resistant; S, susceptible).
Functional effects of amino acid substitutions in Pto:
Since each A− allele was unique, the sequence of each allele was compared in turn to the 11 amino acid sequences of the A+ genes. Substitutions unique to the A− alleles are candidates for mutations that directly affect pathogen recognition and resistance in these individuals or indirectly affect the phenotype by altering protein stability. These substitutions and their positions are summarized in Table 2. The positions of these substitutions on the predicted three-dimensional structure of the Pto protein are illustrated in supplementary Figure S1 (http://www.genetics.org/supplemental/). The chi558 allele has three unique substitutions: two conservative and one nonconservative. The nonconservative substitution at site 135 from a serine (polar) to a phenylalanine (nonpolar) occurs at the junction between domains V and VIa. This region forms an extended chain that connects two α-helices and is important for anchoring the ATP molecule (Hanks and Hunter 1995). A second A− allele, peru592, has nearly the same amino acid sequence as chi558. In addition to the nonconservative change at site 135, this protein has a nonconservative change at site 217 [arginine (+charge) to a glycine (polar)]. This substitution occurs in domain IX along an exposed part of the activation domain. Peru570 has two nonconservative substitutions: site 168 isoleucine (nonpolar) to threonine (polar) and 178 proline (nonpolar) to threonine (polar). The 168 substitution is the first amino acid following the catalytic loop region. This region is involved in phosphoryl-catalysis and transfer. The second substitution occurs in domain VII, which is made up of two β-sheets and an intervening loop. This substitution is embedded in the β-sheet, prior to the activation domain.
TABLE 2.
Summary of unique substitutions in alleles that were A− in the transient expression assay
| Allele | Individual | No. and type of unique substitutions | Position of nonconservative substitutions | Putative function of subdomain in which nonconservative substitution was found |
|---|---|---|---|---|
| chi558 | LA2750 (1) | 2 C,a 1 Nb | S135F: junction of domains V and VIa | Region forms an extended chain between two α-helices, important for anchoring the ATP molecule |
| chi487 | LA2884 (7177) | 1 N | A313Ec: outside domain XI, close to C terminus | The structure and function of this region is undefinede |
| peru592 | LA3636 (7252) | 1 C, 2 N | R217G: domain IX S135Fd | Exposed part of the activation domain |
| peru570 | LA3636 (7258) | 2 N | I168T: catalytic loop region P178T: domain VII in the β-sheet, upstream of activation domain | Phosphoryl-catalysis and transfer |
| hir46f | LA1775 (7219) | 2 C, 1 N | C104F: domain IV | Forms a large hydrophobic β-strand in the small lobe of the protein |
Conservative amino acid substitution.
Nonconservative amino acid substitution.
The introduction of aspartic acid at this position in the functional Pto allele from L. pimpinellifolium results in the loss of HR in the transient assay. Also the gain-of-function phenotype is lost when this substitution is made in the constitutively active form of Pto (L205D).
See chi558 allele.
Yeast-two-hybrid analysis demonstrated that this allele failed to bind the AvrPto ligand in yeast.
The fourth A− allele, chi487, has only one unique substitution relative to the A+ sequences. Alanine 313 (nonpolar) is replaced by a glutamic acid (negative charge). This substitution lies outside the 11 conserved kinase domains close to the C terminus. The structure and function of this region have not been defined (Hanks and Hunter 1995). Site-directed mutagenesis of the wild-type Pto replacing alanine 313 with an aspartic acid (chemically similar to glutamic acid) results in the loss of HR induction in transient assays in planta (Bernal et al. 2005). Also, the gain-of-function phenotype is lost when the same change is made in the constitutively active Pto L205D allele, indicating that a negatively charged amino acid at position 313 either alters protein stability or disrupts downstream signaling rather than recognition of AvrPto per se (Bernal et al. 2005).
