Abstract
Smg1 is a key component of nonsense-mediated decay (NMD) in Caenorhabditis elegans and mammals. Here we report that two nonsense alleles of the ortholog of Smg1 do not affect NMD in Drosophila melanogaster.
NONSENSE-MEDIATED decay (NMD) is an mRNA surveillance mechanism whereby transcripts with premature termination codons (PTC) are degraded (Pulak and Anderson 1993; Wilusz et al. 2001; Maquat 2002). The key genes of NMD in higher eukaryotes were originally defined genetically in three unrelated suppressor screens of Caenorhabditis elegans, which uncovered overlapping subsets of seven genes (Smg 1–7) (Hodgkin et al. 1989). Smg mutations can act as suppressors by allowing transcripts with PTCs to be translated into functional, albeit truncated, proteins. They can also make recessive mutants dominant by allowing the production of truncated dominant-negative proteins whose transcripts would normally be degraded (Cali and Anderson 1998). Orthologs to all Smg genes except Smg7 have been identified in Drosophila melanogaster (Gatfield et al. 2003), although mutations have not previously been documented.
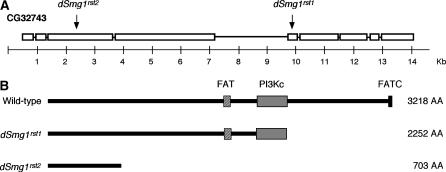
Here we report that two nonsense mutations in the Drosophila ortholog of Smg1 (CG32473) do not affect NMD. Thus Smg1 does not have a role in NMD in Drosophila or its role is redundant in at least some tissues of the adult fly. The mutants were recovered from a screen for cyromazine resistance in flies exposed to ethyl methanesulfonate mutagenesis (Adcock et al. 1993; Daborn et al. 2000). The nonsense mutations were identified by positional cloning of two recessive alleles that failed to complement (Z. Chen, C. Robin and P. Batterham, unpublished results) and would yield proteins truncated at 70% and 22% of the length of the predicted wild-type protein (Figure 1). CG32743 encodes a phosphatidylinositol kinase-like kinase (PIKK) and has been annotated as the ortholog of Smg1 from worms and mammals (FlyBase, http://www.flybase.org; Pulak and Anderson 1993; Yamashita et al. 2001; Gatfield et al. 2003). There is only a low level of amino acid sequence identity between CG32743 and human Smg1 (23%) and C. elegans Smg1 (17%), but it is greater than the level of identity between the human and C. elegans orthologs (16%) and the argument of orthology is supported by phylogenetic analyses of (i) complete sequences of all D. melanogaster, human, and C. elegans PIKKs; (ii) the individual kinase, FAT, and FATC domains; and (iii) regions of similarity outside these domains (not shown). Furthermore, there is no evidence for any other Smg1-like sequence in the D. melanogaster genome or for any of the other Drosophila species genome sequences currently available.
Figure 1.
(A) The genomic structure of the dSmg1 gene, showing the position of the nonsense mutations in dSmg1rst1 (codon 2253 AAA → TAA) and dSmg1rst2 (codon 704 AAA → TAA) alleles. (B) The predicted ORFs of wild-type and mutant dSmg1. The three characteristic domains of PIKKs—FAT, PI3Kc, and FATC—are shown (Bosotti et al. 2000).
Microarray analysis:
We set out to test whether the dSmg1 mutations modified the steady-state levels of other transcripts. Microarrays containing 5849 nonredundant cDNA clones of DGC Release 1.0 were used to compare the transcriptomes of dSmg1rst1 flies and w1118 flies. We performed two replicates of a dye swap experiment (four arrays in total). The overall correlation coefficient between experiments was 0.8 with 72 genes upregulated and 40 genes downregulated in dSmg1rst1 when a B-statistic value of 1 was used as a cutoff (Lonnstedt and Speed 2002). To address whether the differentially regulated transcripts were a result of disruption in NMD, we examined transcripts that would be degraded by NMD if they were in mammals (Nagy and Maquat 1998). Specifically, a bioinformatics analysis of the Drosophila genome uncovered 365 genes with transcripts that had termination codons located >50 bp upstream of the last exon/exon junction (S. Prochnick and N. Harris, personal communication). Although the definition of a PTC is not known in Drosophila, we may still expect to have an overrepresentation of PTC transcripts in this subset. Of these 365 genes, 213 were represented on the microarray, but only 8 of these were considered differentially regulated in Smg1rst1 vs. wild-type flies (i.e., B-statistic >1). Thus the distribution of these 213 PTC-containing genes was not significantly different from that of all other genes (χ2 = 3.4, P > 0.5).
