Abstract
Swi6 associates with Swi4 to activate HO and many other late G1-specific transcripts in budding yeast. Genetic screens for suppressors of SWI6 mutants have been carried out. A total of 112 of these mutants have been identified and most fall into seven complementation groups. Six of these genes have been cloned and identified and they all encode subunits of the mediator complex. These mutants restore transcription to the HO-lacZ reporter in the absence of Swi6 and have variable effects on other Swi6 target genes. Deletions of other nonessential mediator components have been tested directly for suppression of, or genetic interaction with, swi6. Mutations in half of the known subunits of mediator show suppression and/or growth defects in combination with swi6. These phenotypes are highly variable and do not correlate with a specific module of the mediator. Mutations in tail module components sin4 and pgd1 showed both growth defects and suppression when combined with swi6, but a third tail component, gal11, showed neither. A truncated form of the essential Srb7 mediator subunit also suppresses swi6 mutations and shows a defect in recruitment of the tail module components Sin4, Pgd1, and Gal11 to the mediator complex.
APPROXIMATELY 20% of yeast genes are transcribed during the cell cycle such that their mRNAs are present only during a specific phase of the mitotic cycle (Breeden 2003). The largest wave of transcription occurs in late G1, as cells prepare to replicate their DNA and begin the cell division process. More than 300 genes are transcriptionally induced in late G1 (Iyer et al. 2001; Simon et al. 2001) and two promoter elements (SCBs and MCBs) responsible for this regulation have been identified. The transcriptional activators of these late G1-specific elements share a common subunit, Swi6. Swi6 does not bind DNA directly; rather, it associates with either Swi4 or Mbp1, which confer the ability to bind SCBs or MCBs, respectively. Swi4 and Mbp1 are the two founding members of a family of related DNA-binding proteins that mediate switches between mitotic, meiotic, and pseudohyphal growth (Rua et al. 2001; Wittenberg and Flick 2003). As the primary mitotic member of this family, Swi4 is rate limiting for the G1-to-S transition (McInerny et al. 1997) and two of the critical targets of the Swi4/Swi6 complex are the late G1 cyclins CLN1 and CLN2 (Ogas et al. 1991).
Swi4 and Swi6 were identified in screens for genes required for HO expression (Haber and Garvik 1977; Stern et al. 1984; Breeden and Nasmyth 1987). HO encodes the endonuclease involved in mating-type switching. HO transcription is restricted to late G1 and tightly dependent upon Swi4 and Swi6 activity. Early studies showed that cells lacking Swi6 produced an intermediate constitutive level of transcripts for other Swi6 target genes (Dirick et al. 1992; Lowndes et al. 1992), suggesting that Swi6 performs a regulatory function that is both positive and negative within the cell cycle. This hypothesis is supported by the recent discovery of Whi5, which associates with Swi4/Swi6 complexes during early G1 and dissociates when the complex is activated (Jorgensen et al. 2002; Costanzo et al. 2004; de Bruin et al. 2004). Activation of Swi4/Swi6 complexes requires the Cln3 cyclin and the Cdc28 cyclin-dependent kinase, which phosphorylates Whi5 and promotes its dissociation. Release of Whi5 is temporally correlated with activation of Swi4/Swi6 complexes; however, Whi5 is not the only negative regulator of Swi4/Swi6 target promoters because these transcripts retain their late G1-specific regulation in a whi5 mutant. Clearly, there are other negative regulatory components that have not yet been identified.
The HO promoter, because of its tight dependence upon Swi4 and Swi6 for activation, continues to be an important model promoter for studying the temporal order of events that are required to activate late G1-specific transcription. The order of events that lead up to the binding and activation of HO by Swi4/Swi6 complexes involves several other SWI genes (SWI1, -2, -3, and 5), which were identified first for their role in mating-type switching (Haber and Garvik 1977; Stern et al. 1984) and later as activators of HO transcription (Breeden and Nasmyth 1987). During M phase, Swi5 binds to a site ∼1 kb upstream from the translational start site for HO (Stillman et al. 1989). Swi5 recruits the Swi/Snf chromatin-remodeling complex, which in turn enables mediator and the SAGA histone acetyltransferase to bind (Cosma et al. 1999; Bhoite et al. 2001). These large complexes modify and remodel nucleosomal structure within the 1-kb-long promoter and allow Swi4/Swi6 (or SBF) complexed with the Whi5 inhibitor to bind to downstream sites called SCBs. Once bound to SCB elements, Swi4/Swi6/Whi5 recruits mediator, but this takes place well before the complex is activated by Cln3/Cdk. Whi5 dissociates and transcription ensues (Costanzo et al. 2004; de Bruin et al. 2004), with the further recruitment of PolII, TFIIB, and TFIIH (Cosma et al. 2001).
The complex series of events that occur at the HO promoter differs from the order of events at other promoters, including other promoters whose activation is initiated by Swi5 binding (Bhoite et al. 2001). The most striking difference seen at the HO promoter, and at several other Swi4/Swi6 target promoters, is the recruitment of mediator well before transcriptional activation. This was one of the earliest examples that indicated that there is not always a direct correlation between mediator recruitment and initiation of transcription. Moreover, it is not necessarily the gene-specific activator that dictates the timing of mediator recruitment, since this can differ in the context of different promoters.
The mediator is composed of >20 proteins that transduce signals from gene-specific activators to the general transcription machinery (Lewis and Reinberg 2003). This process requires that mediator perform some stereotyped functions to direct the initiation or repression of transcription and some more individual functions tailored to its interaction with gene-specific regulators. Parsing out these functions to specific proteins within each module of the mediator is an ongoing process and few generalities have emerged. Three core modules form the head, middle, and tail of the mediator complex. A fourth, less tightly associated module composed of Srb8, -9, -10, and -11 has also been isolated as a separate complex with kinase activity (Liao et al. 1995; Borggrefe et al. 2002). The head module includes eight tightly associated proteins (Koh et al. 1998) and is thought to interact with the C-terminal domain (CTD) of RNA polymerase II (Lee and Kim 1998) and with two of the general transcription factors (TBP, TFIIB). The middle module with its eight subunits interacts with the CTD and TFIIE and the Srb8–11 module of the mediator (Kang et al. 2001). Genetics suggests that the Srb8–11 kinase module represses activator-dependent transcription (Carlson 1997). The tail module, composed of five proteins, binds to several activators in vitro and mutants in these subunits that are defective in activation and repression have been identified, leading to the view that this module is involved in the recognition of gene-specific regulators (Lewis and Reinberg 2003). However, in vitro studies suggest that this module may also be involved in activator-independent functions of the polymerase (Reeves and Hahn 2003).
Mutations in mediator components have been collected over the last two decades as suppressors of transcriptional activators (Carlson et al. 1984). This article identifies similar genetic interactions between the late G1-specific transcription factor Swi6 and components of the mediator complex. Despite the absolute requirement for Swi6 for HO transcription, mutations in many different components of the mediator complex that activate the HO promoter in the absence of Swi6 have been identified. A total of 112 such mutations have been isolated and characterized in this study. These include temperature-sensitive (ts) alleles of SIN4 and NUT1 and a truncated allele of the essential gene SRB7. In addition, synthetic interactions between swi6 and other nonessential components of the mediator have been investigated.