Hir46 had three unique amino acid substitutions of which one change was nonconservative: at site 104, a cysteine (polar) is changed to a phenylalanine (nonpolar). This substitution is in subdomain IV of the protein kinase, which forms a large hydrophobic β-strand in the small lobe of the protein. The substitution of a bulky nonpolar amino acid at this site may distort the protein structure sufficiently to eliminate function. This allele also has an isoleucine-to-leucine substitution at position 155 and a leucine-to-isoleucine substitution at position 185, but due to the conservative nature of these substitutions, it is less likely that these changes would impair function. Yeast two-hybrid analysis of the hir46 allele demonstrated that this allele failed to bind the AvrPto ligand in yeast. This allele is most similar to hir183, which elicited an AvrPto-specific HR in the transient assay and interacted with AvrPto in the yeast two-hybrid assay. Three of the seven amino acid differences between these two alleles were unique to the nonfunctional hir46 allele compared to the larger data set of Pto alleles. Assuming that recognition of AvrPto has been conserved among alleles of Pto, then the loss of function in hir46 may have been specifically caused by one or more of these substitutions affecting either AvrPto recognition or protein stability rather than downstream signaling.
The presence of polymorphic residues among the A+ alleles indicates that at least some variation is tolerated at these positions. Thirty sites and one indel were polymorphic within the A+ class, indicating that these sites do not affect AvrPto recognition and downstream signaling (Figure 2). Of the 30 polymorphic sites, two codons had three amino acids segregating, while the rest segregated only two amino acids. Eighteen were nonconservative substitutions located throughout the protein (supplementary Figure S1 at http://www.genetics.org/supplemental/).
Pseudogenes of Pto:
Individuals of two different species of Lycopersicon had frameshift mutations in their alleles of Pto (supplementary Table S2 at http://www.genetics.org/supplemental/). The pim114ψ allele from L. pimpinellifolium had a 4-bp deletion at position 168 of the nucleotide alignment, in addition to 14 other unique mutations. Phenotypic assays confirmed that this plant was susceptible to Pst, consistent with this plant lacking a functional allele of Pto. Phylogenetic analysis showed that this allele shared its most recent common ancestor with alleles from L. parviflorum and L. chmielewskii rather than with alleles from other L. pimpinellifolium populations, a result that, taken with the high number of observed unique variants, indicates that this is an ancient pseudogene (Figure 4). A single individual from L. peruvianum population LA 2744 had two distinct pseudogenes (peru602ψ and peru505ψ), each containing a different frameshift mutation. This was the only individual of seven studied from this population from which putative pseudogenes were identified. Pto alleles derived from all other individuals in this population do not contain frameshift or nonsense mutations and the two alleles tested from this population elicited HR in the transient assay. The Pto allele from Solanum ochranthum, a close relative of Lycopersicon, also has a frameshift mutation caused by a 10-bp deletion with respect to the other Pto alleles. This allele did not elicit the HR when tested in transient assays of N. benthamiana. In addition, two alternate approaches to restore the open reading frame using site-directed mutagenesis (one involving an insertion of 2 bp to restore a single missing codon and the other involving the insertion of the 10 bp to restore the four missing codons with respect to the other alleles) failed to restore this HR-inducing activity, indicating that at least one of the other 16 unique amino acid substitutions impairs function.
Loss of the Pto gene:
Attempts were made to amplify Pto alleles from all Lycopersicon species. However, multiple PCRs on some individuals of L. pennellii, L. parviflorum, and L. hirsutum never yielded alleles of Pto. Alleles of paralogs of Pto were amplified and sequenced from the same individuals, indicating that while other members of the gene family were present, the Pto gene may have been specifically absent. The three individuals of L. pennellii from different populations and the L. parviflorum LA247 individual were susceptible to Pst with and without avrPto, which is consistent with the absence of a functional Pto gene. Individuals from LA2133 (L. parviflorum), LA1775 (L. hirsutum), and LA1777 (L. hirsutum) also were missing Pto but were resistant to Pst both with and without avrPto, suggesting that resistance to Pst was not due to Pto in these individuals.
