Abstract
D-type cyclins (cyclins D1, D2, and D3) are key components of cell cycle machinery in mammalian cells. These proteins are believed to drive cell cycle progression by associating with their kinase partners, cyclin-dependent kinases, and by directing phosphorylation of critical cellular substrates. In addition, D-cyclins play a kinase-independent role by sequestering cell cycle inhibitors p27Kip1 and p21Cip1. In the past, we and others generated cyclin D1-deficient mice and have shown that these mice display developmental abnormalities, hypoplastic retinas, and pregnancy-insensitive mammary glands. To test the significance of cyclin D1–p27Kip1 interaction within a living mouse, we crossed cyclin D1-deficient mice with mice lacking p27Kip1, and we generated double-mutant cyclin D1−/−p27−/− animals. Here we report that ablation of p27Kip1 restores essentially normal development in cyclin D1-deficient mice. Our results provide genetic evidence that p27Kip1 functions downstream of cyclin D1.
The progression of mammalian cells through the G1 phase of the cell cycle is governed by two classes of cyclins, namely D-cyclins and cyclin E. The D-cyclins (cyclins D1, D2, and D3) are expressed in an overlapping, redundant fashion in all proliferating cell types. These proteins are rapidly induced after mitogen challenge and hence serve as links between the cell environment and the core cell cycle machinery (1). Consistent with their growth-promoting roles, D-cyclins can act as oncogenes. Indeed, overexpression of D-cyclins is seen in a large number of human cancers (2, 3). For instance, cyclin D1 is overexpressed in the majority of human breast cancers (4, 5).
D-cyclins are thought to drive cell cycle progression by associating with their catalytic partners (termed cyclin-dependent kinases CDK4 and CDK6) and by guiding these kinases to their cellular substrates (1). More recent work indicates that the induction of cyclin D-CDK complexes results in the redistribution of CDK inhibitor p27Kip1 from cyclin E-CDK2 complexes to cyclin D-CDK4/6 complexes, thereby triggering the kinase activity of cyclin E-CDK2 holoenzyme (6). Thus, D-cyclins also control cell cycle progression in a kinase-independent manner, via their interaction with p27Kip1.
In the present work we set out to examine the significance of cyclin D1–p27Kip1 interaction in driving cell proliferation within the context of a living animal. We took advantage of cyclin D1-deficient mice that we and others had previously generated. These cyclin D1−/− mice are viable but show developmental abnormalities and hypoplastic retinas and display mammary glands that fail to undergo normal lobuloalveolar development during pregnancy (7, 8). We crossed cyclin D1-deficient mice with mice lacking p27Kip1 (9–11) and generated double-mutant animals. We reasoned that analyses of these cyclin D1−/−p27−/− animals would provide a stringent test for the significance of cyclin D1–p27Kip1 interaction in controlling the cell proliferation of various cyclin D1-dependent lineages.
Materials and Methods
Generation of Double-Mutant Cyclin D1−/−p27−/− Mice.
Cyclin D1+/− mice (7, 8) were crossed with p27+/− (11) mice. The resulting cyclin D1+/−p27+/− heterozygotes were bred to generate cyclin D1−/−p27−/− mice. All animals were of mixed C57BL/6 × 129/Sv background. Mice were genotyped by PCR as described (7) or by use of the protocol provided by the Jackson Laboratory. To minimize the impact of the mixed genetic background, we used littermates for analyses.
Histopathologic Analyses.
Organs were dissected, fixed in Bouin's fixative (Sigma), and embedded in paraffin. Sections were stained with hematoxylin and eosin. Single-cell suspensions were obtained from freshly dissected thymuses, and the total number of cells was determined.
Superovulation and BrdUrd Staining.
Eight- to 13-week-old female mice were injected i.p. with 10 units of pregnant mare serum gonadotropin (Calbiochem). Forty six hours later mice were injected i.p. with 10 units of human chorionic gonadotropin (Calbiochem) and were mated with stud males. At day 2.5 postcoitum mice were injected i.p. with BrdUrd (Sigma), 100 μg/g of body weight, and sacrificed 3 h later. Ovaries were collected and fixed in 4% paraformaldehyde. Paraffin sections were stained with anti-BrdUrd antibody (Becton Dickinson) followed by detection with a Vectastain ABC kit (Vector Laboratories).