MATERIALS AND METHODS
Strains and growth conditions:
The strains used in this study are listed in Table 1. Standard genetic methods were used for strain construction and tetrad analysis (Sherman et al. 1994). Cells were grown at 30°, unless otherwise specified, in either YEPD medium or synthetic complete medium supplemented with amino acids as appropriate (Sherman et al. 1994). In all cases, the lithium acetate protocol was used for DNA transformation (Ito et al. 1983).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY131 | MATα HO-lacZ46 ura3 his3 leu2-3, -112 trp1-1 can1-100 ade2 met | This study |
| BY134 | MATα HO-lacZ46 ura3 leu2 trp1-1 can1-100 ade2-1 met | This study |
| BY142 | MATα HO-lacZ46 swi6-399 leu2 trp1-1 can1-100 ade2 | This study |
| BY150 | MATα HO-lacZ46 swi6-399 srb9-9 leu2 trp1-1 can1-100 ade2 | This study |
| BY155 | MATα HO-lacZ46 swi6-399 nut1-14 leu2 trp1-1 can1-100 ade2 | This study |
| BY165 | MATα HO-lacZ46 swi6-399 ssx6-24 leu2 trp1-1 can1-100 ade2 | This study |
| BY168 | MATα HO-lacZ46 swi6-399 srb10-27 leu2 trp1-1 can1-100 ade2 | This study |
| BY174 | mat∷LEU2 HO-lacZ46 swi6-399 ura3 trp1-1 can1-100 | This study |
| BY175 | mat∷LEU2 HO-lacZ46 swi6-399 rox3-33 ura3 trp1-1 can1-100 | This study |
| BY178 | mat∷LEU2 HO-lacZ46 swi6-399 srb7-1 ura3 trp1-1 can1-100 | This study |
| BY180 | mat∷LEU2 HO-lacZ46 swi6-399 ssx6-38 ura3 trp1-1 can1-100 | This study |
| BY183 | mat∷LEU2 HO-lacZ46 swi6-399 sb10-41 ura3 trp1-1 can1-100 | This study |
| BY186 | mat∷LEU2 HO-lacZ46 swi6-399 nut1-44 ura3 trp1-1 can1-100 | This study |
| BY296 | mat∷LEU2 HO-lacZ46 swi6-254 his3 leu2-3, -112 trp1-1 can1-100 ade2 met | This study |
| BY338 | MATα HO-lacZ46 swi6-399 srb9-9 ura3 his3 leu2 trp1-1 can1-100 ade2 | This study |
| BY546 | MATaHO-lacZ46 swi6∷LEU2-175 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY547 | MATα HO-lacZ46 swi6∷LEU2-175 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY602 | MATα HO-lacZ46 ura3 his3 leu2-1 leu2-112 trp1-1 can1-100 ade2 met | This study |
| BY603 | MATα HO-lacZ46 swi6∷TRP1-197 ura3 his3 leu2-1 leu2-112 trp1-1 can1-100 ade2 met | This study |
| BY1078 | MATα HO-lacZ46 swi6∷TRP1-197 rox3-102 ura3 his3 leu2-3, -112trp1-1 can1-100 ade2 met | This study |
| BY1087 | MATα HO-lacZ46 swi6∷TRP1-197 sin4-111 ura3 his3 leu2-3, -112 trp1-1 can1-100 ade2 met | This study |
| BY1088 | MATα HO-lacZ46 swi6∷TRP1-197 sin4-112 ura3 his3 leu2-3, -112 trp1-1 can1-100 ade2 met | This study |
| BY1141 | MATα HO-lacZ46 swi6-399 SSX-14 URA his3 trp1-1 | This study |
| BY1152 | MATα HO-lacZ46 swi6∷TRP1 sin4-112 ura3 his3 trp1-1 | This study |
| BY1160 | MATα HO-lacZ46 swi4∷LEU2 ssx3-112 ura3 his3 leu2-3, -112 trp1-1 ade2 | This study |
| DY1430 | MATaHO-lacZ46 swi4∷LEU2 sin4-TRP1 ura3 his3 leu2 trp1 can1-100 ade2 ade6 | D. Stillman |
| BY1974 | MATagic2∷LEU2 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY2125 | MATaura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3883 | MATasrb10∷HIS3MX6 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3885 | MATaswi6∷LEU2 srb10∷HIS3MX6 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3933 | MATaMED7-Flag ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3934 | MATaMED8-Flag ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3935 | MATasrb7-1 MED7-Flag ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY3936 | MATasrb7-1 MED8-Flag ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4010 | MATasrb2∷HIS3MX6 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4011 | MATasrb5∷HIS3MX6 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4018 | MATα HO-lacZ46 swi6∷LEU2-175 srb8∷HIS3MX6 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY4019 | MATα HO-lacZ46 swi6∷LEU2-175 srb11∷HIS3MX6 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY4021 | MATaROX3:HIS3MX6 ybl094c∷HIS3MX6 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4177 | MATα HO-lacZ46 swi6∷LEU2-175 pGAL1-SWI6:URA3 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY4198 | MATasrb7-1 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4199 | MATaswi6∷TRP1 srb7-1 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4200 | MATα swi6∷TRP1 srb7-1 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4201 | MATasrb7-1 srb10∷HIS3MX6 ade2-1 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
| BY4203 | MATaswi6∷TRP1 srb7-1 srb10∷HIS3MX6 ura3 his3-11 his3-15 leu2-3 leu2-112 trp1-1 can1-100 ade2-1 | This study |
| BY4215 | MATasin4∷TRP1 ura3 his3 leu2-3, -112 trp1 can1-100 ade2-1 | This study |
| BY4401 | MATaHO-lacZ46 swi6∷LEU2 sin4∷TRP1 ura3 his3-1 leu2 met15 | This study |
| BY4402 | MATaHO-lacZ46 swi6∷ LEU2 med1∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4406 | MATaHO-lacZ46 swi6∷ LEU2 med9∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4408 | MATaHO-lacZ46 swi6∷ LEU2 gal11∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4410 | MATα HO-lacZ46 swi6∷ LEU2 srb9∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4412 | MATaHO-lacZ46 swi6∷ LEU2 pgd1∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4415 | MATaHO-lacZ46 swi6∷ LEU2 nut1∷KanMX4 ura3 his3-1 leu2 met15 | This study |
| BY4605 | mat∷LEU2 HO-lacZ46 swi6-399 sin4-133ts ura3 his3 trp1-1 ade2 | This study |
| BY4608 | mat∷LEU2 HO-lacZ46 swi6-399 srb9-39 ura3 his3 trp1-1 ade2 | This study |
| DMA688 | MATagal11∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| DMA2548 | MATapgd1∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| DMA3838 | MATamed9∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| DMA3249 | MATamed1∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| DMA4469 | MATasrb9∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| DMA4566 | MATanut1∷KanMX4 ura3-0 his3-1 leu2-0 met15-0 | Winzeler et al. (1999) |
| BY4722 | MATα HO-lacZ46 swi6∷LEU2-175 srb2∷HIS3MX6 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY4723 | MATα swi6∷LEU2-175 srb5∷HIS3MX6 ura3 his3 leu2 trp1-1 can1-100 ade2 met | This study |
| BY4752 | MATα rox3-102 ade2 ura3 his3 leu2-3, -112 trp1-1 met | This study |
| BY4926 | MATaswi6∷LEU2 ura3 his3-11 his3-15 leu2-3, -112 trp1-1 can1-100 ade2-1 | This study |
Mutant isolation:
The screens for suppressors of SWI6 (ssx) mutations were carried out using BY174, BY142, BY296, and BY603. These strains carry one of three different swi6 alleles and the HO promoter driving lacZ expression (HO-lacZ) integrated at the HO locus. In SWI6 cells, β-galactosidase activity expressed from this reporter construct can be observed as blue colonies using the X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) filter assay as described previously (Breeden and Nasmyth 1987). swi6 colonies remain white because Swi6 is absolutely required for HO transcription (Breeden and Nasmyth 1987). Suppressors of the loss of Swi6 activity that could restore lacZ expression in these three different swi6 mutant backgrounds were obtained. Mutagenesis with ethyl methanesulfonate was carried out as previously described (Lycan et al. 1994). Survival rates were between 25 and 50%. Mutagenized cells were plated on YEPD plates and allowed to form colonies at either 30° (BY174, BY142, BY296) or 25° (BY603). Colonies grown at 25° were shifted to 37° for 4 hr before screening. Ten temperature-sensitive suppressors that expressed β-galactosidase activity only at 37° were identified.