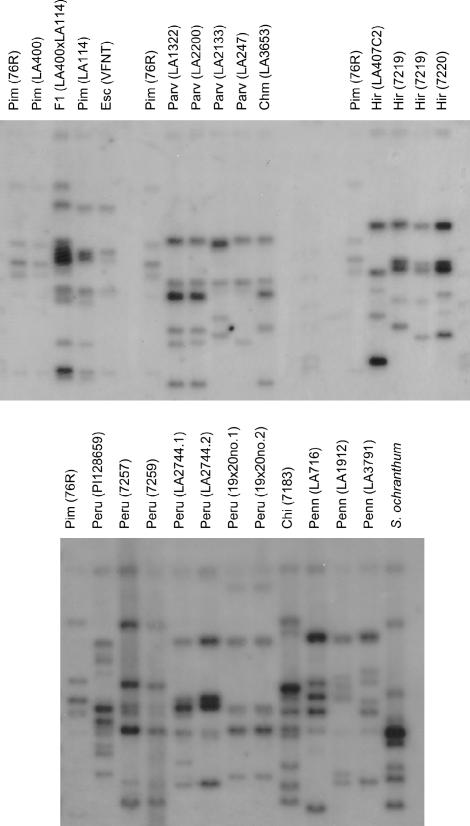
To determine if the failure to amplify Pto from specific individuals was correlated with the absence of specific fragments in genomic Southern hybridizations, banding patterns were compared between individuals within a given species that had Pto to those lacking Pto from the same species (Figure 5; supplementary Table S3 at http://www.genetics.org/supplemental/). In L. parviflorum, there was a correlation between band number and the presence or absence of Pto. The two individuals from LA2200 and LA1322 both had alleles of Pto and had six strongly hybridizing bands. The individual from LA2133 that lacked Pto had five rather than six fragments. The individual from LA247 also lacked Pto and had only four hybridizing bands. Although this species is self-compatible, three different banding patterns were identified in the four individuals from different populations, reinforcing the observation that substantial structural polymorphism exists at this locus, even within the self-compatible species. Southern analysis of L. parviflorum therefore supported the hypothesis that Pto was absent from these genotypes.
Figure 5.
Southern blot hybridization of 26 accessions of Lycopersicon and Solanum species using the entire open reading frame of Pto as a probe. See materials and methods for details. Above each lane is the abbreviated name of the species and the population or individual number of each plant sample. Pim (76R) is the L. esculentum cultivar containing the Pto region introgressed from L. pimpinellifolium. F1 (LA400 × LA114) is an F1 individual of a cross between the LA400 and LA114 individuals.
A correlation between banding pattern and the presence or absence of Pto was not detected in L. hirsutum. All individuals had five bands even though two individuals had Pto and the other two did not. The banding patterns of all three individuals of L. pennellii were surveyed because none of these individuals yielded a copy of Pto and all were susceptible to both strains of Pst. Each banding pattern was different and there were at least six strongly hybridizing bands per individual. Southern hybridizations therefore confirmed that the Pto family is present in these two species. The lack of amplification of a Pto allele could have been due to its absence or sequence divergence at a primer binding site.
DISCUSSION
Up until now, studies of the molecular evolution of resistance genes in plants have been limited to a few model plant species and their closest relatives (Caicedo et al. 1999; Stahl et al. 1999; van der Hoorn et al. 2001; Tian et al. 2002; Mauricio et al. 2003). Here we describe the sequence variability at the Pto locus among seven close relatives of the model plant species L. esculentum. We characterized the phenotypic variability in these plant species and related the amino acid sequence variation at the Pto locus to differences in recognition of a pathogen molecule and activation of downstream resistance response. This is currently one of the most extensive studies of diversity in the function of alleles at a single R-gene across species.