Mammary Epithelial Transplantations.
Mammary fragments were collected from wild-type, p27−/−, or cyclin D1−/−p27−/− females and engrafted into cleared fat pads of B6,129-Rag1tm1Mom recipients (purchased from The Jackson Laboratory) as described (12). The transplantations of cyclin D1−/− epithelium, shown in Fig. 3, were not performed in parallel, but derive from a previous set of experiments. Recipient mice were mated, mammary glands were collected at day 1 postpartum, and whole mounts were prepared and stained with carmine red as described (7).
Figure 3.
Ablation of p27Kip1 rescues the mammary epithelial phenotype of cyclin D1-deficient mice. A wild-type (WT) or mutant mammary epithelial tree 1 day after the delivery of pups. Mammary whole mounts were stained with carmine red. Mammary epithelial transplantations were performed as described in Materials and Methods.
Western Blot and Kinase Assays.
Retinas were microdissected from 1-day-old neonates. Fifty to 150 μg of proteins was separated by 10% SDS/PAGE [or 6% for retinoblastoma protein (pRB) analyses] and transferred to NitroBind membrane (Osmonics, Minnetonka, MN). The immunoblots were probed with the following primary antibodies: anti-cyclin D2 (M-20; Santa Cruz Biotechnology), anti-cyclin D3 (C-16; Santa Cruz Biotechnology), anti-pRB (245; PharMingen), and anti-phospho-Ser780pRB (New England Biolabs). As secondary antibodies, peroxidase-conjugated IgG (Jackson ImmunoResearch) was used, followed by enhanced chemiluminescence detection (Amersham Pharmacia).
For kinase assays, 200 μg of protein lysates was incubated with anti-CDK4 antibody (C-22; Santa Cruz Biotechnology) or with anti-CDK2 antibody (M2; Santa Cruz Biotechnology) conjugated to agarose beads. Immunoprecipitates were subjected to kinase reactions in 15 μl of 50 mM Hepes, (pH 7.5), 10 mM MgCl2, 5 mM MnCl2, 1 mM DTT, 0.013 mM ATP, 2 μCi [γ-32P]ATP, and 1 μg pRB(769) fusion protein (Santa Cruz Biotechnology) for CDK4 activity or 2.5 μg histone H1 for CDK2 activity for 30 min at 30°C.
Results
Appearance of Double-Knockout Mice.
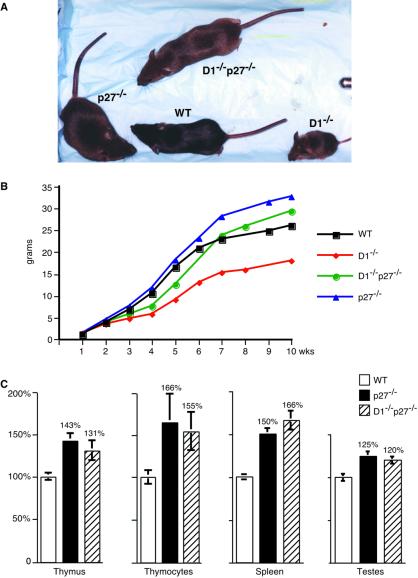
Cyclin D1−/−p27−/− mice, derived from crossing cyclin D1+/−p27+/− heterozygotes, were born with the expected frequency. As reported previously (7, 8), cyclin D1−/− mice displayed grossly reduced body size (Fig. 1 A and B). A significant fraction of these mice died early in life, often within the first 3 weeks (not shown). In contrast, cyclin D1−/−p27−/− mice developed normally and never exhibited premature mortality (Fig. 1 A and B and not shown). Moreover, cyclin D1−/− mice often displayed misaligned teeth, which were likely due to developmental defects in jaw formation. Again, this defect was never seen in double-mutant cyclin D1−/−p27−/− mice (not shown). Thus, ablation of the p27Kip1 gene restored nearly normal development in cyclin D1−/−p27−/− animals.