Mutant identification:
Complementation analysis in this screen is complicated by the fact that HO is repressed in diploid cells. To circumvent this problem, suppressors were isolated in a MAT deletion strain, which mates as an a strain, but does not produce the Mat a1 protein required for diploid-specific repression of HO transcription (Strathern et al. 1981). Suppressors generated in this strain were crossed to suppressors generated in an α-strain. Complementation between two ssx mutants was indicated by the inability to express β-galactosidase driven by HO-lacZ. The complementation analysis was performed at 30° or at 37° for ts alleles. All swi6 ssx mutants were also crossed against the unmutagenized swi6 SSX parent of the opposite mating-type strain to identify dominant suppressors. One dominant suppressor was identified among the 112 suppressors characterized.
To identify these suppressors, representative swi6 ssx HO-lacZ strains were transformed with the genomic DNA 2μ plasmid library (gift from F. Winston) and transformants were selected on SD plates lacking leucine. The colonies were transferred to nitrocellulose filters and assayed for β-galactosidase activity. Transformants that remained white were selected. These plasmids were isolated from the yeast strain, purified, and retransformed to confirm the dependence of the suppression phenotype on the plasmid. Plasmids that prevented suppression of swi6 were sequenced using the Big Dye cycle sequencing method (PE-Biosystems) and the critical open reading frame was identified as indicated.
Strain constructions:
SRB8 and SRB11 were deleted in the α-strain swi6∷LEU2 HO-lacZ (BY547) using pFA6a-HIS3MX6 as described (Longtine et al. 1998). SRB2 and SRB5 were deleted in BY3220 SWI6 ho using the same method. Deletions were verified by polymerase chain reaction (PCR). No srb2 swi6 or srb5 swi6 double mutants could be obtained by crosses, so these crosses were performed with a swi6 strain carrying a galactose-inducible SWI6 gene (BY4177). The double mutants were obtained in this way, and then the plasmid was lost. The resulting strains were viable, but grew very slowly. The ROX3 locus was followed in crosses by disrupting the adjacent YBL094C open reading frame using pFA6a-HIS3MX6 in W303 as above to create BY4021.
To get the double mutants of swi6 and other mediator subunit genes in an HO-lacZ background, we crossed sin4, nut1, pgd1, med1, med9, srb9, and gal11 deletion strains with swi6∷LEU2 HO-lacZ. These were not isogenic crosses, so to minimize the likelihood that the phenotypes that we observed in the triple mutant were due to unknown strain variations, at least five independent segregants bearing all three mutations were analyzed from each cross. The phenotypes reported were consistent among all five isolates and thus unlikely to be caused by unknown background mutations.
RNA measurements:
RNA levels were quantitated with S1 protection assays performed using oligonucleotide probes as described (Mai et al. 2002). The probe sequences are available upon request. mRNA levels were quantified with a PhosphorImager 400A from Molecular Dynamics (Sunnyvale, CA).
Immunoprecipitations:
The C termini of Med7 and Med8 were tagged with the Flag epitope using plasmid pBS-3 Flag-KanMX as described (Gelbart et al. 2001) and insertion of the Flag tag into the native loci was confirmed by PCR. Cells carrying one or the other tagged locus were lysed by vortexing with glass beads five times for 60 sec in lysis buffer (20 mm HEPES pH 7.6, 1 mm EDTA, 10% glycerol, 0.25 KOAC, 0.05% NP40). One milligram of whole cell protein was diluted to 500 μl in lysis buffer and precleared with 20 μl of Protein A coupled to Dynabeads (Dynal Biotech ASA, Oslo) by rotation for 1 hr at 4°. The precleared lysates were mixed with anti-Flag M2 monoclonal antibody (Sigma, St. Louis) at 4° for 2 hr. A total of 30 μl Protein A Dynabeadads were added and incubated at 4° for 2 hr. Beads were pulled down with a magnet and proteins were eluted with 0.1 m sodium citrate pH 2.5 by rotating for 10 min at 4°. Eluate was mixed with SDS loading buffer and boiled for 5 min. Samples were separated by SDS-PAGE, transferred to PVDF membrane (Millipore, Bedford, MA), and immunoblotted with rabbit polyclonal antisera directed to Sin4, Gal11, Pgd1 (all gifts from S. Hahn), or the M2 monoclonal Flag antibody.
RESULTS
Two missense mutants and a deletion of SWI6 were used to screen for suppressors of the requirement for Swi6 at the HO promoter in an HO-lacZ reporter construct. The results of these screens are summarized in Table 2. A total of 112 suppressors of swi6 (ssx) were identified. Most of the ssx mutations fell into seven complementation groups, but 20% of them were either single alleles of other genes or double mutants that were not further characterized. The screen for suppressors of the swi6 deletion yielded alleles of six complementation groups, so mutations in all of these genes can bypass the requirement for Swi6 function at the HO promoter. Representative suppressors from each complementation group were used to clone and identify the complementing open reading frame. The identities of the suppressors are shown in Table 2 and they will be subsequently referred to by their established names. A total of 10 ts suppressors were identified in three different genes (SIN4, NUT1, and SSX6).