Plants of different Pto genotypes show functional differences in their resistance to Pst expressing AvrPto. We observed the cooccurrence of Pto alleles capable of AvrPto recognition, those incapable of AvrPto recognition, and pseudogenes not only within species, but also segregating within single populations (e.g., L. peruvianum and L. chilense). The transient expression of 16 alleles in susceptible plants allowed us to characterize the functional effects of 41 substitutions (i.e., 12.7% of the total protein) that were distributed across the molecule (supplementary Figure S1 at http://www.genetics.org/supplemental/). Amino acid variation in the alleles lacking AvrPto recognition was not localized to regions known to be involved in ligand binding or downstream signaling (e.g., the activation domain) and there were no obvious differences in the patterns of variation between the alleles with and without AvrPto recognition. This lack of differentiation could be due in part to our incomplete understanding of the functional regions of the protein. Although the Pto gene is one of the best-characterized R-genes and previous domain swap, mutational analyses, and DNA shuffling experiments have characterized some of the functional domains of Pto, the precise crystal structure is not known and a thorough molecular dissection of Pto is still underway (Wu et al. 2004, Bernal et al. 2005). However, the observed distribution of the naturally occurring substitutions that affect function suggests that there are several regions critical to function in addition to those previously implicated.
Amino acid substitutions that are restricted to alleles that did not elicit an HR in response to AvrPto are candidates for sites that may alter protein stability, disrupt ligand binding, or interfere with downstream signaling, while polymorphisms among alleles that confer AvrPto recognition identify substitutions that are nonessential for AvrPto-dependent resistance. There was as much variation among Pto alleles that elicit an AvrPto-specific HR (A+ alleles) as there was between this group and the group of alleles that did not recognize AvrPto in planta (A+ vs. A− alleles). The 5 sequences that failed to confer an AvrPto-dependent response appear to be independently derived, each differing by only a few amino acids from one of the 11 different sequences that conferred an AvrPto-dependent HR. Ten substitutions were limited to the class of alleles that lacked AvrPto recognition and downstream signaling, none of which had been identified in previous structure-function studies of Pto utilizing induced and site-directed mutation analyses and domain swaps.
Previous work by Rathjen et al. (1999) demonstrated that specific amino acid substitutions in the P + 1 loop of the activation segment of Pto lead to a constitutive gain-of-function (CGF) phenotype (induction of HR) in the absence of AvrPto. More recently, Wu et al. (2004) used structural modeling of Pto to predict which amino acid residues of the Pto protein are in close proximity to the P + 1 loop and therefore might also play a role in the regulation and function of Pto. Residues in close proximity to the activation loop were systematically mutated to investigate the extent of the negative regulatory patch. In total, Wu et al. mutated 38 residues and tested these molecules for the CGF phenotype and the ability to bind AvrPto and AvrPtoB in yeast. The vast majority of the residues investigated by Wu et al. (33 of 38) were invariant among our 16 alleles derived from natural populations. The high level of protein conservation at these positions can be explained in part by the fact that mutations in these residues are likely to have deleterious effects in nature. In particular, 12 of these mutations lead to the CGF phenotype and substitutions leading to uncontrolled cell death of host tissue are unlikely to be found in natural plant populations. Therefore over one-third of the mutations tested by Wu et al. may be strongly deleterious and would be removed by natural selection.
Among our 16 alleles, we detected natural variation at five sites mutated by Wu et al. However, none of these natural variants showed the same amino acid residue as tested by Wu et al. At three positions, the variation observed was found only among A+ alleles in our study, making these unlikely candidates for affecting AvrPto recognition and signaling of HR in natural populations. Two positions that were predicted to be exposed residues by Wu et al. (isoleucine at position 168 and isoleucine at position 185) showed polymorphisms exclusively in our A− alleles, bolstering the hypothesis that these are functionally important residues; however, the specific mutations studied by Wu et al. did not match those amino acid substitutions that were segregating in our natural populations. The fact that the functionally important residues defined in these two studies do not show greater overlap is perhaps not surprising considering that each work addressed different aspects of Pto function. In our study of natural variants, we anticipated recovering at least some variants that differed in recognition specificity, but we could not predict a priori which types of variants we would recover. In contrast, the Wu et al. study specifically targeted sites predicted to be involved in negative regulation of the protein. Therefore these studies provide complementary data sets for the dissection of the structure and function of resistance proteins.