Figure 1.
Impact of the loss of cyclin D1 and p27Kip1 on animal growth. (A) A photograph of 8-week-old wild-type (WT) and mutant males. (B) Growth curves of wild-type (WT) and mutant males. The mean body weight values were obtained by weekly weighing 6–15 animals per experimental group. An identical analysis was performed for females, and very similar curves were obtained (not shown). (C) Relative organ weight and the number of cells per thymus in wild type (WT) and mutant males. For each genotype, five 10- to 13-week-old males were analyzed. The weight of indicated organs or the total number of cells per thymus (thymocytes) was determined and divided by animal body weight. The mean values shown are expressed relative to the values seen in wild-type mice, which were set at 100%. Error bars indicate SE.
An exception is the “leg-clasping” reflex seen in cyclin D1−/− mice. Thus, when lifted by their tails, cyclin D1-deficient mice invariably draw their limbs toward the trunk, in contrast to wild-type mice, which respond by extending their limbs (7, 8). We found that a moderate leg-clasping reflex persists in cyclin D1−/−p27−/− mice. Because the precise histopathological basis of this defect in cyclin D1−/− mice has not been identified, we can only presume that this defect is partially cured in D1−/−p27−/− animals.
Ablation of p27Kip1 Rescues Retinal Phenotype of Cyclin D1-Deficient Mice.
We and others reported previously that cyclin D1-deficient mice display a dramatic reduction in cell numbers in all layers of the neural retina (7, 8). Regardless of this observation, we found extremely high levels of cyclin D1 but virtually no cyclin D2 or D3 in the developing retinas of wild-type mice (7, 12). We speculated that proliferation of retinal cells is driven by cyclin D1, and, consequently, these cells fail to proliferate normally in the absence of cyclin D1.
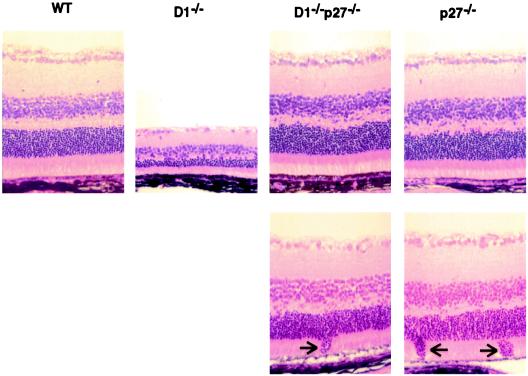
To determine the impact of p27Kip1 loss on retinal development in cyclin D1−/− mice, we examined the histological appearance of retinas derived from double-knockout cyclin D1−/−p27−/− animals and compared them with retinas derived from wild-type and cyclin D1−/− littermates. As reported previously (7, 8), cyclin D1-deficient mice displayed hypoplastic, grossly underdeveloped retinas. In contrast, double-knockout cyclin D1−/−p27−/− mice developed retinas containing normal cell numbers (Fig. 2). Thus, ablation of p27Kip1 in cyclin D1−/− mice fully corrected the phenotype of cyclin D1 deficiency and restored normal retinal development.
Figure 2.
Ablation of p27Kip1 rescues the retinal phenotype of cyclin D1-deficient mice. Histologic sections of retinas derived from wild-type (WT) or mutant mice were stained with hematoxylin and eosin. (Lower) Focal invasions of the outer granular layer into the layer of rods and cones, seen in p27−/− and cyclin D1−/−p27−/− mice, are indicated by arrows.
Ablation of p27Kip1 Rescues Mammary Phenotype of Cyclin D1-Deficient Mice.
As reported previously, cyclin D1-deficient mice display normal mammary glands at the end of sexual maturation. However, these mammary glands fail to undergo normal lobuloalveolar development during pregnancy. As a consequence, cyclin D1-deficient mice display underdeveloped mammary epithelial trees at the end of pregnancy (7, 8). Using a mammary epithelial transplantation approach, we and others have previously shown that this defect is intrinsic to the cyclin D1−/− mammary epithelium, which fails to respond normally to pregnancy-associated morphogenic signals (12, 13).