PTC-bearing Adhn4 transcripts not affected by dSmg1rst1 or dSmgrst2:
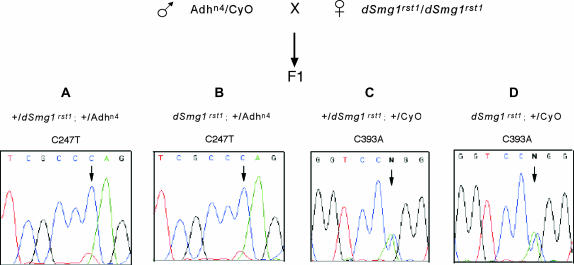
To test more directly the role CG32743 plays in NMD in Drosophila, we measured the effect that the dSmg1rst1 and dSmg1rst2 mutations had on the abundance of specific transcripts thought to be degraded by NMD. The Adhn4 allele contains a premature stop codon located 258 bp upstream from the boundary between exon 3 and exon 4 and Northern data show that it has 7% of the steady-state mRNA of the wild-type allele (Brogna 1999), presumably due to the action of NMD. Adhn4 males were crossed with dSmg1rst1 females and the Adh mRNA levels of F1 offspring were measured. If CG32743 had a role in NMD, then we might expect that dSmg1rst1 flies would have impaired NMD and a greater steady-state abundance of PTC transcripts. We compared the ratio of Adhn4 transcripts to Adh+ transcripts in flies that were heterozygous for these Adh alleles and either hemizygous for dSmg1rst1 (males) or heterozygous for dSmg1rst1 (females). The relative abundance of the Adh transcripts was determined by sequencing cDNA preparations and assessed by examining chromatograms, as each of the Adh alleles under investigation carried specific SNPs. No difference could be seen in the ratio of Adhn4/Adh+ transcripts between males and females. In both cases, the Adhn4 peaks were much smaller than the wild-type Adh+ (Figure 2), even though they are equally abundant in sequence of genomic DNA (not shown). These results were also observed in crosses using dSmgrst2 and using stocks containing Adhn4 in different genetic backgrounds. In 16 experiments on four independent crosses, each with two independent cDNA preps, the chromatogram peak height of the Adhn4 allele ranged from undetectable to ∼10% of the wild-type allele regardless of the dSmg1 allele.
Figure 2.
Genetic crosses between flies harboring the Adhn4 allele (BL-2240 and BL-3410), which has a PTC and is degraded by NMD, and flies carrying Smg1rst1. F1 males do not carry a wild-type Smg1 allele (because Smg1 is on the X chromosome) whereas F1 females do. (A) Sequence trace of Adh cDNA extracted from females heterozygous for Smg1+ and Adhn4 shows that the Adhn4 transcript (T247) is barely detectable, consistent with NMD activity. (B) Sequence trace of Adh cDNA extracted from males hemizygous for Smg1rst1 and heterozygous for Adhn4 also shows that the Adhn4 transcript appears to be degraded by NMD. (C and D) Sequence traces of Adh cDNA extracted from progeny classes that do not contain Adh-PTC but do have a SNP between Adh alleles (C393A), demonstrating that sequencing is quantitative. Total RNA was isolated with the RNeasy mini kit (QIAGEN, Chatsworth, CA). RT-PCR for Adhn4 was performed using the Access RT-PCR System (Promega, Madison, WI). The products of the RT-PCR were directly sequenced with BigDye Terminator V3.1 (Applied Biosystems, Foster City, CA).
Quantitative PCR of Adhfn6 in dSMG1rst2 vs. wild-type flies:
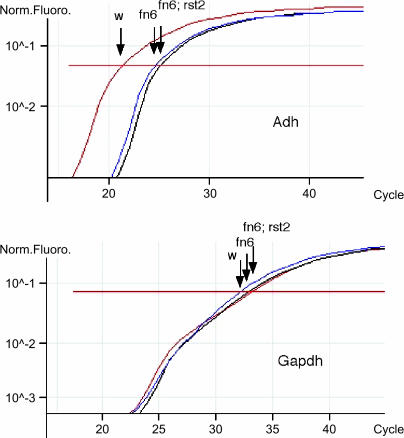
To confirm these results, we investigated the effect of the dSmgrst2 mutation on a different Adh-PTC allele using real-time quantitative PCR. The Adhfn6 allele has a mutation with a sequence repeat and deletion that prevents the splicing of the first intron and leads to a premature stop codon (Benyajati et al. 1983). As in Adhn4 flies, the level of Adhfn6 mRNA is drastically reduced (Brogna 1999). We created a strain (Adhfn6; dSmg1rst2) in which homozygous dSmg1rst2 was placed into a homozygous Adhfn6 background. Quantitative real-time RT-PCR showed that the level of Adhfn6 mRNA is only 6.2% that of wild-type w1118, consistent with previous reports. However, the message level of the Adhfn6 allele was not elevated as a consequence of dSmg1rst2. The transcript expressed in Adhfn6; dSmg1rst2 flies is 6.9% of w1118 and not significantly different from Adhfn6 (P > 0.05) (Figure 3). We conclude from the above results that dSmg1rst1 and dSmg1rst2 do not abolish NMD even though they almost certainly knock out the ortholog of one of the key players in C. elegans and human NMD.