In parallel to the current study, we used DNA shuffling, a PCR-based combinatorial method for generating large numbers of variant progeny molecules from a set of related template molecules in vitro, to investigate functionally important regions of the Pto molecule (Bernal et al. 2005). Pto was shuffled with four of its paralogs and the resulting recombinant molecules were tested for their interaction with AvrPto and AvrPtoB in yeast two-hybrid assays and their activation of HR in an AvrPto- or AvrPtoB-dependent manner in planta. Nine candidate regions important for binding to AvrPto or for signaling downstream were identified by statistical correlations between individual amino acid positions and phenotype. Residues correlated with Pto function were further investigated by site-directed mutational analyses. Since the source of variation among the recombinant molecules was Pto and its paralogs, most of which are expressed and encode potentially functional protein kinases (Chang et al. 2002), we might have expected considerable overlap between the residues identified as functionally important in the study of allelic variation of Pto and the shuffling experiment, assuming that variation in alleles/orthologs of Pto and paralogs of Pto are shaped by similar evolutionary forces. However, nearly all positions implicated and tested in the shuffling experiment were invariant among the 16 alleles of Pto we studied. The one exception was residue 313. This was selected for site-directed mutagenesis because (1) it fell within a large region identified as functionally important by the shuffling experiment, (2) it was the only residue that was polymorphic among our 16 Pto alleles, and (3) it was found exclusively in an A− allele. Therefore, the selection of this site for site-directed mutagenesis was informed by the population study. Due to the greater divergence between Pto and its paralogs as compared to that between alleles of Pto, different subsets of functionally important residues were identified using the shuffling experiment compared to our analysis of alleles and orthologs.
The lack of an AvrPto-dependent elicitation of HR in different accessions was due to a variety of independent genetic events, as indicated by the integration of inoculation, sequence, and transient assay data. Four alleles were putative pseudogenes and no Pto genes could be isolated from seven individuals of different Lycopersicon species. The presence of pseudogenes and null alleles is not uncommon in multigene families including other R-gene loci. At the Rpp5 cluster in A. thaliana, out of a total of 18 genes observed in two ecotypes, only 3 genes encode intact, full-length open reading frames, 1 of which confers resistance to the Peronospora parasitica pathogen (Noel et al. 1999). Several paralogs at the Dm3 locus in lettuce have frameshift mutations and/or are truncated (Meyers et al. 1998). At the Xa21 locus in rice, of seven paralogs, only three are expressed (Wang et al. 1998). In Arabidopsis thaliana, some individuals have a functional RPM1 gene encoding resistance to Ps. pv. maculicola expressing avrRpm1, while all susceptible individuals lacked this gene entirely (Grant et al. 1998; Stahl et al. 1999).
The presence of null alleles at R-gene loci could be due to a number of reasons. If the distribution of the pathogen is heterogeneous in space and time, as is likely, the loss of a resistance gene in a host population could occur by random chance due to drift. In selfing species including several of the Lycopersicon species, stochastic forces such as genetic drift, founder effects, and/or hitchhiking may lead to the fixation of null alleles or pseudogenes in the populations not exposed continuously to the pathogen. However, even within the self-incompatible Lycopersicon species we observed that some individuals within a single population have a functional Pto gene while some lack the Pto gene entirely or have a pseudogene. An alternative explanation is that the loss of function is driven by selection and maintained within populations through negative frequency-dependent selection. This would occur if there were a cost of resistance conferring an advantage to the loss of function. R-genes, although generally expressed at low levels, may be associated with fitness costs (Tian et al. 2003). Such costs need not be strictly metabolic, but could occur because a resistance protein such as Pto could be a target of virulence effectors for some pathogens. These costs would be evident only in the presence of a pathogen strain possessing the virulence effector. However, experiments designed to study costs generally have excluded pathogens and as such would have failed to detect this particular cost.