To examine the impact of p27Kip1 ablation on the mammary epithelial phenotype of cyclin D1−/− mice, we studied mammary development in double-knockout cyclin D1−/−p27−/− females. Because these cyclin D1−/−p27−/− females were found to be sterile (see below), we could not directly analyze the pregnancy-associated mammary development in double-mutant animals. Instead, we again turned to the mammary epithelial transplantation approach. In this approach, a fragment of mammary epithelium is dissected from the donor mouse and grafted into a cleared (i.e., devoid of endogenous epithelium) fat pad of a wild-type recipient. It is well established that transplanted wild-type epithelium behaves like the intact mammary epithelium of host animals and undergoes the same morphological changes during pregnancy (14).
We dissected mammary epithelial fragments from mutant and wild-type mice and transplanted them into cleared fat pads of wild-type recipients. As reported previously (12, 13), cyclin D1-deficient epithelium failed to undergo normal lobuloalveolar development when the recipient mice became pregnant. In contrast, epithelium derived from cyclin D1−/−p27−/− mice underwent full lobuloalveolar development that was indistinguishable from that seen in wild-type animals (Fig. 3). Thus, ablation of p27Kip1 in cyclin D1−/− mammary epithelium restored normal responses to pregnancy-associated signals and afforded normal lobuloalveolar development.
Ablation of Cyclin D1 Does Not Rescue the Phenotypes of p27Kip1-Deficient Mice.
Having shown that deletion of p27Kip1 rescued essentially all of the phenotypic manifestations of cyclin D1 deficiency, we asked whether the converse was true, i.e., whether the loss of cyclin D1 corrected the abnormalities seen in p27−/− mice. As reported previously, p27-deficient mice displayed increased body size (9–11). Double-mutant cyclin D1−/−p27−/− mice also showed increased body mass, although it was lower than the body mass seen in p27-deficient mice (Fig. 1 A and B). Importantly, p27−/− mice displayed enlarged internal organs, most notably spleens, thymuses, and testes (9–11). These internal organs were found to be enlarged to the same extent in double-mutant cyclin D1−/−p27−/− mice (Fig. 1C). Moreover, the thymuses of double-mutant mice showed increased cell numbers, which closely mirrored observations of p27-deficient mice (Fig. 1C). Hence, these phenotypic abnormalities of p27−/− mice were not corrected by the ablation of cyclin D1.
Several p27-deficient mice display focal disorganization of retinal cytoarchitecture. The lesions are composed of the outer granular layer nuclei invading the layer of rods and cones (10, 11). Histopathological examination of retinas derived from cyclin D1−/−p27−/− animals revealed the presence of similar focal lesions in double-mutant animals (Fig. 2 Lower, arrows).
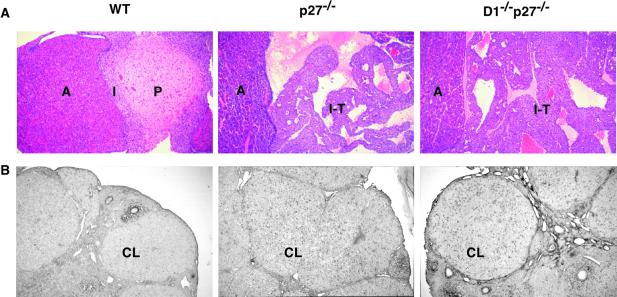
In addition to increased body weight and focal retinal abnormalities, p27-deficient mice are highly prone to pituitary adenomas arising from the intermediate lobe (9–11). Consistent with the previous reports, we found that seven of seven p27−/− mice analyzed that were 10–13 weeks old harbored pituitary tumors. Importantly, all seven age-matched cyclin D1−/−p27−/− mice also developed tumors (Fig. 4A), revealing that the loss of cyclin D1 does not alter the phenotypic manifestations of p27 deficiency in this organ.
Figure 4.