TABLE 2.
ssx alleles obtained with each swi6 mutant
| Complementation group | swi6-399 | swi6-254 | swi6∷TRP1 | Total | Identity of gene |
|---|---|---|---|---|---|
| SSX1 | 1 | 0 | 0 | 1 | SRB7 |
| SSX2 | 9 | 2 | 10 | 21 | ROX3 |
| SSX3 | 1 (1 ts) | 1 | 17 (7 ts) | 19 | SIN4 |
| SSX4 | 4 | 2 | 1 (1 ts) | 7 | NUT1 |
| SSX5 | 8 | 6 | 1 | 15 | SRB9 |
| SSX6 | 9 | 1 | 4 (1 ts) | 14 | ND |
| SSX7 | 7 | 1 | 4 | 12 | SRB10 |
| Unassigned | 5 | 4 | 14 (1 D) | 23 | |
| Total | 44 | 17 | 51 | 112 |
Screens were carried out with BY142 (25% survival), BY174 (25% survival), BY296 (50% survival), and BY603 (42% survival). Temperature-sensitive and dominant alleles obtained are shown in parentheses. ts, temperature sensitive; D, dominant; ND, not determined.
ssx1-1 encodes a truncated allele of the essential gene SRB7:
The library plasmid that suppressed the ssx1-1 phenotype contained the SRB7 gene, which was the obvious candidate suppressor. We sequenced all the mutant's genomic DNA covered by the library plasmid and found only one mutation. The mutation was in the SRB7 locus, causing it to be truncated after codon 89. Since SRB7 is an essential gene, we demonstrated linkage of ssx1-1 to SRB7 by deleting the adjacent gene, GIC2, with a LEU2 marker in a swi6 HO-lacZ strain and crossing this strain to ssx1-1 swi6 HO-lacZ. The suppressor phenotype segregated opposite to the gic2∷LEU2 locus in 19 tetrads, indicating that a mutation linked to GIC2 was responsible for the suppressor phenotype.
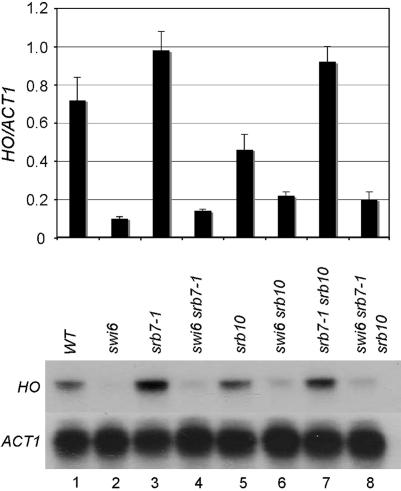
This is the first screen that has uncovered an allele of SRB7 as a suppressor of a defect in transcriptional activation. To further characterize this mutation, the starting strain was backcrossed five times to W303 swi6 ho and tested for the ability to suppress the requirement for Swi6 at the native HO locus. The srb7-1 mutation in SWI6 cells increased transcription of HO by ∼40% above wild-type level; in the swi6 srb7-1 ho strain it also shows a 40% increase above swi6 levels (Figure 1).
Figure 1.
srb7-1 shows a suppression function and a synthetic interaction with srb10 on the requirement for Swi6 at the native HO locus. RNA was collected from exponentially growing cells of the indicated genotypes and the level of HO mRNAs was measured using S1 protection. (Top) Quantification of S1 protection data, normalized to ACT1 loading control. (Bottom) Raw data for HO. Strains used are BY2125, BY4926, BY4198, BY4199, BY3883, BY3885, BY4201, and BY4203.
Truncations of ROX3 also suppress swi6 mutations:
A library plasmid encoding ROX3 suppressed the ssx2-102 (BY1078) phenotype. This DNA encoded six genes, including ROX3. The ROX3 locus was sequenced and a nonsense mutation terminating the protein at codon 135 was identified. Allelism between ssx2-102 and ROX3 was done by crossing swi6∷TRP1 ssx2-102 HO-lacZ with ROX3:HIS3 ho. Among the 26 Trp+ His− segregants, one-half (13) displayed lacZ expression by the filter assay. Since HO-lacZ and ho segregated 2:2, all the segregants carried the suppressor mutation, which indicates that SSX2 and ROX3 are allelic.
sin4 is allelic to ssx3:
Mutations in sin4 were previously identified as suppressors of the requirement for Swi5 and Swi4 at the HO-lacZ locus (Nasmyth et al. 1987; Lycan et al. 1994). Moreover, diploids homozygous for sin4 mutations sporulate inefficiently (Sternberg et al. 1987). We noted the same failure of ssx3 diploids to sporulate, so we transformed a SIN4-containing plasmid (M1387, gift from D. Stillman) into a swi6 ssx3-112 strain (BY1088). These transformants had no detectable lacZ expression, suggesting that ssx3-112 was an allele of SIN4. We then performed crosses between BY1160 swi4 ssx3 and DY1430 swi4 sin4∷TRP1 and observed the same inefficient sporulation that we observed with ssx3 and sin4 homozygotes. Sequencing of two ssx3 alleles confirmed that they contained mutations in the SIN4 locus. Interestingly, we found two mutations in both ssx3 alleles. One of these, a glutamate-to-aspartate substitution at position 612, was shared by both mutants. We then confirmed that this change resides in the parent strain, which had a SIN4 phenotype, and thus it appears to be a neutral substitution in this assay. In addition, we identified mutations at two other sites (see Table 3), which caused a glycine-to-aspartate substitution at position 128 and an alanine-to-threonine substitution at position 241. The latter of these substitutions resulted in a sin4 phenotype only at 37°.
TABLE 3.
Mutations found in representative suppressors
| Allele | Base substitution | Amino acid replacements |
|---|---|---|
| srb7-1 | G265T | E89a |
| rox3-102 | G405A | W135a |
| sin4-111 | G383A | G128D |
| G1836T | E612D | |
| sin4-133ts | G721A | A241T |
| G1836T | E612D | |
| srb9-9 | 2675: deletion of T | L892YFPLSIDa |
| srb10-27 | 768: deletion of G | M257a |
The E612D amino acid substitution identified in both sin4 alleles was traced back to the parent strain, which is phenotypically SIN4.
Termination codon.
NUT1 suppresses the ssx6 mutation, but is linked to ssx4:
ssx6 resisted multiple attempts to identify this gene. Fourteen alleles of ssx6 were isolated, but these mutants all exhibited weak suppressor activity and transformation efficiencies that were 10-fold lower than those of the parent strain. Both of these properties contributed to the difficulties in identifying the mutant gene. Individual transformations of candidate genes showed that ssx6 could not be suppressed or complemented by RGR1, SRB8, SRB11, MED1, MED9, or PGD1. NUT1 suppressed the ssx6 defect, but subsequent crosses that showed that these genes were not allelic were performed between nut1 and ssx6.
ssx4 is also a weak suppressor of swi6, which displayed poor transformation efficiency. Linkage between ssx4 and a nut1 deletion was demonstrated with a cross between swi6 ssx4-14 HO-lacZ (BY155) and swi6∷LEU2 nut1∷KanMX4 HO-lacZ (BY4415), which yielded 4:0 segregation of the suppressor phenotype.
ssx5 and ssx7 identify srb9 and srb10 as suppressors of swi6:
The ssx5-9 mutation (BY338) was suppressed by a library plasmid carrying SRB9. Sequencing of the SRB9 locus of BY338 showed that there was a single base deletion resulting in a reading frameshift terminating the normal coding sequence at codon 892 and replacing the terminal one-third of the protein with seven out-of-frame codons.
SRB10 was encoded on the plasmid that suppressed ssx7-27 (BY168). SRB10 was confirmed to be the critical locus by showing complementation with a plasmid containing only SRB10 (pSH598, gift from Steve Hahn). Sequencing showed that ssx7-27 harbors a single base mutation that causes termination of the protein at codon 257.