Not all resistance observed in this study was determined by Pto. However, nearly all of the individuals that are resistant to Pst expressing avrPto had alleles of Pto that conferred an AvrPto-dependent HR. This is consistent with the conservation of AvrPto recognition conferred by the Pto gene in these species. However, >40% of the individuals tested from these seven species were resistant to the Pst strain lacking avrPto. Presumably other resistance genes in these accessions conferred recognition of additional Pst effector proteins. Previous genetic studies have indicated that resistance to Pst has evolved at several other loci in different Lycopersicon species (Pilowsky and Zutra 1986; Stockinger and Walling 1994). Pseudomonas spp. secrete many different effector molecules into the plant cell; each of these is a potential avirulence protein subject to coevolution with resistance genes (Collmer et al. 2002; Guttman et al. 2002; Deng et al. 2003). Evolution of Pst recognition at independent loci may also make the maintenance of a functional version of Pto redundant. Therefore mutations within this gene or deletion may not be deleterious in all host backgrounds and this could account for the repeated loss-of-function mutations at the Pto locus.
Acknowledgments
We are grateful to H. Akashi, J. Chang, D. Lavelle, T. Long, J. Parsch, J. Rathjen, R. Ree, Y.-S. Tai, and A. Wu for many valuable discussions. We acknowledge C. Rick for help and inspiration during this project. The plant seeds were provided by the C. M. Rick Tomato Genetics Resource Center and the U.S. Department of Agriculture Plant Genetic Resources Unit. We also acknowledge expert technical assistance provided by the University of California (UC) Davis greenhouse and controlled environment facility staff and the Center for Engineering Plants Resistant Against Pathogens (CEPRAP) and UC Davis Division of Biological Sciences DNA sequencing facility staff. This work was supported by a National Science Foundation (NSF) dissertation improvement grant (9902342) and a Jastro Shields graduate research award to L.R. and by NSF Cooperative Agreement (BIR-8920216) to CEPRAP.
References
- Barrett, J. A., 1988. Frequency-dependent selection in plant-fungal interactions. Philos. Trans. R. Soc. Lond. B 319: 473–484. [Google Scholar]
- Bernal, A., Q. Pan, J. Pollack, L. Rose, A. Kozik et al., 2005. Functional analysis of the plant resistance gene Pto using DNA shuffling. J. Biol. Chem. 280: 23073–23083. [DOI] [PubMed] [Google Scholar]
- Bogdanove, A. J., 2002. Pto update: recent progress on an ancient plant defence response signalling pathway. Mol. Plant Pathol. 3: 283–288. [DOI] [PubMed] [Google Scholar]
- Caicedo, A. L., B. A. Schaal and B. N. Kunkel, 1999. Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J. H., Y.-S. Tai, A. J. Bernal, D. T. Lavelle, B. J. Staskawicz et al., 2002. Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol. Plant-Microbe Interact. 15: 281–291. [DOI] [PubMed] [Google Scholar]
- Collmer, A., M. Lindeberg, T. Petnicki-Ocwieja, D. J. Schneider and J. R. Alfano, 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10: 462–469. [DOI] [PubMed] [Google Scholar]
- Dangl, J. L., and J. D. G. Jones, 2001. Plant pathogens and integrated defence responses to infection. Nature 411: 826–833. [DOI] [PubMed] [Google Scholar]
- Deng, W. L., A. H. Rehm, A. O. Charkowski, C. M. Rojas and A. Collmer, 2003. Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J. Bacteriol. 185: 2592–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, J. J., and J. L. Doyle, 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19: 11–15. [Google Scholar]