Ablation of cyclin D1 does not rescue the phenotype of p27Kip1-deficient mice. (A) Histologic sections of pituitaries derived from 10- to 13-week-old wild-type (WT) or mutant mice were stained with hematoxylin and eosin. Note the normal appearance of pituitaries in WT mice and the presence of intermediate lobe tumors (I-T) in p27−/− and cyclin D1−/−p27−/− animals. A, anterior lobe of the pituitary; P, posterior lobe; I, intermediate lobe. (B) Incorporation of BrdUrd into corpora lutea (CL) of wild-type (WT) and mutant ovaries 3 days after superovulation. Mutant p27−/− and cyclin D1−/−p27−/− corpora lutea continue to incorporate BrdUrd, in contrast to wild-type ovaries.
As reported by others (9–11), p27-deficient females mate infrequently and are sterile. Matings of double-mutant cyclin D1−/−p27−/− females with wild-type stud males revealed that these females were also infertile (not shown). The reason for sterility in p27−/− animals is not fully understood and involves both ovarian-dependent and -independent mechanisms (15). The ovarian defect manifests itself in the inability of cells forming corpus luteum to exit the cell cycle in a timely fashion (15). We studied this ovarian phenotype in double-mutant cyclin D1−/−p27−/− mice by injecting females with hormones that cause ovulation, then injecting mice with BrdUrd and analyzing BrdUrd incorporation into corpora lutea 3 days after ovulation. As expected, no BrdUrd incorporation was detected in corpora lutea of wild-type mice. In contrast, BrdUrd incorporation into corpora lutea was evident in both p27−/− and cyclin D1−/−p27−/− females (Fig. 4B), revealing the inability of p27-deficient and double-mutant luteal cells to undergo a timely cell cycle exit. Again, we concluded that the absence of cyclin D1 does not correct the phenotype of p27-deficient mice.
Taken together, phenotypic analyses of double-knockout cyclin D1−/−p27−/− animals revealed that ablation of p27Kip1 restored essentially normal development in cyclin D1-dependent tissues, but the loss of cyclin D1 had virtually no impact on the p27-knockout phenotype.
Molecular Analyses of Developing Retinas.
To study the molecular mechanism of the observed rescue of cyclin D1−/− phenotypes by the ablation of p27Kip1, we performed further analyses on retinas derived from double-knockout cyclin D1−/−p27−/− neonates. We chose retinas for our studies, because this tissue depends on cyclin D1 for its proliferation (7, 8), and it expresses p27Kip1, but no other CDK inhibitors, p21Cip1 and p57Kip2 (16), making it an ideal system for the study of cyclin D1–p27Kip1 interaction at the molecular level. We thus microdissected developing retinas from wild-type and mutant neonates, prepared protein lysates, and subjected them to detailed analyses.
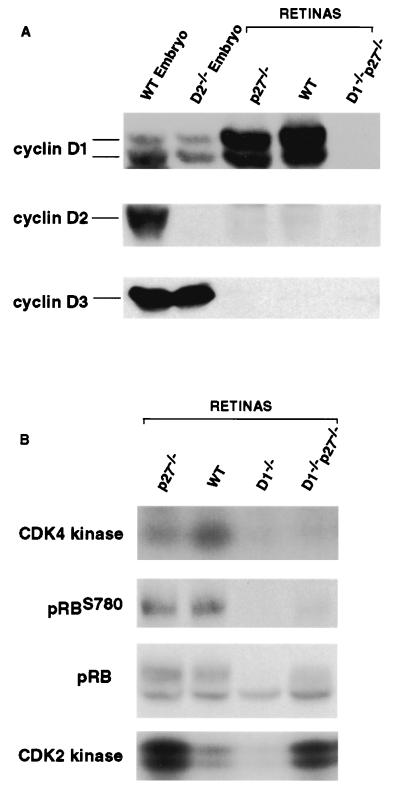
It was possible that the observed restoration of normal morphogenesis in cyclin D1−/−p27−/− retinas was due to derepression of cyclins D2 or D3 in the mutant tissue. To directly address this notion, we determined the expression of D-type cyclins in the retinas of double-knockout mice and compared it with that seen in wild-type animals. As we reported previously (7, 12), developing retinas of wild-type mice expressed very high levels of cyclin D1, no cyclin D2, and virtually no cyclin D3. As expected, the retinas of double-mutant cyclin D1−/−p27−/− mice lacked cyclin D1 protein (Fig. 5A). Importantly, these retinas did not express any detectable cyclin D2 or D3, revealing that ablation of p27Kip1 restored normal retinal morphogenesis in the absence of any detectable levels of D-cyclins (Fig. 5A).