Defects in all components of the kinase module of mediator suppress swi6 mutations:
srb9 and srb10 mutants were readily isolated as suppressors of swi6. However, srb8 and srb11 were not identified. All four of these proteins are subunits of the same module. Moreover, Srb11 is the cyclin that controls the activity of Srb10. To see if srb8 or srb11 mutants could suppress the requirement for Swi6 at HO-lacZ, deletions of these two nonessential genes were generated in the swi6 HO-lacZ strain (BY547) and assayed for β-galactosidase activity. Both deletion mutants were capable of suppressing swi6 (Table 4) to roughly the same extent as srb9 and srb10.
TABLE 4.
Synthetic effects of swi6 and mediator mutants grown at different temperatures
| Growth
|
|||||
|---|---|---|---|---|---|
| 25° | 30° | 37° | Suppression at HO-lacZ
|
||
| swi6: | + | ++ | ++ | ||
| Tail domain | |||||
| SWI6 sin4 | + | + | − | ||
| swi6 sin4 | +/− | +/− | − | + | |
| SWI6 pgd1 | ++ | ++ | ++ | ||
| swi6 pgd1 | (+) | + | (+) | + | |
| SWI6 gal11 | ++ | ++ | ++ | ||
| swi6 gal11 | ++ | ++ | ++ | − | |
| Middle domain | |||||
| SWI6 nut1 | ++ | ++ | ++ | ||
| swi6 nut1 | ++ | ++ | ++ | + | |
| SWI6 med1 | + | ++ | ++ | ||
| swi6 med1 | + | + | (+) | + | |
| SWI6 med9 | ++ | ++ | ++ | ||
| swi6 med9 | (+) | (+) | (+) | + | |
| SWI6 srb7 | ++ | ++ | ++ | ||
| swi6 srb7 | ++ | ++ | (+) | + | |
| Kinase complex | |||||
| SWI6 srb8 | ++ | ++ | ++ | ||
| swi6 srb8 | ++ | ++ | + | + | |
| SWI6 srb9 | ++ | ++ | ++ | ||
| swi6 srb9 | ++ | ++ | + | + | |
| SWI6 srb10 | ++ | ++ | ++ | ||
| swi6 srb10 | ++ | ++ | ++ | + | |
| SWI6 srb11 | ++ | ++ | ++ | ||
| swi6 srb11 | ++ | ++ | + | + | |
| Head domain | |||||
| SWI6 srb2 | ++ | ++ | ++ | ||
| swi6 srb2 | +/− | +/− | +/− | ND | |
| SWI6 srb5 | + | + | + | ||
| swi6 srb5 | +/− | +/− | +/− | ND | |
| SWI6 rox3 | ++ | ++ | ++ | ||
| swi6 rox3 | ++ | ++ | ++ | + | |
The growth phenotypes of mediator subunit mutants in a swi6 background grown on YEPD plates were scored from best to worst as follows: ++, +, (+), +/−, −. ND, not determined. The suppression phenotype of each double mutant is indicated as “+” if the mediator defect restores HO-lacZ expression on the basis of the X-Gal filter assay described in materials and methods.
Mutations in half of the known subunits of mediator either act as suppressors of swi6 or show synthetic growth defects with swi6:
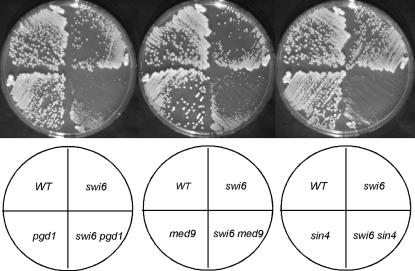
All the suppressors of swi6 mutations that we have identified are subunits of the mediator complex and are scattered throughout all four modules that have been identified. To fully explore the suppressing capacity of mediator mutants, we combined swi6 HO-lacZ with deletion mutations in other subunits of the mediator complex that are known to be nonessential and screened for their ability to suppress. These results are summarized in Table 4. In addition to suppressor activity, we looked for synthetic growth defects between swi6 and these mediator components. Many of the growth defects that we observed were more extreme at either 25° or 37° (Table 4). Mutations in all four proteins in the kinase domain behaved in a similar manner in the swi6 background. However, the other mediator mutations that were tested showed synthetic defects in combination with swi6 to differing extents (Figure 2). srb2 and srb5 showed the most extreme synthetic growth defect with swi6, but another head domain mutant, rox3, showed no such effect. All four middle domain mutations that were made could suppress the swi6 defect in HO-lacZ transcription, but each showed different growth characteristics. Similarly, SIN4 and PGD1 mutants suppress the requirement for Swi6, but a third component of the tail module, gal11, does not suppress. The swi6 pgd1 double also shows a severe growth defect at both 25° and 37°, while the swi6 gal11 strain grows as well as swi6 cells at all three temperatures tested.
Figure 2.
swi6 and mediator mutations show synthetic growth defects. Strains were streaked onto YEP-glucose plates and grown at 25° for 2 days. Strains are from left to right and counterclockwise from wild type (WT): BY134, BY546, BY4412, DMA2548, BY134, BY546, BY4406, DMA3838, BY134, BY546, BY4401, BY4215.
The spectrum of suppression at other swi6 targets is highly variable:
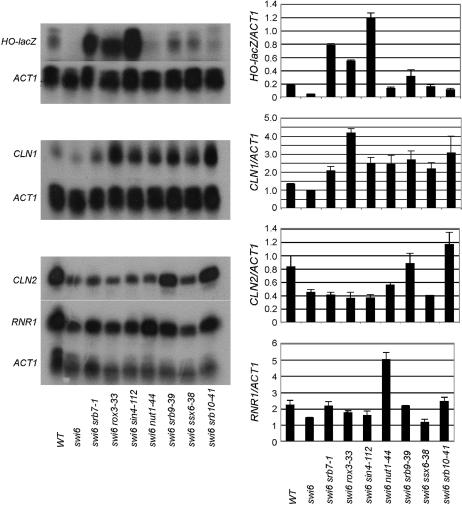
The screen for suppressors of swi6 was originally intended to look for negative regulators that might directly interact with Swi6. However, further characterization of representative alleles of each suppressor showed that these mutants had different effects on the transcription of other Swi6-regulated genes (Figure 3). Only srb10-41 showed modest suppression of the requirement for Swi6 at the CLN2 locus. At the CLN1 locus all the suppressors increased mRNA levels above the SWI6 level but the pattern was completely different from that at HO-lacZ. On the basis of the suppression assay at HO:lacZ, srb7-1, rox3-33, and sin4-112 are the strongest of the suppressors tested, but srb9-39, srb10-41, and ssx6-38 are all stronger suppressors at the CLN1 locus. Interestingly, at RNR1, nut1-44 increased the transcript level more than twofold, while none of the others showed any appreciable effect.
Figure 3.
ssx1–ssx7 show highly variable levels of suppression swi6 at four different Swi6 target promoters. RNA was collected from exponentially growing cells of the indicated genotypes and the levels of HO-LacZ, CLN1, CLN2, and RNR1 mRNAs were measured using S1 protection. (Left) Raw data for HO-lacZ, CLN1, CLN2, and RNR1. (Right) Quantification of S1 protection data, normalized to ACT1 loading control. Strains used are BY602, BY174, BY178, BY175, BY1152, BY186, BY4608, BY180, and BY183.