Figure 3.
Flies containing the dSmg1rst2 mutant do not have impaired nonsense-mediated decay of Adhfn6 null allele transcripts. Real-time quantification of Adh and Gapdh transcripts using dual-labeled fluorogenic gene probes (Adh probe 6-FAM-CTGGCGAAACTGGCCCCCATTA-TAMRA, Gapdh probe TET-CCGCTTGGTTCTCCGCGCC-BHQ) in w1118, Adhfn6, and dSmg1rst2; Adhfn6 flies. Adh transcript levels in Adhfn6 and dSmg1rst2; Adhfn6 flies are similar and much lower than those in w1118. The housekeeper control (Gapdh) transcript levels are the same in the three strains. Each combination of primer set and DNA was done in triplicate.
Discrepancy with dsRNAi in S2 cells:
These results are in contrast with the work of Gatfield et al. (2003) in which reporter genes with PTCs were transfected into S2 cell lines that were then treated with double-stranded RNA interference (dsRNAi) directed against the Drosophila orthologs of Smg genes. Two types of reporter genes were used in these experiments: Adh harboring exactly the same mutation that we used and a GFP transgene containing a PTC. Northern blots were used to determine whether RNAi against the Smg genes increases the abundance of PTC-containing transcripts. While the RNAi against Smg2, -3, -4, -5, and -6 appears to increase the abundance of PTC-containing reporter transcripts 4.6- to 6.9-fold, the effect on RNAi against Smg1 is smaller (2.8-fold) and perhaps less convincing (Maquat 2004). However, it raises the possibility that S2 cells rely more heavily on Smg1's role in NMD than adult male flies do. For instance, there may be a level of redundancy in adult flies caused by some other PIKK fulfilling Smg1's role. Interestingly, nanos PTC transcripts have been shown to be targets for NMD in early embryos but not in later development (Meyer and Gavis 2005). Thus there is independent evidence that the molecular components regulating NMD may change throughout fly development. In conclusion, our data suggest that either Smg1 is not involved in NMD in Drosophila or, if it has a role, it is developmentally limited.
Acknowledgments
We thank Lee Willoughby for assistance with microarrays, Phillip Daborn for discussion, James Wettenhall and Gordon Smyth for their generous help with microarray analysis, and Simon Prochnik and Nomi Harris from the Berkeley Drosophila Genome Project for identifying the PTC transcripts for us. This work was supported by the Australian Research Council through its funding of the Special Research Centre—the Centre of Environmental Research and Adaptation Research.
References
- Adcock, G. J., P. Batterham, L. E. Kelly and J. A. McKenzie, 1993. Cyromazine resistance in Drosophila melanogaster (Diptera, Drosophilidae) generated by ethyl methanesulfonate mutagenesis. J. Econ. Entomol. 86: 1001–1008. [DOI] [PubMed] [Google Scholar]
- Benyajati, C., A. R. Place and W. Sofer, 1983. Formaldehyde mutagenesis in Drosophila molecular analysis of Adh-negative mutants. Mutat. Res. 111: 1–7. [DOI] [PubMed] [Google Scholar]
- Bosotti, R., A. Isacchi and E. L. Sonnhammer, 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25: 225–227. [DOI] [PubMed] [Google Scholar]
- Brogna, S., 1999. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA 5: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali, B. M., and P. Anderson, 1998. mRNA surveillance mitigates genetic dominance in Caenorhabditis elegans. Mol. Gen. Genet. 260: 176–184. [DOI] [PubMed] [Google Scholar]
- Daborn, P. J., J. A. McKenzie and P. Batterham, 2000. A genetic analysis of cyromazine resistance in Drosophila melanogaster (Diptera: Drosophilidae). J. Econ. Entomol. 93: 911–919. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., L. Unterholzner, F. D. Ciccarelli, P. Bork and E. Izaurralde, 2003. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22: 3960–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonnstedt, I., and T. Speed, 2002. Replicated microarray data. Stat. Sin. 12: 31–46. [Google Scholar]
- Maquat, L. E., 2002. Nonsense-mediated mRNA decay. Curr. Biol. 12: R196–R197. [DOI] [PubMed] [Google Scholar]
- Maquat, L. E., 2004. Nonsense-mediated mRNA decay: a comparative analysis of different species. Curr. Genomics 5: 175–190. [Google Scholar]
- Meyer, E. L., and E. R. Gavis, 2005. No nonsense: RNA surveillance in Drosophila melanogaster. Proceedings of the 46th Annual Drosophila Research Conference, San Diego, poster 361A.
- Nagy, E., and L. E. Maquat, 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem. Sci. 23: 198–199. [DOI] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson, 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Wilusz, C. J., W. Wang and S. W. Peltz, 2001. Curbing the nonsense: the activation and regulation of mRNA surveillance. Genes Dev. 15: 2781–2785. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., T. Ohnishi, I. Kashima, Y. Taya and S. Ohno, 2001. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 15: 2215–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]