Figure 5.
Molecular analyses of mutant retinas. Retinal lysates were prepared from mice of indicated genotypes. The levels of D-cyclins and serine 780 phospho-Rb (pRBS780) and total levels of pRB were determined by Western blotting. CDK4-associated or CDK2-associated kinase activity was determined by immunoprecipitation–kinase assays.
To directly gauge the cyclin D-associated kinase activity in mutant retinas, we immunoprecipitated CDK4 from retinal lysates and performed in vitro kinase assays by using pRB as a substrate (CDK4 is the major catalytic partner of D-cyclins in developing retinas; data not shown). Whereas CDK4-associated kinase activity was readily detectable in wild-type and p27-deficient retinas, it was virtually absent in cyclin D1−/− and double-mutant cyclin D1−/−p27−/− tissue (Fig. 5B). Thus, the apparently normal retinal development in cyclin D1−/−p27−/− retinas proceeds in the absence of any detectable cyclin CDK4-associated kinase activity.
This notion was further reinforced by analyses of the phosphorylation status of the pRB in retinal lysates. During cell cycle progression, pRB becomes sequentially phosphorylated by cyclin D-CDK4/6 and later by cyclin E-CDK2 and cyclin A-CDK2 complexes (17). Importantly, D-cyclins were shown to phosphorylate pRB at distinct subsets of sites (18), allowing us to use endogenous pRB phosphorylation as a measure of in vivo cyclin D-associated kinase activity. To this end, we probed immunoblots of retinal lysates with an anti-phospho-pRB antibody specifically recognizing the cyclin D-specific phosphoserine 780 residue of pRB. These analyses revealed that this site was well phosphorylated in wild-type and p27-deficient retinas, but not in cyclin D1−/− or cyclin D1−/−p27−/− tissues (Fig. 5B). Again, we concluded that apparently normal retinal development of double-mutant retinas takes place in the absence of any measurable cyclin D-associated kinase activity.
Last, we determined the levels of CDK2-associated kinase activity in the retinal lysates derived from wild-type and mutant animals. To this end, we immunoprecipitated CDK2 and performed in vitro kinase assays by using histone H1 as a substrate. These analyses revealed a significant reduction of CDK2-associated kinase activity in the retinas derived from cyclin D1−/− mice. In contrast, p27−/− retinas contained elevated levels of CDK2 activity, and these elevated levels persisted in double-mutant tissues (Fig. 5B).
Discussion
D-type cyclins are thought to promote cell proliferation by providing substrate specificity for associated kinases, CDK4 and CDK6. Moreover, induction of D-cyclins causes redistribution of p27Kip1 from cyclin E-CDK to cyclin D-CDK molecules (6).
p27Kip1 was originally regarded as a universal inhibitor of all cyclin-CDK complexes. More recent work revealed that p27Kip1, along with related inhibitor p21Cip1, plays a distinct role with regard to cyclin D-CDK versus cyclin E-CDK2 and cyclin A-CDK2 holoenzymes. Thus, binding of p27Kip1 to cyclin E-CDK2 and cyclin A-CDK2 inhibits their kinase activity. In contrast, at stoichiometric levels, p27Kip1 (and p21Cip1) allows cyclin D to bind CDKs and thus serves as an “assembly” factor for cyclin D-CDK complexes (6, 19, 20). Hence, in addition to their well-documented CDK-dependent role, D-cyclins likely play a kinase-independent function in cell cycle progression by controlling the activity of cyclin E via titration of p27Kip1 (6). However, the significance of cyclin D1–p27Kip1 interaction has never been examined in the context of a living animal.