Synthetic interaction between srb7-1 and srb10:
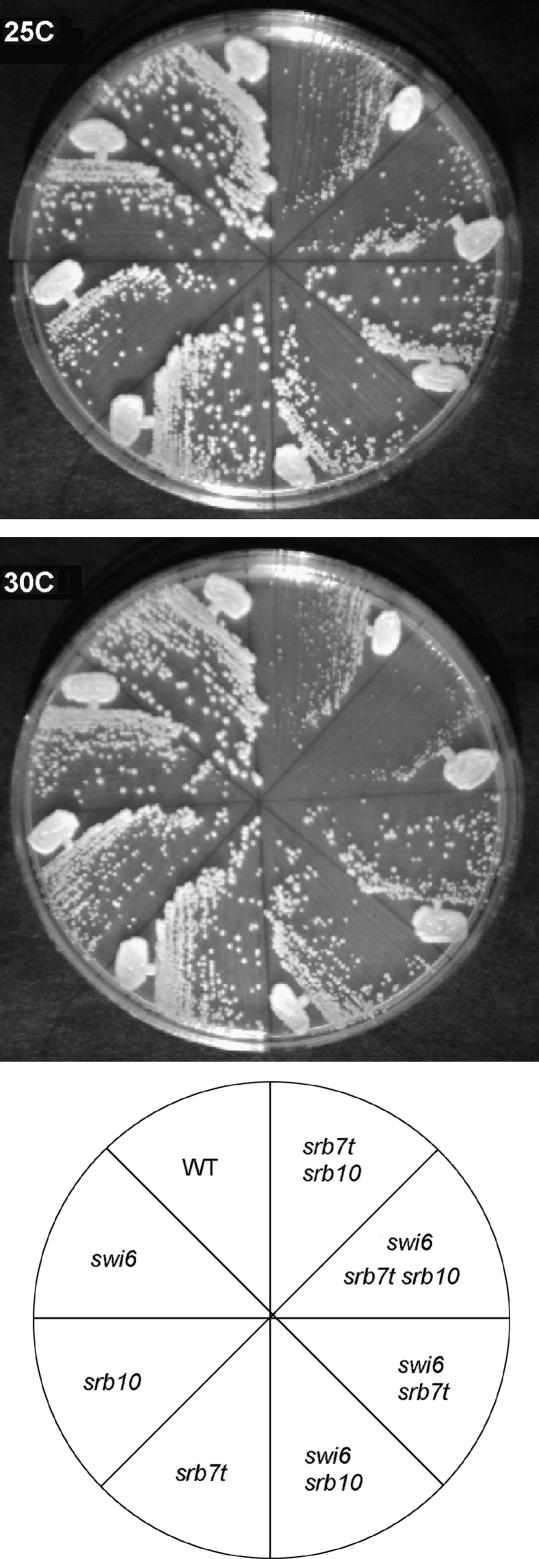
Srb7 is an essential subunit of the middle module of mediator, which interacts with the kinase module (Kang et al. 2001). Since all four components of the kinase module were isolated as suppressors, we wondered if srb7-1 suppressed the requirement for swi6 by preventing association of the kinase module. If this is the case, we would expect that the phenotype of the srb7-1 srb10 deletion would be no more extreme than the srb10 deletion alone. To test this hypothesis, we combined srb7-1 and srb10Δ in the presence or absence of swi6 and looked at their suppression and growth characteristics. In a SWI6 background, srb10 had little or no effect on ho mRNA levels. ho mRNA levels were near wild type in srb10 cells, and both srb7 and srb7 srb10 cells produced 140 ± 10% of the wild-type ho mRNA (Figure 1). In the swi6 background, srb7-1 and srb10 are both weak suppressors and the swi6 srb7-1 srb10 triple is not significantly stronger than the double mutants. This is consistent with a role for the C terminus of Srb7 in interacting with the kinase domain, since the effects of the two suppressors are not additive. However, Figure 4 shows colonies of each genotype, grown at 25° and 30°. Clearly, in both a swi6 and a SWI6 background, the srb7-1 srb10Δ combination is more deleterious than either single mutant. We conclude that Srb10 and the C terminus of Srb7 may function in the same pathway with respect to suppression of swi6 mutants at the ho locus, but there are other critical activities that they do not share.
Figure 4.
Synthetic interactions among swi6, srb7-1, and srb10Δ. Strains of the genotypes indicated were grown to single colonies on YEPD plates at the temperatures indicated. Clockwise from wild type (WT): BY2125, BY4201, BY4203, BY4199, BY3885, BY4198, BY3883, BY1965.
srb7-1 shows reduced association with the tail module:
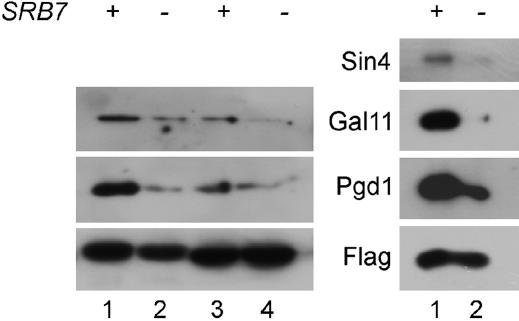
To see if other associations were defective in the srb7-1 strain, we tagged Med7 and Med8, which are subunits of the middle and head domains of mediator, respectively, and looked at the association of the tail subunit components in SRB7 and srb7-1 cells using immunoprecipitation. These results are shown in Figure 5. It is evident that the ability of Med7 and Med8 to co-immunoprecipitate three components of the tail module (Sin4, Gal11, and Pgd1) is reduced in the absence of the C terminus of Srb7.
Figure 5.
The truncation of Srb7 causes reduced association of the tail module with the middle (Med7) and head (Med8) domains of mediator. SRB7 (+) and srb7-1 (−) cells carrying either a Flag-tagged Med7 (left and right, lanes 1 and 2) or a Flag-tagged Med8 (left, lanes 3 and 4) were grown at 30°, harvested, and immunoprecipitated with Anti-Flag M2 monoclonal antibodies. These precipitates were then immunoblotted with Sin4, Gal11, Pgd1, or Flag antibodies as indicated.
DISCUSSION
Swi6 plays a key role in the transition from G1-to-S phase, due to its involvement in both transcription factor complexes that induce late G1-specific transcription. Swi6 interacts with either Swi4 or Mbp1 in >300 genes, but none of these genes have been shown to be as dependent upon Swi6 as the HO gene. Although CLN1 and CLN2 are Swi6-regulated genes on the basis of genetics, promoter dissections, and binding studies, growing cells show less than a twofold drop in CLN1 or CLN2 transcript level in the absence of Swi6 (Figure 3). This difference has not been fully explored; however, several studies have indicated the existence of other regulatory elements in the CLN1 and CLN2 promoters (Stuart and Wittenberg 1994; Partridge et al. 1997).
The uniquely tight requirement for Swi6 at the HO promoter made it possible to identify swi6 mutants in a genetic screen, using the HO promoter driving lacZ as the reporter (Breeden and Nasmyth 1987). This same property enabled us to identify >100 mutations that suppress the requirement for Swi6 at this promoter. Suppressors of Swi6 arose at a frequency of ∼0.1% among the survivors of the mutagenesis. Interestingly, all the mutations identified were components of the mediator complex (Figure 6). Some of these mediator mutations result in a very high level of transcription (Figure 3), causing a dramatic shift from a repressed state to an activated state. Many of the same mediator components were identified as suppressors of swi4 (Lycan et al. 1994; Tabtiang and Herskowitz 1998).