- Flor, H. H., 1956. The complementary genetic systems in flax and flax rust. Adv. Genet. 8: 29–54. [Google Scholar]
- Flor, H. H., 1971. The current status of the gene for gene concept. Annu. Rev. Phytopathol. 9: 275–296. [Google Scholar]
- Frank, S. A., 1992. Models of plant-pathogen coevolution. Trends Genet. 8: 213–219. [DOI] [PubMed] [Google Scholar]
- Frederick, R. D., R. L. Thilmony, G. Sessa and G. B. Martin, 1998. Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2: 241–245. [DOI] [PubMed] [Google Scholar]
- Grant, M. R., J. M. Mcdowell, A. G. Sharpe, Z. M. Detorres, D. J. Lydiate et al., 1998. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 15843–15848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman, D. S., B. A. Vinatzer, S. F. Sarkar, M. V. Ranall, G. Kettler et al., 2002. A functional screen for the type III (Hrp) secretome of the plant pathogen Pseudomonas syringae. Science 295: 1722–1726. [DOI] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1949. Disease and evolution. Ric. Sci. Suppl. 19: 1–11. [Google Scholar]
- Hammond-Kosack, K. E., and J. E. Parker, 2003. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr. Opin. Biotechnol. 14: 177–193. [DOI] [PubMed] [Google Scholar]
- Hanks, S. K., and T. Hunter, 1995. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596. [PubMed] [Google Scholar]
- Innes, R. W., 2004. Guarding the goods. New Insights into the central alarm system of plants. Plant Physiol. 135: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. R., and G. M. E. Mayo, 1990. A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104: 89–100. [Google Scholar]
- Islam, M. R., and K. W. Shepherd, 1991. Present status of genetics of rust resistance in flax. Euphytica 55: 255–268. [Google Scholar]
- Jin, Q., R. Thilmony, J. Zwiesler-Vollick and S.-Y. He, 2003. Type III protein secretion in Pseudomonas syringae. Microbes Infect. 5: 301–310. [DOI] [PubMed] [Google Scholar]
- Kim, Y. J., N.-C. Lin and G. B. Martin, 2002. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598. [DOI] [PubMed] [Google Scholar]
- Kishino, H., and M. Hasegawa, 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data and the branching order in Hominoidea. J. Mol. Evol. 29: 170–179. [DOI] [PubMed] [Google Scholar]
- Leonard, K. J., 1994. Stability of equilibria in a gene-for-gene coevolution model of host-parasite interactions. Phytopathology 84: 70–77. [Google Scholar]
- Loh, Y.-T., and G. B. Martin, 1995. The Pto bacterial resistance gene and the Fen insecticide sensitivity gene encode functional protein kinases with serine/threonine specificity. Plant Physiol. 108: 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. B., M. C. Devicente and S. D. Tanksley, 1993. High-resolution linkage analysis and physical characterization of the Pto bacterial resistance locus in tomato. Mol. Plant Pathol. 6: 26–34. [Google Scholar]
- Martin, G. B., A. Frary, T. Wu, S. Brommonschenkel, J. Chunwongse et al., 1994. A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6: 1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio, R., E. A. Stahl, T. Korves, D. Tian, M. Kreitman et al., 2003. Natural selection for polymorphism in the disease resistance gene Rps2 of Arabidopsis thaliana. Genetics 163: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, R. M., and R. M. Anderson, 1983. Parasite-host coevolution, pp. 186–206 in Coevolution, edited by D. Futuyama and M. Slatkin. Sinauer, Sunderland, MA.