We decided to address this issue by crossing cyclin D1-deficient mice with p27−/− animals. Cyclin D1-deficient mice display focal abnormalities in several organs that likely reflect high levels of cyclin D1 in these tissues of wild-type mice (7, 8).
Our phenotypic analyses of double-mutant cyclin D1−/−p27−/− mice revealed that ablation of p27Kip1 rescued the phenotype of cyclin D1−/− animals and restored virtually normal development. In contrast, the loss of cyclin D1 had essentially no impact on the phenotype of p27 deficiency. These findings provide genetic, in vivo evidence that p27Kip1 functions downstream of cyclin D1. This is a demonstration of the significance of cyclin D1–p27Kip1 interaction within the context of a living mouse.
We realize that the lack of rescue of the p27−/− phenotype by cyclin D1 deletion might reflect a particular expression pattern of cyclin D1 in p27Kip1-dependent tissues. For instance, we do not know whether cyclin D1 is expressed in the cells of the intermediate pituitary lobe that give rise to tumors in p27−/− mice. We note, however, that cyclin D1 ablation failed to rescue the phenotype of p27Kip1 deficiency in the retina, a tissue that expresses solely cyclin D1. Thus, at least in this compartment, the unidirectional rescue of cyclin D1−/− and p27−/− phenotypes cannot be attributed to the expression pattern of D-cyclins but reveals that p27Kip1 functions downstream of cyclin D1.
Our results are consistent with an earlier study of Tsutsui et al. (21), who demonstrated that proliferative defects in in vitro cultured CDK4−/− mouse embryo fibroblasts can be corrected by deletion of p27Kip1. Because CDK4 and CDK6 participate in titrating p27Kip1, by virtue of forming complexes with D-cyclins, we presume that the rescue of the CDK4−/− fibroblast phenotype and the rescue of the developmental phenotype of cyclin D1−/− mice operate on the same mechanistic principle.
Is then the ability of cyclin D1 to neutralize p27Kip1 the only relevant function of cyclin D1 in the cell cycle progression? One simple “extreme” interpretation of our results would be that the kinase-dependent role of cyclin D1 is fully dispensable and that cyclin D-CDK complexes function solely to control the activity of p27Kip1 (and in some tissues likely the activity of p21Cip1). Consistent with this notion are the results of Cheng et al. (22), which revealed that fibroblasts lacking both p27Kip1 and p21Cip1 proliferate in the absence of measurable cyclin D-kinase activity and are resistant to cell cycle arrest induced by D-cyclin inhibitor p16INK4.
The alternative interpretation of our results is that both kinase-dependent and kinase-independent functions of cyclin D1 are required to allow normal cell proliferation. Others have shown that ectopic expression of cyclin E allows a bypass of cyclin D-CDK kinase function with regard to the pRB in in vitro-cultured cells (23–25). Moreover, we have previously shown that replacement of cyclin D1 by cyclin E in the mouse germ line restores normal development in cyclin D1-dependent tissues (12). The deletion of the p27Kip1 gene leads to elevated CDK2-associated kinase activity in p27−/− tissues (10, 11) and these elevated levels persist in cyclin D1−/−p27−/− animals (Fig. 5B). Thus, it is plausible that within the intact, wild-type mouse, both functions of cyclin D1 are required to drive cell proliferation. In contrast, when CDK2 activity is elevated as a result of p27Kip1 deficiency, the CDK4 activity may be expendable. The only definitive way to distinguish between these two explanations is to create a knock-in strain of mice in which the wild-type copies of the cyclin D1 gene will be replaced by the cyclin D1 mutant, which fails to activate CDK4 and CDK6 kinases and yet retains the ability to titrate p27Kip1.
Acknowledgments
We thank B. Alpert, B. Carthon, A. Ciemerych, L. Le Cam, M. Mann, B. Rollins, and L. Zhang for their help. This work was supported by National Institutes of Health Grant CA83688-01, by Department of Defense Breast Cancer Idea Award BC980312, and by Susan G. Komen Breast Cancer Foundation Grant 99-3216 (to P.S.). P.S. is a Barr New Investigator.