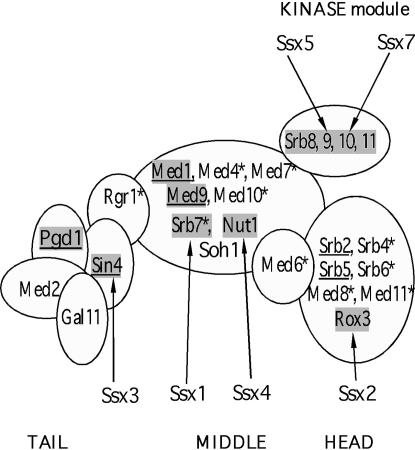
Figure 6.
Mutations throughout the mediator complex suppress the requirement for Swi6 for HO-lacZ transcription. The four mediator subdomains or modules (head, middle, tail, and kinase) are shown and the gene products that are known to be components of each module are labeled. Subunits essential for viability are indicated with asterisks. Subunits in which mutations have been found to confer suppression of swi6 are shaded, and subunit mutants showing synthetic growth defects with swi6 are underlined.
Lycan et al. (1994) analyzed suppressors of swi4 mutations, using the same HO-lacZ reporter and found mutations at roughly one-tenth the frequency of the ssx screen. Three suppressors were identified, only one of which (sin4) is represented in the ssx collection. Tabtiang and Herskowitz (1998) also carried out a search for suppressors of swi4. These investigators used a fragment of the HO promoter with Swi4/Swi6-binding sites sandwiched between the GAL1 upstream activating sequence and a lacZ reporter. Their search identified 14 suppressors, including several that were identified in our screen (srb8–srb11, sin4, rox3). This group also noted, as had previous investigators studying suppressors of Swi5, that suppression was context dependent (Nasmyth et al. 1987; Jiang and Stillman 1992). That is, sin4 could suppress swi4 mutations at their reporter construct, but not at the native ho locus. We observed a similar context dependence in our studies. Looking at three other native Swi6 target promoters, we observed a very different pattern of suppression for the seven classes of suppressor mutants that we identified.
This type of complexity has also been observed on a large scale for Gcn4-activated genes. Gcn4 is required for the full induction of >500 genes (Natarajan et al. 2001). Four native Gcn4 target genes have been analyzed for their inducibility in a panel of 80 deletion mutants, each of which lacked one of the known coactivators or corepressors of transcription (Swanson et al. 2003). Each of the Gcn4 targets displayed a significantly different pattern of dependency, indicating that a different subset of coregulators are recruited by Gcn4 to these promoters. This complexity may be explained, in part, by the fact that Gcn4 target genes are often regulated by more than one transcription factor (Hinnebusch 1992). This appears to be a general phenomenon, because genome-wide location analysis of transcription-factor-binding sites showed that more than one-third of the promoters tested bound more than one transcription factor under the limited set of conditions tested (Lee et al. 2002). These and many other studies indicate that the requirements for transcription of a given promoter depend upon the constellation of transcription factors involved and the features of the surrounding chromatin.
Using the HO-lacZ reporter, we found that mutations in nearly half the proteins that compose the mediator could suppress the requirement for Swi6. As expected, these included all the proteins that make up the kinase domain, which is associated with repression of the transcription of many genes (Carlson et al. 1984; Carlson 1997; Holstege et al. 1998). However, we also observed suppression with mutations in every other module of the mediator, including the essential gene Srb7. This implicates all modules or the mediator as a whole in preventing transcription at HO-lacZ in the absence of its gene-specific activator. Unlike other promoters, mediator is recruited to the HO promoter well before it is activated by Cln3/Cdk, both by Swi5 and by the Swi4/Swi6 complex (Cosma et al. 1999, 2001; Bhoite et al. 2001). It may be this early localization of mediator that facilitates its role as a repressor.
Only one allele of SRB7 was obtained from this screen and no other alleles have been identified in the dozens of such screens that have been carried out. SRB7 is an essential gene in yeast (Hengartner et al. 1995; Lucau-Danila et al. 2000) and mice (Tudor et al. 1999). The srb7-1 allele terminates the protein at position 89. Termination of Srb7 at this position eliminates 40% of the protein, the sequence of which is 40% identical to the human and Drosophila homologs (Gromoller and Lehming 2000). In spite of this, cells carrying srb7-1 grow like wild-type cells. Only when srb7-1 was combined with srb10Δ did we observe a notable drop in growth rate. We have previously shown that BY178 swi6 srb7-1 causes the constitutive transcription of SWI4, which is normally transcribed in late M and early G1 phases of the cell cycle (Breeden and Mikesell 1991). This strain also shows misregulation of YOX1 and YHP1, which are cell-cycle-specific repressors of SWI4 and other M/G1-specific transcripts (our unpublished results). In this study we have shown that the srb7-1 truncation reduces the amount of the three components of the tail module that are associated with mediator. This is the first role ascribed to the C terminus of Srb7. Whether this interaction between Srb7 and the tail module is mediated by a direct or indirect interaction is unknown. For example, Rgr1 is another essential component of the mediator, which has been shown to be important for the association of the tail module (Myers et al. 1998). It is possible that srb7-1 disrupts association with the tail module indirectly through an effect on Rgr1.
In an effort to systematically explore the interactions between Swi6 and mediator, we combined swi6 mutations with mutations in other nonessential subunits of the mediator. Most of these double mutants showed suppression or growth defects or both. Interestingly, with the exception of the kinase module, mutations in the individual components of each module did not behave in the same way with respect to synthetic interactions with swi6. Mutations in the tail module components sin4 and pgd1 showed both growth defects and suppression when combined with swi6, but a third tail component, gal11, showed neither. It has been suggested that GAL11 mutants prevent association of other tail module subunits (Lee et al. 1999; Park et al. 2000). If that were the case, we would expect the gal11 phenotypes to include all the phenotypes associated with loss of Sin4 and Pgd1. In our studies, swi6 sin4 and swi6 pgd1 show strong phenotypes that are undetectable in the swi6 gal11 strain.
This study adds to the ever-growing appreciation of the complexities of transcription. With the number of proteins involved in transcription and the recognition that the unique context of each promoter influences both the sequence of events and the mechanics of the operation, it is not surprising that few generalities have emerged. Many more systematic and comprehensive studies in well-defined settings will be required before the generalities will emerge from the sea of specifics.
Acknowledgments
We thank past and present members of the Breeden lab for their input in this project. Special thanks are also due to Steve Hahn and David Stillman for plasmids, strains, and antibodies. This work was supported by grant GM41073 from the National Institutes of Health to L.B.