- Meyers, B. C., K. A. Shen, P. Rohani, B. S. Gaut and R. W. Michelmore, 1998. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10: 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. C., and S. D. Tanksley, 1990. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor. Appl. Genet. 80: 437–448. [DOI] [PubMed] [Google Scholar]
- Monforte, A. J., and S. D. Tanksley, 2000. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43: 803–813. [PubMed] [Google Scholar]
- Noel, L., T. L. Moores, E. A. Van der biezen, M. Parniske, M. J. Daniels et al., 1999. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Pilowsky, M., and D. Zutra, 1982. Screening wild tomatoes for resistance to bacterial speck pathogen (Pseudomonas tomato). Plant Dis. 66: 46–47. [Google Scholar]
- Pilowsky, M., and D. Zutra, 1986. Reaction of different tomato genotypes to the bacterial speck pathogen (Pseudomonas syringae pv. tomato). Phytoparasitica 14: 39–42. [Google Scholar]
- Pryor, T., 1987. The origin and structure of fungal disease resistance genes in plants. Trends Genet. 3: 157–161. [Google Scholar]
- Rathjen, J. P., J. H. Chang, B. J. Staskawicz and R. W. Michelmore, 1999. Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 18: 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely, B. K, and G. B. Martin, 2001. Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc. Natl. Acad. Sci. USA 98: 2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald, P. C., J. M. Salmeron, F. M. Carland and B. J. Staskawicz, 1992. The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto disease resistance gene. J. Bacteriol. 174: 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield, S. R., C. M. Tobias, J. P. Rathjen, J. H. Chang, D. T. Lavelle et al., 1996. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science 274: 2063–2065. [DOI] [PubMed] [Google Scholar]
- Seger, J., 1988. Dynamics of some simple host-parasite models with more than two genotypes in each species. Philos. Trans. R. Soc. Lond. B 319: 541–556. [DOI] [PubMed] [Google Scholar]
- Sessa, G., and G. B. Martin, 2000. Signal recognition and transduction mediated by the tomato Pto kinase: a paradigm of innate immunity in plants. Microbes Infect. 2: 1591–1597. [DOI] [PubMed] [Google Scholar]
- Sessa, G., M. D'Ascenzo and G. B. Martin, 2000. Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J. 19: 2257–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E. A., G. Dwyer, R. Mauricio, M. Kreitman and J. Bergelson, 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671. [DOI] [PubMed] [Google Scholar]
- Stockinger, E. J., and L. L. Walling, 1994. Pto3 and Pto4: novel genes from Lycopersicon hirsutum var. glabratum that confer resistance to Pseudomonas syringae pv tomato. Theor. Appl. Genet. 89: 879–884. [DOI] [PubMed] [Google Scholar]
- Swofford, D., 1999. PAUP* Version4.0b10. Sinauer Associates, Sunderland, MA.
- Tang, X., R. D. Frederick, J. Zhou, D. A. Halterman, Y. Jia et al., 1996. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 274: 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tanksley, S. D., M. W. Ganal, J. P. Prince, M. C. Devicente, M. W. Bonierbale et al., 1992. High density molecular linkage maps of the tomato and potato genomes. Genetics 132: 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., H. Araki, E. Stahl, J. Bergelson and M. Kreitman, 2002. Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99: 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., M. B. Traw, J. Q. Chen, M. Kreitman and J. Bergelson, 2003. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77. [DOI] [PubMed] [Google Scholar]
- Van der hoorn, R. A. L., M. Kruijt, R. Roth, B. F. Brandwagt, M. H. A. J. Joosten et al., 2001. Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. Proc. Natl. Acad. Sci. USA 98: 10493–10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van ooijen, J. M., J. M. Sandbrink, M. Vrielink, R. Verkerk, P. Zabel et al., 1994. An RFLP linkage map of Lycopersicon peruvianum. Theor. Appl. Genet. 89: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Wang, G.-L., D.-L. Ruan, W.-Y. Song, S. Sideris, L. Chen et al., 1998. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, A., V. Andriotis, M. Durrant and J. P. Rathjen, 2004. A patch of surface-exposed residues mediates negative regulation of immune signaling by tomato Pto kinase. Plant Cell 16: 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., P. Han, I. Lutziger, K. Wang and D. J. Oliver, 1999. A mini binary vector series for plant transformation. Plant Mol. Biol. 40: 711–717. [DOI] [PubMed] [Google Scholar]