Abbreviations
- CDK
cyclin-dependent kinase
- pRB
retinoblastoma protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.011522998.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.011522998
References
- 1.Sherr C J. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 2.Bartkova J, Lukas J, Strauss M, Bartek J. Oncogene. 1995;10:775–778. [PubMed] [Google Scholar]
- 3.Motokura T, Bloom T, Kim H G, Juppner H, Ruderman J V, Kronenberg H M, Arnold A. Nature (London) 1991;350:512–515. doi: 10.1038/350512a0. [DOI] [PubMed] [Google Scholar]
- 4.Dickson C, Fantl V, Gillett C, Brookes S, Bartek J, Smith R, Fisher C, Barnes D, Peters G. Cancer Lett. 1995;90:43–50. doi: 10.1016/0304-3835(94)03676-a. [DOI] [PubMed] [Google Scholar]
- 5.Weinstat-Saslow D, Merino M J, Manrow R E, Lawrence J A, Bluth R F, Wittenbel K D, Simpson J F, Page D L, Steeg P S. Nat Med. 1995;1:1257–1260. doi: 10.1038/nm1295-1257. [DOI] [PubMed] [Google Scholar]
- 6.Sherr C J, Roberts J M. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 7.Sicinski P, Donaher J L, Parker S B, Li T, Fazeli A, Gardner H, Haslam S Z, Bronson R T, Elledge S J, Weinberg R A. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 8.Fantl V, Stamp G, Andrews A, Rosewell I, Dickson C. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 9.Kiyokawa H, Kineman R D, Manova-Todorova K O, Soares V C, Hoffman E S, Ono M, Khanam D, Hayday A C, Frohman L A, Koff A. Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh D Y, Nakayama K. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 11.Fero M L, Rivkin M, Tasch M, Porter P, Carow C E, Firpo E, Polyak K, Tsai L H, Broudy V, Perlmutter R M, et al. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 12.Geng Y, Whoriskey W, Park M Y, Bronson R T, Medema R H, Li T, Weinberg R A, Sicinski P. Cell. 1999;97:767–777. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- 13.Fantl V, Edwards P A, Steel J H, Vonderhaar B K, Dickson C. Dev Biol. 1999;212:1–11. doi: 10.1006/dbio.1999.9329. [DOI] [PubMed] [Google Scholar]
- 14.Robinson G W, Johnson P F, Hennighausen L, Sterneck E. Genes Dev. 1998;12:1907–1916. doi: 10.1101/gad.12.12.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong W, Kiyokawa H, Soos T J, Park M S, Soares V C, Manova K, Pollard J W, Koff A. Cell Growth Differ. 1998;9:787–794. [PubMed] [Google Scholar]
- 16.Zhang P, Wong C, DePinho R A J W H, Elledge S J. Genes Dev. 1998;12:3162–3167. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg R A. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa M, Higashi H, Jung H K, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, et al. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Hannon G J, Beach D. Genes Dev. 1994;8:1750–1758. doi: 10.1101/gad.8.15.1750. [DOI] [PubMed] [Google Scholar]
- 20.La Baer J, Garrett M D, Stevenson L F, Slingerland J M, Sandhu C, Chou H S, Fattaey A, Harlow E. Genes Dev. 1997;11:847–862. doi: 10.1101/gad.11.7.847. [DOI] [PubMed] [Google Scholar]
- 21.Tsutsui T, Hesabi B, Moons D S, Pandolfi P P, Hansel K S, Koff A, Kiyokawa H. Mol Cell Biol. 1999;19:7011–7019. doi: 10.1128/mcb.19.10.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng M, Olivier P, Diehl J A, Fero M, Roussel M F, Roberts J M, Sherr C J. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leng X, Connell-Crowley L, Goodrich D, Harper J W. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 24.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 25.Alevizopoulos K, Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5322–5333. doi: 10.1093/emboj/16.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]