References
- Bhoite, L. T., Y. Yu and D. J. Stillman, 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15: 2457–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst and R. D. Kornberg, 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277: 44202–44207. [DOI] [PubMed] [Google Scholar]
- Breeden, L. L., 2003. Periodic transcription: a cycle within a cycle. Curr. Biol. 13: R31–R38. [DOI] [PubMed] [Google Scholar]
- Breeden, L., and G. Mikesell, 1991. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 5: 1183–1190. [DOI] [PubMed] [Google Scholar]
- Breeden, L., and K. Nasmyth, 1987. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell 48: 389–397. [DOI] [PubMed] [Google Scholar]
- Carlson, M., 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13: 1–23. [DOI] [PubMed] [Google Scholar]
- Carlson, M., B. C. Osmond, L. Neigeborn and D. Botstein, 1984. A suppressor of SNF1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics 107: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma, M. P., T. Tanaka and K. Nasmyth, 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311. [DOI] [PubMed] [Google Scholar]
- Cosma, M. P., S. Panizza and K. Nasmyth, 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7: 1213–1220. [DOI] [PubMed] [Google Scholar]
- Costanzo, M., J. L. Nishikawa, X. Tang, J. S. Millman, O. Schub et al., 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117: 899–913. [DOI] [PubMed] [Google Scholar]
- de Bruin, R. A., W. H. McDonald, T. I. Kalashnikova, J. Yates, III and C. Wittenberg, 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117: 887–898. [DOI] [PubMed] [Google Scholar]
- Dirick, L., T. Moll, H. Auer and K. Nasmyth, 1992. A central role for SWI6 in modulating cell cycle Start-specific transcription in yeast. Nature 357: 508–513. [DOI] [PubMed] [Google Scholar]
- Gelbart, M. E., T. Rechsteiner, T. J. Richmond and T. Tsukiyama, 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21: 2098–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromoller, A., and N. Lehming, 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J. 19: 6845–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., and B. Garvik, 1977. A new gene affecting the efficiency of mating type interconversions in homothallic strains of S. cerevisiae. Genetics 87: 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengartner, C. J., C. M. Thompson, J. Zhang, D. M. Chao, S. M. Liao et al., 1995. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 9: 897–910. [DOI] [PubMed] [Google Scholar]
- Hinnebusch, A. G., 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, pp. 319–414 in The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression, edited by J. R. Broach, E. W. Jones and J. R. Pringle. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner et al., 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- Ito, H., Y. Fukada, K. Murata and A. Kimura, 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder et al., 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409: 533–538. [DOI] [PubMed] [Google Scholar]
- Jiang, H. W., and D. J. Stillman, 1992. Involvement of the SIN4 global transcriptional regulator in the chromatin structure of Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 4503–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz and M. Tyers, 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297: 395–400. [DOI] [PubMed] [Google Scholar]
- Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee et al., 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276: 42003–42010. [DOI] [PubMed] [Google Scholar]
- Koh, S. S., A. Z. Ansari, M. Ptashne and R. A. Young, 1998. An activator target in the RNA polymerase II holoenzyme. Mol. Cell 1: 895–904. [DOI] [PubMed] [Google Scholar]
- Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph et al., 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Lee, Y. C., and Y. J. Kim, 1998. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol. Cell. Biol. 18: 5364–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. C., J. M. Park, S. Min, S. J. Han and Y. J. Kim, 1999. An activator binding module of yeast RNA polymerase II holoenzyme. Mol. Cell. Biol. 19: 2967–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B. A., and D. Reinberg, 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116: 3667–3675. [DOI] [PubMed] [Google Scholar]
- Liao, S. M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson et al., 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374: 193–196. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lowndes, N. F., A. L. Johnson, L. Breeden and L. H. Johnston, 1992. SWI6 protein is required for transcription of the periodically expressed DNA synthesis genes in budding yeast. Nature 357: 505–508. [DOI] [PubMed] [Google Scholar]
- Lucau-Danila, A., R. Wysocki, T. Roganti and F. Foury, 2000. Systematic disruption of 456 ORFs in the yeast Saccharomyces cerevisiae. Yeast 16: 547–552. [DOI] [PubMed] [Google Scholar]
- Lycan, D., G. Mikesell, M. Bunger and L. Breeden, 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in S. cerevisae. Mol. Cell. Biol. 14: 7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, B., S. Miles and L. L. Breeden, 2002. Characterization of the ECB binding complex responsible for the M/G1-specific transcription of CLN3 and SWI4. Mol. Cell. Biol. 22: 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerny, C. J., J. F. Partridge, G. E. Mikesell, D. P. Creemer and L. L. Breeden, 1997. A novel Mcm1-dependent promoter element in the SWI4, CLN3, CDC6 and CDC47 promoters activates M/G1-specific transcription. Genes Dev. 11: 1277–1288. [DOI] [PubMed] [Google Scholar]
- Myers, L. C., C. M. Gustafsson, D. A. Bushnell, M. Lui, H. Erdjument-Bromage et al., 1998. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 12: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth, K. A., D. J. Stillman and D. Kipling, 1987. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating type switching in yeast. Cell 48: 579–587. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts et al., 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., B. J. Andrews and I. Herskowitz, 1991. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell 66: 1015–1026. [DOI] [PubMed] [Google Scholar]
- Park, J. M., H. S. Kim, S. J. Han, M. S. Hwang, Y. C. Lee et al., 2000. In vivo requirement of activator-specific binding targets of mediator. Mol. Cell. Biol. 20: 8709–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge, J. F., G. E. Mikesell and L. L. Breeden, 1997. Cell cycle-dependent transcription of CLN1 involves Swi4 binding to MCB-like elements. J. Biol. Chem. 272: 9071–9077. [DOI] [PubMed] [Google Scholar]
- Reeves, W. M., and S. Hahn, 2003. Activator-independent functions of the yeast mediator sin4 complex in preinitiation complex formation and transcription reinitiation. Mol. Cell. Biol. 23: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua, D., B. T. Tobe and S. J. Kron, 2001. Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4: 720–727. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1994. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi et al., 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106: 697–708. [DOI] [PubMed] [Google Scholar]
- Stern, M., R. Jensen and I. Herskowitz, 1984. Five SW1 genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178: 853–868. [DOI] [PubMed] [Google Scholar]
- Sternberg, P. W., M. J. Stern, I. Clark and I. Herskowitz, 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48: 567–577. [DOI] [PubMed] [Google Scholar]
- Stillman, D. J., A. T. Bankier, A. Seddon, E. G. Groenhout and K. Nasmyth, 1989. Characterization of a transcription factor involved in mother cell-specific transcription of the yeast HO gene. EMBO J. 1: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern, J. N., J. Hicks and I. Herskowitz, 1981. Control of cell type in yeast by the mating type locus. The α1-α2 hypothesis. J. Mol. Biol. 147: 357–372. [DOI] [PubMed] [Google Scholar]
- Stuart, D., and C. Wittenberg, 1994. Cell cycle-dependent transcription of CLN2 is conferred by multiple distinct cis-acting regulatory elements. Mol. Cell. Biol. 14: 4788–4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim et al., 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23: 2800–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabtiang, R. K., and I. Herskowitz, 1998. Nuclear proteins Nut1p and Nut2p cooperate to negatively regulate a Swi4p-dependent lacZ reporter gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 18: 4707–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor, M., P. J. Murray, C. Onufryk, R. Jaenisch and R. A. Young, 1999. Ubiquitous expression and embryonic requirement for RNA polymerase II coactivator subunit Srb7 in mice. Genes Dev. 13: 2365–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wittenberg, C., and K. Flick, 2003. Cell cycle regulation during G1 phase in yeast: decisions, decisions, decisions, pp. 14–39 in G1 Phase Progression, edited by J. Boonstra. Eurekah.com/Kluwer Academic/Plenum Publishers, New York.